Abstract
Background:
Computer vision has promise in image-based cutaneous melanoma diagnosis but clinical utility is uncertain.
Objective:
To determine if computer algorithms from an international melanoma detection challenge can improve dermatologist melanoma diagnostic accuracy.
Methods:
Cross-sectional study using 150 dermoscopy images (50 melanomas, 50 nevi, 50 seborrheic keratoses) from the test dataset of a melanoma detection challenge, along with algorithm results from twenty-three teams. Eight dermatologists and nine dermatology residents classified dermoscopic lesion images in an online reader study and provided their confidence level.
Results:
The top-ranked computer algorithm had a ROC area of 0.87, which was higher than the dermatologists (0.74) and the residents (0.66) (p<0.001 for all comparisons). At the dermatologists’ overall sensitivity in classification of 76.0%, the algorithm had a superior specificity (85.0% vs. 72.6%, p=0.001). Imputation of computer algorithm classifications for dermatologist evaluations with low confidence ratings (26.6% of evaluations) increased dermatologist sensitivity from 76.0% to 80.8% and specificity from 72.6% to 72.8%.
Limitations:
Artificial study setting lacking the full spectrum of skin lesions as well as clinical metadata.
Conclusions:
Accumulating evidence suggests that deep neural networks can classify skin images of melanoma and its benign mimickers with high accuracy and potentially improve human performance.
Keywords: computer vision, machine learning, deep learning, automated melanoma diagnosis, melanoma, reader study, dermatologist, skin cancer, computer algorithm, International Skin Imaging Collaboration, International Symposium on Biomedical Imaging
Capsule Summary
The top-ranked computer algorithm from an international computer vision challenge more accurately classified 150 dermoscopy images of melanoma, nevi, and seborrheic keratoses than dermatologists or dermatology residents
When judiciously applied, use of computer algorithm predictions may improve dermatologist accuracy for melanoma diagnosis
Introduction:
Computer vision has promise in image-based cutaneous melanoma diagnosis.1–7 However, the lack of large public datasets of skin images has restricted the advancement of deep learning algorithms for skin cancer detection; to date, no algorithm has demonstrated clinical utility. The International Skin Imaging Collaboration (ISIC) aims to address these limitations by creating a public archive of images for education and research. Here, we describe results from our second international melanoma detection challenge, which was conducted at the 2017 International Symposium on Biomedical Imaging using dermoscopy images of melanoma and common benign mimickers [i.e., nevi and seborrheic keratoses (SK)]. We (a) compared the diagnostic accuracy of the top-ranked computer algorithm to the performance of dermatologists and residents in a reader study and (b) explored the diagnostic impact of substituting algorithm decisions for dermatologist classifications in instances where reader diagnostic confidence was low.
Methods:
IRB approval was obtained at Memorial Sloan Kettering and the study was conducted in accordance with the Helsinki Declaration. Details of the challenge tasks, evaluation criteria, timeline, and participation are published.8,9 We selected 2,750 high-quality dermoscopy images from the ISIC Archive: 521 (19%) melanomas, 1,843 (67%) melanocytic nevi, and 386 (14%) SK. Images were randomly allocated to training (n=2,000), validation (n=150), and test (n=600) datasets. Twenty-three algorithms were submitted to the melanoma classification challenge, and all used neural networks and deep learning, a form of machine learning that uses multiple processing layers to automatically identify increasingly abstract concepts present in data.10
Algorithms were ranked by area under the receiver operating characteristic (ROC) curve and we chose the top-ranked algorithm for analyses.8,9 A ROC curve is a graphical plot created by plotting sensitivity against the false positive rate (1-specificity) at various threshold settings. The area under the ROC curve is therefore a global measure of the ability of a test to classify whether a specific condition is present or not present; an area under the ROC curve of 0.5 represents a test with no discriminating ability (i.e., no better than chance alone) and an area under the ROC curve of 1.0 represents a test with perfect classification. A ROC curve can be used to determine an appropriate test cut-off but the selection of a test threshold depends on the purpose of the test and the trade-off between sensitivity and specificity in the intended clinical scenario.11
A reader study was performed using 150 images [50 melanomas (15 invasive, 20 in situ, 15 not otherwise specified), 50 nevi, 50 SK] randomly selected from the test set. The median (min-max) Breslow depth for the invasive melanomas was 0.3 (0.15–3.3) mm. Eight dermatologists who specialize in skin cancer diagnosis and management and ten dermatology residents agreed to participate in the study; after beginning evaluations, one resident did not complete the study and was removed. The mean (range) number of years of post-residency clinical experience and use of dermoscopy of the dermatologists was 14 (4–32) and 14.5 (7–28) years, respectively. The dermatologists originated from four countries [United States (n=4), Spain (n=2), Israel (n=1), and Colombia (n=1)] and all the dermatology residents were from the United States. Readers classified the lesions as melanoma, nevus, or SK, indicated a management decision (biopsy or observation), and reported diagnostic confidence on a Likert scale from 0 (extremely unconfident) to 6 (extremely confident). There were 1,200 total image evaluations performed by dermatologists and 1,350 by residents, respectively. Readers were blinded to diagnosis, clinical images, and metadata. There were no time restrictions and participants could complete evaluations over multiple sittings. For comparisons with human readers, algorithm performance metrics were calculated on the same 150 lesions from the reader study.
