Abstract
Absolute quantification of proteins in tissue is important for numerous fields of study. Liquid chromatography–mass spectrometry (LC–MS) methods are the norm but typically involve lengthy sample preparation including tissue homogenization, which results in the loss of information relating to spatial distribution. Here, we propose liquid extraction surface analysis (LESA) mass spectrometry (MS) of stable isotope labeled mimetic tissue models for the spatially resolved quantification of intact ubiquitin in rat and mouse brain tissue. Measured ubiquitin concentrations are in agreement with values found in the literature. Images of rat and mouse brain tissue demonstrate spatial variation in the concentration of ubiquitin and demonstrate the utility of spatially resolved quantitative measurement of proteins in tissue. Although we have focused on ubiquitin, the method has the potential for broader application to the absolute quantitation of any endogenous protein or protein-based drug in tissue.
Proteins are important molecules that play key roles in all life processes. Many proteins serve as biomarkers which distinguish between healthy and diseased tissues; therefore, their quantification may potentially reveal new information about disease state. Relative quantification can be achieved by comparing the abundance of protein signals in different physiological states. Absolute quantification requires comparison to the ion abundance detected from samples of known composition. Liquid chromatography coupled to mass spectrometry (LC–MS) is commonly employed in the quantification of proteins in biological samples. Usually this involves a bottom-up approach in which the protein is digested into proteolytic peptides and tagging of the protein (or its peptides) with a label containing stable isotope(s). The label may be introduced via metabolic, chemical, or enzymatic means.1,2 Commonly used methods include stable isotope labeling by amino acid in cell culture (SILAC), isotope-coded affinity tags (ICAT), isobaric tags for relative and absolute quantification (iTRAQ), and dimethyl labeling.1−3 Sample preparation for such methods is lengthy and there are inherent challenges associated with bottom-up protein identification such as inefficient digestion, failure to identify peptides, and loss of post-translational modifications.4 The alternative to proteolytic digestion is the top-down approach in which intact proteins are analyzed by tandem mass spectrometry.5 Top-down quantification approaches use both labeling and label-free methods; however, labeling has been shown to have its limitations and label-free methods require robust tools for data analysis.6 Top-down protein quantification involves LC–MS separation prior to MS analysis, and LC–MS typically requires sample homogenization. Consequently, spatial information is not retained meaning valuable biological information is lost.7,8
Mass spectrometry imaging (MSI) enables spatial profiling of analytes within thin tissue sections. Ambient MSI methods are particularly suited to quantitative analysis, due to the limited sample preparation required.9 Samples do not undergo any specific sample preparation, such as addition of a matrix compound, resulting in lower ion suppression and enhanced sensitivity.10 A number of examples of quantitative ambient MSI have been reported.11−13 Liquid sampling techniques in which desorption and ionization stages are decoupled are even more attractive because this affords the opportunity for off-line incorporation of an internal standard.14
In 2013, Groseclose and Castellino reported a novel method for spatially resolved quantification of small molecule drugs in tissue via matrix-assisted laser desorption ionization (MALDI) MS imaging (MSI),7 which has since been adopted in conjunction with MALDI and other mass spectrometry imaging techniques.8,12−15 The production of an external calibration sample comprised of tissue homogenates spiked with known amounts of isotopically labeled analyte of interest, termed the “mimetic tissue model”, enabled absolute quantitation. Thin tissue sections of the mimetic model were placed adjacent to sections from experimental samples and imaged under the same conditions. A calibration curve was generated from the mimetic model, and the experimental sample compared against this. Groseclose and Castellino demonstrated that the histology (overall tissue density and distribution of cell nuclei) and mass spectra (total ion currents) were consistent between the mimetic model and intact tissues, i.e., the homogenized and nonhomogenized tissue are comparable as background matrixes. A study by Swales et al. reported quantitative LESA imaging of four drug compounds in rat liver using the mimetic tissue model approach.14
Herein we demonstrate the use of LESA MS for the quantification of intact proteins in biological tissue using the example of ubiquitin. Ubiquitin is a regulatory protein involved in many processes, most notably protein degradation. It is found in every cell type, and it is highly conserved through eukaryotic species.16 It is therefore a suitable model for the quantitative LESA MS approach. In this study, we have incorporated stable isotope labeled (C13, N15) ubiquitin into a rat brain tissue mimetic model for use as a calibration reference standard. We were able to perform quantitative imaging via stable isotope labeled mimetics (SLiM) LESA MS of ubiquitin in rat and mouse brain tissue with a pixel size of 2 mm. Calculated concentration values of ubiquitin across the tissue were compared to those reported in the literature measured via LC–MS techniques and were found to be in good agreement.
