Abstract
Variation and differences of MRSA transmission within and between healthcare settings are not well understood. This variability is critical for understanding the potential impact of infection control interventions and could aid in the evaluation of future intervention strategies. We fit a Bayesian transmission model to detailed individual-level MRSA surveillance data from over 230 Veterans Affairs (VA) hospitals and nursing homes. Our approach disentangles the effects of potential confounders, including length of stay, admission prevalence, and clearance, estimating dynamic transmission model parameters and temporal trends. The median baseline transmission rate in hospitals was approximately four-fold higher than in nursing homes, and declined in 46% of hospitals and 9% of nursing homes, resulting in a median transmission rate reduction of 43% across hospitals and an increase of 2% in nursing homes. For first admissions into an acute care facility, the median (range) importation probability was 10.5% (5.9%–18.4%), and was nearly twice as large, 18.7% (9.2%–37.4%), in nursing homes. This analysis found differences within and between hospitals and nursing homes. The transmission rate declined substantially in hospitals and remained stable in nursing homes, while admission prevalence was considerably higher in nursing homes than in hospitals.
Keywords: MRSA, Transmission dynamic trends, Hospital, Nursing home, Veterans affairs
1. Introduction
Antibiotic-resistant pathogens are major causes of morbidity and mortality in healthcare settings, where Methicillin-resistant Staphylococcus aureus (MRSA), first identified in the 1960s and common in hospitals by the 1980s (McDonald, 2006; Panlilio et al., 1992), increased in prevalence (McDonald, 2006; Klevens et al., 2006) through 2005. Since then, rates of hospital associated MRSA infections and colonization have been on the decline across the United States (Dantes et al., 2013; Landrum et al., 2012).
As with national trends, rates of MRSA healthcare-associated infections in Department of Veterans Affairs (VA) hospitals have declined since late 2007. Some attribute this decline to VA’s MRSA Prevention Initiative which comprised universal nasal surveillance for MRSA colonization, contact precautions for patients who were identified carriers of MRSA, an emphasis on hand hygiene, and an institutional culture change which placed the responsibility of infection control on everyone who had contact with patients. The MRSA Prevention Initiative launched nationally in hospitals October 1, 2007 (Jain et al., 2011), and extended it to all VA nursing homes on January 1, 2009 (Evans et al., 2014). Although VA MRSA infection rates dropped following VA’s MRSA Prevention Initiative, experts debate the extent to which the MRSA Prevention Initiative contributed to those rate reductions (Gurieva et al., 2012; Jones et al., 2014; Lawes and Gould, 2012). This disagreement has prompted further debates about the role of mandated contact precautions (Morgan et al., 2017; Rubin et al., 2018).
The epidemiology of MRSA infection has been studied across a broad range of healthcare settings, yet we have limited understanding of the heterogeneity in MRSA transmission (Kwok et al., 2018). Understanding variation in MRSA transmission is critical for identifying facility and patient characteristics that contribute to transmission, and for informing future interventions to control antibiotic resistant organisms. We conducted an epidemiological analysis of MRSA transmission in more than 230 facilities including acute care and nursing homes. The principal aims of this analysis were to better understand variation in MRSA transmission, within and between healthcare facility types, and to identify trends in the transmission rate immediately following implementation of the VA’s MRSA Prevention Initiative. To our knowledge, this analysis, based on a mechanistic transmission model, represents the largest such analysis to date.
Previous analyses of the MRSA Prevention Initiative focused on MRSA healthcare associated infections but neglected to investigate transmission or account for admission prevalence, length of stay, and decolonization (clearance); important factors underlying the epidemiology of MRSA in healthcare facilities. To provide new and more comprehensive insights into MRSA transmission in VA, we disentangle the effects of length of stay, admission prevalence, transmission rate, clearance rate, and imperfect testing to better understand the variability in MRSA transmission dynamics in VA hospitals and nursing homes. We also estimate trends in the per-capita transmission rate to estimate changes in transmission following implementation of the MRSA Prevention Initiative.
