ABSTRACT
Here, we presented 11 cases with colistin-resistant Pseudomonas aeruginosa infection and co-existence of OXA-48 and NDM-1 in the ST235 high-risk clone. The molecular analyses were performed by Sanger sequencing and RT–PCR. The eight patients (72.7%) had an invasive infection and three (27.3%) had colonization. The 30-day mortality rate was 87.5% (7/8). Three patients (37.5%, 3/8) received colistin therapy before isolation of P. aeruginosa. In the Multilocus sequence typing (MLST) analysis of 11 isolates, eight (72.7%) isolates belonged to P. aeruginosa ST235 clone. All isolates were NDM-1 positive, and nine isolates (81.8%) were found to be positive for both OXA-48 and NDM-1. Sequences of pmrAB and phoPQ revealed numerous insertions and deletions in all isolates. In 10 isolates pmrAB and phoPQ were found to be upregulated. In conclusion, the co-existence of OXA-48 and NDM-1 genes in colistin-resistant P. aeruginosa ST235 high-risk clone indicates the spread of carbapenemases in clinical isolates and highlights need of continuous surveillance for high-risk clones of P. aeruginosa.
KEYWORDS: P. aeruginosa, colistin resistance, high-risk clone, ST235, OXA-48, NDM-1
Pseudomonas aeruginosa (P. aeruginosa) is one of the most common causes of healthcare-related infections [1]. The ST235 high-risk clone of P. aeruginosa has high capacity to acquire antibiotic resistance and is disseminating worldwide. The ST235 clone harbours nearly 39 types of beta-lactamases especially IMP, NDM and VIM type Metallo-β-lactamases (MBLs) [2]. However, up to date, there is no report on co-existence of NDM-1 and OXA-48 in P. aeruginosa. Colistin resistance among P. aeruginosa is rare (<1%) in Europe [3]. However, it was found to be 7.4% in Korea, and 8.8% in Turkey [4,5]. Dissemination of colistin resistance in high-risk clones is concerning because of increased fatality and lack of antimicrobial therapy options. The overexpression of phoPQ and pmrAB two-component regulatory systems contribute colistin resistance by reducing the negative charge of the outer membrane in P. aeruginosa [6].
In this correspondence, we presented 11 cases with colistin-resistant P. aeruginosa infection and reported the presence of OXA-48 along with NDM-1 in the isolates belonging to the ST235 high-risk clone. We also analysed mutations and expressions of phoPQ and pmrAB systems in 11 colistin-resistant P. aeruginosa isolates.
Patients who were diagnosed with colistin-resistant P. aeruginosa infection or colonization in the ICU unit of a Cardio-Pulmonary Surgery Hospital in Istanbul between July 2017 and December 2018 were included in the study. The demographic and clinical data were recorded on a standardized case form. The patients were followed up for clinical outcomes.
The colistin resistance was determined by the broth microdilution method according to EUCAST criteria [7]. In strain typing, Multilocus sequence typing (MLST) was performed by amplifying seven housekeeping genes, according to the protocol on Pseudomonas aeruginosa MLST website (https://pubmlst.org/P.aeruginosa/). Allelic profiles and sequence types (STs) were determined using Applied Math Bionumerics V7.6 software. Clonal relatedness was determined by the repetitive PCR (rep-PCR) (Diversilab, Biomerieux). Isolates that had a similarity index >95% were considered as clonally related. Carbapenem and colistin resistance genes (blaIMP, blaVIM, blaOXA, blaNDM, blaKPC, mcr-1) were screened by PCR using primers as described previously, and the amplicons were confirmed by sequencing (6). For colistin resistance mechanisms, mutations in pmrAB and phoPQ were detected by Sanger Sequencing [6]. Expressions of pmrA and phoP were studied by qRT-PCR [6]. The rpsl was selected for normalization and P. aeruginosa ATCC 27853 was for calibration.