In developing this method, we followed the FDA guidelines for Bioanalytical Method Validation.17 Validation parameters include lower limit of quantification (LLOQ), upper limit of quantification (ULOQ), precision (represented by coefficient of variation), accuracy (% difference between experimental and nominal values), selectivity, sensitivity, reproducibility, and stability of samples over time.
Results and Discussion
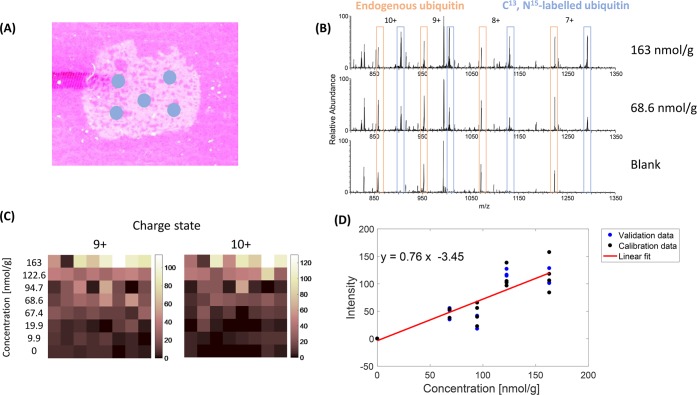
A method was devised to measure the concentration of ubiquitin in rat and mouse brain tissue in a spatially resolved manner. The workflow is summarized in Figure S1 (see the Supporting Information); briefly, a mimetic tissue model was prepared by spiking known quantities of C13, N15-labeled ubiquitin into homogenized rat brain tissue. The labeled ubiquitin is chemically identical to endogenous ubiquitin and is therefore assumed to have the same solubility and desorption/ionization efficiency. The isotopic labeling of ubiquitin with C13 and N15 results in a mass shift of 473.8612 Da between the labeled (9033.4779 Da) and endogenous (8559.6167 Da) proteins, allowing the labeled protein to be detected without interference from the endogenous ubiquitin (which is present in all brain tissues). Thin tissue sections of the mimetic tissue model were placed adjacent to sections of rat or mouse brain tissue on a glass slide, and the whole slide was imaged using LESA MSI. An example section of the tissue mimetic model, labeled with LESA sampling locations, is presented in Figure 1A. Example mass spectra from three mimetics with different concentrations (Figure 1B) show that it was possible to detect both endogenous and labeled ubiquitin simultaneously. Both endogenous and labeled ubiquitin were observed in 7+ through 10+ charge states, with the most abundant charge states being 9+ and 10+. The relative abundance of the labeled ubiquitin increases with mimetic concentration (and none is observed in the blank), whereas the relative abundance of the endogenous ubiquitin remains constant. Figure 1C represents overview intensity maps of the 9+ and 10+ charge states of labeled ubiquitin following LESA MS of the mimetics. Each row comprises eight separate sampling locations for a specific labeled ubiquitin concentration (i.e., one row per mimetic concentration).
Figure 1.
(A) Rat brain tissue mimetic section mounted on a glass slide with LESA sampling points indicated. (B) Mass spectra of the rat brain tissue mimetic models. Top, 163 nmol/g of labeled ubiquitin; middle, 68.6 nmol/g of labeled ubiquitin; bottom, blank mimetic. (C) Ion intensity image comparison of labeled ubiquitin in charge states 9+, m/z 1005.1788 and 10+, m/z 904.7624 in tissue mimetic models. Each horizontal line represents one mimetic concentration in nmol/g. (D) Calibration curve for the 9+ charge state of labeled ubiquitin. Calibration data points are shown in black; validation data points are shown in blue. Least squares fitting was applied to the calibration data.
