Abstract
Native ambient mass spectrometry has the potential for simultaneous analysis of native protein structure and spatial distribution within thin tissue sections. Notwithstanding sensitivity, this information can, in principle, be obtained for any protein present with no requirement for a priori knowledge of protein identity. To date, native ambient mass spectrometry has primarily made use of the liquid extraction surface analysis (LESA) sampling technique. Here, we address a fundamental question: Are the protein structures observed following native liquid extraction surface analysis representative of the protein structures within the substrate, or does the extraction process facilitate refolding (or unfolding)? Specifically, our aim was to determine whether protein–ligand complexes observed following LESA are indicative of complexes present in the substrate, or an artifact of the sampling process. The systems investigated were myoglobin and its noncovalently bound heme cofactor, and the Zn-binding protein carbonic anhydrase and its binding with ethoxzolamide. Charge state distributions, drift time profiles, and collision cross sections were determined by liquid extraction surface analysis ion mobility mass spectrometry of native and denatured proteins and compared with those obtained by direct infusion electrospray. The results show that it was not possible to refold denatured proteins with concomitant ligand binding (neither heme, zinc, nor ethoxzolamide) simply by use of native-like LESA solvents. That is, protein–ligand complexes were only observed by LESA MS when present in the substrate.
Liquid extraction surface analysis (LESA) mass spectrometry (MS) is an ambient, automated surface sampling method that was first reported by Kertesz and Van Berkel.1 LESA makes use of a robotic arm which dispenses a small droplet of solvent onto a substrate of interest and extracts its constituents via formation of a liquid junction. The solvent is reaspirated and the conductive tip introduces the sample to the mass spectrometer by nanoelectrospray ionization. A wide array of applications have emerged for LESA, including the analysis of small molecules,2−6 functionalized nanoparticles,7 lipids,8,9 peptides10−12 and proteins.11,13−18 The majority of these studies used denaturing solvents for extraction purposes, however, more recently it has been shown that despite the inherent challenges, proteins can be sampled under near native-like conditions from solid surfaces15,19or directly from tissue20 with LESA.
The extent to which solution-like protein structures are maintained in the gas-phase is a key question underlying native MS.21−24 It is well-documented that protein conformations influence charge state distributions (CSDs) observed in electrospray MS, with folded proteins displaying a narrow distribution of lower charge states and unfolded proteins a broad distribution of higher charge states.25,26 For protein complexes and assemblies, retention of structure is indicated by stoichiometry (binding of ligands and/or proteins result in mass shifts). Lastly, integration of ion mobility spectrometry, such as traveling wave ion mobility spectrometry (TWIMS),27 provides structural information in the form of drift time profiles or arrival time distributions,28 and estimated rotationally averaged collision cross sections (CCS).29−31
In earlier work, we demonstrated the benefits of TWIMS for native LESA MS of proteins embedded in a complex tissue environment.20 TWIMS not only helps to separate proteins from other classes of compounds such as lipids, carbohydrates, and chemical noise, but also enabled us to calculate the CCS of endogenous protein ions (5+ ions of ubiquitin, 4+ ions of β-thymosin 4, and β-thymosin 10).
A fundamental question remains for native LESA MS: Are the structures of the gas-phase ions observed representative of the structures of the protein in the solid substrate, or does the LESA extraction process facilitate refolding (or unfolding)? Initial attempts at addressing this question by use of a cryo-LESA stage were recently presented by Yan et al.32 Those experiments focused solely on the refolding of ubiquitin and the unfolding of hemoglobin. Ubiquitin is not an ideal model for the study of refolding; its native structure is particularly stable,33 even in solutions with high organic content.34,35 Critically, the study by Yan et al. did not consider protein refolding with concomitant noncovalent binding of ligands. A major potential application of native LESA MS is direct analysis of protein-drug binding from pharmaceutically dosed tissue samples. It is therefore imperative to determine whether protein complexes observed in the gas-phase represent those present in the tissue section or are an artifact of the LESA extraction process. Here, we have performed a detailed investigation of two protein standards associated with noncovalent ligand binding: myoglobin, a holo-protein comprised of apo-myoglobin noncovalently bound to a prosthetic heme group, and the 29 kDa Zn-binding protein carbonic anhydrase (Supporting Information (SI) Figure S1). We also considered the binding of carbonic anhydrase with ethoxzolamide, a sulfonamide derivative. For myoglobin, the presence of the holo-form indicates retention of folding, and the apo-form indicates unfolding. For carbonic anhydrase, the presence of the ethoxzolamide ligand and/or Zn2+ indicates retention of folding and the apo-form indicates unfolding.
Solutions of the proteins were either chemically unfolded (by use of organic solvents) or kept in aqueous ammonium acetate in their native states, before drying onto solid substrates, that is, the dried samples comprised either unfolded or folded proteins. The dried proteins were sampled with either native-like solvents or denaturing solvents and their charge state distributions, arrival time distributions and CCS determined. In this way, the potential capacity of LESA extraction for retaining the structure present in the dried sample, or for refolding or unfolding, could be probed. The results from LESA MS were compared with direct infusion electrospray MS of the folded and unfolded protein solutions.
Experimental Section
Materials
Myoglobin from equine skeletal muscle (M0630), carbonic anhydrase from bovine erythrocytes (C5024), and cytochrome c from equine heart (C2506), 6-ethoxy-2-benzothiazolesulfonamide (ethoxzolamide) (333328) and ammonium hydroxide (A6899) were purchased from Sigma-Aldrich (Dorset, UK). LC/MS grade solvents were purchased from Fisher Scientific (Loughborough, UK), ammonium acetate was purchased from J. T. Baker (Deventer, The Netherlands).
Sample Preparation
Protein stocks were prepared by dissolving the lyophilized powders in LC/MS grade water and adjusting their final concentration to 10 μM in the desired solution based on their molar extinction coefficient (13 940 M–1 cm–1, 1280 M–1 cm–1, 50 070 M–1 cm–1 and 18 600 M–1 cm–1, respectively) using a NanoDrop ND-1000 spectrophotometer at 280 nm. Denatured samples were prepared at 10 μM with 50% acetonitrile, 49% water, and 1% formic acid, under acidic conditions at pH 2.2. Native-like samples were prepared using 25 mM ammonium acetate, pH 7.0. A 135 μM ethoxzolamide stock solution was prepared in 5% methanol and 95% water (v/v) and added at final concentration of 500 nM to 10 μM bovine carbonic anhydrase. The protein–ligand solution contained only residual methanol after mixing and was allowed to incubate for 10 min at room temperature.
1.5 μL of each protein sample was pipetted onto a glass microscope slide purchased from VWR, (Lutterworth, UK) and air-dried for various lengths of times (20 min up to 165 min, as stated in the text). For “liquid LESA”, the protein samples were directly sampled from a droplet pipetted onto the surface of the glass slide without further drying.
