Abstract
Pharmacogenetic testing is a useful and increasingly widespread tool to assist in antidepressant prescription. More than ten antidepressants (including tricyclics, selective serotonin reuptake inhibitors and venlafaxine) have already genetic biomarkers of response/side effects in clinical guidelines and drug labels. These are represented by functional genetic variants in genes coding for cytochrome enzymes (CYP2D6 and CYP2C19). Depending on the predicted metabolic activity, guidelines provide recommendations on drug choice and dosing. Despite not conclusive, the current evidence suggests that testing can be useful in patients who did not respond or tolerate at least one previous pharmacotherapy. However, the current recommendations are based on pharmacokinetic genes only (CYP450 enzymes), while pharmacodynamic genes (modulating antidepressant mechanisms of action in the brain) are still being studied because of their greater complexity. This may be captured by polygenic risk scores, which reflect the cumulative contribution of many genetic variants to a trait, and they may provide future clinical applications of pharmacogenetics. A more extensive use of genotyping in clinical practice may lead to improvement in treatment outcomes thanks to personalized treatments, but possible ethical issues and disparities should be taken into account and prevented.
Keywords: Antidepressant, Major depressive disorder, Genetics, Genome-wide association study, Polygenic risk score, Precision psychiatry
INTRODUCTION
Personalized and precision medicine have become a major area of research and clinical innovation in the post-genomic era. This term refers to the outstanding technological improvements after the sequencing of the first human genome was completed in 2003, exemplified by a more than exponential decrease in time and cost needed for genome sequencing [1]. These advances made possible the extensive study of genetics influence on a number of human complex traits, such as response and side effects to medications, in order to optimize and personalize treatments. In major depressive disorder (MDD), treatment non-response or incomplete symptom remission are common (~50% of cases after the first treatment [2]) and treatment resistant depression (TRD), defined as lack of response to at least two treatments, is observed in ~30% of cases [3]. TRD is associated with higher social and occupational impairment, suicidal thoughts, decline of physical health and increased health care utilization [4]. Another common issue is represented by side effects, which are frequently associated with poor treatment adherence and early treatment discontinuation [5]. Genetic information can provide additional guidance in treatment choice compared to clinical evaluation only and significantly improve these outcomes. A contribution of genetic variants to antidepressant response has been hypothesized since the observation that there is family clustering and the evaluation of family history of treatment response is included among the criteria used to guide drug choice [6]. Pharmacogenetic studies focused on the most plausible candidates for antidepressant response and side effects led to the currently available clinical applications, described in paragraphs 2 and 3, and to the inclusion of pharmacogenetic biomarkers in clinical guidelines and drug labels [7]. This resulted in the flourish of many companies providing commercial kits for pharmacogenetic testing and detailed interpretation of the results [8]. The easy access to these tests and the commonly used direct-to-consumer policy often result in the autonomous decision of the patient to undertake the test and the only subsequent consultation with a psychiatrist, while in other cases the recommendation for testing comes from the treating psychiatrist. In both scenarios, a good knowledge of the available testing options, with their pros and cons, is needed. Pharmacogenetics is also a rapidly advancing field and updates on the most recent developments are required to implement the next wave of clinical applications. The increasing availability of genome-wide or sequence data is leading to the identification of more complex genetic predictors, which are based on the combination of multiple genetic variants into a measure defined as polygenic risk score (PRS). PRSs aim to capture the overall genetic susceptibility to a specific trait, such as treatment response or resistance, and they will probably represent the basis for future clinical applications of pharmacogenetics. Obstacles to the clinical implementation of pharmacogenetic testing and ethical issues will be also discussed, since many practical problems and disparities may emerge as pharmacogenetic testing becomes widespread. Finally, we briefly commented on the possibility to use pharmacogenetic findings to cut time and development costs of new drugs for depression.