2. Methods
2.1. Data
We analyzed electronic health record data from individuals admitted to a VA hospital between October 1, 2007 (when VA launched the initiative in its hospitals) and July 1, 2011; or to a VA nursing home from January 1, 2009 (when VA launched the initiative began in its nursing homes) and December 31, 2010. The different times at which the MRSA Prevention Initiative was implemented in hospitals and nursing homes, together with the completeness of the MRSA surveillance data available in our study cohort led to differing follow-up times between hospitals and nursing homes. Data included facility and patient identifier, date and time of admission and discharge, and MRSA surveillance test results.
There were 145 hospitals with patient data during the study period. Of these, we excluded 7 hospitals that had a mean length of stay > 20 days, 6 additional hospitals with less than 1% of patients having at least one surveillance test, and 3 more hospitals having a mean daily census of under 3 patients per day. Lastly, there were 7 hospitals with incomplete data which were excluded from this analysis, resulting in a total of 23 (16%) hospitals excluded leaving 122 hospitals for this analysis. From 136 nursing homes with patient data during the study period, we excluded 2 nursing homes having mean length of stay <10 days, and 23 nursing homes with incomplete data, resulting in 25 (18%) nursing homes excluded leaving 111 nursing homes in this analysis.
2.2. Model
We extended our recently published dynamic transmission models (Thomas et al., 2015; Khader et al., 2016) to allow for changes in patients’ colonization status, and temporal trends in transmission, and to estimate key epidemiologic parameters. This extended model allowed for patients who acquired and lost colonization with MRSA both during hospitalizations and between consecutive admissions. We assumed there were no false positive surveillance tests, but false negatives were represented by probabilistic uncertainty on the patients’ colonization status at the time of the negative test.
The key parameters (Table 1) include false negative probability, clearance rates both during admissions and between consecutive admissions, and intercept and slope for the log-transformed transmission rate. The intercept parameter corresponds with the baseline transmission rate (at the beginning of the study) while the slope indicates the temporal trend in the transmission rate over time. The in-situ probability is the probability that a patient is colonized at the beginning of the study period, which results from either an acquisition or an importation (MRSA colonized at the time of admission) prior to the beginning of the study. Below, we describe the implementation and the structure of the model in more detail.
Table 1.
Transmission Model Parameters and Markov Chain Monte Carlo Sampling Distribution.
| Parameters | Description | MCMC Update * | Prior distribution |
|---|---|---|---|
| σ | In-situ probability | Gibbs | Beta |
| ν | First-admission importation probability | Metropolis-Hastings | Log-Normal |
| γ | Out-of-unit clearance rate | Metropolis-Hastings | Gamma |
| λI | Transmission rate log-linear intercept | Metropolis-Hastings | Normal |
| λS | Transmission rate log-linear slope | Metropolis-Hastings | Normal |
| η | In-unit clearance rate | Gibbs | Gamma |
| Φ | Surveillance test false negative | Gibbs | Beta |
MCMC = Markov chain Monte Carlo.
The model was implemented within a Bayesian framework, depending on an augmented data set, D, which combined both observed data (i.e. admission, discharge and surveillance test times and results) and unobserved data (i.e. times of acquisition and clearance). We assumed that D was organized as a list of events ordered by their corresponding times, and we represented model parameters by θ. The likelihood of the data, D, given the model parameters is given by
| (1) |
We define te and te− as the times of the events e, and the event prior, e−. The term g(e; θ) represents the contribution to the likelihood of event type e, while h (te, te−; θ) is the probability that no event is observed between times te and te−, given by the formula
| (2) |
The number of uncolonized and colonized individuals at time t are given by Ut and Ct, and Nt = Ut + Ct is the total number of patients at time t. The transmission rate parameter at time t is given by λ (t), and η denotes the constant rate of clearance for colonized patients in the facility.
