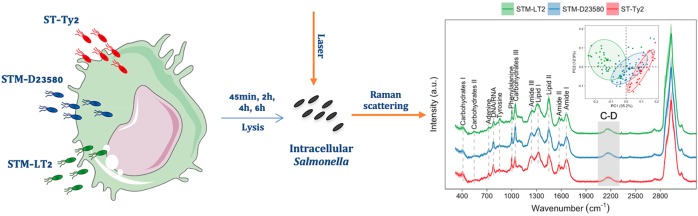
Abstract
The intracellular pathogen Salmonella enterica has evolved an array of traits for propagation and invasion of the intestinal layers. It remains largely elusive how Salmonella adjusts its metabolic states to survive inside immune host cells. In this study, single-cell Raman biotechnology combined with deuterium isotope probing (Raman-DIP) have been applied to reveal metabolic changes of the typhoidal Salmonella Typhi Ty2, the nontyphoidal Salmonella Typhimurium LT2, and a clinical isolate Typhimurium D23580. By initially labeling the Salmonella strains with deuterium, we employed reverse labeling to track their metabolic changes in the time-course infection of THP-1 cell line, human monocyte-derived dendritic cells (MoDCs) and macrophages (Mf). We found that, in comparison with a noninvasive serovar, the invasive Salmonella strains Ty2 and D23580 have downregulated metabolic activity inside human macrophages and dendritic cells and used lipids as alternative carbon source, perhaps a strategy to escape from the host immune response. Proteomic analysis using high sensitivity mass spectrometry validated the findings of Raman-DIP analysis.
Salmonella enterica (S. enterica) is a Gram-negative, facultative anaerobic, intracellular pathogen, responsible for a wide range of diseases. Salmonella accounts for 16 million cases of typhoid fever and 1.3 billion cases of gastroenteritis per year.1 In humans, the outcome of S. enterica infection mostly depends on the infecting serovar. Infection with human-restricted typhoidal serovars, such as S. enterica serovar Typhi (S. Typhi) and Paratyphi (S. Paratyphi), results in typhoid fever; while nontyphoidal broad host range serovars, such as S. enterica serovar Typhimurium (S. Typhimurium), are usually responsible of localized self-limiting gastroenteritis.2
Nevertheless, highly invasive multidrug resistant S. Typhimurium serovars have emerged in sub-Saharan Africa, as a major cause of lethal bacteremia in malnourished anemic children and individuals affected by underlying diseases such as malaria or HIV.3−8S. Typhimurium isolates from Kenya and Malawi are predominantly of a novel multilocus sequence type (ST) named ST313.7 The S. Typhimurium STS313 pathovar D23580 (STM-D23580) is a representative blood-stream clinical isolate which demonstrates genome degradation, resembling that of the human restricted pathogen S. Typhi.7 STM-D23580 has been largely used in studies of host–pathogen interactions to gain deeper insights into its increased pathogenicity, named as invasive nontyphoidal salmonellosis (iNTS).
In order to survive and replicate within host cells, Salmonella undergoes dramatic metabolic changes to utilize the available intracellular nutrients and to overcome adverse endosomal conditions.9,10 These adaptions, alongside with genetic variances, contribute to the different disease outcomes upon Salmonella infections. Technological perspectives have emerged to obtain a comprehensive picture of intracellular pathogen lifestyle. Transcriptomic11 and proteomic studies12,13 give insights into the mechanisms adopted by Salmonella to regulate its metabolic pathways in response to host cell environment. However, these technologies rely on highly expressed genes and proteins, thus overseeing subtle metabolic changes and a crucial fraction of nonenzymatic metabolic reactions.14 Most of the investigations provide a bulk average measurement of the total bacterial population, but phenotypes of bacteria will adapt when invading human cells.15−17
Single-cell Raman microspectroscopy (SCRM) offers phenotypic analysis at the single-cell resolution. Compared with other single-cell techniques, such as flow cytometry and nanosecondary ion mass spectrometry (NanoSIMS), SCRM measures intrinsic chemical profile of a cell in a nondestructive and relatively cheap manner.18−20 Single cell Raman spectrum (SCRS) provides a biochemical “fingerprint” of individual cell containing the molecular vibrational modes of all biomolecules, reflecting physiological and metabolic states of the cell.21,22 Raman-deuterium isotope labeling (Raman-DIP) is able to identify and measure general metabolic activity of microbes by detecting a C–D signature band.23 When cells are cultured in heavy water (D2O), NADPD (originally NADPH) is formed due to an exchange of H/D and facilitates synthesis of deuterated biomolecules, leading to the formation of carbon-deuterium (C–D) bonds that display a distinguishable Raman band in an otherwise silent region (2070–2300 cm–1) in SCRS.
In this study, we applied SCRS, Raman-DIP, and reverse labeling24 to obtain comprehensive and time-resolved profiles of the intracellular metabolism of three Salmonella strains responsible for different clinical outcomes: the gastroenteritis-associated S. Typhimurium LT2 (STM-LT2), the nontyphoidal invasive S. Typhimurium D23580 (STM-D23580), and the typhoidal S. Typhi Ty2 (ST-Ty2). The results from single-cell Raman analysis were further validated by proteomics using high sensitivity mass spectrometry.
Experimental Section
Bacterial Strains and Culture Conditions
Salmonella enterica serovar Typhimurium (STM) strain LT2 (ATCC 700220) and the clinical isolate STM-D23580, were used as representative nontyphoidal Salmonella sequence type 19 (ST19) and type 313 (ST313), respectively. Strain D23580 was isolated from the blood of an HIV-negative Malawian child with malaria and anemia. Salmonella enterica serovar Typhi (ST) strain Ty2 (ATCC 700931) was used as representative typhoidal serovar. Escherichia coli strain DH5α (EC-DH5α) (ATCC 67878) was used as an external reference for comparison with Salmonella strains. Details of the growth conditions for infection experiments are in the Supporting Information.
Infection
Generation of eukaryotic cells is detailed in the Supporting Information. Two hours before infection, the medium was replaced with fresh complete medium without antibiotics. THP-1 cells, monocyte-derived dendritic cells (MoDCs), and macrophages (Mf) were infected with D-labeled STM-LT2, STM-D23580, or ST-Ty2 at a MOI of 30 bacteria per cell (THP-1), 10 bacteria per cell (MoDCs), or 15 bacteria per cell (Mf). After 30 min incubation at 37 °C (THP-1 and Mf) or 45 min at 37 °C (MoDCs), extracellular bacteria were removed by washing cells twice with phosphate-buffered saline (PBS) and further incubated for 30 min in RPMI supplemented with 10% FBS, 2 mM l-glutamine, and 100 μg/mL of gentamycin (MP Biomedicals) to kill extracellular bacteria. After that, the gentamycin concentration was reduced to 30 μg/mL for the rest of the experiment. At each time point, eukaryotic cells were lysed by addition of 500 μL of saponin 2% (w/v) (Sigma) in PBS, followed by 5 min incubation at 37 °C. Cell lysates were used for SCRS or were serially diluted 10-fold in PBS and plated onto LB Lennox Broth agar (Sigma) to count CFU.
