Abstract
Staphylococcus aureus can cause severe life‐threatening illnesses such as sepsis and endocarditis. Although S. aureus has been isolated from marine water and intertidal beach sand, only a few studies have been conducted to assess prevalence of S. aureus at freshwater recreational beaches. As such, we aimed to determine prevalence and molecular characteristics of S. aureus in water and sand at 10 freshwater recreational beaches in Northeast Ohio, USA. Samples were analyzed using standard microbiology methods, and resulting isolates were typed by spa typing and multilocus sequence typing. The overall prevalence of S. aureus in sand and water samples was 22.8% (64/280). The prevalence of methicillin‐resistant S. aureus (MRSA) was 8.2% (23/280). The highest prevalence was observed in summer (45.8%; 55/120) compared to fall (4.2%; 5/120) and spring (10.0%; 4/40). The overall prevalence of Panton‐Valentine leukocidin genes among S. aureus isolates was 21.4% (15/70), and 27 different spa types were identified. The results of this study indicate that beach sand and freshwater of Northeast Ohio were contaminated with S. aureus, including MRSA. The high prevalence of S. aureus in summer months and presence of human‐associated strains may indicate the possibility of role of human activity in S. aureus contamination of beach water and sand. While there are several possible routes for S. aureus contamination, S. aureus prevalence was higher in sites with wastewater treatment plants proximal to the beaches.
Keywords: freshwater beaches, S. aureus, MRSA, antibiotic resistance, wastewater treatment
Key Points
A total of 280 samples was collected from 10 beaches on Northeast Ohio lakes; distances to sources of treated sewage effluent were examined
Overall prevalence of S. aureus was 22.8% (64/280); prevalence was higher in sites with wastewater treatment plants proximal to the beaches
More than one fifth of the positive S. aureus isolates harbored PVL genes, and almost one third of the isolates were ST 8
1. Introduction
Staphylococcus aureus is a ubiquitous bacterium that causes a wide range of infections, from superficial skin infections to severe, and potentially fatal, invasive disease (Kadariya et al., 2014). S. aureus infections have become increasingly difficult to treat due to the emergence of antibiotic resistance, including resistance to the antibiotic methicillin (methicillin‐resistant S. aureus, MRSA). Approximately 30% and 1.5% of the U.S. population is colonized with methicillin‐susceptible S. aureus (MSSA) and MRSA, respectively (Graham et al., 2006) with the most important site for colonization being the anterior nares (nostrils) (Wertheim et al., 2005). While colonization itself does not harm the host, it is a risk factor for developing subsequent symptomatic infections (Fritz et al., 2009; Graham et al., 2006). S. aureus may also be acquired via contact with fomites contaminated with the organism.
The strains causing health care‐associated MRSA infections (HA‐MRSA) have been distinct from the strains causing community‐associated MRSA (CA‐MRSA) infections: HA‐MRSA typically are resistant to many classes of antibiotics and carry the methicillin resistance gene, mecA, on the staphylococcal cassette chromosome (SCC) of type II (SCCmec type II), while CAMRSA tend to be resistant to fewer classes of antibiotics, carry the Panton‐Valentine leukocidin (PVL) genes, and carry SCCmec type IV. A third group, livestock‐associated MRSA (LA‐MRSA), has recently been identified and has typically been associated with swine or cattle (Smith & Pearson, 2010; Wulf & Voss, 2008). LA‐MRSA include strains such as ST398 and ST9, often carry SCCmec type V, are typically PVL negative, and, like HA‐MRSA, tend to be resistant to multiple classes of antibiotics. Lines are blurring between the strains, as both CA‐MRSA and LA‐MRSA have caused outbreaks or nosocomial infections in hospitals (Fanoy et al., 2009; Jenkins et al., 2009; Kourbatova et al., 2005; Seybold et al., 2006; Tattevin et al., 2009; van Rijen et al., 2008, 2009; Wulf et al., 2008).
A recent report has suggested that MRSA caused over 80,000 invasive infections and more than 11,000 deaths in 2011 (Dantes et al., 2013); the additional burden of MSSA infections has not been quantified. Most invasive MRSA infections were associated with healthcare exposure; however, this trend is changing, as the CA‐MRSA strain USA300 was recently identified in 29% of invasive MRSA isolates collected from U.S. metropolitan areas (Klevens et al., 2007). These invasive infections are on top of millions of more minor skin and soft tissue infections, which are the primary infections that S. aureus causes in the United States.