A total of seven mimetics with nonzero concentrations were prepared and analyzed. In the initial experiment, the most abundant charge state observed in the mass spectra was 10+. Therefore, the signal intensity of the most abundant isotopomer of the 10+ charge state was plotted against the mimetic ubiquitin concentration (see Figure S2). FDA guidance for the validation of analytical and bioanalytical methods17 states that the analyte response at the lower limit of quantification (LLOQ) should be at least 5 times that of the blank response. Results for the three lowest concentration mimetics did not meet this criterion, where the LLOQ was 68.6 nmol/g, and were excluded from the data set. The upper limit of quantification (ULOQ) for these experiments was 163 nmol/g (% CV 7.55%).
Subsequent experiments made use of five mimetic concentrations (blank, 68.6 nmol/g, 94.7 nmol/g, 122.6 nmol/g, 163 nmol/g). For each concentration, seven locations were sampled. Replicate data acquired for each calibration sample were randomly split into 4 calibration and 3 validation (or quality control) data points in accordance with FDA guidance.17 In these experiments, the most abundant charge state was observed to be 9+ instead of 10+, and therefore the calibration data points were used to generate a calibration curve from the 9+ charge state (see Figure 1D). The data were analyzed by linear regression and a line was fitted with an associated R of 0.948 (R2 = 0.898). Individual data points acquired from the same concentration sample showed a fairly high level of variation (see Figure 1C,D). Variation can be introduced during LESA sampling and analysis via several routes including variable spreading of the sampling droplet resulting in variation in area sampled, variation in proportion of sampling solvent recovered from the surface during reaspiration, and in the case of sampling a tissue mimetic, spatial variation in concentration resulting from incomplete mixing of the homogenate. An assessment of homogenate mixing was performed by taking multiple sections at various depths of the mimetic volume, see Figure S3. The level of variability in the labeled and endogenous protein signal was comparable; therefore, the source of variation is more likely a function of the sampling process. The incorporation of an internal standard (at fixed concentration) into the sampling solvent or coated as a homogeneous layer over the sample could address this challenge; however, a second labeled version of the protein would be required for each separate internal standard, considerably increasing the cost of the experiment. This will be considered as a future step toward improving the reproducibility of the method.
The three validation data points (at each of the mimetic concentrations) were used to provide assessment of the method according to the guidance provided by the FDA for the validation of analytical and bioanalytical methods.17 The mean experimental concentration was calculated and compared with the nominal concentration of the mimetic (see Table 1). The coefficient of variation (% CV) between the experimental concentration and the nominal concentration of each mimetic sample was then calculated and meets the FDA precision requirements (<15%, and <20% for the LLOQ) for all mimetic samples. To measure the accuracy of the method, the % difference between back-calculated and nominal concentrations were calculated. To meet accuracy requirements, concentrations must fall within 15% deviation of the mean, except for LLOQ where 20% deviation is acceptable. Two of the three samples meet this requirement.
Table 1. Nominal and Calculated Concentration of Validation/Quality Control (QC) Data Points with Standard Deviation, Coefficient of Variation (% CV), and Calculated % Difference.
| QC sample | nominal concentration [nmol/g] | mean of the calculated concentration [nmol/g] | standard deviation | % CV | % difference |
|---|---|---|---|---|---|
| high QC | 163.0 | 150.68 | 16.66 | 11.09 | 7.55 |
| middle QC | 122.6 | 162.32 | 7.29 | 4.49 | 32.40 |
| low QC | 68.6 | 67.01 | 11.78 | 17.58 | 2.32 |
To demonstrate the selectivity of our method, we calculated the mean concentration in the blank mimetic sample which was 1.3 nmol/g and below our LLOQ. Sensitivity is defined as the LLOQ as long as it fulfills the accuracy and precision requirements; therefore, the sensitivity of the method is 68.6 nmol/g.