Mf and differentiated THP-1 cells are specialized phagocytic cells that quickly uptake and clear pathogens by engulfing them. Across ex-vivo settings, incubation times of 30 min or even less, guarantee successful internalization of Salmonella by human macrophages. The phagocytic activity of dendritic cells is less pronounced.25 After sensing pathogens via receptors or via inflammatory stimuli, dendritic cells switch on the expression of chemokine receptors, migrate to draining lymphoid organs and undergo a process of maturation. Unlike Mf, the major task of the dendritic cell is not to clear pathogens but to present antigens to T lymphocytes. Pathogen uptake, phagosomal degradation, and acidification are much lower in dendritic cells than in macrophages. Hence, to maximize the phagocytic activity of MoDCs, Salmonella strains were sampled at different time points, reflecting effector functions of each cell type. This sampling strategy is consistent to the previous reports.26−28
Single-Cell Raman Spectra (SCRS) Measurements and Analysis
At specific time point p.i., bacteria grown in RPMI and cell lysates of THP-1, MoDCs, and Mf were washed with PBS, fixed in 4% paraformaldehyde/PBS (PFA, EMS) (v/v) at RT for 15 min, and washed twice with Milli-Q water. SCRS were acquired for at least 30 bacterial cells per each condition using an HR Evolution confocal Raman microscope (Horiba Jobin-Yvon, France). Parameters of acquisition are in the Supporting Information.
Raman Data Preprocessing and Analysis
All spectra were recorded and preprocessed by comic ray correction and polyline baseline fitting using LabSpec 6 (Horiba Scientific, France). Spectral normalization was done by vector normalization of the entire spectral region. Data analysis, statistics and visualization were done under an R environment using in-house scripts. Quantification of intracellular biomolecule and D content was done by integrating corresponding Raman bands in SCRS (details in the Supporting Information). The ratio of C–D/(C–D + C–H) was used to indicate the extent of deuterium incorporation and this ratio of three Salmonella strains was mean-normalized at t = 0 h. Donor-wise centering has been done in all analysis. A machine learning classification model was built based on the fingerprint region of SCRS of inoculum STM-LT2, STM-D23580 and ST-Ty2, based on k-Nearest-Neighbor (kNN) algorithm. Leave-one-out-cross-validation (LOOCV) was used to evaluate the performance of the model and performance measures were computed as sensitivity for each strain as well as an overall accuracy rate.
LC-MS/MS Measurements and Analysis
The preparation of bacteria from MoDCs for proteomic experiments is described in the Supporting Information. LC-MS/MS analyses of tryptic peptides were performed using an UltiMate 3000 RSLC coupled to a Q Exactive plus mass spectrometer via a Nanospray Flex Ion Source (Thermo-Fisher Scientific Idstein, Germany; Table S1). Raw data files from the LC-MS/MS runs were analyzed using MaxQuant version 1.5.3.8. Protein identification for STM-LT2 and STM-D23580 was based upon a Salmonella FASTA database file which contained 4,547 target entries (S. Typhimurium strain LT2/SGSC1412/ATCC 700720, UniProt; proteome ID UP000001014; November 28, 2017). A FASTA database with 4,295 Salmonella typhi entries (S. Typhi strain Ty2/ATCC 700931, UniProt, proteome ID UP000002622; February 27, 2018) was applied for the ST-Ty2MS data. Details in MaxQuant analyses are in the Supporting Information. Log2 transformed label-free quantification (LFQ) intensities were calculated and pairwise comparisons of p.i. 0.75, 2, 4, and 6 h (6 pairs in total) were done using empirical Bayes approach in LIMMA package. Proteins with p < 0.05 (moderated t test) at any pairwise comparison were determined as differentially expressed proteins (DEPs). DEPs were then functionally annotated using Biocyc database.
Due to the very low number of bacteria isolated, a classical prenormalization by total protein concentration determination could not be applied. Similar to previous in vivo studies on S. aureus,29 all available bacteria cells were used to get a maximum of proteins per sample. In order to get equal amounts in MS, we used a 2 μg ZipTip bed to roughly equalize the peptide amounts before loading them on HPLC-column. In addition, a postmeasurement statistical normalization was further done by default in LFQ calculation in MaxQuant,30 assuming that most proteins do not change between different samples of the same organism. Supporting evidence of this assumption is that 602 proteins were found in all three analyzed conditions.
Results and Discussion
Phenotypic Profiles Based on Single-Cell Raman Spectra Distinguished Closely Related Salmonella Strains
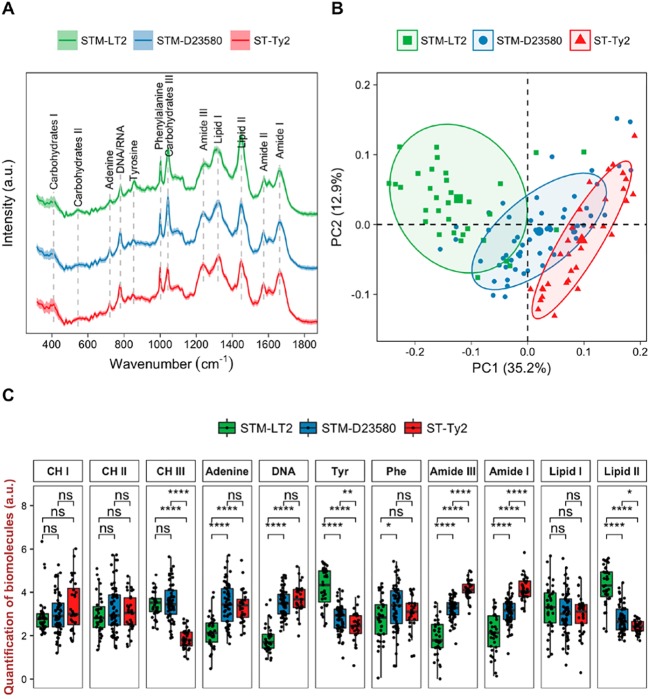
In order to understand whether the genetically close Salmonella strains (STM-LT2, STM-D23580, and ST-Ty2), yet responsible of distinct clinical manifestations in humans, can be discriminated by their metabolic profiles, SCRS of bacterial cultures were obtained and analyzed. Figure 1A shows the SCRS of STM-LT2, STM-D23580, and ST-Ty2, averaged from 30 to 50 single-cell measurements of each strain. Wavenumbers and corresponding vibrational modes of spectral features assigned to nucleic acids, proteins, lipids, and carbohydrates are summarized in Table S2. Unsupervised principal component analysis (PCA) was used to reduce the high dimensional Raman data set and reveal key information responsible for single-cell variances. PCA plot along the first two principal components (PC1 and PC2) separates the three bacterial strains, in which the invasive, nontyphoidal STM-D23580 sits in between the noninvasive, nontyphoidal STM-LT2 and the invasive, typhoidal ST-Ty2 (Figure 1B). It indicates that the three Salmonella strains (STM-LT2, STM-D23580 and ST-Ty2) have distinctive phenotypic profiles.