Recently, MRSA have been recovered from water and sand at public beaches in several areas in the United States (Soge et al., 2009; Yamahara et al., 2012). Domestically and internationally, antibiotic‐resistant S. aureus has also been isolated from municipal (Borjesson et al., 2009; Porrero et al., 2014; Rosenberg Goldstein et al., 2012; Wan & Chou, 2014), hospital (Thompson et al., 2013), and agricultural wastewaters/sewage (Brooks et al., 2014; Wan & Chou, 2014), representing potential sources of human environmental contamination. The primary sources of antibiotic resistance genes or bacteria derived from human sewage in the aquatic environment are (1) treated effluent from municipal wastewater treatment plants (WWTP) and (2) untreated sewage from combined sewer overflows (CSOs) (Tchobanoglous et al., 2003). In municipalities, most sewage is treated at WWTPs, but in 772 U.S. cities, combined storm water and sanitary sewers result in overflows that discharge untreated sewage into lakes and rivers during wet weather (Environmental Protection Agency, 2015). In areas with low population density, sewage treatment occurs by home septic systems. When properly functioning, such septic systems should not impact surface water quality, but failure of these systems, with attendant impacts on water quality, is common (Ohio Department of Health, 2013). Streams, lakes, and beaches may receive water affected by one or more of these sources, but some streams and small lakes have no identified sources of human wastewater discharge. MRSA are also able to survive for extended periods of time in sea and river water, though they are killed within 24 h in properly chlorinated pool water (Tolba et al., 2008). Taken together, these studies suggest that S. aureus in the environment represent a reservoir of bacteria that may result in colonization or infection of individuals who may come into contact with them.
As MRSA becomes increasingly common in nonhospitalized human and animal populations, the potential for environmental contamination and therefore additional acquisition of these strains by the general population similarly increases. Indeed, the ability to survive for extended periods in the environment has been suggested to be a factor in the generation or selection of strains capable of causing outbreaks (Wagenvoort et al., 2000). As such, novel studies of S. aureus in the environment and the community are critically needed. The objective of this study was to determine the prevalence and molecular characteristics of S. aureus at freshwater recreational beaches in Northeast Ohio.
2. Methods
2.1. Sample Collection
A convenience sampling method was used to collect sand and water samples from 10 Northeast Ohio recreational beaches (Figure 1). These beaches were selected to represent both rural and urban beaches as well as inland and Lake Erie beaches. Water (n = 30) and beach sand (n = 90) samples were collected from these beaches between 24 June and 15 July 2014; a second sampling was carried out with identical sample numbers between 8 October and 5 November 2014. For each sampling time period (summer and fall), three dates were sampled with an approximately 2 week interval between samples. One additional sampling was done to collect water (n = 10) and beach sand (n = 30) between 22 and 30 April 2015. During each location visit, three wet sand samples were collected at the tide line along with one 500 mL sample of freshwater. Water sample was collected using sterile methods via gloved hand immersion of 250 mL autoclaved wide‐mouth bottles with screw caps. All samples were processed immediately upon arrival to the laboratory or within 24 h of collection.
Figure 1.
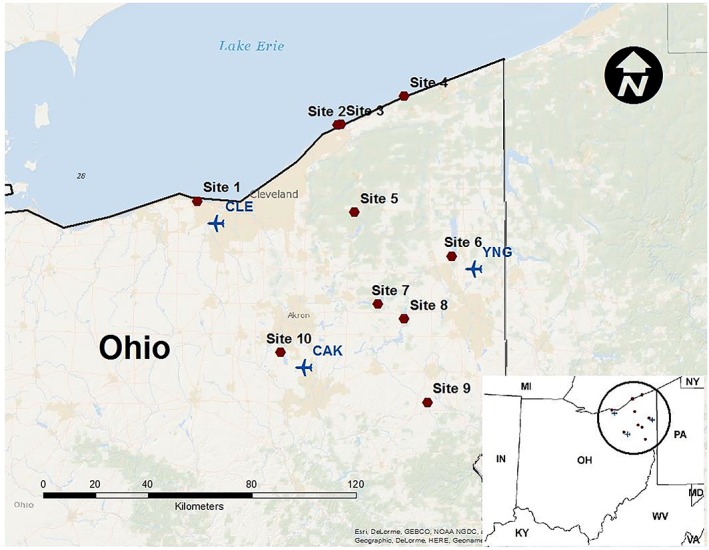
Sampling sites. CAK, Akron‐Canton; CLE, Cleveland; and YNG, Youngstown‐Warren.