CURRENT CLINICAL APPLICATIONS: CYTOCHROME GENES
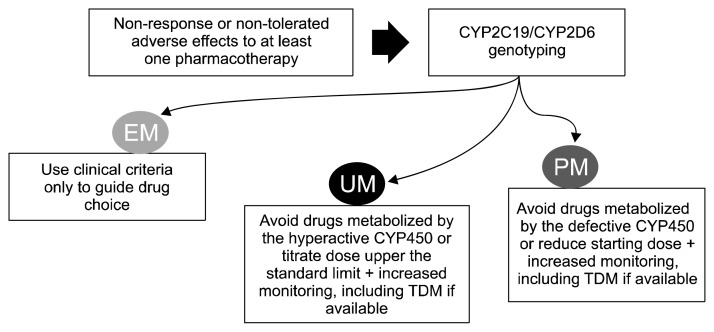
Candidate genes involved in antidepressant metabolism (pharmacokinetics) have sufficiently robust scientific evidence for clinical applications, as documented in clinical guidelines such as those curated by the Clinical Pharmacogenetics Implementation Consortium (CPIC) and Dutch Pharmacogenetic Working Group (DPWG) [9,10]. These genes are those coding for cytochrome P450 (CYP450) enzymes responsible for antidepressant metabolism, namely CYP2D6 and CYP2C19. Functional genetic variants within these genes are common in the population and they result in significant variation in the enzymatic activity, which can be classified in four main groups (poor metabolizers [PMs], intermediate metabolizers [IMs], extensive metabolizers [EMs] and ultrarapid metabolizers [UMs]). These metabolizing groups were associated with pharmacokinetic parameters (e.g., drug and metabolites plasma concentration) for several antidepressants, demonstrating an impact on drug metabolism [11]. There is however weaker evidence in terms of association between metabolizing groups and clinical outcomes (response/side effects), as well as unclear relationship between drug plasma concentration and clinical outcomes, except for tricyclic antidepressants (TCAs), es(citalopram) and venlafaxine [12–14]. Selective serotonin reuptake inhibitors (SSRIs) have probably a mostly flat plasma concentration-response curve, with relevant changes only for very low and very high plasma concentrations [14]. This means that significant differences in clinical outcomes may be observed in patients with severely compromised or increased enzymatic activity (PMs and UMs, respectively), but there was poor evidence in the literature due to the relatively rarity of these groups in the population. Recent findings from a larger sample demonstrate that CYP2C19 PMs have an increased risk of side effects (gastrointestinal, neurological and sexual) during (es) citalopram treatment, as well as higher symptom improvement and better chances of symptom remission [15]. These results are in line with the recommendations provided by clinical guidelines, confirming that the starting dose should be reduced to 50% of the standard dose and increased clinical monitoring should be performed in CYP2C19 PMs, but they suggested that being CYP2C19 PMs does not contraindicate treatment with (es) citalopram. Recommendations provided for other antidepressants (mostly TCAs and SSRIs) follow the same schema: in PMs and UMs consider avoiding drugs mostly metabolized by the defective or hyperactive enzyme, if there is a clinical indication for one of those drugs adjust the dose accordingly and monitor closer. An overview of recommendations based on CYP2D6/CYP2C19 metabolizing activity is reported in Table 1 and summarized in Figure 1. In total, 13 antidepressant drugs have pharmacogenetic biomarkers provided by CPIC and/or DPWG. Of particular interest, guidelines do not provide indications on when and to whom pharmacogenetic testing should be recommended, since there is still no conclusive information on the groups which are expected to benefit more from testing. According the best current evidence, pharmacogenetic testing should be recommended in patients who had not responded or tolerated at least one previous pharmacotherapy [16].
Table 1.