Figure 1.
(A) Average SCRS of overnight cultures of STM-LT2, STM-D23580, and ST-Ty2 from 30 to 50 single-cell measurements. The shaded area in SCRS represents the standard deviation. (B) PCA plot based on the fingerprint region of the SCRS. (C) Boxplots show semiquantification of selected biomolecules by integrating relevant Raman bands. Statistics (Welch’s t test) indicates pairwise differences (ns: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****; p < 0.0001). “CH” represents “carbohydrates”; “DNA” represents “DNA/RNA”; “Tyr” represents “tyrosine”; “Phe” represents “phenylalanine”.
We then performed quantification of intracellular biomolecules by integrating relevant Raman bands to highlight the phenotypic differences among the three strains (Figure 1C and Table S2). The most significant differences were observed in carbohydrates (carbohydrates III), nucleic acids (adenine and DNA/RNA), lipids (lipid II), amino acids, and proteins (tyrosine, amide III, and amide I). These differences correlate with the Raman contributions along PC1 of the PCA, which accounts for the highest variances among single bacterial cells (Figure S1).
A number of traits in the Raman metabolic profiles indicate a closer congruity between the invasive STM-D23580 and ST-Ty2, than to the noninvasive STM-LT2 (Figure 1C). The most significant changes in peak intensity between the two invasive strains and STM-LT2 were observed in Raman wavenumbers at 722 and 780 cm–1, corresponding to nucleic acids (adenine and DNA/RNA) and at 1574 and 1662 cm–1, corresponding to proteins (amide III and amide I; Figure 1A and Table S2). An increased DNA/RNA intensity could be due to different DNA states, transitioning from a condensate form in STM-LT2 to an uncoiled form in STM-D23580 and ST-Ty2, ready for more transcription activities in these invasive strains. The relaxed states of DNA facilitate efficient transcription, which may also account for the high intensity of the amide peaks, and thus more protein synthesis in STM-D23580 and ST-Ty2.
It was previously reported that S. Typhi no longer retains the pathway for tyrosine utilization31 because this serovar does not remain long enough in the gut to utilize the aromatic compounds produced by the human intestinal flora.32 In agreement, the tyrosine peak (853 cm–1) is significantly lower in ST-Ty2 and STM-D23580 as compared to STM-LT2 (Figure 1C). Surface lipids and proteins are often antigenic and represent the first line of contact with the host immune system. Lipid II at 1450 cm–1 which refers to saturated lipids that are abundant in bacterial cell wall is significantly higher in STM-LT2 as compared to STM-D23580 and ST-Ty2. The development of a robust outer membrane with increased level of saturated lipids confers resistance to antimicrobial peptides.33 This characteristic might be of relevance to STM-LT2, which primarily resides in the gut but can be lost in those pathovars that do not remain long enough in the gut, such as ST-Ty2 and STM-D23580.
The carbohydrates III (1043 cm–1) quantification, on the other hand, indicates dissimilarities between the typhoidal and the typhimurium strains, with ST-Ty2 exhibiting a significantly lower content. The human-restricted serovars S. Typhi and S. Paratyphi A harbor pseudogenes related to the transport of d-glucarate into the cell, suggesting that the utilization of this carbon source may not be advantageous in causing invasive infection.31
While feature extraction provides invaluable biochemical information on the phenotypic differences, machine learning algorithms can be used to resolve complex Raman information, classify bacterial species and benefit diagnostic purposes. A classification model based on k-nearest-neighbor (kNN) algorithm was applied to analyze SCRS of STM-LT2, STM-D23580, and ST-Ty2. A leave-one-out-cross-validation evaluated the kNN model, achieving an overall accuracy rate of 92.4% (Table S3). This classification model correctly classifies 106 out of 116 STM-LT2 spectra (sensitivity 91.4%), 135 out of 152 STM-D23580 spectra (sensitivity 88.8%), and 122 out of 125 ST-Ty2 spectra (sensitivity 97.6%). The highest sensitivity rate is achieved in classifying ST-Ty2 indicating that substantial differences emerge between the typhoidal and the nontyphoidal strains. Notably, all incorrect assignments of STM-LT2 and ST-Ty2 solely belong to the class of STM-D23580, while STM-D23580 is incorrectly classified as both STM-LT2 and ST-Ty2. This is consistent with both results from the PCA and quantification of biomolecules (Figure 1B,C). While STM-LT2 and ST-Ty2 bear distant resemblance, STM-D23580 is closely related to STM-LT2 at a taxonomic level but features many of the ST-Ty2 traits.
Intracellular Salmonella Shows Distinctive Metabolic Activities Revealed by Raman-DIP
We then sought to implement Raman-DIP and reverse labeling to detect the metabolic rates of Salmonella serovars. With the presence of D2O, SCRS of STM-LT2 showed a distinguishable C–D band at 2170 cm–1, similar to Escherichia coli DH5α (EC-DH5α), due to an incorporation of D via NADPH during active metabolism (Figure S2). Hence, the D incorporation in cells can be used as a universal indicator of metabolic activity applicable to Salmonella spp..
