Abstract
Purpose
CD44 isoforms are highly expressed in cancer stem cells, initiating tumor growth and sustaining tumor self-renewal. Among these isoforms, CD44 variant 9 (CD44v9) is overexpressed in chronic inflammation-induced cancer. CD44 and the mesenchymal-to-epithelial transition (MET) receptor tyrosine kinase are coactivated in some gastric cancers (GCs). In this study, we characterized MET and CD44 expression and signaling in human GC cell lines and analyzed differences in the susceptibility of these lines to foretinib.
Patients and Methods
We analyzed cell viability and the rate of apoptotic cells using MTS assays and flow cytometry, respectively. Gene and protein expression were assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and immunoblotting, respectively.
Results
Foretinib treatment resulted in dose-dependent inhibition of growth in c-MET-amplified MKN45 and SNU620 cells with concomitant induction of apoptosis, but not in c-MET-reduced MKN28 and AGS cells. Foretinib treatment also significantly reduced phosphor-c-MET, phosphor-AKT, beta-catenin, and COX-2 protein expression in MKN45 and SNU620 cells. Interestingly, foretinib significantly reduced CD44, CD44v9, COX-2, OCT3/4, CCND1, c-MYC, VEGFA, and HIF-1a gene expression in CD44 and MET coactivated MKN45 cells and increased CD44s gene expression; in contrast, these drugs were only slightly active against SNU620 cells.
Conclusion
The results of this study indicate that foretinib could be a therapeutic agent for the prevention or treatment of GCs positive for CD44v9 and c-MET.
Keywords: c-MET, CD44v9, foretinib, gastric cancer, OCT3/4
Introduction
The rate of gastric cancer (GC) incidence is high in East Asia, Eastern Europe, and South America.1–3 Mortality rates have decreased markedly in recent years;4 however, GC remains the third most common cancer, causing 12% of all cancer-related deaths every year.5,6 To develop and identify new drug candidates with the aim of reducing GC mortality rates, it is important to determine its tumor characteristics and treatment parameters. The MET proto-oncogene encode the receptor tyrosine kinase (RTK) c-MET. In GC, such activation of MET has been attributed to gene amplification.7 MET amplification occurred in 2–20% of GC patients, however, only 7% of advanced GC patients overexpressed MET.8,9 In gastric cancer tissue, MET overexpression has been observed in gastric epithelium harboring dysregulated MET signaling, either as a result of high level focal amplification of the MET gene (>12 copies) or via HGF autocrine activation of MET.10 Previous study MET amplification was not detected in the cases with a gene copy number of <4. These study identified MET amplification at a frequency of 1.5% (4 out of 266 cases) in GC.11 Many MET tyrosine kinase inhibitors (MET-TKIs) were discontinued in clinical trials for multiple cancer types, including GC, because they were unable to determine the amplification cutoff that was MET for patient enrollment. The issue of how to determine the cutoff value for MET amplification in such cases has not yet been resolved.
HGF/c-MET targeting inhibitors are TKIs (e.g. ATP-competitive MET kinase inhibitors, non-ATP-competitive MET kinase inhibitors and multi-targeted inhibitors of MET and other kinases), anti-HGF neutralizing antibodies and anti-MET neutralizing antibodies used for various cancer which is known to overexpress MET.12,13 Foretinib, ATP-competitive MET kinase inhibitors, it inhibits HGF-mediated MET phosphorylation, migration and invasion of MET-amplified human gastric cancer cell lines.14 Foretinib failed to meet its primary end point of an objective response rate >25% in Phase II GC clinical trial. In this study, only 3patients had MET amplification, one of whom had disease stabilization.15 Other research patients based on germline or acquired MET mutation, germline MET mutations were a strong predictor of response to foretinib compared to acquired MET mutations.16 In GC cancer, gain-of-function mutation of MET are rare, with MET activation having been attributed mostly to gene amplification.7,17,18 Previous studies MET gene amplification rather than protein overexpression as a true oncogenic driver and a predictive marker for MET TKIs in GC.11,19,20 Interestingly though, other study revealed a strong association between p-MET expression and MET amplification in GC.9 Moreover, c-MET amplification is a frequent molecular abnormality in GC.21,22