Prescribing recommendations provided by clinical guidelines based on pharmacogenetic biomarkers
| Drug | Gene(s) | CPIC | DPWG | FDA | Recommendations |
|---|---|---|---|---|---|
| Amitriptyline | CYP2D6, CYP2C19 | × | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication |
| Citalopram | CYP2C19 | × | × | × | Consider an alternative drug in UMs or titrate dose to a maximum of 150% and a 50% reduction of the standard starting dose in PMs |
| Clomipramine | CYP2D6, CYP2C19 | × | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication |
| Desipramine | CYP2D6 | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication | |
| Doxepin | CYP2D6, CYP2C19 | × | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication |
| Escitalopram | CYP2C19 | × | × | × | Consider an alternative drug in UMs or titrate dose to a maximum of 150% and a 50% reduction of the standard starting dose in PMs |
| Fluvoxamine | CYP2D6 | × | × | Consider a 25–50% reduction of recommended starting dose in PMs | |
| Imipramine | CYP2D6, CYP2C19 | × | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication |
| Nortriptyline | CYP2D6 | × | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication |
| Paroxetine | CYP2D6 | × | × | Select an alternative drug in UMs, consider alternative drug or 50% reduction of the standard starting dose in PMs | |
| Sertraline | CYP2C19 | × | × | 50% reduction of the standard starting dose or alternative drug in PMs | |
| Trimipramine | CYP2D6, CYP2C19 | × | × | Avoid drug in PMs and UMs or consider dose adjustments if clinical indication | |
| Venlafaxine | CYP2D6 | × | × | Select alternative drug in PMs and IMs or adjust dose, titrate dose to a maximum of 150% of the normal dose or select alternative drug in UMs |
In the column Food and Drug Administration (FDA), we reported if the drug has an actionable pharmacogenetic biomarker also in drug label according to the FDA.
CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetic Working Group; CYP2D6, cytochrome 2D6; CYP2C19, cytochrome 2C19; PMs, poor metabolizers; UMs, ultrarapid metabolizers; IMs, intermediate metabolizers.
Fig. 1.
Schema of CYP2D6/CYP2C19 genetic testing implementation in clinical practice.
EM, extensive metabolizer; UM, ultrarapid metabolizer; PM, poor metabolizer; TDM, therapeutic drug monitoring.
Polymorphisms in other CYP450 genes (e.g., CYP1A2, CYP3A4/A5, CYP2B6) have probably not clinically relevant consequences on drug metabolism, because their level of activity is much more dependent from environmental factors (e.g., smoking, diet, concomitant drugs) than genetic factors [17]. Therefore, polymorphisms in these genes should not be included in pharmacogenetic testing.
COMMERCIAL PHARMACOGENETIC TESTS
The inclusion of pharmacogenetic biomarkers in clinical guidelines and drug labels provided by the main drug regulatory agencies have led to the rapid development of many commercial pharmacogenetic tests which are available on the Internet and in drug stores in some countries. These tests can be a useful option when testing is not provided by the national health care system (greatest part of cases currently, see paragraph 4) or when there is no nearby laboratory having a certification for pharmacogenetic testing for clinical use. When considering the use of a commercial pharmacogenetic test, a few points should be kept in mind. Only some of the available tests were evaluated in clinical trials for potential benefits compared to standard care, and only six randomized controlled trials (RCTs) evaluating five different commercial tests are published [18–23], while dozens of different tests exist and have no published evidence from RCTs or non-randomized case control studies. The available studies, including RCTs, had some methodological limitations, for example limited sample size and imperfect blinding, but the meta-analytic evidence coming from RCTs shows that there is probably a benefit in terms of symptom remission compared to standard care, particularly in patients who did not respond to at least one previous pharmacotherapy [24]. The last key point to consider when evaluating commercial tests is which genes and genetic variants are genotyped and how the prescribing recommendations are formulated based on the genotyping results. All the commercial tests are indeed multi-gene and multi-variant tests, and they typically include also genetic variants which are not endorsed by clinical guidelines and not cited in drug labels [8]. The interpretative report should explain on which genetic variants the provided recommendations are based, in a way that it is possible to select recommendations based on genes with the strongest scientific evidence and having clinical usefulness according to guidelines.