Sagittal sections of rat and mouse brain (ubiquitin is highly conserved through eukaryotic species, i.e., the protein sequence is the same in both organisms) were imaged using LESA MS under the same sampling parameters as the calibration samples to quantify endogenous ubiquitin, see Figure S4. The average concentration of ubiquitin detected across the whole rat brain section was 139.641 ± 95.403 nmol/g and in mouse brain was 90.233 ± 51.512 nmol/g. Assuming an average rat brain mass of 2 g and mouse brain of 0.4 g,18 these values equate to ∼2.392 ±1.634 mg ubiquitin per rat brain and ∼0.309 ± 0.177 mg per mouse brain. Kaiser et al. used a protein standard absolute quantification (PSAQ) method which uses LC–MS to quantify ubiquitin in mouse brain tissue and found that the total concentration of ubiquitin was ∼121 nmol/g tissue.16 This equates to ∼0.416 (±0.017) mg ubiquitin per brain. To date and to our knowledge, there is no information about the total ubiquitin content in the rat brain available; therefore, it was not possible to compare the calculated results to any reference value. Considering the size of the rat and mouse brain, these numbers are in approximate agreement. The LESA images in Figure S4 also suggest there might be a variability in the ubiquitin distribution across the brain. The mouse tissue shows higher variability compared to the rat tissue.
A complete repeat of the same experiment was performed 1 year after the initial experiment (see Figure S5), using the same mimetic samples and quantifying ubiquitin in the rat brain by using the most abundant charge state in those experiments (10+). The average concentration of ubiquitin in the rat brain was measured at 129.358 ± 98.401 nmol/g corresponding to ∼2.216 ± 1.686 mg of ubiquitin per rat brain. The percentage difference between this value and the value from the repeated experiment is within 7.4%. FDA guidelines suggest that the variation ±15% is acceptable, and therefore this result demonstrates the stability of the samples and reproducibility of the method.
Conclusions
We report quantitative mass spectrometry imaging of ubiquitin in tissue for the first time. The method passes validation criteria for LLOQ, ULOQ, selectivity, sensitivity, precision, reproducibility, and stability according to FDA guidelines. Absolute quantification of ubiquitin was performed in a pixel-wise fashion and demonstrated variability of ubiquitin concentration in different regions of the mouse and rat brain. Results obtained from the two most abundant charge states 9+ and 10+ were in good agreement. Future work will focus on incorporation of an internal standard into the workflow to improve the variability related to extraction efficiency, accuracy, and signal intensity observed in LESA sampling. Although the focus here was on ubiquitin, the method could in principle be adapted, with appropriate validation, to measure the absolute concentration of any protein in tissue including endogenous proteins and antibodies or protein-based therapeutic compounds.
Acknowledgments
H.J.C. is an EPSRC Established Career Fellow (Grants EP/L023490/1 and EP/S002979/1). E.C.R. was financially supported from EPSRC through PSIBS Doctoral Training Centre (Grant EP/F50053X/1) with additional funding from NiCE-MSI and AstraZeneca. J.H. is funded by the EPSRC Physical Sciences for Health Doctoral Training Centre (Grant EP/L016346/1). R.L.G. was funded by EPSRC (Grant EP/L023490/1). The Advion Triversa Nanomate and Thermo Fisher Orbitrap Elite mass spectrometer used in this research were funded through Birmingham Science City Translational Medicine, Experimental Medicine Network of Excellence Project with support from Advantage West Midlands.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b04148.
Materials and methods; Figure S1, workflow describing the preparation of tissue mimetic models; Figure S2, data obtained from LESA MS of mimetic tissue; Figure S3, schematic describing section locations within mimetic, locations selected for LESA sampling and LESA MS images of ions detected at m/z 904.6543 and 857.3632; Figure S4, LESA MS imaging of brain tissue sections from mouse and rat; and Figure S5, LESA MS imaging of rat brain (PDF)
Author Contributions
+ J.H. and E.C.R. contributed equally.