LESA Traveling Wave Ion Mobility Mass Spectrometry
An image of the glass slides containing the protein samples was acquired using an Epson Perfection V300 flatbed photo scanner. Both the dried or wet droplet locations were selected with the aid of LESA Points software (Advion). Samples were introduced for ion mobility mass spectrometry with an Advion TriVersa NanoMate (Ithaca, NY) using an electroconductive tip coupled to a chip either by direct infusion from a 96 well microtiter plate or by LESA from the surface of a glass slide. LESA sampling solvents were 25 mM ammonium acetate, pH 7.0; 50% acetonitrile, 1% formic acid and 49% water, pH 2.2; or 2.5% ammonium hydroxide and 97.5% water (v/v), pH 11.7, as stated in the text. The robot was operated with the ChipSoft 8.3.3 software at 1.4 kV capillary voltage and 40 bar pressure, aspirating 3 μL solvent, dispensing 3 μL and reaspirating 3.5 μL from the surface of the glass.
The Synapt G2 S (Waters Corp., Wilmslow, UK) mass spectrometer was operated in positive electrospray ionization mode, at a helium flow rate of 180 mL/min and nitrogen flow rate of 90 mL/min. Detailed parameters for each protein are given in the Supporting Information.
External mass calibration was performed with CsI clusters in the corresponding mass ranges. All measurements were performed in triplicate, and at least 80–100 scans were accumulated with a 2 s scan time. CCS calibration was performed with myoglobin and cytochrome c following the protocol described by Ruotolo et al.29 CCS are reported as TWCCSN2→He, that is, helium reference values have been used to calibrate TWIMS measurements made in nitrogen.36 Reference CCS values were obtained from the Bush database30 (myoglobin) and the Clemmer database24 (cytochrome c). Mass spectra and drift times were not smoothed, except for the mass spectra of carbonic anhydrase to which Savitzky Golay function was applied with a smooth window of three channels and five cycles. All data were analyzed in MassLynx V.4.1 (Waters, Wilmslow, UK) and exported for plotting in either Prism GraphPad 6.01 or Origin 2016. Chimera (UCSF, San Francisco, CA)37 was used for viewing pdb structures of horse heart myoglobin (1DWR), and bovine carbonic anhydrase (1 V9E).
Results and Discussion
For the proteins studied, dried protein spots were generated from either neutral solutions or solutions in which the proteins were chemically unfolded through use of acidified organic solvents. Hereafter, these samples are referred to as native protein spots or denatured protein spots.
Myoglobin
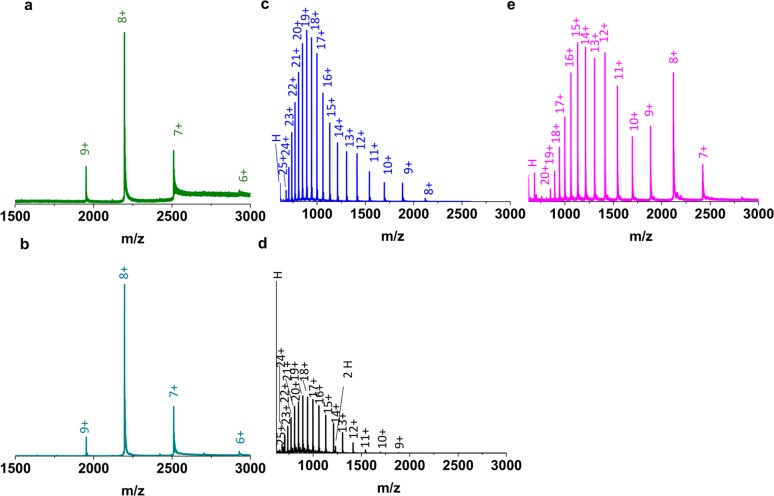
Myoglobin is a well-characterized globular protein which in the condensed-phase comprises eight helices, β turns and a heme group.38−40 The prosthetic heme group41,42 is noncovalently bound in its hydrophobic pocket in a particular orientation, forming the 17 567 Da holo-form. Figure 1a shows a representative LESA mass spectrum obtained from a native myoglobin spot sampled under native conditions, at 60 s dwell time, neutral pH. Figure 1b shows the direct infusion electrospray mass spectrum of myoglobin under native conditions. Both display a narrow charge state distribution from 6+ to 9+ indicating a compact, folded structure. (The relative intensity of the 6+ ions was very low in LESA mode, thus it is excluded from further discussion). By contrast, it is widely accepted that under acidic conditions myoglobin undergoes unfolding.43 The unfolding results in the loss of its prosthetic group to form apo-myoglobin (16 952 Da) and leads to altered interactions between the multiple helices.44Figure 1c and d show representative mass spectra following LESA of a denatured myoglobin spot using denaturing solvents under acidic conditions, and direct infusion electrospray of myoglobin under denaturing conditions, respectively. In both cases, a broad charge state distribution, from 8+ to 25+, of apo-myoglobin was observed, indicating an elongated conformation accommodating a large number of charges on its surface.26,45 Moreover, an abundant peak corresponding to the heme group dominated the mass spectra, especially in direct infusion electrospray mode. (A heme dimer was also seen in the direct infusion mass spectrum that is most likely a product of aggregation46,47).
Figure 1.
(a) LESA mass spectrum of a native myoglobin (holo-) protein spot sampled under native conditions at 60 s dwell time. (b) Direct infusion electrospray mass spectrum of myoglobin (holo-) under native conditions. (c) LESA mass spectrum of a denatured myoglobin (apo-) protein spot sampled under denaturing conditions with LESA at 60 s dwell time. (d) Direct infusion electrospray mass spectrum of myoglobin (apo-) under denaturing conditions. (e) LESA mass spectrum of a denatured myoglobin protein spot sampled under native conditions at 60 s dwell time.
The results above show that for both native and denaturing conditions, the LESA mass spectra mirror the direct infusion mass spectra. To determine whether it is possible to refold unfolded myoglobin during the LESA extraction, denatured protein spots were sampled with native LESA solvent at neutral pH (Figure 1e). The appearance of a multimodal spectrum26 with a CSD of 7+ to 20+ suggests that a number of folded conformers of apo-myoglobin were present; however, refolding of myoglobin with incorporation of heme, that is, reconstitution of holo-myoglobin, was not observed. Feng and Konishi48 showed that acid denatured myoglobin could be reconstituted with the heme group when the pH was increased from 2.2 to pH 6.0–8.0 upon addition of ammonium hydroxide. Similarly, Lee et al.47 showed that myoglobin could be refolded to its holo-form after short-term acid exposure by the subsequent addition of ammonium hydroxide to raise the pH to 7.5 or pH 11.47 Both of these studies47,48 showed that refolding of denatured myoglobin follows a pathway in which apo-myoglobin first folds into a compact form and second binds the heme. We also sampled a denatured myoglobin spot with a LESA solvent comprising 2.5% ammonium hydroxide in water (SI Figure S2). Some refolding (∼3%) of holo-myoglobin was observed.