We assumed that acquisitions in the hospital at time t per uncolonized individual occur at a rate of λ(t)·Ct/Nt, corresponding to a frequency-dependent transmission model. Since we were interested in estimating temporal trends in the transmission rate parameter, and because we require λ(t) ≥ 0, we assumed that λ(t) = exp(λI + λSt), where λI and λS are the intercept and slope respectively of the log-transmission rate. Consequently, the contribution of an acquisition event e to the likelihood is given by g(e; θ) = λ(te)·Cte−/Nte−.
At the time of admission into the facility, patients were assumed to be either colonized or uncolonized. The probability of colonization at admission, or importation, depended on whether the admission was a first-admission, in which case the probability was represented by the first-admission importation probability, or whether the patient had been previously admitted. Colonization status between consecutive admissions was modeled as a two-state continuous-time Markov chain (CTMC). In particular, uncolonized patients were colonized at rate α and colonized individuals lost colonization at rate γ. From these two parameters, we defined first-admission importation probability as the steady-state fraction of individuals who were colonized between consecutive admissions ν = α/(α + γ).
We explicitly modeled first-admission importation ν and outpatient clearance γ, from which estimates of α were uniquely determined. Therefore, for a first-admission event, the contribution to the likelihood was given by
| (3) |
where s(e) indicates that the patient corresponding with event e was colonized at time te. For readmissions, the importation probability depended on patients’ colonization status at prior discharge, denoted ed, and the time since prior discharge Δt = te – ted. Computing the probability of importation for readmissions required computing transition probabilities for the out-of-facility CTMC, which resulted in the contribution of a readmission to the likelihood given by
| (4) |
The contribution to the likelihood for admission, discharge, surveillance test results and the in-situ event are summarized in Table 2.
Table 2.
Contribution of Event-Specific Probabilities to the Transmission Model Likelihood.
| Event | Contribution to likelihood - g(e; θ) |
|---|---|
| In-situ | σs(e)(1 – σ)1–s(e) |
| Clearance | η |
| Negative surveillance test | Φ s(e) |
| Positive surveillance test | 1 – Φs(e) |
| Admission and discharge | 1 |
2.3. MCMC updates
Parameters and augmented data were updated using Markov chain Monte Carlo (MCMC). Estimation within each iteration of the MCMC consisted of generating a new sample of both the augmented data and the model parameters.
To sample the augmented data, we sequentially stepped through each patient and proposed an initial colonization status and a sequence of times that the patient switched between being colonized and uncolonized. The proposal for the new augmented data depended on the transmission rates, clearance rates, and probability of importation. The newly proposed augmented data was then accepted according to standard Metropolis acceptance probability (Hastings, 1970). Once the augmented data were updated, parameter values were proposed conditional on the observed data and the new augmented data. The parameter updates were accomplished via both Gibbs (Geman and Geman, 1984) and Metropolis-Hastings samples (Table 1) and updated sequentially. The process of updating the augmented data and parameter values was iterated and resulted in the posterior distribution for the parameters, which formed the basis for the analysis.
2.4. Analysis
We computed facility-specific estimates of posterior means for the model parameters, ranges and quartiles (Q1 – Q3), as well as 95% credible intervals (95% CI), a Bayesian analogue to the confidence interval. Additionally, we reported median, quartiles and range for parameter estimates across the facilities. In addition to the parameter estimates obtained from the dynamic transmission model, we derived empirical estimates of admission and discharge prevalence to compare with the corresponding model-derived estimates. We defined empirical estimates of admission (discharge) prevalence as the percentage of admission (discharge) surveillance tests that were positive, where an admission (discharge) surveillance test occurred within 1 day of admission (discharge). In contrast to empirical estimates of prevalence, model-based estimates of prevalence were based on the estimated proportion of individuals colonized according to the underlying transmission model, and accounted for the imperfect nature of surveillance tests.