Crosstalk between HGF/c-MET signaling pathway, PI3K/AKT signaling pathway and Wnt/β-catenin signaling pathway has been implicated in numerous cancers.23,24 The HGF/c-MET interact and cooperate with tyrosine kinases, can stimulate various downstream PI3K/AKT and Wnt/β-catenin signaling pathway in tumor cells. Prominent Wnt/β-catenin target genes include the CD44 and proto-oncogene c-MYC.25,26 Especially, cell adhesion molecule CD44 promotes stimulates inflammation, M2 macrophages accumulation and ECM deposition.27 M2 macrophages exert pro-tumoral functions by enhancing invasion and angiogenesis, immunosuppression and drug resistance through high expression of cyclooxygenase-2 (COX-2), VEGF and MMPs.28 The hypoxic tumor microenvironment induce phenotypic changes by hypoxic-inducible factor 1α (HIF-1α) that promote EMT and the stem-like properties of gastric cancer cells.29,30 HIF-1α increases CD44 expression levels and the number of CD44-positive cells.31 HIF-1α regulates CD44 may be associated with the Wnt/β-catenin signaling pathway in gastric cancer.32 CD44-positive GC cells exhibit the cancer stem cell (CSC) phenotype, which exhibits tumor growth initiation and self-renewal.34 Other study has demonstrated a positive correlation between NANOG levels and CD44 positive phenotype.33 CD44 also modulates interaction, movement, and metastatic potential.35 The standard isoform CD44s is generally expressed in normal epithelial cells; CD44v, the variant isoform, is expressed in epithelial-type carcinomas.36 CD44v9 is particularly overexpressed in GC caused by chronic inflammation.37,38 COX-2 stimulates cancer stem cell development in the inflammatory tumor microenvironment39 and promotes tumor growth by inducing proliferation, invasion, apoptosis inhibition, and angiogenesis induction.40,41
In the present study, we evaluated the suppressive effects of the c-MET inhibitor foretinib on GC proliferation, apoptosis and cancer stemness.
Materials and Methods
Cell Culture and Reagents
GC cell lines SNU620, MKN45, MKN28, and AGS were obtained from the Korean Cell Line Bank (Seoul, Korea) and maintained in RPMI1640 supplemented with 10% fetal bovine serum. Cells were cultured at 100% humidity and 5% CO2 at 37°C. The c-MET inhibitor foretinib (GSK1363089) was purchased from Selleck Chemicals (Houston, TX, USA). The Annexin V-APC/propidium iodide (PI) Apoptosis Detection Kit (Thermo Fisher Scientific, Rockford, IL, USA) was used to determine apoptosis rates.
Growth Inhibition Assays
IC50 values for foretinib in SNU620 and MKN45 cells were measured using the MTS assay for foretinib concentrations of 10, 1, 0.1, 0.05, 0.0025, 0.00125, 0.001, 0.0001, 0.00001, and 0.000001 µM for 48 h. On the day of the proliferation assay, the medium was removed, and 200 µL fresh medium was added to each well in 96-well plates, followed by 20 µL of MTS solution. The plates were then incubated at 37°C for 1 h in a humidified environment with 5% CO2. Absorbance was read at 490 nm using a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek). IC50 values were determined after fitting growth inhibition curves to dose–response curves using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA, USA).
Apoptosis Analysis
SNU620, MKN45, MKN28, and AGS cells seeded into 6-well plates at a density of 5 × 104 cells/mL were treated with foretinib IC50 values. Cell death was determined using the Annexin V-APC/PI Apoptosis Detection Kit (Thermo Fisher Scientific) using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, USA). Percentages of intact and apoptotic cells were calculated using the CytExpert software (Beckman Coulter).