Descriptive statistics were used to explore the distributions of reader and algorithm results by lesion diagnostic classification and reader confidence. Summary measures of diagnostic accuracy were estimated for lesion classification and management for readers. Two sample tests for proportions were used to assess differences in diagnostic accuracy measures between sample subgroups. Where applicable, variance estimates were inflated to address clustering of responses within readers. Algorithm diagnostic accuracy was assessed for lesion classification. ROC curves were calculated for algorithms, reader, and reader subgroups. Comparisons of ROC area between algorithms and human readers used a non-parametric approach.12,13
Reader results were imputed with algorithm responses when reader confidence in classification of the lesion was low (confidence classification: 0–3). This was accomplished by dichotomizing the algorithm using a pre-determined sensitivity threshold of 90%. After imputation, diagnostic accuracy measures were recalculated. The alpha level for analyses was 5% and tests were two-sided. Analyses were performed using Stata v.14.2, Stata Corporation, College Station, TX.
Results:
The overall sensitivity, specificity, and ROC area of the dermatologists for melanoma classification was 76.0% (95% CI:71.5–80.1), 72.6% (95% CI:69.4–75.7) and 0.74 (95% CI:0.72–0.77), respectively. The overall sensitivity, specificity, and ROC area of the residents for melanoma classification was 56.0% (95% CI:51.3–60.6), 76.3% (95% CI:73.4–79.1) and 0.66 (95% CI:0.6–0.69), respectively. The ROC area of the top-ranked algorithm in melanoma classification was 0.8685 (Figure 1), which was greater than the overall ROC areas in classification and management of 0.74 and 0.70 for the dermatologists and 0.66 and 0.67 for the residents (p<0.001 for all comparisons).
Figure 1. Diagnostic accuracy of the top-ranked algorithm, dermatologists, and residents for melanoma on the 150-image dataset.
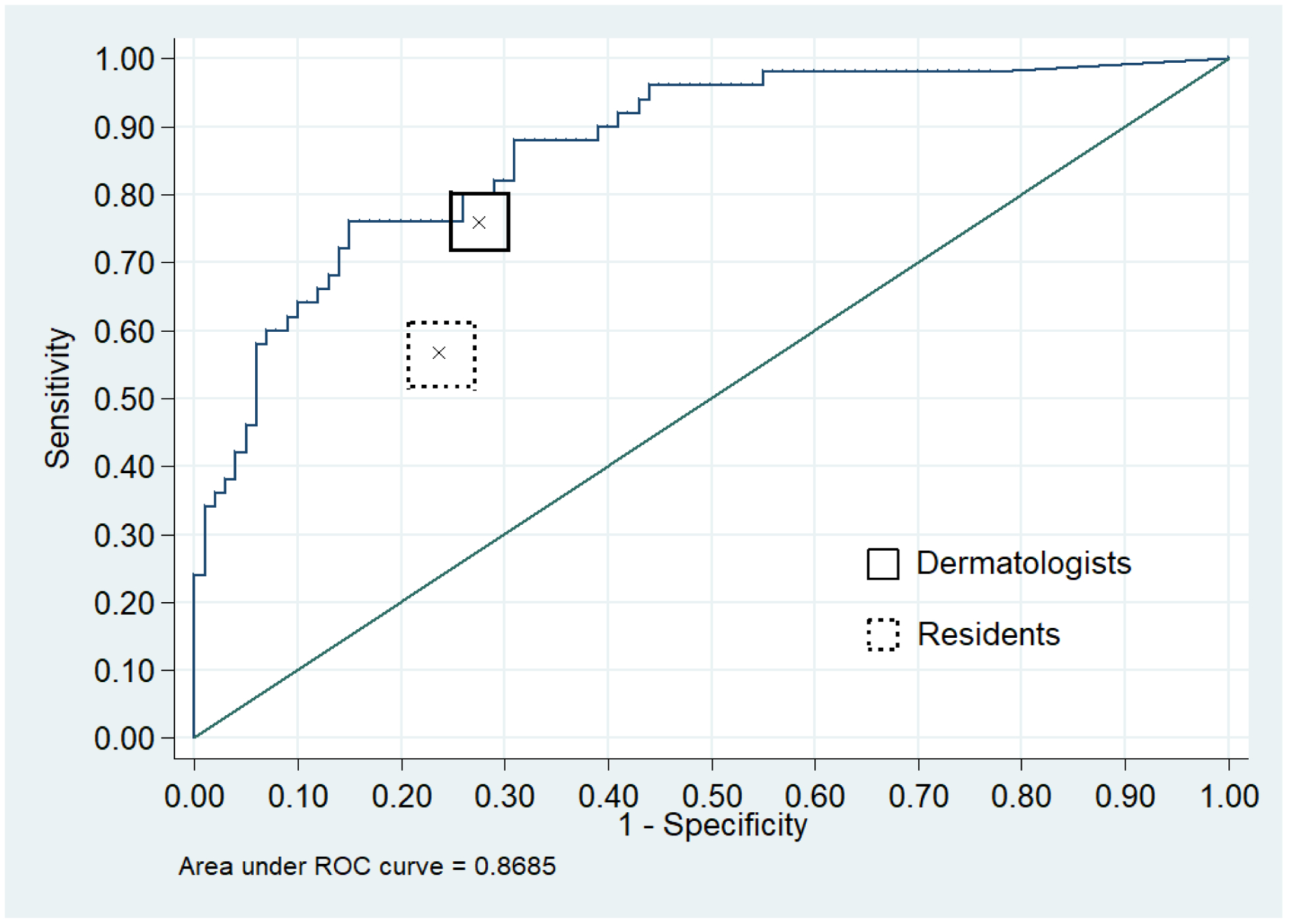
Receiver operating characteristic curve demonstrating sensitivity and specificity for melanoma of the top-ranked algorithm from the 2017 ISIC melanoma detection challenge (blue curve). Solid black box indicates the overall performance of 8 dermatologists (center “x”) along with 95% confidence band (rectangular bounding box). Dashed gray box indicates the overall performance of 9 residents (center “x”) along with 95% confidence intervals (rectangular bounding box).