3. Results
3.1. Facility patient summaries
We analyzed 122 VA hospitals with 971,412 patients having 1,857,328 admissions and 3,525,837 MRSA surveillance tests, resulting in approximately 1.9 surveillance tests per admission. Additionally, we analyzed 111 nursing homes with 63,838 residents having 95,549 admissions and 188,425 MRSA surveillance tests corresponding with approximately 2.0 surveillance tests per admission. The median length of stay across all facilities was 5.1 days in hospitals and 45 days in nursing homes, and a high proportion of admission and discharge surveillance samples were taken from all admissions (Table 3). The median time between consecutive tests in hospitals was 2.5 days during an admission and 72 days between consecutive admissions (from discharge to readmission). In nursing homes, the median time between consecutive tests was 17 days during an admission and 10 days between consecutive admissions. The facilities included in this analysis represent a geographically diverse collection of facilities across the US.
Table 3.
Summaries of Hospitals, Nursing Homes and Admissions from Veterans Affairs Facilities in the United States.
| Factor | Acute Care | Long-term care |
|---|---|---|
| Mean LOS (days) median (Q1-Q3) | 5.1 (4.3–6.0) | 45 (38–63) |
| Mean # patients median (Q1-Q3) | 62 (29–99) | 71 (50–104) |
| Fraction admission tests median (Q1-Q3) | 0.91 (0.86–0.94) | 0.80 (0.71–0.86) |
| Fraction discharge tests median (Q1-Q3) | 0.85 (0.76–0.89) | 0.76 (0.66–0.83) |
| Tests per admission median (Q1-Q3) | 1.9 (1.8–2.1) | 2.0 (1.8–2.1) |
| Percent of admissions that are readmissions median (Q1-Q3) | 46.7% (42.5%–49.6%) | 31.7% (26.3%–37.3%) |
| Region no. (%) | 122 (100%) | 111 (100%) |
| West | 23 (19%) | 19 (17%) |
| Midwest | 30 (25%) | 27 (24%) |
| South | 48 (39%) | 41 (37%) |
| Northeast | 21 (17%) | 24 (22%) |
3.2. Transmission
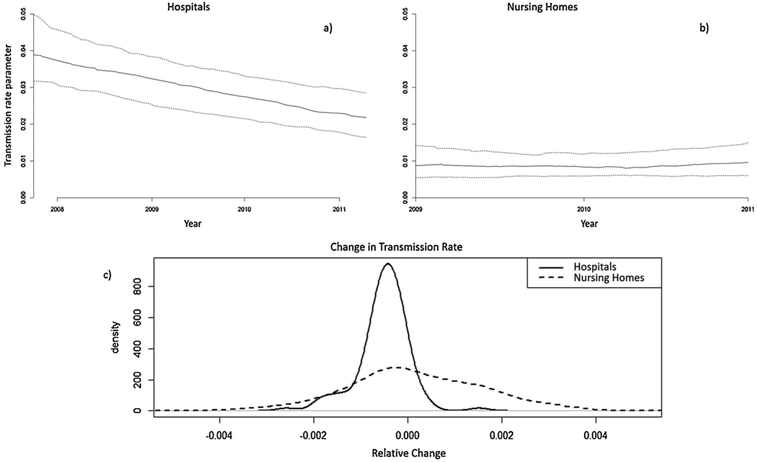
The median baseline transmission rate, or transmission rate at the beginning of the study in hospitals was 0.039 per day (Q1 – Q3; 0.032 – 0.050) and declined to 0.022 (0.016 – 0.029) at the end of the study period, a reduction of 43.1% during approximately 3.5 years (Fig. 1a). The majority of hospitals demonstrated a decreasing transmission rate parameter (Fig. 1c); the 95% CI of the declining temporal trend term did not include zero in 59 hospitals (46%). In two (1.7%) facilities, the temporal trend term was increasing, with the lower bound of the 95% credible interval above zero.
Fig. 1.
The top panel shows the estimated temporal trend of the median (—quartiles) for the transmission rate parameter in a) hospitals and b) nursing homes during the study period, The bottom panel shows c) the distribution of the estimated relative change in transmission in hospitals and nursing homes.
Among nursing homes, the median baseline transmission rate was 0.009 per day (Q1 – Q3; 0.005 – 0.014) and remained stable at 0.010 (0.006 – 0.015) after 2 years (Fig. 1b). The temporal trend parameter for nursing homes was centered on zero, with a much flatter distribution than for hospitals. Inspection of the 95% credible intervals demonstrated that 10 (8.5%) had decreasing transmission and 7 (6.0%) increasing transmission.
