Abstract
There is much interest in using nanosensors to monitor biologically relevant species such as glucose, or cellular pH, as these often become dysregulated in diseases such as cancer. This information is often inaccessible at depth in biological tissue, due to the highly scattering nature of tissue. Here we show that gold nanoparticles labeled with pH-sensitive reporter molecules can monitor pH at depth in biological tissues. This was achieved using deep Raman spectroscopy (spatially offset Raman and transmission Raman) in combination with surface-enhanced Raman spectroscopy, allowing chemical information to be retrieved significantly deeper than conventional Raman spectroscopy permits. Combining these approaches with chemometrics enabled pH changes to be monitored with an error of ±∼0.1 pH units noninvasively through 22 mm of soft tissue. This development opens the opportunity for the next generation of light-based medical diagnostic methods, such as monitoring of cancers, known to significantly alter pH levels.
The deep Raman techniques, which mainly comprise spatially offset Raman spectroscopy (SORS)1,2 and transmission Raman spectroscopy (TRS),3 have been demonstrated to be versatile tools for the analysis of complex diffusely scattering (turbid) samples at depth.4−6 These techniques are based around separating the laser illumination and Raman collection zones from each other on the sample surface. This allows signal retrieval on a scale of 2 orders of magnitude deeper within turbid samples compared to traditional Raman approaches. These developments have opened new applications of Raman spectroscopy in a number of areas such as pharmaceutical drug analysis in quality control,3 airport security screening,7 and in healthcare.8,9 Recent advances in the deep Raman techniques have also established that not only can specific chemical information that the Raman spectra inform upon be retrieved, but also the physical properties of a sample such as temperature. We previously named this approach temperature-SORS or T-SORS.10 This opens up further areas where this emerging technology can play a disruptive role such as process control and food manufacturing. It has also been previously demonstrated that combining SORS with surface enhanced Raman spectroscopy (SESORS) can allow monitoring of chemical signals at depth.11−13 Elsewhere, much work using traditional SERS has recently focused on creating stable SERS nanosensors that can detect and monitor a number of biologically relevant moieties or processes, such as pH14−16 and redox levels,17 among many others.18,19 In this context, the ability to accurately monitor in vivo pH levels at a depth that is noninvasive is of particular relevance to a number of diseases, where disruption to the usually tightly regulated pH value is a possible indicator of an associated disease state such as cancer20−22 and is known to be an important factor in wound healing and infections.23 For example, in healthy breast tissue, the average intracellular pH (pHi) is ∼7.2 and the extracellular pH (pHe) is ∼7.4; however, in advanced invasive breast cancer, a reversal of the pH gradient is observed with a pHi and pHe of 7.2 and 6.7, respectively.24 To date, most of the studies using SERS nanosensors have been performed in transparent samples, that is, optical vials, and the SERS enhancement was used as a way to reduce measurement times and increase the sensitivity of analyte detection. Pioneering experiments by Campbell et al. have demonstrated the possibility of measuring subsurface pH levels using SERS nanoparticles from zones within cell culture spheroids at depths of 0.5–1 mm using conventional Raman microscopy.25 Here we demonstrate the feasibility of monitoring pH levels noninvasively in scattering tissue at depths by more than an order of magnitude greater using combined labeled nanoparticles and the deep Raman approaches (SESORS). This new capability opens exciting prospects in medical sciences in photonic methods of diagnosis, for example, for early stage noninvasive cancer diagnosis (e.g., breast cancer), where local pH levels are significantly lowered from biological normal levels by tumor driven hypoxia.17,22,25
Experimental Section