At the dermatologists’ overall sensitivity in classification of 76.0%, the computer algorithm had a specificity of 85.0%, which was higher than the dermatologists’ specificity of 72.6% (p=0.001). At the dermatologists’ overall sensitivity in management of 89.0%, the algorithm specificity was 61%, which was higher than the dermatologists’ specificity of 51.1% (p=0.02).
To explore the feasibility of algorithms aiding lesion classification, we imputed algorithm classifications for reader evaluations with low confidence scores (range 0–3), constituting 51% of resident and 26.6% of dermatologist evaluations, respectively. After imputation, sensitivity of resident evaluations increased from 56.0% to 72.9%, with a decrease in specificity from 76.3% to 72.6%. The proportion of the 1,350 evaluations correctly classified by residents increased from 69.4% (n=939) to 72.6% (n=981). The sensitivity of dermatologist classifications increased from 76.0% to 80.8% and the specificity increased from 72.6% to 72.8%. The proportion of evaluations correctly classified by dermatologists increased from 73.8% (n=885) to 75.4% (n=905).
Discussion:
These results and others2–5 demonstrate that deep neural networks can classify skin images of melanoma with high accuracy. Compared to our 2016 challenge,1 we observed an increase in the relative diagnostic performance of the top-ranking algorithm compared to the same eight dermatologist readers. This suggests that the performance of algorithms is improving, possibly due to availability of larger training datasets or advances in algorithm development.
Although studies have demonstrated that algorithms can identify melanoma with diagnostic accuracy superior to dermatologists in reader studies, their clinical applicability remains uncertain. To examine the feasibility of an algorithm augmenting physician performance, we imputed algorithm classifications for lesions in which the physician reported low diagnostic confidence. We hypothesized that this would represent the most likely circumstance in which a physician would seek/use diagnostic help in a clinical setting. In this analysis, we found that the sensitivity and overall proportion of correct responses by readers increased by imputing algorithm classifications. Further studies are required to determine the optimal algorithm thresholds that would benefit physicians in a range of clinical settings and scenarios.