3.3. Prevalence
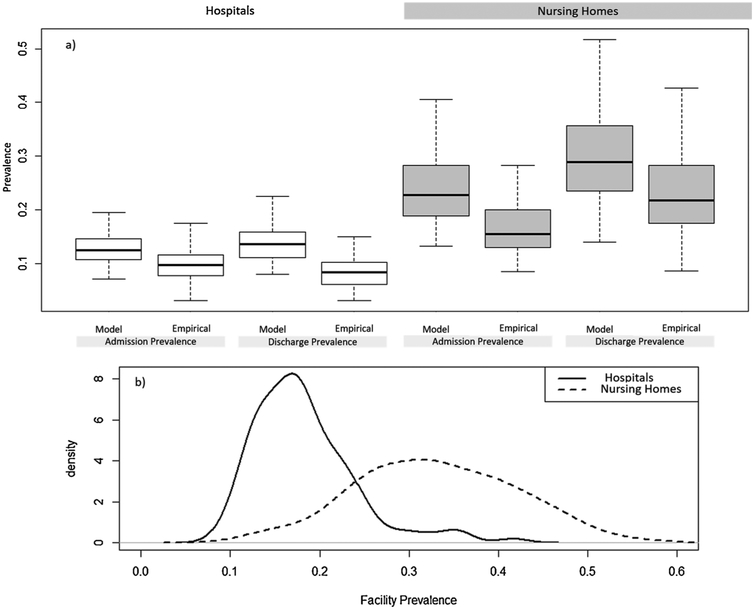
All prevalence measures were substantially higher in nursing homes than in hospitals. The first-time admission prevalence, our “importation” parameter, had a median (range) of 10.5% (5.9%–18.4%) in hospitals and 18.7% (9.2%–37.4%) in nursing homes. Admission prevalence, which includes readmissions, was 12.5% (range: 7.1%–21.9%) in hospitals and 22.8% (13.2%–40.5%) in nursing homes. The average point prevalence was particularly high in nursing homes relative to hospitals, and exhibited a broader distribution (Fig. 2b); median point prevalence (range) was 33.0% (12.0%–56.1%) and 17.1% (9.2%–41.6%), respectively, in the two types of facilities.
Fig. 2.
Shows a) comparison of distributions for admission prevalence and discharge prevalence in hospitals, and nursing homes, shaded gray. Comparison is between those estimates derived from the Bayesian transmission model and those estimated empirically, based on the proportion of positive admission or discharge surveillance tests, and b) the variation and differences in the distribution of facility prevalence within and between hospitals and nursing homes.
Model based estimates of admission prevalence and discharge prevalence were considerably higher than empirical estimates of admission and discharge prevalence in both hospitals and nursing homes (Fig. 2a). The relative difference between empirical and model-based estimates ranged from 18% to 36%.
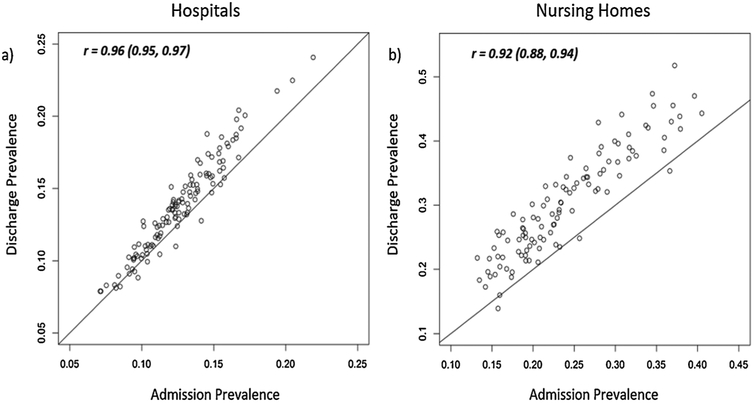
Admission prevalence was highly predictive of discharge prevalence in both hospitals and nursing homes (Figs. 3a and 3b; correlation coefficients were 0.96 and 0.92, respectively). The mean difference between discharge prevalence and admission prevalence was higher in nursing homes than in hospitals: 6.1% and 1.1% respectively.