qRT-PCR Analysis
To quantitate mRNA expression, total RNA from each sample was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR was performed using the Power SYBR Green PCR Master Mix and a LightCycler 96 instrument (Roche Applied Science, Indianapolis, IN, USA). Transcript levels of glyceraldehydes-3-phosphate dehydrogenase (GAPDH) were used for sample normalization. The primer sequences were as follows: c-MET (FW 5ʹ-AAG AGG GCA TTT TGG TTG TG-3ʹ; RW 5ʹ-GAT GAT TCC CTC GGT CAG AA-3ʹ), POU5F1 (OCT3/4, FW 5ʹ-TTC AGC CAA ACG ACC ATC TG-3ʹ; RW 5ʹ-GAA CCA CAC TCG GAC CAC ATC-3ʹ), NANOG (FW 5ʹ-CAC CAG TCC CAA AGG CAA AC-3ʹ; RW 5ʹ-GCC TTC TGC GTC ACA CCA TT-3ʹ), CCND1 (FW 5ʹ-GAT CAA GTG TGA CCC GGA CT-3ʹ; RW 5ʹ-TCC TCC TCT TCC TCC TCC TC-3ʹ), VEGFA (FW 5ʹ-AGG CCA GCA CAT AGG AGA GA-3ʹ; RW 5ʹ-TTT CTT GCG CTT TCG TTT TT-3ʹ), CD44 (FW 5ʹ-AGC ATC GGA TTT GAG ACC TG-3ʹ; RW 5ʹ-GTT GTT TGC TGC ACA GAT GG-3ʹ), CD44s (FW 5ʹ-AAA GGA GCA GCA CTT CAG GA-3ʹ; RW 5ʹ-TGT GTC TTG GTC TCT GGT AGC-3ʹ), CD44v9 (FW 5ʹ-ACC ATC CAA CAA CTT CTA CTC TGA CA-3ʹ; RW 5ʹ-CCT TCA GAA TGA TTT GGG TCT CTT-3ʹ), ECAD (FW 5ʹ-TGG GCC AGG AAA TCA CAT CC-3ʹ; RW 5ʹ-GGC ACC AGT GTC CGG ATT AA-3ʹ), COX-2 (FW 5ʹ-TGA GCA TCT ACG GTT TGC TG-3ʹ; RW 5ʹ-AAC TGC TCA TCA CCC CAT TC-3ʹ), HIF-1α (FW 5ʹ-CCA CCT ATG ACC TGC TTG GT-3ʹ; RW 5ʹ-TAT CCA GGC TGT GTC GAC TG-3ʹ), c-MYC (FW 5ʹ-TCA AGA GGC GAA CAC ACA AC-3ʹ; RW 5ʹ-GGC CTT TTC ATT GTT TTC CA-3ʹ), and GAPDH (FW 5ʹ-TTC ACC ACC ATG GAG AAG GC-3ʹ; RW 5ʹ-GGC ATG GAC TGT GGT CAT GA-3ʹ).
Immunoblot Analysis
Immunoblot analysis was performed using standard procedures. Commercially available primary antibodies were directed against anti-phospho-c-MET (Tyr1234/1235; 1:1000; #3077; Cell Signaling Technology, Danvers, MA, USA), anti-c-MET (1:1000; #4560; Cell Signaling Technology), anti-phospho-AKT (1:1000; #4060; Cell Signaling Technology), anti-AKT (1:1000; #1085-1; Epitomics), anti-β-catenin (1:1000; #610153; BD Biosciences), anti-COX-2 (1:1000; sc1745; Santa Cruz Biotechnology), anti-β-actin (1:1000; sc47778; Santa Cruz Biotechnology), and anti-α-tubulin (1:4000; # 05–829; Millipore).
Statistical Analysis
Data were analyzed using the Prism 5 (GraphPad Software Inc.). Values are means ± standard deviation (S.D.). Statistical significance was determined using one-way analysis of variance (ANOVA); significance was determined at a level of P < 0.05.
Results
Determining the Effective Dose of Foretinib in c-MET-Positive Cells
We tested the dose-dependent inhibitory effects of foretinib in SNU620 and MKN45 cells (Figure 1). Cells were treated with different concentrations of foretinib for 48 h, and the optimal dose was determined by evaluating cell viability using MTS assays. Treatment with foretinib decreased cell viability in a dose-dependent manner in c-MET-amplified SNU620 and MKN45 cells (n = 3) (Figure 1). Non-linear regression analysis revealed foretinib IC50 values of 13.4 nM for MKN45 cells and 21.9 nM for SNU620 cells.
Figure 1.
Effect of foretinib on gastric cancer (GC) cells positive for c-MET amplification. (A) SNU620 and MKN45 cells were treated with various concentrations of foretinib for 48 h. (B) Immunodetection of endogenous c-MET and phosphor c-MET (pY1234/1235) in GC cell lines.