4-Mercaptobenzoic acid (MBA) labeled 100 nm gold nanoparticles were produced following a previously described method.16 A total of 1 mL of the gold nanoparticles (nanoComposix) were mixed for 5 min with 100 μL of 1 mM MBA. This was followed by centrifugation at 3000 rpm for 10 min. The supernatant was removed from the pellet of nanoparticles, which were then resuspended in 1 mL of 0.1 M phosphate buffered saline (PBS) solution. The pH of the nanoparticle suspensions was adjusted with HCl and NaOH solutions (0.5 M), and the pH was measured using a VWR pH110 pH meter. Porcine tissue was purchased locally. For transmission Raman experiments, a 35 × 35 × 35 mm3 cube of porcine tissue was used with a central cavity, perpendicular to the optical axis, allowing placement of a quartz cell (10 × 10 × 48 mm3) containing the labeled nanoparticles. For the inverse SORS measurements, the hole was cut off center at set distances from the side walls to provide multiple depths (5, 8, and 12 mm), depending on the orientation of the block of tissue with respect to the illumination beam. Meat cores were pH-adjusted through overnight soaking in 50 mL of 10–50 mM arginine solutions. The meat cores were briefly rinsed with distilled water prior to introduction of nanoparticles and homogenized after experiments, and the pH was confirmed. Raman measurements were gathered on a previously described home-built spatially offset Raman system in Exeter.5 This system comprises two spatially offset modalities, transmission Raman (TRS), where the Raman illumination zone and collection zone are on the opposite sides of the sample (Scheme S1A). Transmission Raman measurements provide a signal that is a composite of all constituents present within the sample volume, that is, bulk analysis. The second deep Raman modality is inverse SORS, where an axicon lens is used to create a variable diameter ring-shaped laser illumination zone on the sample surface, and Raman spectra are collected from the center of this zone, with the radius of the illumination zone approximately corresponding to the SORS spatial offset, Δs (Scheme S1B). As the values of Δs are increased, in the inverse SORS modality, the larger the relative contribution of the subsurface constituents to the surface ones is observed in the collected Raman spectra. Due to this, SORS enables sensitive depth discrimination, making it ideal in layer discrimination studies. A laser with an 830 nm excitation wavelength was used (Innovative Photonic Solutions: I0830MM0350MF-EM), the laser beam was filtered using two 830 nm laser line filters (Semrock) to provide a spectrally clean laser profile. The resulting laser power at the sample surface was around 350 mW. All Raman spectra were collected using an Andor iDus 420A – BR-DD deep depletion CCD, which was coupled to a Kaiser spectrometer (Holospec 1.8i) with an f-number of 1.8. Raman spectra of suspended nanoparticles within the vial were measured for 2.5 s × 24 (=60 s), while nanoparticles once buried in tissue were measured for 5 s × 120 (=600 s). All Raman spectra were processed using Matlab 2014a; in summary, the data was baseline corrected with a linear fit under the region of interest (1550–1620 cm–1), all data was intensity normalized [0, 1] and the x-axis was interpolated to 0.1 cm–1 increments. Principal component analysis (PCA) was used to create a scores model of MBA sensitivity to pH, and new Raman data collected from nanoparticles buried in meat was projected onto this model and a pH estimate calculated.
Results and Discussion
Our measurements were performed on porcine tissue (Figure 1A) with a thickness of 35 mm. The collected Raman spectra remained stable throughout the experiment, with only a slight drop in the background fluorescence signal being observed (Figure 1B). MBA-NP’s have a number of active Raman bands (Figure 1C), with several of them being sensitive to the local pH levels.14−16
Figure 1.
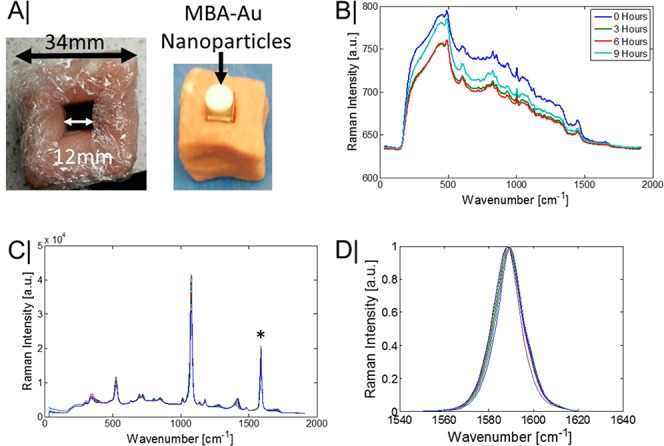
(A) Porcine tissue used to conceal a quartz vial containing MBA labeled 100 nm diameter gold nanoparticles. (B) Pure transmission Raman spectra of porcine tissue at set time intervals. (C) Pure transmission Raman spectra of the MBA-NP solution. (D) Transmission Raman spectra of the pH position sensitive benzene ring stretching mode of MBA at ∼1585 cm–1.