Fig. 3.
Scatterplots indicate the strength of the relationship between model-based estimates of admission prevalence and discharge prevalence in a) hospitals, and b) nursing homes. Model-based prevalence estimates are the estimated proportion of patients (residents) who are colonized at admission and discharge.
3.4. Clearance rates and false negative
The median (Q1-Q3) of the estimated mean time until loss of colonization (inverse of the clearance rate) was 40.1 (30.1–54.5) days during an acute care facility hospitalization and 274.0 (237.7–330.0) days during the period between consecutive stays. For long-term care facility residents, median estimated time to clearance was 215.4 (142.0–279.1) days during their stay, and 108.1 (44.3–191.0) days in the period between consecutive stays. We estimated the median (Q1-Q3) false negative probability to be 23.5% (19.5%–26.6%) among hospitals and 21.8% (16.6%–25.9%) among nursing homes.
4. Discussion
We fit Bayesian models to VA health system data to estimate the key mechanistic parameters that govern the epidemiology of resistant bacteria in healthcare settings. Our findings demonstrate that transmission dynamics in nursing homes and hospitals are distinctly different. The transmission rate parameter, which represents the linear relationship between colonization prevalence and rate of new colonizations, was substantially lower in nursing homes than in hospitals. Stated a different way, MRSA cases in nursing homes were less infectious per day than those in hospitals, a finding that has potential implications for design and implementation of control policies (Slayton et al., 2015). Interventions that address the cumulative risk of transmission across longer time scales may be particularly beneficial in nursing homes. Heterogeneity in the transmission rate parameter across facilities may also serve as a useful facility-level surrogate for quality of infection control practices. However, the transmission rate parameter may also be affected by patient factors, such as antibiotic use, that influence susceptibility of acquisition as well as bacterial shedding.
One of the key goals of this study was to improve understanding of the impact of VA’s MRSA prevention initiative. Our results demonstrate that the median transmission rate parameter progressively declined in VA hospitals but not in VA nursing homes. Moreover, within the population of hospitals, the temporal trends in transmission rate parameters were highly heterogeneous. Further studies to identify which facility characteristics predict transmission trends over time may shed light on the factors that influence success of the MRSA prevention initiative.
Another finding was that the estimated MRSA clearance rate was much lower in nursing homes than in hospitals. The apparent reduction in rate of MRSA clearance in nursing homes may partially be due to the decreased intensity of exposure to anti-MRSA antibiotics (e.g., vancomycin) compared to hospitals (unpublished data). In order to estimate clearance, collection of surveillance tests after a positive test is necessary. In general, since the model denotes that negative tests after a positive test represent either clearance or a false negative, the higher the fraction of such tests that are positive, the lower the estimated clearance rate. It is important to acknowledge that the effect of antibiotics may be to suppress positive test results, without necessarily eradicating MRSA, a difference that our model cannot distinguish.
Admission prevalence, discharge prevalence, and average prevalence are three types of prevalence that are relevant to healthcare epidemiology. Typically, prevalence is estimated directly from the proportion of surveillance tests that are positive, within a time window defined according to the type of prevalence (e.g., for admission prevalence, tests collected within 24 h of admission). Because of false negativity, this approach yields an underestimate of true prevalence. Bias may also arise because of sample selection and test timing. For example, a test obtained 24 h after admission may not reflect true admission status; similarly, tests obtained prior to discharge may not reflect true discharge status.
Our methods addressed these biases in estimation of these quantities and again demonstrated important distinctions between hospitals and nursing homes. In both types of facilities, admission prevalence was a very strong predictor of discharge prevalence. However, admission prevalence was much higher in nursing homes than hospitals. We note that a much higher proportion of long-term care facility patients had prior healthcare facility exposure. Moreover, the difference between discharge prevalence and admission prevalence was substantially higher in nursing homes. The combination of high admission prevalence, reduced clearance, and increased length of stay elevated MRSA prevalence in nursing homes relative to hospitals, compensating for the comparatively lower transmission rate parameter.