Among 11 patients, eight (72.7%) had an invasive infection and three (27.3%) had colonization with colistin-resistant P. aeruginosa. Eight (72.7%) patients stayed in ICU and six (54.5%) had lung transplantation. Sepsis was diagnosed in three (37.5%) of eight patients. The 30-day mortality rate was 88% (7/8). The overall mortality of P. aeruginosa infections in the hospital was 46% and it was 68% for carbapenem-resistant colistin susceptible P. aeruginosa infections. In a recent report from Spain, the overall 30-day mortality of bacteremia cases caused by ST235 was found to be 82%, however, it was 42.2% in other clones [10]. In our study, the eight of 11 (72.7%) isolates belonged to P. aeruginosa ST235 high-risk clone. The three isolates were identified as a novel allele (ST3078) referred by Pasteur Institutes MLST database (https://pubmlst.org/paeruginosa/)(Table 1). In Germany, the mortality rate of P. aeruginosa bacteremia was reported as 26% in 937 ICU units [8]. The increased mortality rates are associated with multidrug resistance [9]. The ST235 is associated with MDR or PDR profile and fatal infections [2]. In genotyping K741-K748 and K752-K753 were found to be clonally related (>95%). Other isolates belong to different clone.
Table 1. Clinical and laboratory characteristics of study population.
| Code | ST | Source | Colistin MIC | Meropenem MIC | Carbapenemase | Empirical Therapy | Pre-exposure time to colistin | Active Therapy | Duration of Active Colistin Therapy | Survival | 30-day Mortality | Clonal Relatedness |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K704 | 235 | Catheter | >64 | 16 | OXA-48, NDM-1 | Meropenem Vancomycin | 0 | 0 | 0 | Ex | 1 | |
| K741 | 235 | BAL | 16 | 16 | OXA-48, NDM-1 | Moxifloxacin | 0 | Meropenem Colistin Ertapenem | 1 | Ex | 2 | * |
| K740 | 3078 | BAL | >64 | 8 | OXA-48, NDM-1 | Piperacillin Tazobactam | 0 | Meropenem Colistin Ertapenem | 27 | Ex | 30 | |
| K748 | 235 | DTA | 64 | 16 | OXA-48, NDM-1 | 0 | 0 | 0 | 0 | Discharge | Discharge | * |
| K752 | 3078 | DTA | 4 | 16 | OXA-48, NDM-1 | 0 | 0 | 0 | 0 | Discharge | Discharge | ** |
| K753 | 3078 | BAL | 4 | 8 | OXA-48, NDM-1 | Piperacillin Tazobactam | 0 | Meropenem Colistin | 14 | Discharge | Discharge | ** |
| K783 | 235 | BAL | 16 | 16 | OXA-48, NDM-1 | Meropenem Colistin | 10 | Meropenem Colistin | 10 | Ex | 7 | |
| K970 | 235 | Catheter | 32 | 16 | NDM-1 | Levofloxacin Colistin | 21 days | 0 | 0 | Ex | 10 | |
| K982 | 235 | BAL | 16 | 16 | OXA-48, NDM-1 | Colistin (inhalation) | 25 days | 0 | 0 | Ex | 1 | |
| K989 | 235 | Catheter | >64 | 16 | NDM-1 | Meropenem | 0 | Meropenem Colistin | 38 | Ex | 42 | |
| K1009 | 235 | DTA | 16 | 16 | OXA-48, NDM-1 | Colistin (inhalation) | 8 days | 0 | 0 | Ex | 19 |
Notes: BAL: Bronchoalveolar lavage; DTA: Deep tracheal aspirate.
*clone 1, **clone 2.
All isolates were found to be carbapenem-resistant. All of them were NDM-1 positive, and nine (81.8%) harbour both OXA-48 and NDM-1 beta-lactamases. Carbapenem-resistant OXA-48 positive P. aeruginosa was isolated in Sudan and India [11] and NDM-1 positive P. aeruginosa was detected in Serbia [12]. However, this is the first report of co-existence of OXA-48 and NDM-1 producing P. aeruginosa isolation in Turkey and Europe.