2.2. Bacterial Isolation and Identification
One hundred milliliters of water was added with 50 mL Baird Parker broth (2 times concentrations) with tellurite enrichment (Sigma products—Sigma‐Aldrich, St. Louis, MO) in a 250 mL sterile screw cap jar and incubated for 18–24 h at 37°C. Five grams of sand was added to 40 mL Baird Parker broth (2 times concentrations) with tellurite enrichment in a sterile 50 mL conical centrifuge tube (Thermo Fisher Scientific, Pittsburg, PA) and vortexed for approximately 30 s. The supernatant liquid from each sample was then transferred into a sterile, 250 mL screw cap jar and incubated at 37°C for 18–24 h. After 24 h of incubation, 25 mL of broth was inoculated onto Baird Parker agar (BPA) with EY tellurite enrichment (BD Difco ™, Becton, Dickinson and Company, Sparks, MD) and selective MRSA agar plates (BBL CHROMagar MRSA, Becton, Dickinson and Company, Sparks, MD) and incubated 24–48 h at 37°C and examined for bacterial growth. Presumptive S. aureus (black colonies with clear halos on BPA) and presumptive MRSA (mauve colonies on CHROMagar) were subcultured onto Columbia colistin‐nalidixic acid agar (with 5% sheep blood) plates (BD) and incubated at 37°C for 24 h. S. aureus was confirmed by doing the catalase test, the slide coagulase test, and the S. aureus latex agglutination assay (Pastorex Staph‐plus, Bio‐Rad, Richmond, CA). S. aureus isolates were stored at −80°C.
2.3. Antimicrobial Susceptibility Testing
All S. aureus isolates were tested for antibiotic susceptibility with the VITEK 2 system (version R06.01) according to the manufacturer's instructions. Antimicrobial susceptibility testing (AST)‐GP71 cards of the VITEK 2 system were inoculated with a bacterial suspension prepared in 0.45% saline equal to the turbidity of a 0.5–0.63 McFarland standard with the DensiCHEK™ Plus (bioMérieux, Durham, NC). S. aureus isolates were tested for susceptibility to benzylpenicillin, oxacillin, tetracycline, erythromycin, ciprofloxacin, moxifloxacin, minocycline, clindamycin, trimethoprim‐sulfamethoxazole, quinupristin/dalfopristin, gentamicin, levofloxacin, linezolide, daptomycin, vancomycin, and rifampin, tigecycline, and nitrofurantoin per Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute, 2012). Isolates showing intermediate levels of susceptibility were classified as resistant. Isolates that were resistant to three or more classes of antimicrobials or that were resistant to oxacillin were considered multidrug resistant (MDR) (Magiorakos et al., 2012).
2.4. Molecular Testing
Genomic DNA was extracted using the Wizard Genomic DNA preparation kit (Promega, Madison, WI). Polymerase chain reaction (PCR) was performed on all isolates. The detection of mecA and PVL (lukS, lukF) genes in the S. aureus isolates were determined by PCR (Bosgelmez‐Tinaz et al., 2006; Lina et al., 1999). The Staphylococcus protein A (spa) gene was amplified as previously described (Koreen et al., 2004; Shopsin et al., 1999). spa types were assigned using Ridom® StaphType software (version2.2.1; Ridom GmbH, Wṻrzburg, Germany). The Based Upon Repeat Pattern (BURP) algorithm was applied to spa types to group S. aureus isolates into genetic clusters, according to their genetic proximity (Mellmann et al., 2007). All isolates were tested for multilocus sequence typing (MLST) as described previously (Enright et al., 2000). Sequence types (STs) were assigned using organism specific MLST database (http://saureus.mlst.net/). PHYLOViZ software v2.0 was used to analyze Global optimal eBURST of STs and to draw minimum spanning tree and relatedness of STs as described by Francisco et al. (2009, 2012) (Figure 2). Positive (USA300) and negative controls (reaction mixture without DNA template) were used in mecA, PVL, and spa PCR.
Figure 2.