Our study design and analytical approach had notable strengths. Fitting dynamic statistical models to MRSA surveillance data make it feasible to explore epidemiological mechanisms that influence disease spread. A key limitation was the simplifying assumption that there were only two infection states, namely colonized and uncolonized. This assumption meant we were unable to differentiate between patients who were truly uncolonized, and those who were exposed or colonized but undetectable due to low-level colonization. An additional limitation is that we were unable to explicitly incorporate information on adherence to contact precautions, antimicrobial exposure and other factors that may have influenced transmission. For example, adherence to contact precautions may be correlated with surveillance test compliance. If so, less compliant facilities could have implemented contact precautions with lower adherence. Additionally, adherence to contact precautions could have changed over time during the study, either improving or waning, due to an initial resistance to the policy or an increasing complacency over time. Not explicitly accounting for adherence to contact precautions could lead to some bias. Finally, both the VA patient population and the VA Healthcare System may not be representative of other US hospitals and settings, and the findings of this study with respect to trend and variation in epidemiological dynamics may not be generalizable to the entire United States. A fundamental challenge in designing and implementing infection control interventions is their high resource utilization. Modeling and forward simulation can provide preliminary evidence on the effectiveness of interventions while accounting for indirect effects, and may identify important limitations or optimal implementation strategies earlier, leading to more cost-effective intervention implementations (Halloran et al., 2017). In order for forward simulations to reliably predict the impact of intervention strategies, it is critical that their predictions be consistent with observed data, and in particular that the parameter value inputs for the simulation are precisely estimated from data. We have estimated the variability of key mechanistic parameters across a broad range of facilities, and these parameters can be used to test the sensitivity of intervention effects across a wide range of realistic settings.
A facility connected to other facilities via patient transfer or readmission may serve as a source of antibiotic resistance, sustaining transmission across a region (Toth et al., 2017). Our future extensions of this work will allow for tracking readmissions and transfers across multiple facilities and improve our understanding of key factors for controlling regional transmission of antibiotic resistance. Additionally, we plan to incorporate detailed patient and facility characteristics and to test mechanistic hypotheses to better understand specific mechanisms and factors, including progression to clinical infection, that are most important for driving transmission and infection within and across a network of healthcare facilities.
Acknowledgements
We would like to thank Dr. Mary-Claire Roghmann for her review and thoughts on this manuscript. Additionally, we would like to thank Ms. Carrie Edlund for her many iterations reviewing and editing this manuscript.
Fundings
This work was supported by the Department of Veterans Affairs, Veterans Health Administration, Health Services Research and Development Service project REA-08-264 and COIN grant CIN 13-414. This work was supported using resources and facilities at the VA Salt Lake City Health Care System with funding support from the Centers for Disease Control Prevention’s Epicenter ProgramU54CK000456-01 and the Centers for Disease Control and Prevention Cooperative Agreement RFA-CK-17-001-Modeling Infectious Diseases in Healthcare Program (MInD-Healthcare). This investigation was supported by the University of Utah Study Design & Biostatistics Center, with funding in part from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 5UL1TR001067-02 (formerly 8UL1TR000105 and UL1RR025764).
Footnotes
Disclaimer
The views expressed in this article are those of the authors and do not necessarily represent the position or policy of the Department of Veterans Affairs, the United States government, or any of the affiliated institutions.
Declarations of interest
None.