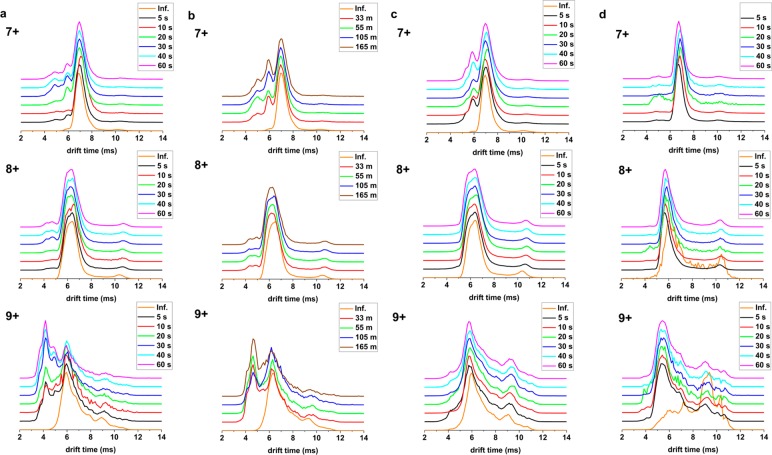
The correlation between CSDs and observed m/z values (apo vs holo) for LESA and direct infusion electrospray, and the fact that LESA extraction did not result in reconstitution of denatured myoglobin suggest that the mass spectra observed following LESA are a true representation of the folding state of the protein on the glass slide. To further investigate, we considered the drift time profiles observed by TWIMS. In particular, we investigated the effect of dwell time (that is, length of extraction) on observed conformers. (The protein spot drying time was kept constant at 20 min in each case). Figure 2a shows the drift time profiles of three charge states of holo-myoglobin (7+, 8+, and 9+) observed following LESA of native protein spots with native solvents at various dwell times together with those for direct infusion electrospray of myoglobin under native conditions. The drift time profile for the 8+ charge state acquired in LESA was in agreement with that from direct infusion performed under native conditions. For the 7+ and 9+ charge states, however, the LESA drift time profiles revealed peaks to the left of the main peak, indicating the presence of a collapsed conformer(s). Counterintuitively, the intensity of these collapsed conformers increased at longer dwell times. We hypothesized that the collapsed conformers are a consequence of drying of the protein onto the glass substrate and that the observed increase in intensity with increasing dwell time is due to improved extraction efficiency of the dried (and collapsed) conformers. That is, the longer the dwell time, the more likely that the collapsed conformers adjacent to the glass substrate will be extracted.
Figure 2.
(a) Mean TWIMS drift time profiles obtained following LESA of native myoglobin (holo-) protein spots sampled under native conditions for various dwell times and mean TWIMS drift time profiles obtained following direct infusion electrospray of myoglobin (holo-) under native conditions (orange line) for 7+, 8+, and 9+ charge states. (b) Mean TWIMS drift time profiles of native myoglobin (holo-) protein spots sampled under native conditions following different drying times at 40 s dwell time and mean TWIMS drift time profiles obtained following direct infusion electrospray of holo-myoglobin (orange line) for 7+, 8+, and 9+ charge states. (c) Mean TWIMS drift time profiles of native myoglobin (holo-) droplets sampled with “liquid LESA” under native conditions at different dwell times and mean TWIMS drift time profiles obtained following direct infusion electrospray of native myoglobin (holo-) (orange line) for 7+, 8+, and 9+ charge states.) (d) Mean TWIMS drift time profiles of denatured myoglobin (apo-) protein spots sampled under native conditions for various dwell times and mean TWIMS drift time profiles obtained following direct infusion electrospray of myoglobin (apo-) under denaturing conditions (orange line) for 7+, 8+, and 9+ charge states.
To investigate the effect of drying prior to sampling by LESA, we varied the drying times of the native myoglobin spots from 33 to 165 min and kept the dwell time fixed at 40 s. Figure 2b shows the drift time profiles obtained for the 7+, 8+, and 9+ charge states. Collapsed features were observed for both the 7+ and 9+ charge states, and the relative intensities of these features increased with increased drying time. For the 9+ charge state, a peak corresponding to a single dominant collapsed conformer was observed, whereas for the 7+ charge state, multiple collapsed conformers were observed. The role of water in mediating the hydrogen-bonding networks and electrostatic interactions central to protein structure is well-established.49 Our results suggest that these interactions are disrupted when the protein is dried onto the glass slide giving rise to the appearance of a minor, more compact population. Next, we deposited a droplet of myoglobin in native solvent onto the glass substrate but did not allow it to dry. The droplet was subsequently sampled by LESA (“liquid LESA”) using various dwell times (Figure 2c). For the 9+ charge state, no collapsed conformers were observed regardless of dwell time, and the liquid LESA drift profiles mirrored that obtained following direct infusion electrospray. For the 7+ charge state, the extent of collapse was reduced to a single dominant conformer whose relative intensity did not differ significantly with dwell time; nevertheless, some collapse was observed in comparison with the direct infusion electrospray results. The reason for this collapse is not clear. It may be that a surface interaction between the myoglobin droplet and the glass slide is responsible.
Returning to the question of LESA-induced refolding, the results shown in Figure 1e suggest that LESA sampling of denatured myoglobin spots with native solvents results in some refolding of the apo-myoglobin but without rebinding of the heme group to form holo-myoglobin. The aggregation of heme has been reported previously46,47 and that may have prevented refolding of holo-myoglobin here. To gain insight into the refolding of the apo-myoglobin, we compared the drift time profiles obtained by TWIMS. Figure 2d shows the drift time profiles of the 7+, 8+, and 9+ charge states of apo-myoglobin following native LESA of denatured protein spots and the drift time profiles obtained from direct infusion of denatured myoglobin (note that the 7+ charge state was not observed in the latter). The remaining charge states (10+ through 18+) are shown in SI Figure S3. The LESA drift time profiles agreed well with the denatured direct infusion samples for most charge states (8+, and 13+ to 18+), but differences were observed for the intermediate charge states (9+ to 12+). Multiple shoulders were observed to the left of the apex of the main peak indicating collapsed conformers. For comparison, we looked at the drift time profiles obtained for apo-myoglobin ions observed following LESA of denatured protein spots with denaturing solvents, see SI Figure S4. Again, for most charge states the drift time profiles were in agreement with those obtained following direct infusion electrospray of denatured myoglobin; however, multiple collapsed conformers were seen for intermediate charge states, for example, 9+ charge state. Interestingly, the distribution of observed conformers in the 9+ charge state differed for the native LESA and the denaturing LESA of the denatured spot, the former demonstrating a skew toward more compact structures. It is well-established that myoglobin acquires a number of conformations at the intermediate and low charge states under denaturing conditions,25,38,40 the existence of which is further supported by solution state measurements.50−52 That is, their distribution heterogeneity has been recognized, and it is plausible that subtle differences occur among these intermediate forms which are observed in LESA and direct infusion electrospray.