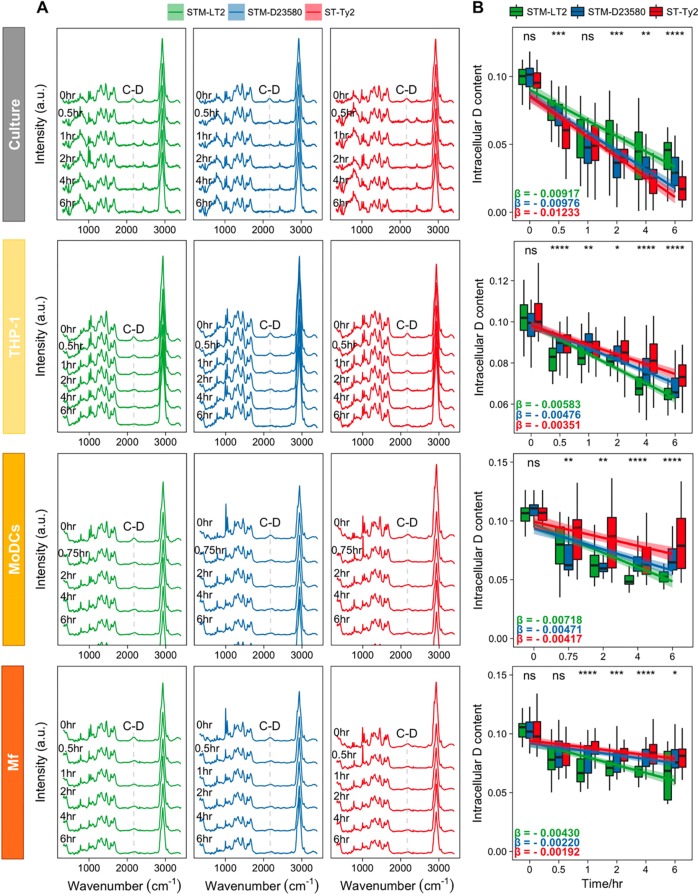
To better understand the metabolic features that underlie Salmonella’s survival and replication within the host cells, we initially labeled the three Salmonella strains with deuterium and employed reverse labeling24 to trace the metabolic changes over time during the interaction of Salmonella with human host cells. We obtained SCRS of STM-LT2, STM-D23580 and ST-Ty2 recovered at different time points post infection (p.i.) of THP-1 cell line, human MoDCs and human Mf. Three strains growing in liquid culture were used as extracellular controls. At each time point, the cellular D content was quantified by calculating the ratio of C–D/(C–D + C–H) in SCRS of single bacterial cells. A greater gradient (β) of D loss represents a faster replacement of C–H band during active metabolism, thus a higher bacterial metabolic rate.
At free-living condition in the growth medium, SCRS of extracellular Salmonella illustrate a gradually decreasing C–D band over time in all three Salmonella strains (Figure 2A). ST-Ty2 displays the fastest D loss (−0.01233), compared to STM-LT2 (−0.00917) and STM-D23580 (−0.00976), indicating that ST-Ty2 is the most active and fastest duplicative strain in liquid culture, consistent with the colony forming unit (CFU) counts (Figure S3A). In support of these results, the Raman phenotypic quantification (Figure 1C) also shows that in a nutrient-rich environment, the higher energy requirements of ST-Ty2, necessary for its replication, is fulfilled by higher consumption of carbohydrate stocks (Carbohydrates III). The extensive growth is accompanied by a higher content of nucleic acids (DNA/RNA) ready for transcription and increased protein production (amide III and amide I; Figure 1C).
Figure 2.
(A) SCRS of Salmonella serovars under four experimental conditions: extracellular growth in liquid culture and intracellular metabolism during infection of a differentiated THP-1 cell line, primary MoDCs, and primary Mf (the band at ∼2500 cm–1 in the “culture” panel is instrumental artifact and it has been corrected in later measurements). (B) Boxplots quantifying D content at different time points. Statistics was done with one-way ANOVA (ns: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****; p < 0.0001). The absolute value of linear regression slope β represents the metabolism speed.
A decrease of the C–D band in the recovered intracellular Salmonella was also observed in all strains, during time-course infection of THP-1 cells, MoDCs and Mf (Figure 2A). In contrast to the observation in the liquid growth medium, all infection models show that ST-Ty2 was the strain with the slowest metabolic rate (Figure 2B). In THP-1 cells, at 6 h p.i., a visible amount of the C–D band is still present in the spectra of ST-Ty2, as opposed to the complete absence of the C–D band in STM-LT2 and STM-D23580 (Figure 2A). Quantification of D content (Figure 2B) confirms that ST-Ty2 has the slowest metabolic rate (−0.00351), comparing to STM-LT2 (−0.00583) and STM-D23580 (−0.00476). A similar spectral pattern is also observed in human MoDCs and Mf (Figure 2), where ST-Ty2 has the slowest metabolism (−0.00417 in MoDCs, −0.00192 in Mf), followed by STM-D23580 (−0.00471 in MoDCs, −0.00220 in Mf) and ST-LT2 (0.00718 in MoDCs, −0.00430 in Mf). The slowest metabolic rate of ST-Ty2 during infection in THP-1, MoDCs and Mf also correlates with the CFU counting of the three Salmonella strains (Figure S3B–D). Therefore, in terms of Salmonella metabolic activity in all infection models, the invasive ST-Ty2 has the slowest metabolic rate, and the noninvasive STM-LT2 has the fastest metabolic rate, while the activity of the nontyphoidal but invasive clinical strain STM-D23580 is in between. It is known that the invasive host-adapted Salmonella replicates slowly and elicits weaker pro-inflammatory responses2 in order to avoid immune-surveillance and to disseminate inside host cells. Our results highlight a plausible strategy adopted also by STM-D23580 to slow down its central metabolism in an effort to survive intracellularly while avoiding immune recognition.
Overall, all three Salmonella strains exhibited a slower metabolism in intracellular conditions than in free-living, liquid growth medium. The metabolic activity was particularly reduced in Mf as compared to THP-1 and MoDCs, most likely as a result of the hostile acidic phagosomal environment.34 Likewise, Mf exhibited increased killing ability compared to THP-1 and MoDCs (Figure S3).
SCRS Map Dynamic Phenotypic Profiles of Intracellular Salmonella within Primary Cells
As SCRS provides a snapshot of bacterial phenotypic profiles, we analyzed the SCRS of intracellular cells for each Salmonella strain by quantifying relevant Raman bands at different time points p.i., thus providing a phenotypic comparison across strains. Time-resolved Raman profiles are summarized in Figure S4, while characteristic bands during primary cell infections are emphasized in Figure 3. The detailed analysis is shown as follows.
Figure 3.