Effects of Foretinib on Cell Apoptosis
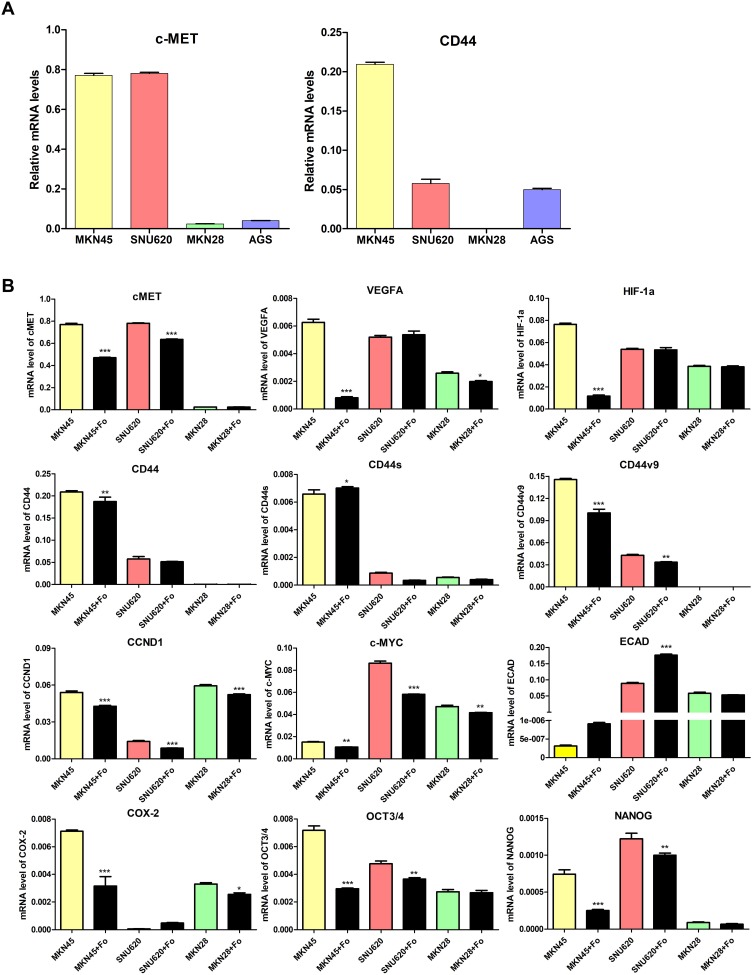
To evaluate the effects of foretinib on cell death in SNU620, MKN45, MKN28, and AGS cells, apoptosis was examined by staining with Annexin V-APC/PI, followed by flow cytometry (Figure 2). Cells were stained with Annexin V-APC and PI, which assess early apoptotic and late apoptotic, and necrotic cell populations, respectively. Foretinib showed the best cell death rates in SNU620 and MKN45 cells, whereas apoptosis was seldom observed in MKN28 and AGS cells (Figure 2A and B), with apoptotic cell percentages of 23.02 and 12.7%, respectively, after exposure to foretinib for 48 h (Figure 2A). SNU620 and MKN45 were high-c-MET expressors, whereas others such as MKN28 and AGS belonged to the low-c-MET expressor subtype. Notably, MKN45 cells were a high-CD44 expressor subtype (Figure 3A).
Figure 2.
Apoptotic activity of foretinib in (A) c-MET-positive SNU620 and MKN45 cells and (B) c-MET-negative MKN28 and AGS cells. Flow cytometric assay of GC cells treated with 30 nM foretinib for 48 h. Data are means ± S.D.
Figure 3.
Effect of foretinib on carcinogenesis-related genes in GC cells. (A) c-MET and CD44 gene expression in gastric cancer cells and (B) mRNA levels of c-MET, HIF-1a, VEGFA, CD44, CD44s, CD44v9, CCND1, COX-2, and ECAD in MKN45, SNU620, MKN28, and AGS cells were determined by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis after treatment with 30 nM foretinib for 48 h. Data are means ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 (one-way analysis of variance [ANOVA]).