Carbonic Anhydrase
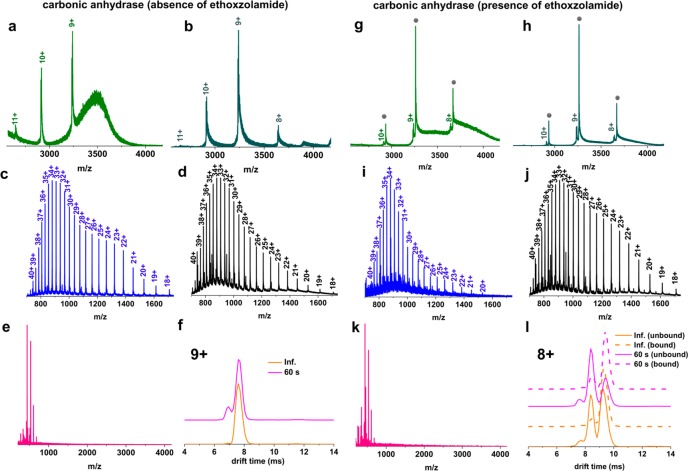
To investigate the effects of LESA on a slightly larger protein, we chose carbonic anhydrase, a β-sheet rich, N-terminally acetylated, zinc-containing metalloenzyme53 with 259 amino acid residues54,55 and its well-characterized binding with the ligand ethoxzolamide. Ethoxzolamide, a sulfonamide derivative, has been shown to bind to both human56,57 and bovine carbonic anhydrases58 by MS and other biophysical tools.56,59−61 Initial experiments were performed in the absence of ligand. We determined the mass of zinc-bound carbonic anhydrase (holo-) to be 29 089 Da in close agreement with the observed mass reported by Gudiksen et al.62 In addition to sodium and ammonium adducts, we also observed the holo-protein bound to a bicarbonate ion (+62 Da) as described previously63 (SI Figure S5). At neutral pH, native LESA of the native protein spot resulted in a narrow CSD from 9+ to 11+, with the 9+ ions being the most dominant (Figure 3a), while direct infusion electrospray under native conditions additionally revealed a low intensity peak corresponding to the 8+ charge state (Figure 3b). Yin and Loo64 observed charge states 9+ to 11+, with the 10+ charge states being the most abundant, at pH 6.8 in the absence of a supercharging reagent. Under denaturing conditions at pH 2.2, carbonic anhydrase displayed a CSD of 18+ to 40+ (Figure 3c, d) with the release of Zn2+. A bimodal distribution appears in the denatured mass spectra that suggest an expanded, flexible structure, potentially with multiple conformers. Nabuchi et al.65 noted that at pH 3.8 the mass spectrum of carbonic anhydrase shows a bimodal distribution, comprised of the apoprotein with a CSD of 11+ to 38+ and the holo (zinc-bound)-protein with a CSD of 10+ to 20+. In a follow-up study,66 these researchers demonstrated that a drop in pH from 5.0 to 3.6 resulted in the loss of Zn2+ and in significant unfolding of the N-terminus. We attempted refolding of denatured bovine carbonic anhydrase by sampling with native LESA solvent (Figure 3e), however, peaks corresponding to well-resolved charge states were not observed presumably due to aggregation of the protein, an occurrence that has been documented with various alcohols.67Figure 3f shows the drift time profile of the 9+ charge state of the zinc-bound holo-protein obtained at neutral pH following either direct infusion or LESA under native conditions. The LESA drift time profiles revealed the presence of a minor, more compact conformer in addition to the dominant conformer. This minor conformer is likely due to drying of the sample rather than LESA itself as described above. The LESA drift time profiles of the 10+ and 11+ charge states acquired at 60 s dwell time are in excellent agreement with those observed following direct infusion electrospray (SI Figure S6).
Figure 3.
(a) LESA mass spectrum of native bovine carbonic anhydrase protein spot acquired under native conditions. (b) Direct infusion electrospray mass spectrum of carbonic anhydrase acquired under native conditions. (c) LESA mass spectrum of denatured bovine carbonic anhydrase protein spot acquired under denaturing conditions at 60 s dwell time. (d) Direct infusion electrospray mass spectrum of carbonic anhydrase acquired under denaturing conditions. (e) LESA mass spectrum of a denatured bovine carbonic anhydrase protein spot sampled under native conditions at 60 s dwell time. (f) Mean TWIMS drift time profiles obtained from native carbonic anhydrase spots with LESA at 60 s dwell time under native conditions (pink line) and following direct infusion electrospray (orange line) for the 9+ charge state ions. (g) LESA mass spectrum of a 10 μM native bovine carbonic anhydrase/500 nM ethoxzolamide protein spot acquired under native conditions at 60 s dwell time. Gray circles indicate ligand-bound protein peaks. (h) Direct infusion electrospray mass spectrum of a 10 μM native bovine carbonic anhydrase/500 nM ethoxzolamide acquired under native conditions. (i) LESA mass spectrum of a denatured 10 μM bovine carbonic anhydrase/500 nM ethoxzolamide protein spot acquired under denaturing conditions. (j) Direct infusion electrospray mass spectrum of 10 μM carbonic anhydrase/500 nM ethoxzolamide acquired under denaturing conditions. (k) LESA mass spectrum of a 10 μM denatured bovine carbonic anhydrase/500 nM ethoxzolamide protein spot sampled under native conditions at 60 s dwell time. (l) Mean TWIMS drift time profiles obtained from native, unbound (pink continuous line) or ligand bound (pink dotted line) carbonic anhydrase sampled with LESA at 60 s dwell time, and from unbound (orange continuous line) or ligand bound (orange dotted line) carbonic anhydrase obtained with direct infusion under native conditions, for the 8+ ions.
We subsequently probed the binding of carbonic anhydrase with one of its well-characterized ligands, ethoxzolamide.60,61 Sulfonamide derivatives, for example, ethoxzolamide, are thought to interact with carbonic anhydrase via direct association with the Zn2+ in its active site and key amino acid residues in the hydrophobic pocket.68 Here, 10 μM bovine carbonic anhydrase and 500 nM ethoxzolamide showed a strong interaction, as revealed by a 258.3 Da mass shift, with the majority of the protein (86%) being in a ligand bound form. Under native conditions, for both LESA and direct infusion electrospray, the charge state distribution was narrow, spanning charge states from 8+ to 10+ (Figure 3g and h). Under denaturing conditions, a CSD of 20+ to 40+ was observed following both LESA MS and direct infusion electrospray MS (Figure 3i and j), both of which revealed the loss of Zn2+ and the ligand. LESA sampling of native protein/ethoxzolamide spots under denaturing conditions yielded a similar mass spectrum with a wide charge state distribution with 19+ to 40+ ions and without Zn2+ or ligand bound (SI Figure S7). This broad CSD demonstrates that the tertiary structure of bovine carbonic anhydrase had been disrupted, leading to major structural rearrangements in its active site, hence dissociation of both Zn2+ and the ligand. Significantly, native LESA sampling of denatured protein/ethoxzolamide spots did not result in detection of the protein–ligand complex (Figure 3k). Neither did inclusion of 2.5% ammonium hydroxide in the LESA sampling solvent result in reconstitution of the protein–ligand complex (SI Figure S8). Inspection of drift time profiles for the 9+ and 10+ ions in both the unbound and bound forms showed excellent agreement between LESA MS and direct infusion electrospray MS (Figure S9). For the 8+ charge state we observed good agreement between drift times associated with the ligand bound ions for both LESA MS and direct infusion electrospray MS (Figure 3l). A small discrepancy was observed, however, in the relative intensity of two different unbound conformational populations. For LESA MS, the dominant conformer has drift time 8.4 ms, whereas for direct infusion electrospray MS, the conformer with drift time 9.4 ms is dominant (Figure 3l). (Both conformers are observed in each case). We attribute this deviation to the drying process as discussed above, and point out that the relative abundance of these ions was particularly low in the mass spectrum.
CCS Measurements of Myoglobin and Carbonic Anhydrase
For the proteins studied, CCSs were calculated from TWIMS measurements for the conformers, including minor conformers, observed in charge states under native LESA conditions (of native protein spots) (SI Figure S10). Experimental CCS values were compared with CCSs calculated from the results of direct infusion electrospray under native conditions (Table 1). In all cases, there was good agreement between the CCS calculated following LESA and direct infusion electrospray.