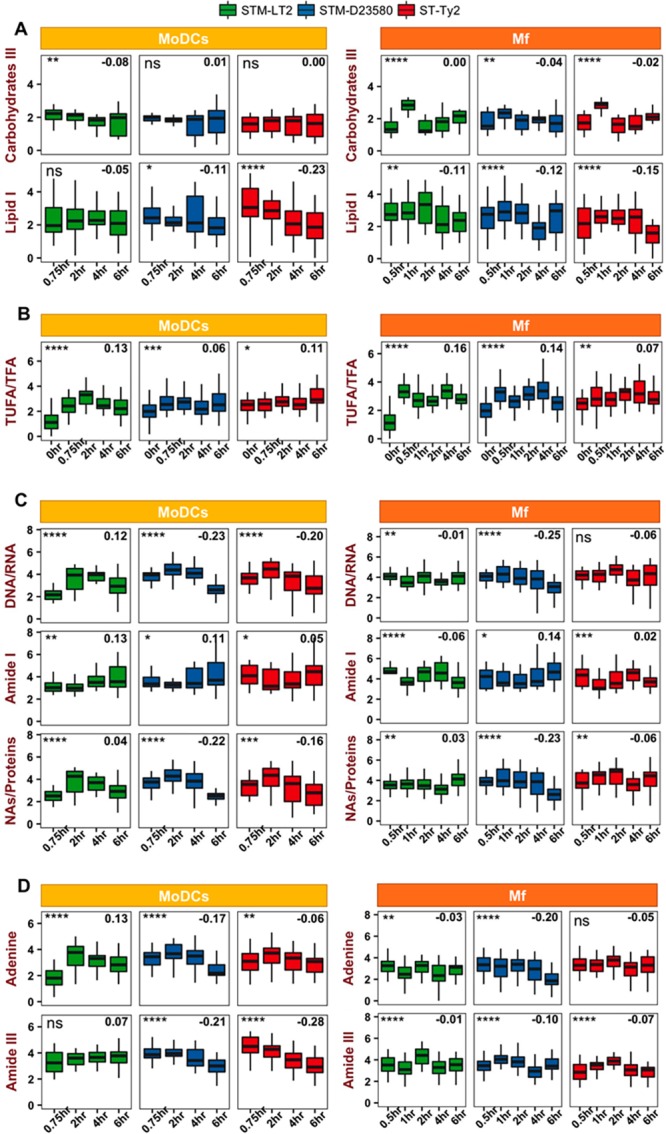
Raman profiling of biomolecules in intracellular bacteria. Raman bands of (A) carbohydrates III and lipid I, (B) the ratio of total unsaturated fatty acids to total fatty acids (TUFA/TFA), (C) DNA/RNA, amide I and the ratio of nucleic acids to proteins, and (D) adenine and amide III were quantified in SCRS of STM-LT2, STM-D23580, and ST-Ty2 at different time point of infection in MoDCs and Mf. Statistics was done by using one-way ANOVA with a global p-value to indicate general differences among time points (ns: p > 0.05; *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****; p < 0.0001). Linear regression was fitted to compare the time-wise changes of one specific quantification in each strain and the slope was labeled at the top right corner, where a negative value indicates a decrease over time and a positive value indicates an increase over time.
Carbohydrates and Lipids
In MoDCs, an increased consumption of carbohydrates III in STM-LT2 (−0.08) is observed compared with STM-D23580 (0.01) and ST-Ty2 (0.00), which indicates a greater utilization of carbohydrate stocks by the noninvasive STM-LT2 during infection (Figure 3A). On the contrary, a decrease of lipid I (saturated long chain fatty acids) at 1320 cm–1 is observed in STM-D23580 (−0.11 in MoDCs, −0.12 in Mf) and ST-Ty2 (−0.23 in MoDCs, −0.15 in Mf), compared with in STM-LT2 (−0.05 in MoDCs, −0.11 in Mf). The results suggest that the invasive ST-Ty2 and STM-D23580 should adopt a switch from carbohydrate consumption to lipid consumption. In line with a previous study,35 our results suggest that invasive Salmonella appears to switch off the glycolytic pathways, using fatty acids as carbon sources instead. The contribution of lipid metabolism to the virulence and survival of Salmonella in the intravacuolar environment has been highlighted in a number of studies.35,36
To further assess the modulation of Salmonella lipidic phenotype during its intracellular life, we analyzed the higher wavenumber region of the SCRS (2700–3300 cm–1). The band centered at 3008 cm–1 in SCRS is representative of unsaturated =CH stretching and the one at 2930 cm–1 represents all CH, CH2, and CH3 stretching in the cell. The ratio of total unsaturated fatty acids/total fatty acids (TUFA/TFA) is commonly used in Raman studies to characterize lipid membrane profile as an index of bacterial adaption and response to oxidative stress.37 Increased unsaturation of lipids causes fluidizing of the membrane and allows membrane modifications that enhance the tolerance of bacteria under stress conditions. As bacterial membrane modification is particularly upon internalization, we compared the TUFA/TFA ration between the intracellular and the extracellular conditions (Figure 3B). The TUFA/TFA ratio in all conditions show a marked upregulation in STM-LT2 (0.13 in MoDCs, 0.16 in Mf) compared to STM-D23580 (0.06 in MoDCs, 0.14 in Mf) and ST-Ty2 (0.11 in MoDCs, 0.07 in Mf), highlighting the differences in membrane alteration between the invasive and noninvasive strains. Notably, the TUFA/TFA ratio in all strains increased dramatically from 0 to 0.5 h in Mf (t test, **** in STM-LT2, *** in STM-D23580 and * in ST-Ty2), suggesting a rearrangement of the surface moieties upon phagocytosis necessary to resist to the hostile intramacrophage environment.
Nucleic Acids, Proteins, and Aromatic AAs
A variation in DNA/RNA is detected during MoDC infection for all bacterial strains, with a marked upregulation at 2 h p.i. (t test, * in STM-LT2, *** in STM-D23580 and ** in ST-Ty2), likely due to increased DNA synthesis and transcriptions (Figure 3C). A decrease was observed in the invasive STM-D23580 (−0.23) and ST-Ty2 (−0.20), as compared to STM-LT2 (0.12). Amide I, a representative peak of C=O stretching in proteins (Table S2), showed different time-wise modulation as compared to DNA/RNA. We calculated the ratio of nucleic acids/protein for all three Salmonella strains during infection as an indicator of intracellular bacterial replication. Following a short increase of the ratio, a significant decrease is observed in the invasive STM-D23580 (−0.22) and ST-Ty2 (−0.16) compared to STM-LT2 (−0.04; Figure 3C). This result suggests that the intracellular growth of the invasive strains may be arrested in response to the adverse intracellular environment.