However, to be suitable for monitoring pH at depths in turbid media, ideally the chosen reporter band should have a large Raman cross-section and, preferably, be nonoverlapping with the Raman signal of the surrounding medium, thus, simplifying analysis and increasing the maximal depth a signal can be recovered from. With these considerations in mind, the benzene ring stretching mode of MBA was chosen as a pH reporter due to exhibiting a measurable peak shift with changes in pH levels (Figure 1D) and minimally overlapping with the tissue Raman signal. Once the MBA-NP are inserted into the porcine tissue, the overall observable MBA signal in both modalities is considerably diminished (Figure 2A,B), with only the generally invariant ring breathing mode at ∼1078 cm–1 and the ring stretching mode ∼1585 cm–1 still being identifiable in the spectrum by the naked eye.
Figure 2.
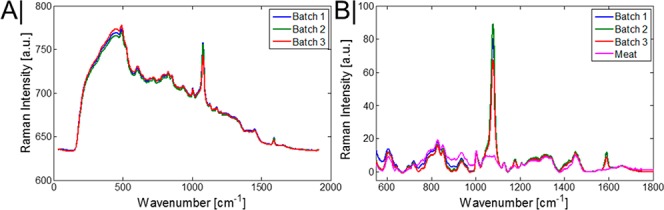
(A) Raw transmission Raman spectra of the average MBA-NP signal when in the center of the porcine tissue (tissue thickness = 35 mm). (B) Raman spectra of MBA-NP and porcine tissue alone, following baseline correction and data scaling.
The approach explored here prohibits basic ratio-metric (univariate) analysis that has been previously reported in the literature as the Raman bands that this analysis is based on are still readily overwhelmed by the tissue signal. For this reason we have resorted to multivariate analysis to extract maximum informational content from data. Specifically, we used principal component analysis (PCA) in this study. For nonburied NPs, in the pH range that was explored (∼6.2–7.8), a strong correlation was present between the peak position of the ring stretching mode of MBA-NP (nonburied) and pH (R2 ∼ 0.92; Figure 3A). The spectral region of 1520–1620 cm–1 was used to create a PCA model of the spectral variation. In this model, 99% of the variance was explained in PC1, and an improved R2 value was observed (R2 0.95) plotting the intensity scores of PC1 versus pH levels (Figure 3B). Furthermore, a root-mean-square error (RMSE) of 0.11 pH units was achieved in a leave one out cross validation of this model. Using the PCA model constructed of the MBA-NP measured alone (i.e., outside tissue), it was possible to project new measurements, that is, of the MBA-NP once enclosed in the porcine tissue. As is observable, the loadings of PC1 for the model of nanoparticles matches that of the data set that is surrounded with porcine tissue (i.e., buried NP’s), albeit with a lower signal-to-noise ratio (Figure 3C). Furthermore, it was possible to predict the pH level based on this approach, with an RMSEP of 0.13 pH units achieved, which is only a slight increase in error of prediction compared with the original nonburied model, validating the robustness of this approach (Figure 3D).
Figure 3.