Foretinib Inhibits c-MET Activation and Cancer Stemness in GC Cells
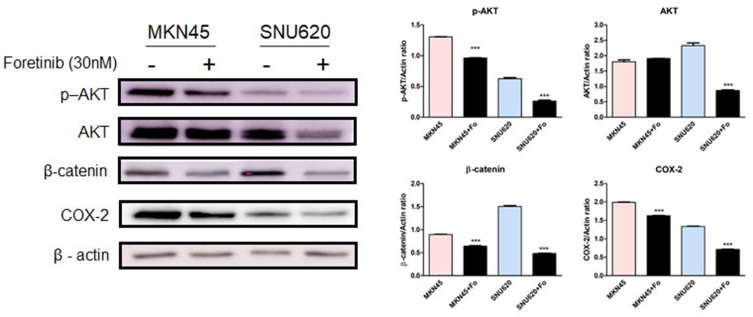
To examine the inhibitory effects of foretinib on GC cells (high-c-MET/high-CD44 [MKN45], high-c-MET/low-CD44 [SNU620], and low-c-MET/low-CD44 [MKN28]), oncogenic pathways were examined by analyzing gene and protein expression. Following treatment with foretinib, levels of c-MET, HIF-1α, VEGFA, CD44, CD44v9, CCND1, c-MYC, COX-2, and OCT3/4 mRNA decreased in MKN45 cells, whereas CD44s expression increased. In contrast, these drugs were only slightly active against SNU620 cells (Figure 3B). However, phosphor-c-MET, phosphor-AKT, β-catenin, and COX-2 protein expression decreased in MKN45 and SNU620 cells (Figure 4).
Figure 4.
Effect of foretinib on p-AKT, AKT, b-catenin, and COX-2 protein expression in GC cells. Protein levels of p-AKT, AKT, b-catenin, and COX-2 in MKN45 and SNU620 cells were determined by Western blot analysis after treatment with 30 nM foretinib for 48 h. Data are means ± S.D. ***P < 0.001 (one-way ANOVA).
Discussion
Carcinogenesis is complex process whereby malignant transformation occurs through genome instability and inflammation.42 Several studies have reported that COX-2 activation may be involved in inflammation-mediated CSC proliferation and differentiation.43,44 Many solid tumors possess CSCs, which share several characteristics with stem cells.45–47 Presently, CD44 is considered the most useful marker of gastric CSCs and a number of other solid tumors.34,48–51 CD44 is well known as a downstream target of Wnt/ß-catenin signaling.25 CD44 affect ß-catenin phosphorylation and nuclear accumulation in myeloid leukemia cells.52 Other study reveals that CD44 regulates Wnt signaling at the level of membrane receptors.53 In addition, CD44 controls signaling through interactions with TRKs, including c-Met and VEGFR2.54 Therefore, we examined CD44 downregulated drugs in MKN45 cells, which have a high expression of CD44 among c-MET-amplified cell lines. In a previous study, CD44 inhibitor drugs were screened using qRT-PCR analysis of MKN45 cells with CSCs to determine their suitability as therapeutic agents in GC treatment. Among the six c-MET inhibitor drugs tested, only foretinib downregulated the CD44 gene; therefore, foretinib was selected for further study.
Protein kinase mutations, overexpression, and dysregulation play an important role in the pathogenesis of many diseases including cancer.55–57 Therefore, this enzyme family (e.g. ALK, EGFR, ERBBs, VEGFR2, FGFR, PDGFR, c-MET, etc.) has become one of the important drug targets during the past 20 years.58 Foretinib is multi-kinase inhibitor targeting to c-MET and VEGFR2.14 In the present study, foretinib showed higher inhibition rates but lower apoptosis rates in MKN45 cells than in SNU620 cells. CD44-positive GC cells showed increased resistance for chemotherapy-induced cell death.34 Alternative splicing results in several CD44 isoforms with diverse functions. Notably, CD44v9 expression was higher in cancer cells from cancer patients than in normal tissues.37,38,44 CD44v9 is contained in the v6-10, v7-10, and v8-10 CD44 combinations.59 Among these, CD44v8-10 acts as a cancer stem cell in cancer development.60 Stem cell markers include OCT3/4, Nanog, and CD44v9, which are involved in the inflammatory microenvironment during cancer development.61 When MKN45 cells were treated with foretinib, levels of CD44, CD44v9, OCT3/4, NANOG, and COX-2 genes/proteins decreased. In contrast, those of CD44s increased. Previous study, attenuated ECAD expression, upregulation of β-catenin and enhanced ovarian cancer cell migration.62 Other study has reported that suppressed EMT through attenuation of AKT phosphorylation and β-catenin.63 Our results also indicated that foretinib may suppress β-catenin and AKT phosphorylation through increased ECAD expression. Indeed, foretinib inhibited angiogenesis, Wnt/β-catenin signaling, and the PI3K/Akt pathway in this study by inhibiting VEGFA, HIF-1α, c-MET, and AKT phosphorylation, as well as CCND1, c-MYC, and β-catenin in c-MET-positive GC. Our in vitro study strongly supports the clinical evaluation of foretinib, which prevents cancer stemness and c-MET-associated GC.