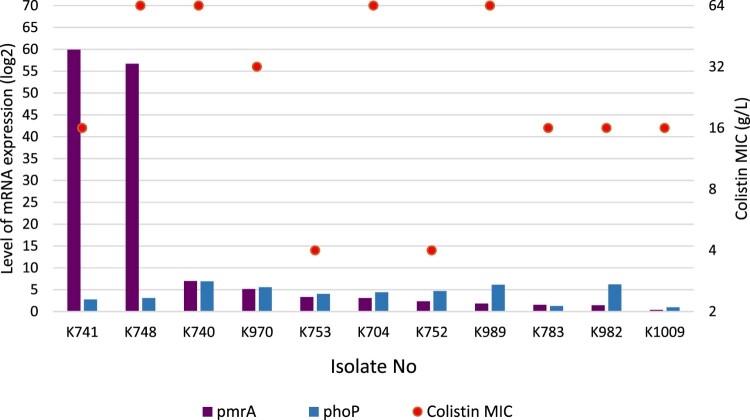
Colistin use is one of the major factors responsible for the development of colistin resistance. In our study, only four patients (36,3%) received colistin therapy before the isolation of P. aeruginosa (Table 1). The MICs for colistin were between 4 and >64 mg/L. In all isolates, sequences of pmrAB and phoPQ revealed numerous insertions and deletions. In the ten of them, pmrAB and phoPQ were found to be upregulated. Relative expressions of pmrA and phoQ genes were between 0.3–59.9-fold (mean 12.9-fold) and 0.9-6.9-fold (mean 4.15-fold), respectively. In nine isolates, colistin MICs and pmrAB-phoPQ expressions were found to be related (Figure 1). These results suggested that there could be additional mechanisms contributing to colistin resistance in P. aeruginosa.
Figure 1.
PmrA and PhoP expressions of the colistin-resistant P. aeruginosa in correlation with colistin MIC values.
In conclusion, colistin resistance is emerging in P. aeruginosa ST235 global high-risk clone. The co-existence of OXA-48 and NDM-1 genes in colistin-resistant P. aeruginosa ST235 high-risk clone indicates the spread of carbapenemases in clinical isolates and highlights need of continuous surveillance for high-risk clones of P. aeruginosa.
Acknowledgements
Koç University Institutional Review Board approved the study by the number of 2015.048.IRB1.008.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Moradali MF, Ghods S, Rehm BH.. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7(39):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver A, Mulet X, Lopez-Causape C, et al. . The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Update. 2015 Jul–Aug;21-22:41–59. [DOI] [PubMed] [Google Scholar]
- 3.European Centre of Diseases Prevention and Control. Surveillence of antimicrobial resistance in Europe . (2016). Anual Report of the European Antimicrobial Resistance Surveillence Network (EARS-NET). Stocholm: ECDC; 2016.
- 4.Aydin M, Ergonul O, Azap A, et al. . Rapid emergence of colistin resistance and its impact on fatality among healthcare-associated infections. J Hosp Infect. 2018 Mar;98(3):260–263. [DOI] [PubMed] [Google Scholar]
- 5.Wi YM, Choi JY, Lee JY, et al. . Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int J Antimicrob Agents. 2017 Jun;49(6):767–769. [DOI] [PubMed] [Google Scholar]
- 6.Lee JY, Ko KS.. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis. 2014 Mar;78(3):271–276. [DOI] [PubMed] [Google Scholar]
- 7.Muggeo A, Guillard T, Klein F, et al. . Spread of Klebsiella pneumoniae ST395 non-susceptible to carbapenems and resistant to fluoroquinolones in North-Eastern France. J Glob Antimicrob Re. 2018 Jun;13:98–103. [DOI] [PubMed] [Google Scholar]
- 8.Schwab F, Geffers C, Behnke M, et al. . ICU mortality following ICU-acquired primary bloodstream infections according to the type of pathogen: a prospective cohort study in 937 Germany ICUs (2006–2015). PLoS One. 2018;13(3):1–13. e0194210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandra S, Tseng KK, Arora A, et al. . The mortality burden of multidrug-resistant pathogens in India: a retrospective observational study. Clin Infect Dis. 2018;69(4):563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Recio R, Villa J, Viedma E, et al. . Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents. 2018 Aug;52(2):172–179. [DOI] [PubMed] [Google Scholar]
- 11.Mohamed SER, Alobied A, Hussien WM, et al. . blaOXA-48 carbapenem resistant Pseudomonas aeruginosa clinical isolates in Sudan. Journal of Advances in Microbiology. 2018;10(4):1–5. [Google Scholar]
- 12.Jovcic B, Lepsanovic Z, Suljagic V, et al. . Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob Agents Chemother. 2011 Aug;55(8):3929–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]