A transient accumulation of adenine is observed for all of the bacterial strains during the early stage of MoDC infection (Figure 3D). This result can be explained by the fact that bacteria might have sufficient purine reserves to sustain only the initial host cell attack. In Mf, significant alterations of the adenine peak are detected for the Typhimurium strains only (Figure 3D). Purine nucleotides are critical for intramacrophage survival of Salmonella. Under purine-limiting environments like the macrophage phagosome, Salmonella is unable to repair the DNA damage, which ultimately results in bacterial death.
Significant variations in the amide III peak were also detected (Figure 3D) except for STM-LT2 in MoDCs, possibly due to either intracellular stress that changes the properties of bacterial proteins and enzymes, or protein denaturation as a result of bacteria being killed. Indeed, significant changes in the amide III peak are noticeable during Mf infection, in agreement with the decreased bacterial survival (Figure S3) and metabolic activity (Figure 2) previously described. Additionally, a decrease can be observed in STM-D23580 and ST-Ty2 during infection of MoDCs as compared to STM-LT2, which might contribute to invasive characteristics of these strains in MoDCs.
Single-Cell Raman Analysis Resolves Phenotypic Heterogeneity in Isogenic Populations
Salmonella displayed significant cell-to-cell variation resulting in distinct subpopulations with different metabolic phenotypes. A t-distributed stochastic neighbor embedding (t-SNE) of all bacterial cells in MoDCs and Mf at 2 h p.i. was used to exemplify and visualize the multimodal behaviors of intracellular bacteria (Figure S5A). Three subpopulations were clustered for bacteria in MoDCs (MoDC1, MoDC2, and MoDC3) and Mf (Mf1, Mf2, and Mf3).
Bacteria in the MoDC3 cluster was enriched in lipid content while displaying low levels of carbohydrates (Figure S5B). This metabolic phenotype is compatible with that of bacteria restricted to phagosomal compartments, in which glucose is less available and alternative carbon sources are required.38 The NO produced in the SCV targets the pyruvate and α-ketoglutarate dehydrogenase complexes, which would strongly affect glycolysis, glycogenesis, and the TCA cycle.39 The higher lipid content, on the other hand, suggests the employment of alternative energy pathways.
MoDC3 and Mf3 clusters also had a low TUFA/TFA ratio. When exposed to acidic pH, such as in the SCV, Salmonella induces a profound rearrangement of lipid membranes by increasing the unsaturated fatty acids content.40 On the contrary, the higher TUFA/TFA ratio, observed for the bacteria in the clusters MoDC2 and Mf2, could suggest continuous internal turnover, damage, and/or loss to the environment, which has previously been proposed as a potential strategy adopted by the persisters.41
Diversity in proliferation is also observed in different bacterial groups. A low DNA/RNA contents and NAs/protein ratio was observed for the bacteria of the “intraphagosomal” clusters MoDC3 and Mf3. In fact, under particular stress conditions, such as the presence of NO, Salmonella cell division is reduced. High protein-specific peak intensities indicate an increase in protein synthesis, which may be required for the correct assembly of the type II secretion system (T2SS) machinery.42 On the other hand, bacteria in the clusters of MoDC1, MoDC2, Mf1, and Mf2 exhibit a different metabolic profile characterized by high content of carbohydrates and nucleic acids concomitant with a reduced TUFA/TFA ratio.
The Mf3 population is bigger than MoDC3, suggesting that a more efficient compartmentalization of Salmonella in the SCV could be achieved by Mf. Previous studies have indicated that the intracellular lifestyle of Salmonella in MoDCs differs from that in Mf,34,43 implying that Salmonella’s gene expression required for survival inside MoDCs is not the same as the one needed for survival in Mf. Diverse adaptations and isogeneic heterogeneity within different host compartments in cell lines and primary human cells highlights the importance of choosing single-cell tools and different cell infection models for the study of intracellular pathogens.
Intracellular Raman Profiles Are Consistent with the Proteomic Analysis
In order to validate the Raman observations, we performed time-course proteomics experiments of intracellular Salmonella recovered from MoDCs, which are the most important antigen-presenting cells. We identified 687 proteins in STM-LT2, 1277 proteins in STM-D23580, and 1408 proteins in ST-Ty2 (Figure S6). For each bacterial strain, differentially expressed proteins (DEPs; p < 0.05) were determined across the time points. Those DEPs were then functionally annotated to bacterial pathways related to either energy metabolism (Figures S7A and S8A-C) or biosynthesis (Figures S7B and S8D-F).
Carbon metabolism and energy utilization are key to all bacterial metabolic events. We therefore hypothesize that regulation in energy pathways could support the observation that the three Salmonella strains demonstrate distinctive metabolism rates by Raman-DIP. In Figure S7A, most of the proteins related to bacterial energy pathways were upregulated during the intracellular life of STM-LT2 except for those responsible for glycolysis and fermentation. STM-D23580 exhibited a mixture of upregulated and downregulated pathways. All pathways related to energy metabolism had a decreasing trend in ST-Ty2. This is in a good agreement with the Raman-DIP results, indicating that the invasive typhoidal ST-Ty2 had the lowest metabolic activity among three Salmonella pathovars (Figure 2), by slowing down most metabolic reactions to obtain energy and nutrients.
Overall, a downregulation of many metabolic enzymes in the TCA cycle is observed in all Salmonella strains (Figure S8A-C), in good agreement with a previous report,44 including the aminotransferase AspC, the NADP-specific glutamate dehydrogenase GdhA, the succinyl-CoA synthetases SucC and SucD, and AcnA and AcnB. The inactivation of the TCA cycle increases the ability of Salmonella to survive within resting and activated murine macrophages,45 which might explain the prominent disruption of TCA cycle and aerobic respiration in STM-D23580 and ST-Ty2 (Figure S7A). This result supports the dynamic Raman profiling in MoDCs, where a significant consumption of carbon stock (carbohydrates III) was observed in STM-LT2, while higher consumption of lipids occurred in STM-D23580 and ST-Ty2 (Figure 3A). These results highlight a possible strategy adopted by the invasive strains to survive intracellularly: switching energy sources from aerobic respiration to lipid-driven metabolisms. This conclusion is also supported by a decrease in lipid biosynthesis pathway in STM-LT2, STM-D23580, and ST-Ty2 (Figure S7B).