Table 1. Mean TWCCSN2→He Values of Myoglobin and Carbonic Anhydrase (In the Presence or Absence of Ethoxzolamide) With ±1 STD Acquired on a Synapt G2 S Instrument in 25 mM Ammonium Acetate, pH 7.0, (n = 3)a.
| protein | charge | mean TWCCSN2→He ± STD inf. (Å2) | mean TWCCSN2→He ± STD LESA (Å2) | ||
|---|---|---|---|---|---|
| myoglobin | 7+ | 1620 ± 62* | 1630 ± 68* | ||
| 1460 ± 55 | |||||
| 1260 ± 54 | |||||
| 8+ | 2430 ± 95 | 2480 ± 97 | |||
| 1750 ± 46* | 1730 ± 66* | ||||
| 1420 ± 60 | |||||
| 1320 ± 43 | |||||
| 9+ | 2460 ± 107 | 2530 ± 68 | |||
| 1890 ± 66* | 1890 ± 79* | ||||
| 1490 ± 64 | |||||
| carbonic anhydrase (absence of ethoxzolamide) | 8+ | 2310 ± 46* | |||
| 9+ | 2200 ± 107* | 2270 ± 44* | |||
| 2140 ± 77 | |||||
| 10+ | 2220 ± 55* | 2230 ± 24* | |||
| 11+ | 2230 ± 38* | 2250 ± 49* | |||
| carbonic anhydrase (presence of ethoxzolamide) | 8+ | unbound | bound | unbound | bound |
| 2280 ± 19* | 2290 ± 8* | 2320 ± 16 | 2320 ± 16* | ||
| 2130 ± 6 | 2150 ± 6 | 2150 ± 15* | 2150 ± 6 | ||
| 2010 ± 13 | 1990 ± 24 | ||||
| 9+ | 2230 ± 18* | 2250 ± 5* | 2250 ± 5* | 2270 ± 6* | |
| 2100 ± 2 | 2110 ± 11 | ||||
| 10+ | 2220 ± 19* | 2230 ± 4* | 2250 ± 18* | 2260 ± 4* | |
The asterisk denotes the most abundant conformer.
Literature CCS values for each protein in different charge states are shown in SI Tables S1 and S2. A range of values have been reported, and these can be explained by the use of different buffer gases and experimental conditions,38,40,69,70 and by the presence of multiple conformers populated for a given charge state in the gas-phase.38,40 These observations highlight the importance of reporting instrumental parameters that influence the mobility of ions, such as the type of drift gas, temperature, pressure, and electric field, and the method of sample preparation (solvent, any additives and pH) to enable comparison of experimentally derived CCSs between different laboratories.36 In all cases, our results obtained via LESA or direct infusion electrospray fall within the CCSHe or CCSN2→He ranges reported in the literature, or are within 4% (myoglobin, 8+). As discussed above, we attribute the presence of additional, lower abundance, more compact features in the LESA TWIMS of holo-myoglobin and carbonic anhydrase to dehydration of the proteins during drying onto the glass slide, and not the LESA extraction process itself.
Conclusions
Proteins and peptides are essential components of a myriad of cellular functions. Investigation of changes in their conformation can provide important insights into the breakdown of biochemical pathways and disease states. One approach to probe conformation of proteins is the use native MS. Combining native MS with mass spectrometry imaging (MSI) has the potential to provide not just structural information but also information relating to the spatial distribution of proteins in tissue. To this end, we have already begun exploring native MS imaging of proteins from thin tissue sections, using TWIMS coupled to LESA, and reporting respective TWCCSN2→He.20 In this study, we sought to address a fundamental question: to what extent do the protein structures, specifically protein–ligand complexes, observed after LESA extraction correlate with those present in the solid substrate? We selected two well-characterized, protein standards: a medium sized α-helix rich protein with a cofactor (holo-myoglobin, 17.5 kDa) and a larger metal-bound β-sheet rich protein (carbonic anhydrase, 29 kDa) and its well-characterized ligand. Both proteins displayed well-preserved tertiary structures, maintaining the prosthetic group or metal ion/ligand in the active center, following native LESA of native protein spots. It was not possible, however, to refold denatured myoglobin with incorporation of the heme group simply by use of native-like LESA solvents. Addition of a strong base to the LESA solvent elicited some (3%) refolding. Neither was it possible to refold denatured carbonic anhydrase with incorporation of Zn2+ or its ligand ethoxzolamide. That is, our results suggest that observation of protein complexes in native LESA is indicative of their presence in the substrate and not simply an artifact of the sampling process.
Interestingly, for some charge states, the drift time profiles obtained following LESA of the protein spots revealed the presence of more compact conformers which were not observed following direct infusion electrospray. The abundance of these conformers increased with both drying time and dwell time, suggesting that they are the result of dehydration at the surface of the glass substrate. (The longer the dwell time, the more efficient the extraction of the lower reaches of the protein spots). Happily, when considering the question of the relationship between LESA-sampled structures and those present in the substrate, this phenomenon is more of a challenge for purified protein standards in aqueous solution, as is the case here, than for endogenous proteins in thin tissue sections which are not dried but are in their physiological environment. An alternative and exciting consideration is that the effects of dehydration on structure are of great interest to protein scientists. Our results suggest that native LESA MS may provide a tool for interrogation of the effects of water removal and further work in this area is warranted.
Acknowledgments
E.I.T. and H.J.C. are funded by EPSRC (EP/R018367/1). H.J.C. is an EPSRC Established Career Fellow (EP/L023490/1 and EP/S002979/1). We thank Emma Sisley and Dr Chi Tsang for technical help and Dr Aneika Leney for helpful discussions. We also thank Dr Klaudia I. Kocurek for assistance in preparing the abstract figure. Supplementary data supporting this research is openly available from the University of Birmingham data archive at DOI: 10.25500/edata.bham.00000375.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b02075.