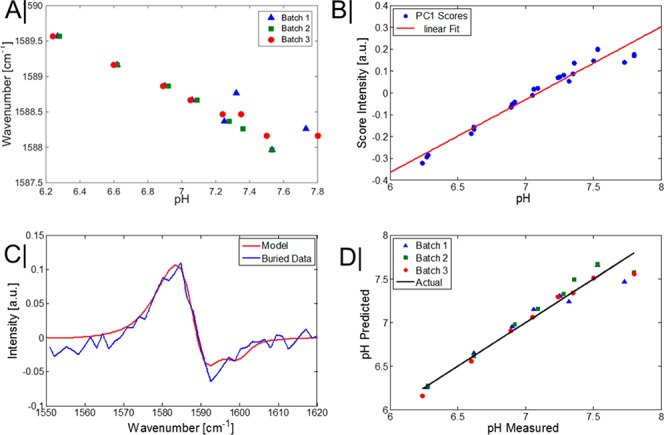
(A) Wavenumber position of the benzene ring breathing mode of MBA-NP at different pH levels for nonburied particles using TRS. (B) Score intensities of principal component one versus pH level. (C) Loadings of principal component (PC) 1 of MBA-NP solution alone (Model) and the loadings of PC1 of MBA-NP when buried in porcine tissue (tissue thickness within optical path = 22 mm). (D) Prediction of pH level using leave one out cross validation from the scores intensities.
Due to the sample size/access requirements of TRS it has more limited applications in vivo, for example, measuring through the hand or breast tissue. Therefore, in addition to the TRS measurements of the MBA-NP in porcine tissue, the nanoparticles were also measured using the inverse SORS modality. This mode is beneficial in situations where access to the other side of sample is not possible, or the sample size is prohibitively too large to be measured in transmission. However, the penetration depths achievable with SORS are approximately half of those achievable with TRS. For these experiments the nanoparticles were inserted in an off-center hole (Figure 4A), thereby providing multiple depths (5, 8, and 12 mm) from different tissue side walls, where the depth was set by the orientation of the tissue block with respect to the laser illumination axis.
Figure 4.
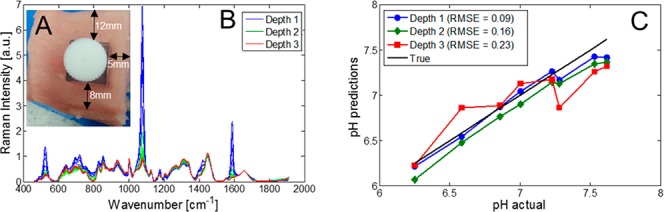
(A) Porcine tissue with an off-center hole for the quartz vial, thereby providing multiple depths (5, 8, and 12 mm) of signal retrieval depending on orientation with the laser beam for SORS measurements. (B) Raman spectra of MBA, as measured in the SORS modality, at three different depths, “1”, “2”, and “3” (5, 8, and 12 mm, respectively), when buried in porcine tissue. (C) Predicted pH levels using SORS vs actual pH levels using the PCA scores model.
As to be expected, the depth of sample has a large influence on the observed Raman intensity of the nanoparticles. At the most superficial depth (5 mm), the MBA-NP dominates that of the meat signal (Figure 4B), while at 12 mm depth, the MBA-NP signal is comparable to the tissue intensity. The same approach was taken as with the TRS data, that being a PCA model of the nonburied MBA-NPs was first created, then new data from buried NPs was projected onto this. An apparent trend is observed that as the signal-to-noise of the data decreases, that is, is recovered from deeper within the porcine tissue, there is a worsening of RMSEP of pH (from 0.09 to 0.23 pH units). The explored pH levels achieved accuracy in a biologically important range, as cancerous lesions are known to exhibit lower extracellular pH levels (6.7) compared with healthy biological tissue (7.4).24 As such, cancer diagnosis based around pH levels alone, or in conjunction with other chemical markers derived from SORS signals, could be used to identify the presence of such lesions and monitor the efficacy of treatments. Other potential applications include the monitoring of the presence of infections and their healing, the monitoring of pH levels in industrial manufacture in process and quality control (e.g., biopharmaceutical manufacture) or in the food industry where pH levels could signify, for example, the spoilage or maturity of food products: meat, dairy products, and so on. Finally, to demonstrate the overall robustness of pH monitoring using MBA labeled nanoparticles, they were inserted directly into porcine tissue (Figure 5A). Due to the continuation of anaerobic respiration in animal muscle tissue postslaughter and a subsequent buildup of lactic acid, tissue of “high quality” for retail has a pH between 5.4 and 5.8. Due to this being lower than what is expected in living organisms, the tissue pH was adjusted in a series of meat cores with a known range of pH from ∼6 to 7.5. With the nanoparticles directly inserted into tissue, it was still possible to accurately monitor the pH of the tissue cores, monitoring the position of the SERS Raman reporter allowed prediction of pH with an RMSEP of ∼0.2 pH units.