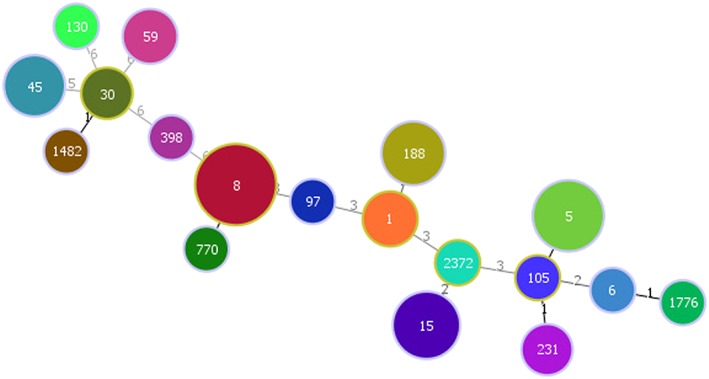
Minimum spanning tree demonstrating diversity of sequence types identified (constructed by the goeBURST algorithm using PHYLOViZ software v2.0).
2.5. Environmental Analysis
Data from Global Historical Climatology Network stations at three regional airports were used to compare the seasonal differences across sampling periods, as well as the spatial variability within the study area (Menne, Durre, Korzeniewski et al., 2012a; Menne, Durre, Vose et al., 2012b). Average daily precipitation for the duration of each sampling period, plus 2 weeks prior to the first sampling date, was calculated to indicate overall wetness. Average daily temperature was calculated for the same periods.
Potential exposure of water and beach sand to untreated or treated sewage effluent was qualitatively assessed by examining distances to (a) municipal WWTPs as identified by the National Pollutant Discharge Elimination System permits issued discharge into the lake and tributary streams or rivers, using the online mapping application available at http://wwwapp.epa.ohio.gov/dsw/gis/npdes/index.php and (b) CSO outfalls, via an Ohio Environmental Protection Agency map available at http://wwwapp.epa.ohio.gov/dsw/maps/cso/index.php. Potential influence of septic systems was qualitatively assessed based on whether residential areas surrouding the beach and tributary streams and rivers were served by WWTPs or not. For the four sites on Lake Erie, we also considered the patterns of lake currents, both generally (Bai et al., 2013) and for approximately weekly snapshots during the summer and fall sampling periods based on Great Lakes Coastal Forecasting system (GLCFS), as archived by the Experimental Lake Erie Harmful Algal Bloom Bulletin (https://www.glerl.noaa.gov/res/HABs_and_Hypoxia/lakeErieHABArchive/). These snapshots are not available for April 2015, and no information on currents was available for the inland lakes.
2.6. Statistical Analysis
Frequency distribution and proportions were calculated for the categorical variables. One‐way analysis of variance with least significant difference posthoc tests was conducted to assess the differences among group means. Statistical significance was assessed at α = 0.05 level. All statistical analyses were conducted using SAS statistical software (version 9.3, SAS Institute Inc., Cary, NC).
3. Results
3.1. Prevalence of S. aureus
A total of 280 environmental samples (fresh beach water and sand) was collected from 10 public freshwater recreational beaches in Northeast Ohio. A total of 70 isolates was chosen from 64 S. aureus‐positive samples: one from each positive BPA plate (n = 64) and one from each positive CHROMagar plate (n = 6). The overall prevalence of S. aureus was 22.8% (64/280; 95% confidence interval (CI) 17.9%–27.7%), with a prevalence of 14.6% (41 /280; 95% CI 10.5%–18.8%) and 8.2% (23/280; 95% CI 4.9%–11.4%) for MSSA and MRSA, respectively. Table 1 shows the distribution of S. aureus in sand and water across 10 sampling sites. The prevalence of S. aureus was significantly higher in Sites 6, 7, and 10 (p value 0.028, 0.001, and 0.027, respectively). The overall presence of S. aureus in sand and water was 20.5% (43/210) and 30% (21/70), respectively. The overall prevalence of S. aureus in summer, fall, and spring was 45.8% (55/120; 95% CI 37.0–54.7), 4.2% (5/120; 95% CI 0.6–7.7), and 10% (4/40; 95% CI 0.7–19.3), respectively. Similarly, the overall prevalence of MRSA in summer, fall, and spring was 15.8% (19/120; 95% CI 0.1–0.2), 0.8% (1/120; 95% CI 0–0.02), and 7.5% (3/40; 95% CI 0–0.2), respectively. The prevalence of S. aureus was higher in water (63.3%; 19/30; 95% CI 46.1–80.6) than in sand (40%; 36/90; 95% CI 29.9–50.1) in summer and in fall (6.7%; 2/30; 95% CI 0–0.15 versus 3.3%; 3/90; 95% CI 0–0.07). However, the difference was significant only in summer with p value = 0.03 (odds ratio (OR) 0.38 95% CI 0.164–0.906) and not significant in the fall (p value = 0.59, OR 0.48 95% CI 0.076–3.037).