Conclusion
The results of this study indicate that c-MET and CD44v9 are differentially expressed in GCs and that foretinib exhibits significant inhibitory activity in c-MET- and CD44v9-expressed GC. Our in vitro study strongly supports the clinical evaluation of foretinib, which prevents cancer stemness and c-MET-associated GC.
Acknowledgments
This research was supported by the National R & D Program for Cancer Control, Ministry of Health and Welfare (HA17C0054), the National Research Foundation of Korea (grant NRF-2017R1A2B4005055, funded by the Korean Ministry of Science and ICT), and the Ministry of Food and Drug Safety of Korea (18183MFDS491; 2018), and the Hallym University Medical Center Research Fund. No funding body had any role in the study design, data collection, analysis, interpretation or writing of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Peleteiro B, Severo M, La Vecchia C, et al. Model-based patterns in stomach cancer mortality worldwide. Eur J Cancer Prev. 2014;23:524–531. doi: 10.1097/CEJ.0b013e328364f2b6 [DOI] [PubMed] [Google Scholar]
- 2.Peleteiro B, Barros S, Castro C, et al. Worldwide burden of gastric cancer in 2010 attributable to high sodium intake in 1990 and predicted attributable burden for 2030 based on exposures in 2010. Br J Nutri. 2016;116:728–733. doi: 10.1017/S0007114516002518 [DOI] [PubMed] [Google Scholar]
- 3.Ferro A, Peleteiro B, Malvezzi M, et al. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 4.Patru CL, Surlin V, Georgescu I, et al. Current issues in gastric cancer epidemiology. Rev Med Chir Soc Med Nat Iasi. 2013;117:199–204. [PubMed] [Google Scholar]
- 5.Jung KW, Won YJ, Kong HJ, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2011. Cancer Res Treat. 2014;46:109–123. doi: 10.4143/crt.2014.46.2.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999 [DOI] [PubMed] [Google Scholar]
- 7.Nakajima M, Sawada H, Yamada Y, et al. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer. 1999;85:1894–1902. [DOI] [PubMed] [Google Scholar]
- 8.Janjigian YY, Tang LH, Coit DG, et al. MET expression and amplification in patients with localized gastric cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:1021–1027. doi: 10.1158/1055-9965.EPI-10-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Z, Li Z, Gao J, et al. Tumor MET expression and gene amplification in chinese patients with locally advanced or metastatic gastric or gastroesophageal junction cancer. Mol Cancer Ther. 2015;14:2634–2641. doi: 10.1158/1535-7163.MCT-15-0108 [DOI] [PubMed] [Google Scholar]
- 10.Hughes PE, Rex K, Caenepeel S, et al. In vitro and in vivo activity of AMG 337, a potent and selective MET kinase inhibitor, in MET-dependent cancer models. Mol Cancer Ther. 2016;15:1568–1579. doi: 10.1158/1535-7163.MCT-15-0871 [DOI] [PubMed] [Google Scholar]
- 11.Kawakami H, Okamoto I, Arao T, et al. MET amplification as a potential therapeutic target in gastric cancer. Oncotarget. 2013;4:9–17. doi: 10.18632/oncotarget.v4i1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh RA, Wang P, Beumer JH, et al. The potential roles of hepatocyte growth factor (HGF)-MET pathway inhibitors in cancer treatment. Onco Targets Ther. 2014;7:969–983. doi: 10.2147/OTT.S40241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, Chen H, Huang X, et al. ARQ-197 enhances the antitumor effect of sorafenib in hepatocellular carcinoma cells via decelerating its intracellular clearance. Onco Targets Ther. 2019;12:1629–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian F, Engst S, Yamaguchi K, et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009;69:8009–8016. doi: 10.1158/0008-5472.CAN-08-4889 [DOI] [PubMed] [Google Scholar]
- 15.Shah MA, Wainberg ZA, Catenacci DV, et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PLoS One. 2013;8:e54014. doi: 10.1371/journal.pone.0054014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eder JP, Vande Woude GF, Boerner SA, et al. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009;15:2207–2214. doi: 10.1158/1078-0432.CCR-08-1306 [DOI] [PubMed] [Google Scholar]
- 17.Hara T, Ooi A, Kobayashi M, et al. Amplification of c-myc, K-sam, and c-met in gastric cancers: detection by fluorescence in situ hybridization. Lab Invest. 1998;78:1143–1153. [PubMed] [Google Scholar]