Figure 5.
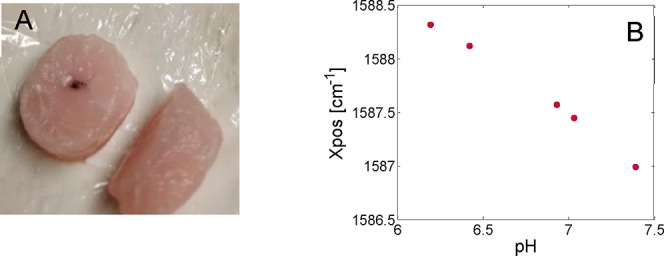
(A) pH-adjusted porcine tissue core, with MBA-labeled nanoparticles deposited in the middle of the core. (B) Wavenumber position of MBA SERS as a function of pH while embedded in porcine tissue with RMSE ∼ 0.2 pH units.
Conclusions
The feasibility of monitoring pH at a significant depth in porcine tissue, using labeled nanoparticles, has been demonstrated using both of the two principal deep Raman approaches (TRS and SORS). A biologically relevant range of pH levels were explored (6.2–7.8 pH units). Accurate reporting was easily achievable in both deep Raman modalities with a best RMSEP ∼ 0.1 pH units, with transmission Raman yielding a deeper retrieval (22 mm tissue thickness in optical path) of pH-sensitive information compared to inverse SORS (8 mm).
The approach holds promise for future noninvasive photon-based disease diagnosis in situations where local pH levels are altered (e.g., breast cancer).
Acknowledgments
EPSRC Grants (EP/K020374/1, EP/P012442/1, and EP/R020965/1) partly funded the work presented here.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b01015.
Schematic diagrams of the deep Raman spectroscopy illumination schemes utilized: SORS and TRS (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Matousek P. Appl. Spectrosc. 2006, 60 (11), 1341–1347. 10.1366/000370206778999102. [DOI] [PubMed] [Google Scholar]
- Matousek P.; Clark I. P.; Draper E. R. C.; Morris M. D.; Goodship A. E.; Everall N.; Towrie M.; Finney W. F.; Parker a W. Appl. Spectrosc. 2005, 59 (4), 393–400. 10.1366/0003702053641450. [DOI] [PubMed] [Google Scholar]
- Matousek P.; Parker A. W. J. Raman Spectrosc. 2007, 38 (5), 563–567. 10.1002/jrs.1688. [DOI] [Google Scholar]
- Matousek P.; Conti C.; Realini M.; Colombo C. Analyst 2016, 141 (3), 731–739. 10.1039/C5AN02129D. [DOI] [PubMed] [Google Scholar]
- Vardaki M. Z.; Gardner B.; Stone N.; Matousek P. Analyst 2015, 140 (15), 5112–5119. 10.1039/C5AN01118C. [DOI] [PubMed] [Google Scholar]
- Liao Z.; Sinjab F.; Gibson G.; Padgett M.; Notingher I. Opt. Express 2016, 24 (12), 12701–12712. 10.1364/OE.24.012701. [DOI] [PubMed] [Google Scholar]
- Eliasson C.; Macleod N. A.; Matousek P. Anal. Chem. 2007, 79 (21), 8185–8189. 10.1021/ac071383n. [DOI] [PubMed] [Google Scholar]
- Matousek P.; Stone N. Chem. Soc. Rev. 2016, 45 (7), 1794–1802. 10.1039/C5CS00466G. [DOI] [PubMed] [Google Scholar]
- Stone N.; Baker R.; Rogers K.; Parker A. W.; Matousek P. Analyst 2007, 132 (9), 899–905. 10.1039/b705029a. [DOI] [PubMed] [Google Scholar]