References
- Dantes R, Mu Y, Belflower R, et al. , 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern. Med 173 (21), 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ME, Kralovic SM, Simbartl LA, et al. , 2014. Nationwide reduction of health care-associated methicillin-resistant Staphylococcus aureus infections in Veterans Affairs long-term care facilities. Am. J. Infect. Control 42 (1), 60–62. [DOI] [PubMed] [Google Scholar]
- Geman S, Geman D, 1984. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell PAMI-6 (6), 721–741. [DOI] [PubMed] [Google Scholar]
- Gurieva T, Bootsma MCJ, Bonten MJM, 2012. Successful veterans affairs initiative to prevent methicillin-resistant staphylococcus aureus infections revisited. Clin. Infect. Dis 54 (11), 1618–1620. [DOI] [PubMed] [Google Scholar]
- Halloran ME, Auranen K, Baird S, et al. , 2017. Simulations for designing and interpreting intervention trials in infectious diseases. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings WK, 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57 (1), 97–109. [Google Scholar]
- Jain R, Kralovic SM, Evans ME, et al. , 2011. Veterans affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med 364 (15), 1419–1430. [DOI] [PubMed] [Google Scholar]
- Jones M, Ying J, Huttner B, et al. , 2014. Relationships between the importation, transmission, and nosocomial infections of methicillin-resistant staphylococcus aureus: an observational study of 112 veterans affairs medical centers. Clin. Infect. Dis 58 (1), 32–39. [DOI] [PubMed] [Google Scholar]
- Khader K, Thomas A, Huskins WC, et al. , 2016. A dynamic transmission model to evaluate the effectiveness of infection control strategies. Open Forum Infect. Dis 195 (1), ofw247 Oxford University Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R, 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin. Infect. Dis 42 (3), 389–391. [DOI] [PubMed] [Google Scholar]
- Kwok KO, Read JM, Tang A, Chen H, Riley S, Kam KM, 2018. A systematic review of transmission dynamic studies of methicillin-resistant Staphylococcus aureus in non-hospital residential facilities. BMC Infect. Dis [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum ML, Neumann C, Cook C, et al. , 2012. Epidemiology of Staphylococcus aureus blood and skin and soft tissue infections in the US Military Health System, 2005-2010. JAMA – J. Am. Med. Assoc 308 (1), 50–59 American Medical Association. [DOI] [PubMed] [Google Scholar]
- Lawes TG, Gould IM, 2012. Dissecting a multi-intervention methicillin-resistant staphylococcus aureus prevention bundle may miss emergent properties. Clin. Infect. Dis 55 (7), 1027–1028. [DOI] [PubMed] [Google Scholar]
- McDonald LC, 2006. Trends in antimicrobial resistance in health care-associated pathogens and effect on treatment. Clin. Infect. Dis 42 (Supplement 2), S65–S71 Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Wenzel RP, Bearman G, 2017. Contact precautions for endemic MRSA and VRE: time to retire legal mandates. JAMA – J. Am. Med. Assoc 318 (4), 329–330 American Medical Association. [DOI] [PubMed] [Google Scholar]
- Panlilio AL, Culver DH, Gaynes RP, et al. , 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol 13 (10), 582–586. [DOI] [PubMed] [Google Scholar]
- Rubin MA, Samore MH, Harris AD, 2018. The importance of contact precautions for endemic methicillin-resistant staphylococcus aureus and vancomycin-resistant enterococci. JAMA – J. Am. Med. Assoc 319 (9), 863–864 American Medical Association. [DOI] [PubMed] [Google Scholar]
- Slayton RB, Toth D, Lee BY, et al. , 2015. Vital signs: estimated effects of a coordinated approach for action to reduce antibiotic-resistant infections in health care facilities - United States. Am. J. Transplant 15 (11), 3002–3007. [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Redd A, Khader K, Leecaster M, Greene T, Samore M, 2015. Efficient parameter estimation for models of healthcare-associated pathogen transmission in discrete and continuous time. Math. Med. Biol 32 (1), 79–98. [DOI] [PubMed] [Google Scholar]
- Toth DJA, Khader K, Slayton RB, et al. , 2017. The potential for interventions in a long-term acute care hospital to reduce transmission of carbapenem-resistant Enterobacteriaceae in affiliated healthcare facilities. Clin. Infect. Dis 65 (4), 581–587. [DOI] [PubMed] [Google Scholar]