There are notable limitations to our study.1 Our test dataset did not include the full spectrum of skin lesions, particularly banal lesions and less common presentations of melanoma, and the setting was artificial as physicians did not have access to data used when evaluating patients (e.g., age, personal/family history of melanoma, lesion symptoms). We did not perform external validity analyses, which are important for demonstrating algorithm generalizability.14 Comparisons of skin cancer diagnostic accuracy of dermatologists and computer algorithms through reader studies should be cautiously interpreted. One device approved by the US Food and Drug Administration that used multispectral digital skin lesion analysis had been shown to have high melanoma sensitivity15 and to improve both the sensitivity and specificity of dermatologists after clinical and dermoscopic examination of suspicious skin lesions via reader studies16; despite these apparent strengths, the device was discontinued in 2017.
Unlike other studies2–5 examining the diagnostic accuracy of automated systems for skin cancer diagnosis, our study used a dataset that is publicly available for external use and future benchmarking. We further compared dermatologist accuracy to the top-ranked algorithm from a computer vision challenge, suggesting that the performance of the classifier is reflective of the current state-of-the-art in machine learning. Our annual ISIC melanoma detection challenges17 are the largest comparative studies of computerized skin cancer diagnosis to date and have attracted global participation. As our ISIC image archive expands, we anticipate hosting continuous public challenges with larger and more varied datasets with clinically relevant metadata.
In conclusion, the top-ranked algorithm from an international melanoma detection challenge exceeded the diagnostic accuracy of both dermatologists and residents in an artificial study setting. The sensitivity and overall proportion of correct evaluations by readers improved when imputing algorithm classifications for lesions in which the physician reported low diagnostic confidence, suggesting that augmented human classification is feasible. Future studies demonstrating clinical utility in a real-world setting are needed.
Table 1.
Measures of diagnostic accuracy for lesion classification by reported confidence in the diagnosis.
| Residents | |||||
|---|---|---|---|---|---|
| Confidence Level | Freq. (%) | Sensitivity (95% CI) | ptrend | Specificity (95% CI) | ptrend |
| 0 | 7 (0.5) | 100.0 (2.5–100.0) | 0.54 | 16.7 (0.4–64.1) | <0.001 |
| 1 | 160 (11.8) | 57.6 (44.1–70.4) | 61.4 (51.2–70.9) | ||
| 2 | 238 (17.6) | 48.6 (36.9–60.6) | 73.2 (65.7–79.8) | ||
| 3 | 289 (21.4) | 53.6 (43.2–63.8) | 70.3 (63.3–76.7) | ||
| 4 | 397 (29.4) | 51.8 (43.1–60.4) | 81.9 (76.7–86.4) | ||
| 5 | 204 (15.1) | 63.1 (50.2–74.7) | 87.8 (81.1–92.7) | ||
| 6 | 55 (4.1) | 100.0 (80.5–100.0) | 89.5 (75.2–97.1) | ||
| Dermatologists | |||||
| Confidence Level | Freq. | Sensitivity (95% CI) | ptrend | Specificity (95% CI) | ptrend |
| 0 | 26 (2.2) | 75.0 (34.9–96.8) | 0.002 | 61.1 (35.7–82.7) | <0.001 |
| 1 | 65 (5.4) | 62.5 (40.6–81.2) | 68.3 (51.9–81.9) | ||
| 2 | 97 (8.1) | 52.0 (31.3–69.8) | 58.3 (46.1–69.8) | ||
| 3 | 131 (10.9) | 67.3 (52.9–79.7) | 63.3 (51.7–73.9) | ||
| 4 | 301 (25.1) | 74.3 (64.8–82.3) | 64.8 (57.7–71.5) | ||
| 5 | 342 (28.5) | 79.5 (70.8–86.5) | 76.1 (70.0–81.4) | ||
| 6 | 238 (19.8) | 91.9 (83.2–97.0) | 90.2 (84.6–94.3) | ||
Readers reported a mean confidence of 3.7 (SD=1.51). Dermatologists had higher confidence than residents (4.2 vs. 3.3, p<0.001).
Acknowledgements:
This study reports the efforts of the International Skin Imaging Collaboration (ISIC). We thank the leadership of the ISIC collaboration: H. Peter Soyer, MD, Dermatology Research Centre, The University of Queensland, Brisbane, Australia (Technique Working Group Co-Leader); Clara Curiel-Lewandrowski, MD, University of Arizona Cancer Center, Tucson, AZ, USA (Technique Working Group Co-Leader); Harald Kittler, MD, Department of Dermatology, Medical University of Vienna, Austria (Terminology Working Group Leader); Liam Caffery, PhD, The University of Queensland, Brisbane, Australia (Metadata Working Group Leader); Josep Malvehy, MD; Hospital Clinic of Barcelona, Spain (Technology Working Group Leader); Rainer Hofmann Wellenhof, MD, Medical University of Graz, Austria (Archive Group Leader).