BARRIERS TO THE CLINICAL IMPLEMENTATION OF PHARMACOGENETICS
Pharmacogenetics is still not implemented in routine clinical practice in the most part of countries, with some exceptions (for example, Switzerland). The main barrier consists in the need to invest resources to create standard procedures for test prescription and genotyping, but also for standard interpretation of the genotyping results in prescriptive recommendations. This would mean educational programs for clinicians and other health care professionals, costs for sample transport and genotyping, integration of Clinical Decision Support (CDS) tools in clinical settings, in other words: time and money. CDS tools can be optimized to integrate different types of information: genetic information, medical history from electronic health records (e.g., hospitalizations and diagnoses, drug prescriptions, blood tests and other tests), standard assessment scales for symptom evaluation and disease diagnosis, prescription recommendations from guidelines. This would of course require relevant investments to create and harmonize a common CDS, accessible from different health care services within a country, but it could provide major improvements in the quality of health care. This and the other steps needed for routine clinical application of pharmacogenetic testing require robust cost-effectiveness evidence before implementation. A medical intervention is defined as cost-effective when it costs more than standard care, but the extra-cost is considered acceptable based on the improvements in clinical outcomes, according to what is usually defined as willingness to pay threshold [25]. There are a few published studies looking at the cost-effectiveness of pharmacogenetic testing in guiding antidepressant prescription compared to standard care [26–28]. They reported evidence of cost-effectiveness compared to standard care, even though they were based on simulated clinical outcomes using input data from short-term clinical trials, while results coming from longer follow-up non-sponsored studies are lacking. Large collaborative projects such as the European consortium Ubiquitous Pharmacogenetics (U-PGx) will be able to answer in a more definitive way to the cost-effectiveness question, using large RCTs comparing treatment as usual with pharmacogenetic-guided treatment according to the available guidelines [29].
FUTURE CLINICAL APPLICATIONS: POLYGENIC RISK SCORES
Current recommendations for guiding antidepressant prescription are based on pharmacokinetic genes only, while the mechanisms responsible for drug action (pharmacodynamics) are not taken into account. This approach is clearly an oversimplification, but it comes from the fact that antidepressant pharmacodynamics is far more complex than pharmacokinetics and it is only partially understood. Many commercial pharmacogenetic tests include also variants in pharmacodynamic genes, such as the serotonin transporter, but the role of these genes in treatment outcomes is quite controversial and they are not endorsed by guidelines [8]. On a pharmacodynamic level, we expect that many different genes are involved and they interact among each other. A popular approach to consider this polygenic and complex architecture is represented by PRSs, which reflect the cumulative contribution of a number of genetic variants associated with a trait [30]. PRSs can also be described as a measure of the genetic predisposition of an individual towards the development of a certain trait (e.g., a disease or treatment outcome). Their calculation is relatively easy in terms of costs and time, since genome-wide genotyping can be performed for 50 US dollars or less and the results can be available within few days. Among complex traits, there is already convincing evidence supporting a strong effect of a PRS estimating the risk of cardiovascular disease, which was shown to outperform traditional risk factors in risk prediction [31]. Very promising results were reported also for a PRS of type 2 diabetes, which improved 5-year risk prediction of 32% [32]. In traits related to mental health, results are currently not as much close to clinical application, probably the difficulty in collecting large and well clinically characterized samples and the relative low standardization of psychopathological measures have significantly contributed to the slower progress in this field. The most consistently replicated results were obtained for a PRS reflecting the risk of schizophrenia [33], while non-significant results were obtained for antidepressant response PRSs [34]. In addition to the discussed limitations of PRSs in predicting traits related to mental health, another potential issue is represented by the fact that the standard PRS approach is based on common genetic variants only (which are usually defined as those observed in > 1% of the population). Rare variants may contribute to the inter-individual variability in antidepressant response, but they are more difficult to genotype and analyze, since genome sequencing rather than genome-wide genotyping is needed (all the DNA basis have to be determined, not just a pool of selected common variants) and larger samples are required in theory to perform standard statistical tests. The creation of polygenic risk scores including both rare and common variants could potentially improve the prediction of antidepressant response and other similar traits.
PHARMACOGENETIC-GUIDED DRUG DISCOVERY AND REPOSITIONING
Another interesting implication of antidepressant pharmacogenetic research at clinical level is the possibility to guide the development of new drugs for depression. Standard drug discovery and development is a very long and expensive process, usually between 12 and 16 years with a cost of 1–2 billion US dollars [35]. Pharmacogenetics can help in identifying new pharmacological targets and speed up the first phase of drug development, but more interestingly it can guide the repurposing of existing drugs, approved or in the experimental phase for other conditions, leading to even larger time and cost savings since the first phases of drug development would be skipped. In practical terms, genes targeted by existing drugs can be tested for enrichment in genes associated with antidepressant response. This approach identified some possible pharmacological modes of action for new antidepressants which include the modulation of sex hormones, calcium channels and GABA-A receptor [36]. This last mode of action is in line with that of a new generation of antidepressant drugs which has been recently developed, represented by positive allosteric modulators at GABA-A receptor subunit [37]. The first drug of this new class (brexanolone) has been recently approved by the Food and Drug Administration for the treatment of post-partum depression, while other compounds with analogous mechanisms of action are currently in phase II or III for MDD treatment [37]. Drugs developed using pharmacogenetic information may be targeted on specific patient groups (i.e., the ones carrying genetic variants in specific genes or pathways), similarly to what we commonly observe for anti-cancer drugs. This can be an advantage in terms of treatment personalization, but it could limit the benefits of new drugs to restricted groups of patients.