Figure S1, pdb structure of myoglobin and carbonic anhydrase. Figure S2, mass spectrum of denatured myoglobin (apo-) protein spot sampled with 2.5% ammonium hydroxide. Resultant deconvoluted mass of apo- and holo-myoglobin. Figure S3, mean TWIMS drift time profiles of charge states 10+ to 18+ for denatured myoglobin (apo-) obtained following native LESA. Figure S4, mean TWIMS drift time profiles of denatured myoglobin (apo-) obtained following LESA under denaturing conditions or direct infusion electrospray. Figure S5, deconvoluted mass spectra of carbonic anhydrase. Figure S6, mean TWIMS drift time profiles of native carbonic anhydrase for 10+ and 11+ charge state ions. Figure S7, mass spectrum of carbonic anhydrase deposited under native conditions and sampled with LESA under denaturing conditions. Figure S8, LESA mass spectrum of denatured 10 μM carbonic anhydrase/500 nM ethoxzolamide protein spot sampled with 2.5% ammonium hydroxide. Figure S9. mean TWIMS drift time profiles of native carbonic anhydrase/500 nM ethoxzolamide for 9+ and 10+ charge state ions. Figure S10, TWCCSN2→He profiles of native myoglobin and carbonic anhydrase in the presence or absence of ethoxzolamide. Table S1. mean experimental TWCCSN2→He ± 1 STD of myoglobin acquired by direct electrospray ionization or native LESA MS compared with previously published literature values. Table S2, mean experimental TWCCSN2→He ± 1 STD of carbonic anhydrase acquired by direct electrospray ionization- or native LESA MS compared with previously published literature values. Detailed experimental section (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kertesz V.; Van Berkel G. J. Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform. J. Mass Spectrom. 2010, 45 (3), 252–260. 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- Parson W. B.; Koeniger S. L.; Johnson R. W.; Erickson J.; Tian Y.; Stedman C.; Schwartz A.; Tarcsa E.; Cole R.; Van Berkel G. J. Analysis of chloroquine and metabolites directly from whole-body animal tissue sections by liquid extraction surface analysis (LESA) and tandem mass spectrometry. J. Mass Spectrom. 2012, 47 (11), 1420–1428. 10.1002/jms.3068. [DOI] [PubMed] [Google Scholar]
- Swales J. G.; Tucker J. W.; Spreadborough M. J.; Iverson S. L.; Clench M. R.; Webborn P. J.; Goodwin R. J. Mapping drug distribution in brain tissue using liquid extraction surface analysis mass spectrometry imaging. Anal. Chem. 2015, 87 (19), 10146–10152. 10.1021/acs.analchem.5b02998. [DOI] [PubMed] [Google Scholar]
- Paine M. R.; Barker P. J.; Maclauglin S. A.; Mitchell T. W.; Blanksby S. J. Direct detection of additives and degradation products from polymers by liquid extraction surface analysis employing chip-based nanospray mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26 (4), 412–418. 10.1002/rcm.6116. [DOI] [PubMed] [Google Scholar]
- Kim A. J.; Basu S.; Glass C.; Ross E. L.; Agar N.; He Q.; Calligaris D. Unique Intradural Inflammatory Mass Containing Precipitated Morphine: Confirmatory Analysis by LESA-MS and MALDI-MS. Pain Pract 2018, 18 (7), 889–894. 10.1111/papr.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M.; Gonzalez I.; Genilloud O.; Singh S. B.; Svatos A. Direct mass spectrometric screening of antibiotics from bacterial surfaces using liquid extraction surface analysis. Rapid Commun. Mass Spectrom. 2012, 26 (20), 2477–2482. 10.1002/rcm.6365. [DOI] [PubMed] [Google Scholar]
- Yan X.; Xu L.; Bi C.; Duan D.; Chu L.; Yu X.; Wu Z.; Wang A.; Sun K. Lactoferrin-modified rotigotine nanoparticles for enhanced nose-to-brain delivery: LESA-MS/MS-based drug biodistribution, pharmacodynamics, and neuroprotective effects. Int. J. Nanomed. 2018, 13, 273–281. 10.2147/IJN.S151475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelsbach M.; Varesio E.; Hopfgartner G. Liquid extraction surface analysis (LESA) of hydrophobic TLC plates coupled to chip-based nanoelectrospray high-resolution mass spectrometry. Chimia 2014, 68 (3), 150–154. 10.2533/chimia.2014.150. [DOI] [PubMed] [Google Scholar]
- Hall Z.; Chu Y.; Griffin J. L. Liquid Extraction Surface Analysis Mass Spectrometry Method for Identifying the Presence and Severity of Nonalcoholic Fatty Liver Disease. Anal. Chem. 2017, 89 (9), 5161–5170. 10.1021/acs.analchem.7b01097. [DOI] [PubMed] [Google Scholar]
- Montowska M.; Alexander M. R.; Tucker G. A.; Barrett D. A. Rapid detection of peptide markers for authentication purposes in raw and cooked meat using ambient liquid extraction surface analysis mass spectrometry. Anal. Chem. 2014, 86 (20), 10257–10265. 10.1021/ac502449w. [DOI] [PubMed] [Google Scholar]
- Ryan D. J.; Nei D.; Prentice B. M.; Rose K. L.; Caprioli R. M.; Spraggins J. M. Protein identification in imaging mass spectrometry through spatially targeted liquid micro-extractions. Rapid Commun. Mass Spectrom. 2018, 32 (5), 442–450. 10.1002/rcm.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont L.; Baumert M.; Ogrinc Potocnik N.; Allen M.; Vreeken R.; Heeren R. M. A.; Porta T. Integration of Ion Mobility MS(E) after Fully Automated, Online, High-Resolution Liquid Extraction Surface Analysis Micro-Liquid Chromatography. Anal. Chem. 2017, 89 (20), 11143–11150. 10.1021/acs.analchem.7b03512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. L.; Creese A. J.; Baumert M.; Griffiths P.; Bunch J.; Cooper H. J. Hemoglobin variant analysis via direct surface sampling of dried blood spots coupled with high-resolution mass spectrometry. Anal. Chem. 2011, 83 (6), 2265–2270. 10.1021/ac1030804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. J.; Bunch J.; Cooper H. J. Dried blood spot proteomics: surface extraction of endogenous proteins coupled with automated sample preparation and mass spectrometry analysis. J. Am. Soc. Mass Spectrom. 2013, 24 (8), 1242–1249. 10.1007/s13361-013-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. J.; Griffiths R. L.; Edwards R. L.; Cooper H. J. Native Liquid Extraction Surface Analysis Mass Spectrometry: Analysis of Noncovalent Protein Complexes Directly from Dried Substrates. J. Am. Soc. Mass Spectrom. 2015, 26 (8), 1320–1327. 10.1007/s13361-015-1152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall E. C.; Bunch J.; Cooper H. J. Direct analysis of intact proteins from Escherichia coli colonies by liquid extraction surface analysis mass spectrometry. Anal. Chem. 2014, 86 (21), 10504–10510. 10.1021/ac503349d. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L.; Cooper H. J. Direct Tissue Profiling of Protein Complexes: Toward Native Mass Spectrometry Imaging. Anal. Chem. 2016, 88 (1), 606–609. 10.1021/acs.analchem.5b03993. [DOI] [PubMed] [Google Scholar]