The authors declare the following competing financial interest(s): J.G.S. and R.J.A.G. are employees of AstraZeneca.
Notes
Supplementary data supporting this research is openly available from the University of Birmingham data archive at DOI: 10.25500/edata.bham.00000391.
Supplementary Material
References
- Yates J. R.; Ruse C. I.; Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009, 11, 49–79. 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- Bantscheff M.; Schirle M.; Sweetman G.; Rick J.; Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Boersema P. J.; Raijmakers R.; Lemeer S.; Mohammed S.; Heck A. J. R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484. 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Cui W.; Rohrs H. W.; Gross M. L. Top-down mass spectrometry: recent developments, applications and perspectives. Analyst 2011, 136, 3854–3864. 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X.; Aslanian A.; Yates J. R. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toby T. K.; Fornelli L.; Kelleher N. L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseclose M. R.; Castellino S. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal. Chem. 2013, 85, 10099–10106. 10.1021/ac400892z. [DOI] [PubMed] [Google Scholar]
- Rzagalinski I.; Volmer D. A. Quantification of low molecular weight compounds by MALDI imaging mass spectrometry - A tutorial review. Biochim. Biophys. Acta, Proteins Proteomics 2017, 1865, 726–739. 10.1016/j.bbapap.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Takáts Z.; Wiseman J. M.; Cooks R. G. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J. Mass Spectrom. 2005, 40, 1261–1275. 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- Randall E. C.; Race A. M.; Cooper H. J.; Bunch J. MALDI Imaging of Liquid Extraction Surface Analysis Sampled Tissue. Anal. Chem. 2016, 88, 8433–8440. 10.1021/acs.analchem.5b04281. [DOI] [PubMed] [Google Scholar]
- Chen W.; Wang L.; Van Berkel G. J.; Kertesz V.; Gan J. Quantitation of repaglinide and metabolites in mouse whole-body thin tissue sections using droplet-based liquid microjunction surface sampling-high-performance liquid chromatography-electrospray ionization tandem mass spectrometry. J. Chromatogr. 2016, 1439, 137–143. 10.1016/j.chroma.2015.10.093. [DOI] [PubMed] [Google Scholar]
- Bergman H.-M.; Lundin E.; Andersson M.; Lanekoff I. Quantitative mass spectrometry imaging of small-molecule neurotransmitters in rat brain tissue sections using nanospray desorption electrospray ionization. Analyst 2016, 141, 3686–3695. 10.1039/C5AN02620B. [DOI] [PubMed] [Google Scholar]
- Almeida R.; Berzina Z.; Arnspang E. C.; Baumgart J.; Vogt J.; Nitsch R.; Ejsing C. S. Quantitative Spatial Analysis of the Mouse Brain Lipidome by Pressurized Liquid Extraction Surface Analysis. Anal. Chem. 2015, 87, 1749–1756. 10.1021/ac503627z. [DOI] [PubMed] [Google Scholar]
- Swales J. G.; Strittmatter N.; Tucker J. W.; Clench M. R.; Webborn P. J. H.; Goodwin R. J. A. Spatial Quantitation of Drugs in tissues using Liquid Extraction Surface Analysis Mass Spectrometry Imaging. Sci. Rep. 2016, 6, 37648. 10.1038/srep37648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadoul L.; Longuespée R.; Noël A.; De Pauw E. A spiked tissue-based approach for quantification of phosphatidylcholines in brain section by MALDI mass spectrometry imaging. Anal. Bioanal. Chem. 2015, 407, 2095–2106. 10.1007/s00216-014-8232-7. [DOI] [PubMed] [Google Scholar]
- Kaiser S. E.; Riley B. E.; Shaler T. A.; Trevino R. S.; Becker C. H.; Schulman H.; Kopito R. R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 8, 691. 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . Guidance for Industry–Bioanalytical Method Validation, 2013.
- Bishop K. M.; Wahlsten D. Sex and Species Differences in Mouse and Rat Forebrain Commisures Depend on the Method of Adjusting for Brain Size. Brain Res. 1999, 815, 358–366. 10.1016/S0006-8993(98)01088-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.