ETHICAL CONSIDERATIONS AND POTENTIAL DISPARITIES
In a hypothetical future when genetic information is routinely collected and used to provide personalized medical interventions, a number of ethical issues may become relevant and appropriate management should be planned in advance. One major problem could be how to manage genetic information predicting poor clinical outcomes with the available treatments. This may result in the impossibility to have health insurance coverage in some countries and have a negative impact on the patient’s perception of treatment (nocebo effect). Newer and more expensive treatments may not be available or reimbursable. Incidental findings would also be possible, including the genetic diagnosis of severe and fatal disorders for which there could not be available treatments, or genetic variants with unclear clinical consequences. In these scenarios, the communication of the results of testing to the patient may have extremely negative consequences on his/her quality of life and lead to no benefits, but some patients may prefer to be informed anyway. There are also consequences in terms of risks for the offspring which should be carefully evaluated. The manipulation of the human genome to protect the offspring from developing severe diseases have already become a reality and it is object of great perplexity and concerns [38]. Another relevant issue would be represented by disparities in the quality and quantity of knowledge about the clinical consequences of genetic variants in different ethnic groups. The frequency of many genetic variants widely changes among ethnic groups, including for example many functional variants within CYP2D6 and CYP2C19 genes with consequent different prevalence of the corresponding metabolizing groups [39,40] (Fig. 2). In 2017, 88% of patients included in genome-wide association studies were of European ancestry, meaning that almost all findings may be poorly translatable in another ethnic groups [41]. This issue already led to striking cases of misdiagnosis, such as in the case of seven patients of African ancestry who were treated for hypertrophic cardiomyopathy diagnosed based on genetic variants which have high frequency differences between European and African populations and were eventually shown to be benign [42].
Fig. 2.
Examples of prevalence of CYP2D6 and CYP2C19 metabolizing groups based on functional genetic variants across different ethnic groups.
These differences have clinical implications in terms of relevance of certain variants and metabolizing groups across ethnicities.
PM, poor metabolizer; UM, ultrarapid metabolizer.
DISCUSSION
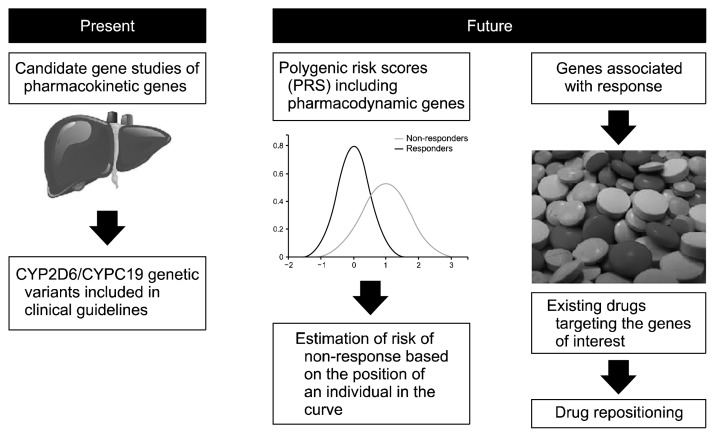
Pharmacogenetics already represents an available tool in assisting antidepressant prescription. Despite not definitive consensus on when and to whom testing should be recommended, the available evidence supports that testing can be useful in patients who did not respond or tolerate at least one previous pharmacological treatment [16]. The current recommendations are based on variants in the genes coding for CYP2D6 and CYP2C19, the main cytochrome enzymes responsible for antidepressant metabolism [7]. Indications in terms of drug choice and dosing are based on the predicted level of CYP2D6/CYP2C19 enzymatic activity. Variants in these genes are usually combined with variants in pharmacodynamic genes in many commercial pharmacogenetic tests [8]. Prescription based on variants in pharmacodynamic genes with inconclusive level of evidence for association with treatment outcomes may lead to prescriptive errors, such as exemplified in a recently described case report [43]. While candidate gene studies were able to identify meaningful findings for pharmacokinetic genes, the complexity of antidepressant mechanisms of action probably requires a different approach (Fig. 3). PRSs or analogous approaches are probably more suitable to capture the polygenic contribution of pharmacodynamic genes to antidepressant efficacy, despite clinical applications using this methodology are still not available. Other future developments in the field include the evaluation of the role of rare variants and more robust estimations of pharmacogenetic testing cost-effectiveness in clinical practice. A comprehensive predictive model including clinical predictors may the then be hypothesized as a valuable support tool for clinicians [44], ideally integrated in electronic health records [45]. Routine clinical implementation, particularly when more extensive genotyping will be performed, will however have to face a number of management and ethical issues. In any case, genetics should be seen as an additional tool for guiding prescription, and not as a replacement of a complete clinical evaluation.