- 18.Tsugawa K, Yonemura Y, Hirono Y, et al. Amplification of the c-met, c-erbB-2 and epidermal growth factor receptor gene in human gastric cancers: correlation to clinical features. Oncology. 1998;55:475–481. doi: 10.1159/000011898 [DOI] [PubMed] [Google Scholar]
- 19.Lin X, Peng Z, Wang X, et al. Targeting autophagy potentiates antitumor activity of Met-TKIs against Met-amplified gastric cancer. Cell Death Dis. 2019;10:139. doi: 10.1038/s41419-019-1314-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak EL, Ahronian LG, Siravegna G, et al. Molecular heterogeneity and receptor coamplification drive resistance to targeted therapy in MET-amplified esophagogastric cancer. Cancer Discov. 2015;5:1271–1281. doi: 10.1158/2159-8290.CD-15-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuniyasu H, Yasui W, Kitadai Y, et al. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun. 1992;189:227–232. doi: 10.1016/0006-291X(92)91548-5 [DOI] [PubMed] [Google Scholar]
- 22.Nessling M, Solinas-Toldo S, Wilgenbus KK, et al. Mapping of chromosomal imbalances in gastric adenocarcinoma revealed amplified protooncogenes MYCN, MET, WNT2, and ERBB2. Genes Chromosomes Cancer. 1998;23:307–316. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Zhu GY, Gao HY, et al. Expression of tissue levels of matrix metalloproteinases and tissue inhibitors of metalloproteinases in gastric adenocarcinoma. J Surg Oncol. 2011;103:243–247. doi: 10.1002/jso.21824 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Xia M, Jin K, et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45. doi: 10.1186/s12943-018-0796-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wielenga VJ, Smits R, Korinek V, et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509 [DOI] [PubMed] [Google Scholar]
- 27.Kuwahara G, Hashimoto T, Tsuneki M, et al. CD44 promotes inflammation and extracellular matrix production during arteriovenous fistula maturation. Arterioscler Thromb Vasc Biol. 2017;37:1147–1156. doi: 10.1161/ATVBAHA.117.309385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang X, Gong K, Zhang X, et al. Osteopontin as a multifaceted driver of bone metastasis and drug resistance. Pharmacol Res. 2019;144:235–244. doi: 10.1016/j.phrs.2019.04.030 [DOI] [PubMed] [Google Scholar]
- 29.Guo J, Wang B, Fu Z, et al. Hypoxic microenvironment induces EMT and upgrades stem-like properties of gastric cancer cells. Technol Cancer Res Treat. 2016;15:60–68. doi: 10.1177/1533034614566413 [DOI] [PubMed] [Google Scholar]
- 30.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- 31.Krishnamachary B, Penet MF, Nimmagadda S, et al. Hypoxia regulates CD44 and its variant isoforms through HIF-1alpha in triple negative breast cancer. PLoS One. 2012;7:e44078. doi: 10.1371/journal.pone.0044078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G, Li S, Du W, et al. Hypoxia regulates CD44 expression via hypoxia-inducible factor-1alpha in human gastric cancer cells. Oncol Lett. 2017;13:967–972. doi: 10.3892/ol.2016.5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeter CR, Yang T, Wang J, et al. Concise review: NANOG in cancer stem cells and tumor development: an update and outstanding questions. Stem Cells. 2015;33:2381–2390. doi: 10.1002/stem.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takaishi S, Okumura T, Tu S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27:1006–1020. doi: 10.1002/stem.v27:5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasui W, Oue N, Aung PP, et al. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0 [DOI] [PubMed] [Google Scholar]
- 36.Ishimoto T, Nagano O, Yae T, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 37.Go SI, Ko GH, Lee WS, et al. CD44 variant 9 serves as a poor prognostic marker in early gastric cancer, but not in advanced gastric cancer. Cancer Res Treat. 2016;48:142–152. doi: 10.4143/crt.2014.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirata K, Suzuki H, Imaeda H, et al. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br J Cancer. 2013;109:379–386. doi: 10.1038/bjc.2013.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumder M, Xin X, Liu L, et al. COX-2 induces breast cancer stem cells via EP4/PI3K/AKT/NOTCH/WNT axis. Stem Cells. 2016;34:2290–2305. doi: 10.1002/stem.2426 [DOI] [PubMed] [Google Scholar]