- Kocurek K. I.; Stones L.; Bunch J.; May R. C.; Cooper H. J. Top-Down LESA Mass Spectrometry Protein Analysis of Gram-Positive and Gram-Negative Bacteria. J. Am. Soc. Mass Spectrom. 2017, 28 (10), 2066–2077. 10.1007/s13361-017-1718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov V. A.; Griffiths R. L.; Cooper H. J. Liquid extraction surface analysis for native mass spectrometry: Protein complexes and ligand binding. Int. J. Mass Spectrom. 2017, 420, 43–50. 10.1016/j.ijms.2016.09.011. [DOI] [Google Scholar]
- Griffiths R. L.; Sisley E. K.; Lopez-Clavijo A. F.; Simmonds A. L.; Styles I. B.; Cooper H. J. Native mass spectrometry imaging of intact proteins and protein complexes in thin tissue sections. Int. J. Mass Spectrom. 2019, 437, 23–29. 10.1016/j.ijms.2017.10.009. [DOI] [Google Scholar]
- McLafferty F. W.; Castro S.; Breuker K. Multi-step evolution of protein conformation on electrospray into the gas phase. Eur. J. Mass Spectrom. 2010, 16 (3), 437–42. 10.1255/ejms.1058. [DOI] [PubMed] [Google Scholar]
- Bakhtiari M.; Konermann L. Protein Ions Generated by Native Electrospray Ionization: Comparison of Gas Phase, Solution, and Crystal Structures. J. Phys. Chem. B 2019, 123 (8), 1784–1796. 10.1021/acs.jpcb.8b12173. [DOI] [PubMed] [Google Scholar]
- Hamdy O. M.; Julian R. R. Reflections on charge state distributions, protein structure, and the mystical mechanism of electrospray ionization. J. Am. Soc. Mass Spectrom. 2012, 23 (1), 1–6. 10.1007/s13361-011-0284-8. [DOI] [PubMed] [Google Scholar]
- Shelimov K. B.; Clemmer D. E.; Hudgins R. R.; Jarrold M. F. Protein Structure in Vacuo: Gas-Phase Conformations of BPTI and Cytochrome c. J. Am. Chem. Soc. 1997, 119 (9), 2240–2248. 10.1021/ja9619059. [DOI] [Google Scholar]
- Mohimen A.; Dobo A.; Hoerner J. K.; Kaltashov I. A. A chemometric approach to detection and characterization of multiple protein conformers in solution using electrospray ionization mass spectrometry. Anal. Chem. 2003, 75 (16), 4139–4147. 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- Grandori R.; Santambrogio C.; Brocca S.; Invernizzi G.; Lotti M. Electrospray-ionization mass spectrometry as a tool for fast screening of protein structural properties. Biotechnol. J. 2009, 4 (1), 73–87. 10.1002/biot.200800250. [DOI] [PubMed] [Google Scholar]
- Giles K.; Wildgoose J. L.; Langridge D. J.; Campuzano I. A method for direct measurement of ion mobilities using a travelling wave ion guide. Int. J. Mass Spectrom. 2010, 298 (1), 10–16. 10.1016/j.ijms.2009.10.008. [DOI] [Google Scholar]
- Marchand A.; Livet S.; Rosu F.; Gabelica V. Drift Tube Ion Mobility: How to Reconstruct Collision Cross Section Distributions from Arrival Time Distributions?. Anal. Chem. 2017, 89 (23), 12674–12681. 10.1021/acs.analchem.7b01736. [DOI] [PubMed] [Google Scholar]
- Ruotolo B. T.; Benesch J. L.; Sandercock A. M.; Hyung S. J.; Robinson C. V. Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3 (7), 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- Bush M. F.; Hall Z.; Giles K.; Hoyes J.; Robinson C. V.; Ruotolo B. T. Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem. 2010, 82 (22), 9557–9565. 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- Smith D. P.; Knapman T. W.; Campuzano I.; Malham R. W.; Berryman J. T.; Radford S. E.; Ashcroft A. E. Deciphering drift time measurements from travelling wave ion mobility spectrometry-mass spectrometry studies. Eur. J. Mass Spectrom. 2009, 15 (2), 113–130. 10.1255/ejms.947. [DOI] [PubMed] [Google Scholar]
- Yan B.; Taylor A. J.; Bunch J. Cryo-LESA Mass Spectrometry—a Step Towards Truly Native Surface Sampling of Proteins. J. Am. Soc. Mass Spectrom. 2019, 30 (7), 1179–1189. 10.1007/s13361-019-02178-7. [DOI] [PubMed] [Google Scholar]
- Wyttenbach T.; Bowers M. T. Structural stability from solution to the gas phase: native solution structure of ubiquitin survives analysis in a solvent-free ion mobility-mass spectrometry environment. J. Phys. Chem. B 2011, 115 (42), 12266–12275. 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- Shi H.; Clemmer D. E. Evidence for two new solution states of ubiquitin by IMS-MS analysis. J. Phys. Chem. B 2014, 118 (13), 3498–3506. 10.1021/jp4097327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H.; Atlasevich N.; Merenbloom S. I.; Clemmer D. E. Solution dependence of the collisional activation of ubiquitin [M + 7H](7+) ions. J. Am. Soc. Mass Spectrom. 2014, 25 (12), 2000–2008. 10.1007/s13361-014-0834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabelica V.; Shvartsburg A. A.; Afonso C.; Barran P.; Benesch J. L. P.; Bleiholder C.; Bowers M. T.; Bilbao A.; Bush M. F.; Campbell J. L.; Campuzano I. D. G.; Causon T.; Clowers B. H.; Creaser C. S.; De Pauw E.; Far J.; Fernandez-Lima F.; Fjeldsted J. C.; Giles K.; Groessl M.; Hogan C. J. Jr.; Hann S.; Kim H. I.; Kurulugama R. T.; May J. C.; McLean J. A.; Pagel K.; Richardson K.; Ridgeway M. E.; Rosu F.; Sobott F.; Thalassinos K.; Valentine S. J.; Wyttenbach T. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom. Rev. 2019, 38 (3), 291–320. 10.1002/mas.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25 (13), 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Schenk E. R.; Almeida R.; Miksovska J.; Ridgeway M. E.; Park M. A.; Fernandez-Lima F. Kinetic intermediates of holo- and apo-myoglobin studied using HDX-TIMS-MS and molecular dynamic simulations. J. Am. Soc. Mass Spectrom. 2015, 26 (4), 555–563. 10.1007/s13361-014-1067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.; Hoffmann W.; Warnke S.; Bowers M. T.; Pagel K.; von Helden G. Retention of Native Protein Structures in the Absence of Solvent: A Coupled Ion Mobility and Spectroscopic Study. Angew. Chem., Int. Ed. 2016, 55 (45), 14173–14176. 10.1002/anie.201606029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J. C.; Jurneczko E.; Stow S. M.; Kratochvil I.; Kalkhof S.; McLean J. A. Conformational Landscapes of Ubiquitin, Cytochrome c, and Myoglobin: Uniform Field Ion Mobility Measurements in Helium and Nitrogen Drift Gas. Int. J. Mass Spectrom. 2018, 427, 79–90. 10.1016/j.ijms.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. V.; Brayer G. D. High-resolution study of the three-dimensional structure of horse heart metmyoglobin. J. Mol. Biol. 1990, 213 (4), 885–897. 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- Santucci R.; Ascoli F.; La Mar G. N.; Parish D. W.; Smith K. M. Horse heart myoglobin reconstituted with a symmetrical heme. A circular dichroism study. Biophys. Chem. 1990, 37 (1–3), 251–255. 10.1016/0301-4622(90)88024-M. [DOI] [PubMed] [Google Scholar]
- Puett D. The equilibrium unfolding parameters of horse and sperm whale myoglobin. Effects of guanidine hydrochloride, urea, and acid. J. Biol. Chem. 1973, 248 (13), 4623–4634. [PubMed] [Google Scholar]