Fig. 3.
Current and future clinical applications of antidepressant pharmacogenetics.
Footnotes
Conflicts of Interest
Prof. Alessandro Serretti is or has been consultant/speaker for: Abbott, Abbvie, Angelini, Astra Zeneca, Clinical Data, Boheringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, Innovapharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polifarma, Sanofi, Servier.
Chiara Fabbri has no potential conflicts of interest.
Author Contributions
Chiara Fabbri performed the literature search relevant to this review and wrote the first draft; Alessandro Serretti developed the general framework of the review and comments that led to the final draft.
REFERENCES
- 1.National Human Genome Research Institute. DNA sequencing costs: data [Internet] Bethesda: National Institute of Health; 2017. [cited at 2017 Nov 27]. Available from: https://www.genome.gov/sequencingcostsdata/ [Google Scholar]
- 2.Thomas L, Kessler D, Campbell J, Morrison J, Peters TJ, Williams C, et al. Prevalence of treatment-resistant depression in primary care: cross-sectional data. Br J Gen Pract. 2013;63:e852–e858. doi: 10.3399/bjgp13X675430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souery D, Amsterdam J, de Montigny C, Lecrubier Y, Montgomery S, Lipp O, et al. Treatment resistant depression: methodological overview and operational criteria. Eur Neuropsychopharmacol. 1999;9:83–91. doi: 10.1016/S0924-977X(98)00004-2. [DOI] [PubMed] [Google Scholar]
- 4.Mrazek DA, Hornberger JC, Altar CA, Degtiar I. A review of the clinical, economic, and societal burden of treatment-resistant depression: 1996–2013. Psychiatr Serv. 2014;65:977–987. doi: 10.1176/appi.ps.201300059. [DOI] [PubMed] [Google Scholar]
- 5.Ho SC, Jacob SA, Tangiisuran B. Barriers and facilitators of adherence to antidepressants among outpatients with major depressive disorder: a qualitative study. PLoS One. 2017;12:e0179290. doi: 10.1371/journal.pone.0179290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetin M, Hoepner CT, Bjornson L. Rational antidepressant selection: applying evidence-based medicine to complex real-world patients. Psychopharmacol Bull. 2006;39:38–104. [PubMed] [Google Scholar]
- 7.PharmGKB®. Drug labels and clinical guideline annotations [Internet] Stanford: PharmGKB®; 2019. [cited at 2019 Mar 11]. Available from: https://www.pharmgkb.org. [Google Scholar]
- 8.Fabbri C, Zohar J, Serretti A. Pharmacogenetic tests to guide drug treatment in depression: comparison of the available testing kits and clinical trials. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:36–44. doi: 10.1016/j.pnpbp.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Clinical Pharmacogenetics Implementation Consortium (CPIC®) Clinical Pharmacogenetics Implementation Consortium guidelines [Internet] Clinical Pharmacogenetics Implementation Consortium (CPIC®) 2019 [cited at 2019 Nov 7]. Available from: https://cpicpgx.org/guidelines/
- 10.KNMP. Dutch Pharmacogenetic Working Group guidelines [Internet] The Hague: KNMP; 2019. [cited at 2019 Jul 11]. Available from: https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1/pharmacogenetics. [Google Scholar]
- 11.Porcelli S, Fabbri C, Spina E, Serretti A, De Ronchi D. Genetic polymorphisms of cytochrome P450 enzymes and antidepressant metabolism. Expert Opin Drug Metab Toxicol. 2011;7:1101–1115. doi: 10.1517/17425255.2011.597740. [DOI] [PubMed] [Google Scholar]