- 40.Sun WH, Sun YL, Fang RN, et al. Expression of cyclooxygenase-2 and matrix metalloproteinase-9 in gastric carcinoma and its correlation with angiogenesis. Jpn J Clin Oncol. 2005;35:707–713. doi: 10.1093/jjco/hyi196 [DOI] [PubMed] [Google Scholar]
- 41.Lazar D, Taban S, Ardeleanu C, et al. Immunohistochemical expression of the cyclooxygenase-2 (COX-2) in gastric cancer. The correlations with the tumor angiogenesis and patients’ survival. Rom J Morphol Embryol. 2008;49:371–379. [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 43.Thanan R, Murata M, Ma N, et al. Nuclear localization of COX-2 in relation to the expression of stemness markers in urinary bladder cancer. Mediators Inflamm. 2012;2012:165879. doi: 10.1155/2012/165879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suwannakul N, Ma N, Thanan R, et al. Overexpression of CD44 variant 9: a novel cancer stem cell marker in human cholangiocarcinoma in relation to inflammation. Mediators Inflamm. 2018;2018:4867234. doi: 10.1155/2018/4867234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobo NA, Shimono Y, Qian D, et al. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154 [DOI] [PubMed] [Google Scholar]
- 46.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells–perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126 [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen L, Sprick MR, Kemper K, et al. Cancer stem cells–old concepts, new insights. Cell Death Differ. 2008;15:947–958. doi: 10.1038/cdd.2008.20 [DOI] [PubMed] [Google Scholar]
- 48.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018 [DOI] [PubMed] [Google Scholar]
- 50.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 52.Chang G, Zhang H, Wang J, et al. CD44 targets Wnt/beta-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int. 2013;13:117. doi: 10.1186/1475-2867-13-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt M, Metzger M, Gradl D, et al. CD44 functions in Wnt signaling by regulating LRP6 localization and activation. Cell Death Differ. 2015;22:677–689. doi: 10.1038/cdd.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orian-Rousseau V. CD44, a therapeutic target for metastasising tumours. Eur J Cancer. 2010;46:1271–1277. doi: 10.1016/j.ejca.2010.02.024 [DOI] [PubMed] [Google Scholar]
- 55.Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol Res. 2019;144:19–50. doi: 10.1016/j.phrs.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 56.Roskoski R Jr. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;129:65–83. doi: 10.1016/j.phrs.2018.01.021 [DOI] [PubMed] [Google Scholar]
- 57.Roskoski R Jr. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol Res. 2019;139:395–411. doi: 10.1016/j.phrs.2018.11.014 [DOI] [PubMed] [Google Scholar]
- 58.Cohen P. Protein kinases–the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773 [DOI] [PubMed] [Google Scholar]
- 59.Chen C, Zhao S, Karnad A, et al. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thanee M, Loilome W, Techasen A, et al. CD44 variant-dependent redox status regulation in liver fluke-associated cholangiocarcinoma: a target for cholangiocarcinoma treatment. Cancer Sci. 2016;107:991–1000. doi: 10.1111/cas.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawanishi S, Ohnishi S, Ma N, et al. Nitrative and oxidative DNA damage in infection-related carcinogenesis in relation to cancer stem cells. Genes Environ. 2016;38:26. doi: 10.1186/s41021-016-0055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin J, Zeng F, Wu N, et al. Interleukin-8 promotes human ovarian cancer cell migration by epithelial-mesenchymal transition induction in vitro. Clin Transl Oncol. 2015;17:365–370. doi: 10.1007/s12094-014-1240-4 [DOI] [PubMed] [Google Scholar]
- 63.Sohn EJ, Won G, Lee J, et al. Blockage of epithelial to mesenchymal transition and upregulation of let 7b are critically involved in ursolic acid induced apoptosis in malignant mesothelioma cell. Int J Biol Sci. 2016;12:1279–1288. doi: 10.7150/ijbs.13453 [DOI] [PMC free article] [PubMed] [Google Scholar]