- Barrick D.; Hughson F. M.; Baldwin R. L. Molecular mechanisms of acid denaturation. The role of histidine residues in the partial unfolding of apomyoglobin. J. Mol. Biol. 1994, 237 (5), 588–601. 10.1006/jmbi.1994.1257. [DOI] [PubMed] [Google Scholar]
- Dobo A.; Kaltashov I. A. Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESI MS. Anal. Chem. 2001, 73 (20), 4763–4773. 10.1021/ac010713f. [DOI] [PubMed] [Google Scholar]
- Adams P. A. The kinetics and mechanism of the recombination reaction between apomyoglobin and haemin. Biochem. J. 1976, 159 (2), 371–376. 10.1042/bj1590371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. W.; Chen Y. L.; Konermann L. Reconstitution of acid-denatured holomyoglobin studied by time-resolved electrospray ionization mass spectrometry. Anal. Chem. 1999, 71 (19), 4154–4159. 10.1021/ac9904664. [DOI] [PubMed] [Google Scholar]
- Feng R.; Konishi Y. Stepwise refolding of acid-denatured myoglobin: evidence from electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 1993, 4 (8), 638–645. 10.1016/1044-0305(93)85028-V. [DOI] [PubMed] [Google Scholar]
- Levy Y.; Onuchic J. N. Water and proteins: a love-hate relationship. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (10), 3325–3326. 10.1073/pnas.0400157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmanshin R.; Gulotta M.; Dyer R. B.; Callender R. H. Structures of apomyoglobin’s various acid-destabilized forms. Biochemistry 2001, 40 (17), 5127–5136. 10.1021/bi002255v. [DOI] [PubMed] [Google Scholar]
- Gulotta M.; Gilmanshin R.; Buscher T. C.; Callender R. H.; Dyer R. B. Core Formation in Apomyoglobin: Probing the Upper Reaches of the Folding Energy Landscape. Biochemistry 2001, 40 (17), 5137–5143. 10.1021/bi002256n. [DOI] [PubMed] [Google Scholar]
- Fink A. L.; Oberg K. A.; Seshadri S. Discrete intermediates versus molten globule models for protein folding: characterization of partially folded intermediates of apomyoglobin. Folding Des. 1998, 3 (1), 19–25. 10.1016/S1359-0278(98)00005-4. [DOI] [PubMed] [Google Scholar]
- Lindskog S.; Coleman J. E. The catalytic mechanism of carbonic anhydrase. Proc. Natl. Acad. Sci. U. S. A. 1973, 70 (9), 2505–2508. 10.1073/pnas.70.9.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R.; Sato T.; Ikai A.; Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 A resolution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60 (Pt 4), 792–795. 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- Eriksson A. E.; Liljas A. Refined structure of bovine carbonic anhydrase III at 2.0 A resolution. Proteins: Struct., Funct., Genet. 1993, 16 (1), 29–42. 10.1002/prot.340160104. [DOI] [PubMed] [Google Scholar]
- Jecklin M. C.; Schauer S.; Dumelin C. E.; Zenobi R. Label-free determination of protein-ligand binding constants using mass spectrometry and validation using surface plasmon resonance and isothermal titration calorimetry. J. Mol. Recognit. 2009, 22 (4), 319–29. 10.1002/jmr.951. [DOI] [PubMed] [Google Scholar]
- Nguyen G. T. H.; Tran T. N.; Podgorski M. N.; Bell S. G.; Supuran C. T.; Donald W. A. Nanoscale Ion Emitters in Native Mass Spectrometry for Measuring Ligand-Protein Binding Affinities. ACS Cent. Sci. 2019, 5 (2), 308–318. 10.1021/acscentsci.8b00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu H.; Pham N. B.; Quinn R. J. Direct screening of natural product extracts using mass spectrometry. J. Biomol. Screening 2008, 13 (4), 265–75. 10.1177/1087057108315739. [DOI] [PubMed] [Google Scholar]
- Iyer R.; Barrese A. A. 3rd; Parakh S.; Parker C. N.; Tripp B. C. Inhibition profiling of human carbonic anhydrase II by high-throughput screening of structurally diverse, biologically active compounds. J. Biomol. Screening 2006, 11 (7), 782–91. 10.1177/1087057106289403. [DOI] [PubMed] [Google Scholar]
- Conroy C. W.; Maren T. H. The effect of temperature on the binding of sulfonamides to carbonic anhydrase isoenzymes I, II, and IV. Mol. Pharmacol. 1995, 48 (3), 486–91. [PubMed] [Google Scholar]
- Di Fiore A.; Pedone C.; Antel J.; Waldeck H.; Witte A.; Wurl M.; Scozzafava A.; Supuran C. T.; De Simone G. Carbonic anhydrase inhibitors: the X-ray crystal structure of ethoxzolamide complexed to human isoform II reveals the importance of thr200 and gln92 for obtaining tight-binding inhibitors. Bioorg. Med. Chem. Lett. 2008, 18 (8), 2669–74. 10.1016/j.bmcl.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Gudiksen K. L.; Urbach A. R.; Gitlin I.; Yang J.; Vazquez J. A.; Costello C. E.; Whitesides G. M. Influence of the Zn(II) Cofactor on the Refolding of Bovine Carbonic Anhydrase after Denaturation with Sodium Dodecyl Sulfate. Anal. Chem. 2004, 76 (24), 7151–7161. 10.1021/ac0488560. [DOI] [PubMed] [Google Scholar]
- Schachner L. F.; Ives A. N.; McGee J. P.; Melani R. D.; Kafader J. O.; Compton P. D.; Patrie S. M.; Kelleher N. L. Standard Proteoforms and Their Complexes for Native Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30 (7), 1190–1198. 10.1007/s13361-019-02191-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S.; Loo J. A. Top-down mass spectrometry of supercharged native protein–ligand complexes. Int. J. Mass Spectrom. 2011, 300 (2), 118–122. 10.1016/j.ijms.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabuchi Y.; Hirose K.; Takayama M. pH Dependence of the Number of Discrete Conformers of Carbonic Anhydrase 2, as Evaluated from Collision Cross-Section Using Ion Mobility Coupled with Electrospray Ionization. Mass Spectrom. 2018, 7 (1), A0064. 10.5702/massspectrometry.A0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabuchi Y.; Hirose K.; Takayama M. Ion mobility and collision-induced dissociation analysis of carbonic anhydrase 2. Anal. Chem. 2010, 82 (21), 8890–8896. 10.1021/ac101482b. [DOI] [PubMed] [Google Scholar]
- Es-haghi A.; Ebrahim-Habibi A.; Sabbaghian M.; Nemat-Gorgani M. Amyloid-like aggregates formation by bovine apo-carbonic anhydrase in various alcohols: A comparative study. Int. J. Biol. Macromol. 2016, 92, 573–580. 10.1016/j.ijbiomac.2016.07.083. [DOI] [PubMed] [Google Scholar]
- Supuran C. T. How many carbonic anhydrase inhibition mechanisms exist?. J. Enzyme Inhib. Med. Chem. 2016, 31 (3), 345–360. 10.3109/14756366.2015.1122001. [DOI] [PubMed] [Google Scholar]
- Jurneczko E.; Kalapothakis J.; Campuzano I. D. G.; Morris M.; Barran P. E. Effects of Drift Gas on Collision Cross Sections of a Protein Standard in Linear Drift Tube and Traveling Wave Ion Mobility Mass Spectrometry. Anal. Chem. 2012, 84 (20), 8524–8531. 10.1021/ac301260d. [DOI] [PubMed] [Google Scholar]
- Harrison J. A.; Kelso C.; Pukala T. L.; Beck J. L. Conditions for Analysis of Native Protein Structures Using Uniform Field Drift Tube Ion Mobility Mass Spectrometry and Characterization of Stable Calibrants for TWIM-MS. J. Am. Soc. Mass Spectrom. 2019, 30 (2), 256–267. 10.1007/s13361-018-2074-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.