- 12.Perry PJ, Zeilmann C, Arndt S. Tricyclic antidepressant concentrations in plasma: an estimate of their sensitivity and specificity as a predictor of response. J Clin Psychopharmacol. 1994;14:230–240. doi: 10.1097/00004714-199408000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Charlier C, Pinto E, Ansseau M, Plomteux G. Venlafaxine: the relationship between dose, plasma concentration and clinical response in depressive patients. J Psychopharmacol. 2002;16:369–372. doi: 10.1177/026988110201600413. [DOI] [PubMed] [Google Scholar]
- 14.Florio V, Porcelli S, Saria A, Serretti A, Conca A. Escitalopram plasma levels and antidepressant response. Eur Neuropsychopharmacol. 2017;27:940–944. doi: 10.1016/j.euroneuro.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur Neuropsychopharmacol. 2018;28:945–954. doi: 10.1016/j.euroneuro.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 16.International Society of Psychiatric Genetics. Genetic testing statement [Internet] Brentwood: International Society of Psychiatric Genetics; 2019. [cited at 2019 Mar 14]. Available from: https://ispg.net/genetic-testing-statement/ [Google Scholar]
- 17.Spina E, de Leon J. Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna) 2015;122:5–28. doi: 10.1007/s00702-014-1300-5. [DOI] [PubMed] [Google Scholar]
- 18.Elliott LS, Henderson JC, Neradilek MB, Moyer NA, Ashcraft KC, Thirumaran RK. Clinical impact of pharmacogenetic profiling with a clinical decision support tool in polypharmacy home health patients: A prospective pilot randomized controlled trial. PLoS One. 2017;12:e0170905. doi: 10.1371/journal.pone.0170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley P, Shiekh M, Mehra V, Vrbicky K, Layle S, Olson MC, et al. Improved efficacy with targeted pharmacogenetic-guided treatment of patients with depression and anxiety: a randomized clinical trial demonstrating clinical utility. J Psychiatr Res. 2018;96:100–107. doi: 10.1016/j.jpsychires.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Pérez V, Salavert A, Espadaler J, Tuson M, Saiz-Ruiz J, Sáez-Navarro C, et al. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double-blind clinical trial. BMC Psychiatry. 2017;17:250. doi: 10.1186/s12888-017-1412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin Psychopharmacol Neurosci. 2015;13:150–156. doi: 10.9758/cpn.2015.13.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16:219–227. [PubMed] [Google Scholar]
- 23.Greden JF, Parikh SV, Rothschild AJ, Thase ME, Dunlop BW, DeBattista C, et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: a large, patient- and rater-blinded, randomized, controlled study. J Psychiatr Res. 2019;111:59–67. doi: 10.1016/j.jpsychires.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Bousman CA, Arandjelovic K, Mancuso SG, Eyre HA, Dunlop BW. Pharmacogenetic tests and depressive symptom remission: a meta-analysis of randomized controlled trials. Pharmacogenomics. 2019;20:37–47. doi: 10.2217/pgs-2018-0142. [DOI] [PubMed] [Google Scholar]
- 25.Verbelen M, Weale ME, Lewis CM. Cost-effectiveness of pharmacogenetic-guided treatment: are we there yet? Pharmacogenomics J. 2017;17:395–402. doi: 10.1038/tpj.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornberger J, Li Q, Quinn B. Cost-effectiveness of combinatorial pharmacogenomic testing for treatment-resistant major depressive disorder patients. Am J Manag Care. 2015;21:e357–e365. [PubMed] [Google Scholar]
- 27.Groessl EJ, Tally SR, Hillery N, Maciel A, Garces JA. Cost-effectiveness of a pharmacogenetic test to guide treatment for major depressive disorder. J Manag Care Spec Pharm. 2018;24:726–734. doi: 10.18553/jmcp.2018.24.8.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciel A, Cullors A, Lukowiak AA, Garces J. Estimating cost savings of pharmacogenetic testing for depression in real-world clinical settings. Neuropsychiatr Dis Treat. 2018;14:225–230. doi: 10.2147/NDT.S145046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.U-PGx. Ubiquitous Pharmacogenomics (U-PGx) [Internet] Ubiquitous Pharmacogenomics (U-PGx); 2019. [cited at 2019 Mar 14]. Available from: http://upgx.eu. [Google Scholar]