Biosynthesis pathways related to amino acids (AA), amine, and polyamine were found to be differentially regulated (Figure S7B). While proteins associated with glutamine and lysine synthesis were upregulated at final time points in STM-LT2 (STM1795 and DapD), proteins associated with these pathways (e.g., STM1795, Gln A, ThrA, IlvC, IlvD, DapA, and DapD) were detected at significantly lower level at 6 h, as compared to the initial stage of the infection, in STM-D23580 and ST-Ty2 (Figure S8D-F). Overall, AA biosynthesis was more repressed in STM-D23580 and ST-Ty2 during the infection, as a significant decrease of amide III was observed in the invasive strains while nonsignificant changes were detected in STM-LT2 (Figure 3B). Similarly, more evidently reduced abundance of nucleosides and nucleotides (NN) synthesis proteins was observed in STM-D23580 and ST-Ty2 (Figure S7B), similar to marked decreasing DNA/RNA bands in the Raman profile of the invasive strains (Figure 3C).
The Fad-family proteins are known to be the key players in the metabolism of saturated and unsaturated fatty acids and are indicative of a modulation of bacterial membrane fluidity in response to environmental stress.46 While all Fab proteins were downregulated in STM-D23580 and ST-Ty2, STM-LT2 exhibited an increased abundance of FabB and FabG, responsible for the de novo synthesis of UFA via the fabA-fabB pathway46,47(Figure S8D-F). This suggests variability in the membrane composition between the invasive strains and the noninvasive strain, verified by the markedly increased Raman TUFA/TFA ratio in STM-LT2 (Figure 3B).
Conclusions
In this study, we employed a single-cell Raman technique to detect the different metabolic profiles of typhoidal S. typhi Ty2 and the nontyphoidal S. Typhimurium LT2 and D23580. By using Raman-DIP and reverse labeling, we were able to characterize the metabolic activity of the three different Salmonella strains during infection of human cell lines and primary monocyte-derived dendritic cells (MoDCs) and macrophages (Mf). In a paradox, ST-Ty2 had the fastest extracellular metabolic rate but the slowest metabolic rate inside immune cells. Our study also revealed a decreased uptake of carbohydrate stocks but increased lipid consumption in both invasive STM-D23580 and ST-Ty2.
In summary, we have demonstrated that single-cell Raman-DIP technologies are capable of revealing metabolic changes, highlighting molecular heterogeneity and cell-to-cell variations among invasive Salmonella serovars. Through comprehensive profiling and comparison of three different Salmonella pathovars, this study sheds light into bacterial metabolic traits and adaptations inside human host cells, which might contribute to the distinct pathogenicity of Salmonella. As several aspects of metabolism can influence antimicrobial susceptibility and tolerance, a better understanding of the Salmonella-host metabolic landscape might open new perspectives for antimicrobial development.
Acknowledgments
WEH acknowledges finance and instrumentation support from EPSRC (EP/M002403/1, EP/M02833X/1) and NERC (NE/M002934/1).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b01010.
Supporting methods; tables of Raman assignment, model performance, and LC-MS/MS details; figures of PCA loading plots, SCRS of STM-LT2 and E. coli DH5α, CFU during infection, complete Raman profiling, t-SNE plots, proteomics Venn diagram, dotplots, and heatmaps of DEPs (PDF)
Author Contributions
# L.P. and A.A. have contributed equally to this study.
The authors declare no competing financial interest.
Supplementary Material
References
- Pui C. F.; Wong W. C.; Chai L. C.; Tunung R.; Jeyaletchumi P.; Noor Hidayah M. S.; Ubong A.; Farinazleen M. G.; Cheah Y. K.; Son R. Int. Food Res. J. 2011, 18, 465–473. [Google Scholar]
- Dougan G.; Baker S. Annu. Rev. Microbiol. 2014, 68, 317–336. 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- Gordon M. A. J. Infect. 2008, 56, 413–422. 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Graham S. M. Curr. Opin. Infect. Dis. 2010, 23, 409–414. 10.1097/QCO.0b013e32833dd25d. [DOI] [PubMed] [Google Scholar]
- Hohmann E. L. Clin. Infect. Dis. 2001, 32, 263–269. 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- Gordon M. A.; Graham S. M.; Walsh A. L.; Wilson L.; Phiri A.; Molyneux E.; Zijlstra E. E.; Heyderman R. S.; Hart C. A.; Molyneux M. E. Clin. Infect. Dis. 2008, 46, 963–969. 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- Kingsley R. A.; Msefula C. L.; Thomson N. R.; Kariuki S.; Holt K. E.; Gordon M. A.; Harris D.; Clarke L.; Whitehead S.; Sangal V.; Marsh K.; Achtman M.; Molyneux M. E.; Cormican M.; Parkhill J.; MacLennan C. A.; Heyderman R. S.; Dougan G. Genome Res. 2009, 19, 2279–2287. 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leekitcharoenphon P.; Friis C.; Zankari E.; Svendsen C. A.; Price L. B.; Rahmani M.; Herrero-Fresno A.; Fashae K.; Vandenberg O.; Aarestrup F. M.; Hendriksen R. S. J. Infect. Dev. Countries 2013, 7, 696–706. 10.3855/jidc.3328. [DOI] [PubMed] [Google Scholar]
- Bumann D.; Schothorst J.. Cell Microbiol 2017, 19. [DOI] [PubMed] [Google Scholar]
- Thompson A.; Fulde M.; Tedin K. Environ. Microbiol. Rep. 2018, 10, 140–154. 10.1111/1758-2229.12628. [DOI] [PubMed] [Google Scholar]