- Gardner B.; Matousek P.; Stone N. Anal. Chem. 2016, 88 (1), 832–837. 10.1021/acs.analchem.5b03360. [DOI] [PubMed] [Google Scholar]
- Stone N.; Faulds K.; Graham D.; Matousek P. Anal. Chem. 2010, 82 (10), 3969–3973. 10.1021/ac100039c. [DOI] [PubMed] [Google Scholar]
- Gardner B.; Stone N.; Matousek P. Faraday Discuss. 2016, 187 (0), 329–339. 10.1039/C5FD00154D. [DOI] [PubMed] [Google Scholar]
- Ma K.; Yuen J. M.; Shah N. C.; Walsh J. T.; Glucksberg M. R.; Van Duyne R. P. Anal. Chem. 2011, 83 (23), 9146–9152. 10.1021/ac202343e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.; Flynn K. J.; Xia Z.; Dunstan P. R. J. Raman Spectrosc. 2016, 47 (7), 819–827. 10.1002/jrs.4910. [DOI] [Google Scholar]
- Zheng X. S.; Hu P.; Cui Y.; Zong C.; Feng J. M.; Wang X.; Ren B. Anal. Chem. 2014, 86 (24), 12250–12257. 10.1021/ac503404u. [DOI] [PubMed] [Google Scholar]
- Jaworska A.; Jamieson L. E.; Malek K.; Campbell C. J.; Choo J.; Chlopicki S.; Baranska M. Analyst 2015, 140, 2321–2329. 10.1039/C4AN01988A. [DOI] [PubMed] [Google Scholar]
- Jamieson L. E.; Jaworska A.; Jiang J.; Baranska M.; Harrison D. J.; Campbell C. J. Analyst 2015, 140 (7), 2330–2335. 10.1039/C4AN02365J. [DOI] [PubMed] [Google Scholar]
- McNay G.; Eustace D.; Smith W. E.; Faulds K.; Graham D. Appl. Spectrosc. 2011, 65 (8), 825–837. 10.1366/11-06365. [DOI] [PubMed] [Google Scholar]
- McAughtrie S.; Faulds K.; Graham D. J. Photochem. Photobiol., C 2014, 21, 40–53. 10.1016/j.jphotochemrev.2014.09.002. [DOI] [Google Scholar]
- Johnsson M. S.; Nancollas G. H. Crit. Rev. Oral Biol. Med. 1992, 3 (1–2), 61–82. 10.1177/10454411920030010601. [DOI] [PubMed] [Google Scholar]
- Scott R.; Stone N.; Kendall C.; Geraki K.; Rogers K. npj Breast Cancer 2016, 2 (1), 1–6. 10.1038/npjbcancer.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Kim B.; Sohn W. B.; Byun K. M.; Lee S. Y. Proc. SPIE 2017, 1005813–1005816. 10.1117/12.2249622. [DOI] [Google Scholar]
- Percival S. L.; McCarty S.; Hunt J. A.; Woods E. J. Wound Repair Regen 2014, 22 (2), 174–186. 10.1111/wrr.12125. [DOI] [PubMed] [Google Scholar]
- Damaghi M.; Wojtkowiak J. W.; Gillies R. J. Front. Physiol. 2013, 4, 1–10. 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson L. E.; Camus V. L.; Bagnaninchi P. O.; Fisher K. M.; Stewart G. D.; Nailon W. H.; McLaren D. B.; Harrison D. J.; Campbell C. J. Nanoscale 2016, 8 (37), 16710–16718. 10.1039/C6NR06031E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