- 30.Anderson JS, Shade J, DiBlasi E, Shabalin AA, Docherty AR. Polygenic risk scoring and prediction of mental health outcomes. Curr Opin Psychol. 2019;27:77–81. doi: 10.1016/j.copsyc.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles JW, Ashley EA. Cardiovascular disease: the rise of the genetic risk score. PLoS Med. 2018;15:e1002546. doi: 10.1371/journal.pmed.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Läll K, Mägi R, Morris A, Metspalu A, Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19:322–329. doi: 10.1038/gim.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonas KG, Lencz T, Li K, Malhotra AK, Perlman G, Fochtmann LJ, et al. Schizophrenia polygenic risk score and 20-year course of illness in psychotic disorders. 2019 Mar 18; doi: 10.1038/s41398-019-0612-5. BioRxiv 581579 [Preprint] [cited 2019 Jul 12]. Available from: https://www.biorxiv.org/content/10.1101/581579v1. [DOI] [PMC free article] [PubMed]
- 34.García-González J, Tansey KE, Hauser J, Henigsberg N, Maier W, Mors O, et al. Pharmacogenetics of antidepressant response: a polygenic approach. Prog Neuropsychopharmacol Biol Psychiatry. 2017;75:128–134. doi: 10.1016/j.pnpbp.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 36.Gaspar HA, Gerring Z, Hübel C, Middeldorp CM, Derks EM, Breen G Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Using genetic drug-target networks to develop new drug hypotheses for major depressive disorder. Transl Psychiatry. 2019;9:117. doi: 10.1038/s41398-019-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson ST, Sanacora G. A new generation of antidepressants: an update on the pharmaceutical pipeline for novel and rapid-acting therapeutics in mood disorders based on glutamate/GABA neurotransmitter systems. Drug Discov Today. 2019;24:606–615. doi: 10.1016/j.drudis.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasson K, Darnovsky M. Gene-edited babies: no one has the moral warrant to go it alone [Internet] The Guardian; 2018. Nov 27, [cited at 2019 Jul 12]; Available from: https://www.theguardian.com/science/2018/nov/27/gene-edited-babies-no-one-has-moral-warrant-go-it-alone. [Google Scholar]
- 39.Bousman C, Maruf AA, Müller DJ. Towards the integration of pharmacogenetics in psychiatry: a minimum, evidence-based genetic testing panel. Curr Opin Psychiatry. 2019;32:7–15. doi: 10.1097/YCO.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 40.PharmVar. Genes [Internet] PharmVar; 2019. [cited at 2019 Jul 12]. Available from: https://www.pharmvar.org/genes. [Google Scholar]
- 41.Mills MC, Rahal C. A scientometric review of genome-wide association studies. Commun Biol. 2019;2:9. doi: 10.1038/s42003-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman T, Ash DM, Lauriello J, Rawlani R. Misleading guidance from pharmacogenomic Testing. Am J Psychiatry. 2017;174:922–924. doi: 10.1176/appi.ajp.2017.16121353. [DOI] [PubMed] [Google Scholar]
- 44.Serretti A. The present and future of precision medicine in psychiatry: focus on clinical psychopharmacology of antidepressants. Clin Psychopharmacol Neurosci. 2018;16:1–6. doi: 10.9758/cpn.2018.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maron E, Baldwin DS, Balõtšev R, Fabbri C, Gaur V, Hidalgo-Mazzei D, et al. Manifesto for an international digital mental health network. Digit Psychiatry. 2019;2:14–24. doi: 10.1080/2575517X.2019.1617575. [DOI] [Google Scholar]