The authors also thank the organizing committee of the 2017 International Symposium on Biomedical Imaging (ISBI), the chairs of the 2017 ISBI Grand Challenges: Bram van Ginneken, Radboud University Medical Center, NL; Adriënne Mendrik, Utrecht University, NL; Stephen Aylward, Kitware Inc., USA, the participants of the 2017 ISBI Challenge “Skin Lesion Analysis towards Melanoma Detection, and the participants of the reader study: Cristina Carrera, MD, PhD; Jennifer L. DeFazio, MD; Natalia Jaimes, MD; Ashfaq A. Marghoob, MD; Elizabeth Quigley, MD; Alon Scope, MD; Oriol Yelamos, MD; Allan C. Halpern, MD; Caren Waintraub, MD; Meryl Rosen, MD; Sarah Jawed, MD; Priyanka Gumaste, MD; Miriam R. Lieberman, MD; Silvia Mancebo, MD; Christine Totri, MD; Corey Georgesen, MD; Freya Van Driessche, MD; Maira Fonseca, MD.
Funding/Support: This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
Financial Disclosure of the Authors: Dr. Codella is an employee of IBM and an IBM stockholder. Dr. Halpern is a consultant for Canfield Scientific Inc, Caliber I.D., and SciBase. Drs. Marchetti, Liopyris, Kalloo, Gutman, Helba, and Dusza have no financial disclosures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None declared.
Statement on prior presentation: Preliminary study data was presented at the World Congress of Melanoma in Brisbane, Australia in October 2017.
IRB Statement: This research received IRB approval at Memorial Sloan Kettering Cancer Center.
References
- 1.Marchetti MA, Codella NCF, Dusza SW, et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(2):270–277.e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tschandl P, Rosendahl C, Akay BN, et al. Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks. JAMA Dermatol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8):1836–1842. [DOI] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J Invest Dermatol. 2018;138(7):1529–1538. [DOI] [PubMed] [Google Scholar]
- 6.Codella N, Lin CC, Halpern A, Hind M, Feris R, Smith JR. Collaborative Human-AI (CHAI): Evidence-Based Interpretable Melanoma Classification in Dermoscopic Images. 2018; https://arxiv.org/pdf/1805.12234v3.pdf. Accessed December 7, 2018.
- 7.Codella N, Nguyen QB, Pankanti S, et al. Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images. IBM Journal of Research and Development. 2017;61(4/5). [Google Scholar]
- 8.ISIC 2017: Skin Lesion Analysis Towards Melanoma Detection. https://challenge.kitware.com/#challenge/n/ISIC_2017%3A_Skin_Lesion_Analysis_Towards_Melanoma_Detection. Accessed December 21, 2016.
- 9.Codella NCF, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, Halpern A. . Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). 2017; https://arxiv.org/pdf/1710.05006.pdf. Accessed December 12, 2018.
- 10.Suzuki K Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10(3):257–273. [DOI] [PubMed] [Google Scholar]
- 11.Hoo ZH, Candlish J, Teare D. What is an ROC curve? Emerg Med J. 2017;34(6):357–359. [DOI] [PubMed] [Google Scholar]
- 12.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 13.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. New York: Oxford University Press; 2003. [Google Scholar]
- 14.Navarrete-Dechent C, Dusza SW, Liopyris K, Marghoob AA, Halpern AC, Marchetti MA. Automated Dermatological Diagnosis: Hype or Reality? J Invest Dermatol. 2018;138(10):2277–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monheit G, Cognetta AB, Ferris L, et al. The performance of MelaFind: a prospective multicenter study. Arch Dermatol. 2011;147(2):188–194. [DOI] [PubMed] [Google Scholar]
- 16.Farberg AS, Glazer AM, Winkelmann RR, Tucker N, White R, Rigel DS. Enhanced melanoma diagnosis with multispectral digital skin lesion analysis. Cutis. 2018;101(5):338–340. [PubMed] [Google Scholar]
- 17.ISIC Melanoma Challenges. https://www.isic-archive.com/#!/topWithHeader/tightContentTop/challenges. Accessed January 11, 2019.