- Faucher S. P.; Porwollik S.; Dozois C. M.; McClelland M.; Daigle F. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 1906–1911. 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zhang Q.; Hu M.; Yu K.; Fu J.; Zhou F.; Liu X. Infect. Immun. 2015, 83, 2897–2906. 10.1128/IAI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.; Adkins J. N.; Coleman J. R.; Schepmoes A. A.; Dohnkova A.; Mottaz H. M.; Norbeck A. D.; Purvine S. O.; Manes N. P.; Smallwood H. S.; Wang H.; Forbes J.; Gros P.; Uzzau S.; Rodland K. D.; Heffron F.; Smith R. D.; Squier T. C. J. Biol. Chem. 2006, 281, 29131–29140. 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- Keller M. A.; Piedrafita G.; Ralser M. Curr. Opin. Biotechnol. 2015, 34, 153–161. 10.1016/j.copbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumann D. Cell Host Microbe 2015, 17, 13–19. 10.1016/j.chom.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Knodler L. A. Curr. Opin. Microbiol. 2015, 23, 23–31. 10.1016/j.mib.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Helaine S.; Thompson J. A.; Watson K. G.; Liu M.; Boyle C.; Holden D. W. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 3746–3751. 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Q.; Xu J.; Romero-Gonzalez M.; Banwart S. A.; Huang W. E. Curr. Opin. Biotechnol. 2012, 23, 56–63. 10.1016/j.copbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Wagner M. Annu. Rev. Microbiol. 2009, 63, 411–429. 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Huang W. E.; Cui L.; Wagner M. Curr. Opin. Biotechnol. 2016, 41, 34–42. 10.1016/j.copbio.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Huang W. E.; Griffiths R. I.; Thompson I. P.; Bailey M. J.; Whiteley A. S. Anal. Chem. 2004, 76, 4452–4458. 10.1021/ac049753k. [DOI] [PubMed] [Google Scholar]
- Schie I. W.; Huser T. Appl. Spectrosc. 2013, 67, 813–828. 10.1366/12-06971. [DOI] [PubMed] [Google Scholar]
- Berry D.; Mader E.; Lee T. K.; Woebken D.; Wang Y.; Zhu D.; Palatinszky M.; Schintlmeister A.; Schmid M. C.; Hanson B. T.; Shterzer N.; Mizrahi I.; Rauch I.; Decker T.; Bocklitz T.; Popp J.; Gibson C. M.; Fowler P. W.; Huang W. E.; Wagner M. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E194–203. 10.1073/pnas.1420406112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Song Y.; Tao Y.; Muhamadali H.; Goodacre R.; Zhou N. Y.; Preston G. M.; Xu J.; Huang W. E. Anal. Chem. 2016, 88, 9443–9450. 10.1021/acs.analchem.6b01602. [DOI] [PubMed] [Google Scholar]
- Nagl M.; Kacani L.; Mullauer B.; Lemberger E. M.; Stoiber H.; Sprinzl G. M.; Schennach H.; Dierich M. P. Clin. Diagn. Lab. Immunol. 2002, 9, 1165–1168. 10.1128/CDLI.9.6.1165-1168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno S. M.; Gonzalez P. A.; Carreno L. J.; Tobar J. A.; Mora G. C.; Pereda C. J.; Salazar-Onfray F.; Kalergis A. M. Immunology 2008, 124, 522–533. 10.1111/j.1365-2567.2008.02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheminay C.; Mohlenbrink A.; Hensel M. J. Immunol. 2005, 174, 2892–2899. 10.4049/jimmunol.174.5.2892. [DOI] [PubMed] [Google Scholar]
- Fu G.; Wijburg O. L. C.; Cameron P. U.; Price J. D.; Strugnell R. A. Infect. Immun. 2005, 73, 1714–1722. 10.1128/IAI.73.3.1714-1722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F.; Scharf S. S.; Hildebrandt P.; Burian M.; Bernhardt J.; Dhople V.; Kalinka J.; Gutjahr M.; Hammer E.; Volker U. Proteomics 2010, 10, 2801–2811. 10.1002/pmic.201000045. [DOI] [PubMed] [Google Scholar]
- Cox J.; Hein M. Y.; Luber C. A.; Paron I.; Nagaraj N.; Mann M. Mol. Cell. Proteomics 2014, 13, 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C.; Fookes M.; Connor T. R.; Feltwell T.; Feasey N.; Parsons B. N.; Seth-Smith H. M.; Barquist L.; Stedman A.; Humphrey T.; Wigley P.; Peters S. E.; Maskell D. J.; Corander J.; Chabalgoity J. A.; Barrow P.; Parkhill J.; Dougan G.; Thomson N. R. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 863–868. 10.1073/pnas.1416707112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matono T.; Morita M.; Yahara K.; Lee K. I.; Izumiya H.; Kaku M.; Ohnishi M. Open Forum Infect Dis 2017, 4, ofx230. 10.1093/ofid/ofx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.; Lim K. B.; Poduje C. M.; Daniel M.; Gunn J. S.; Hackett M.; Miller S. I. Cell 1998, 95, 189–198. 10.1016/S0092-8674(00)81750-X. [DOI] [PubMed] [Google Scholar]
- Swart A. L.; Hensel M. Virulence 2012, 3, 660–667. 10.4161/viru.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L.; Lorenzi L.; Tomassetti M.; Meresse S.; Gramajo H. Virulence 2017, 8, 975–992. 10.1080/21505594.2016.1270493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro C. K.; Barquist L.; Connor T. R.; Harris S. R.; Clare S.; Stevens M. P.; Arends M. J.; Hale C.; Kane L.; Pickard D. J.; Hill J.; Harcourt K.; Parkhill J.; Dougan G.; Kingsley R. A. PLoS Neglected Trop. Dis. 2015, 9, e0003611. 10.1371/journal.pntd.0003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J. B.; Rock C. O. Prog. Lipid Res. 2013, 52, 249–276. 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger M.; Kasper L.; Hensel M.; Hube B. Int. J. Med. Microbiol. 2018, 308, 215. 10.1016/j.ijmm.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Dandekar T.; Fieselmann A.; Fischer E.; Popp J.; Hensel M.; Noster J. Front. Cell. Infect. Microbiol. 2015, 4, 191. 10.3389/fcimb.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ordonez A.; Fernandez A.; Lopez M.; Arenas R.; Bernardo A. Int. J. Food Microbiol. 2008, 123, 212–219. 10.1016/j.ijfoodmicro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Hurdle J. G.; O’Neill A. J.; Chopra I.; Lee R. E. Nat. Rev. Microbiol. 2011, 9, 62–75. 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta A. L.; Lassak J.; Jung K.; Wilson D. N. FEMS Microbiol Rev. 2014, 38, 1172–1201. 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Del Portillo F.; Jungnitz H.; Rohde M.; Guzman C. A. Infect. Immun. 2000, 68, 2985–2991. 10.1128/IAI.68.5.2985-2991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gutierrez E.; Chidlaw A. C.; Le Gall G.; Bowden S. D.; Tedin K.; Kelly D. J.; Thompson A. PLoS One 2016, 11, e0150687. 10.1371/journal.pone.0150687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden S. D.; Ramachandran V. K.; Knudsen G. M.; Hinton J. C.; Thompson A. PLoS One 2010, 5, e13871. 10.1371/journal.pone.0013871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino A.; Pendleton S.; Zhang N.; Chen W.; Critzer F.; Hanning I. Poult. Sci. 2014, 93, 441–447. 10.3382/ps.2013-03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Cronan J. E. J. Biol. Chem. 2009, 284, 29526–29535. 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.