Table 1.
Distribution of S. aureus in Environmental Samples
| Sites (N) | Total samples N = 280 (%) | P value | ||||
|---|---|---|---|---|---|---|
| Sand, N = 210 | Water, N = 70 | Total S. aureus | ||||
| MRSA | MSSA | MRSA | MSSA | |||
| 1a | 0 | 0 | 0 | 0 | 0 | |
| 2a | 0 | 3(14.3) | 1(14.3) | 1(14.3) | 5(17.9) | 0.483 |
| 3a | 3(14.3) | 1(4.8) | 0 | 3(42.8) | 7(25.0) | 0.145 |
| 4a | 1 (4.8) | 4(19.0) | 0 | 1(14.3) | 6(21.4) | 0.281 |
| 5b | 1(4.8) | 0 | 2(28.6) | 1(14.3) | 4(14.3) | 0.725 |
| 6b | 3(14.3) | 4(19.0) | 1(14.3) | 1(14.3) | 9(32.1) | 0.028c |
| 7b | 2(9.6) | 7(33.3) | 1(14.3) | 2(28.6) | 12(42.8) | 0.001c |
| 8b | 2(9.6) | 1(4.8) | 0 | 2(28.6) | 5(17.8) | 0.483 |
| 9b | 2(9.6) | 3(14.3) | 2 (28.6) | 0 | 7(25.0) | 0.145 |
| 10b | 1(4.8) | 5(23.8) | 1(14.3) | 2(28.6) | 9(32.1) | 0.027c |
| Total | 15 (7.1) | 28 (13.3) | 8 (11.4) | 13 (18.6) | 64(22.8) | |
Note. Same number of samples was taken from each site. Percentage of positive isolates resulting from each sampling site is shown in parentheses.
Lake Erie sites.
Inland sites.
Statistically significant.
No significant difference was found in S. aureus or MRSA prevalence between Lake Erie and inland lakes (p value for S. aureus 0.19, OR 1.5; 95% CI: 0.82–2.7; for MRSA, p value 0.07, OR 0.38; 95% CI 0.14–1.08).
3.2. Characterization of S. aueus Isolates
spa typing was carried out on all positive isolates. A total of 27 spa types was detected from 70 S. aureus isolates. t008 was the most common spa type (18; 25.7%), followed by t346 (5; 7.1%), t189 (6; 8.6%), t688 (4; 5.7%), t002 (4; 5.7%), t726 (3; 4.3%), t216 (3; 4.3%), t2143 (3; 4.3%), t021 (2; 2.9%), t548 (2; 2.9%), t6713 (2; 2.9%), t14316 (2; 2.9), and t304 (2; 2.9) (Table 2). All other spa types were less than 1% of S. aureus isolates.
Table 2.
Molecular Characteristics of S. aureus
| Isolate ID | Source | mecA | PVL | spa | AST | MLST |
|---|---|---|---|---|---|---|
| 062406S3 | Sand | − | − | t548 | P and E | ST5 |
| 062409S1 | Sand | − | − | t065 | P, E, and C | ST45 |
| 062409S2 | Sand | − | − | t371 | P | ST45 |
| 062409S3 | Sand | + | − | t216 | P and O | ST59 |
| 062410S1 | Sand | + | + | t008 | P, O, E, and Cl | ST8 |
| 062410S3 | Sand | − | − | t688 | P | ST5 |
| 062406W | Water | + | + | t008 | P, O, and E | ST8 |
| 062408W | Water | − | − | t267 | P | ST97 |
| 062409W | Water | − | − | t304 | P and E | ST6 |
| 062410W | Water | + | − | t008 | P, O, and E | ST231 |
| 062602S1 | Sand | − | − | t2049 | P and T | ST5 |
| 062603S2 | Sand | + | + | t008 | P, O, and E | ST8 |
| 062604S2 | Sand | − | + | t346 | P | ST15 |
| 062602W | Water | + | + | t1178 | P, O, and E | ST8 |
| 062605W | Water | + | + | t008 | P, O, E, T, Q, and R | ST8 |
| 070503S2 | Sand | + | − | t304 | P, O, and E | ST1776 |
| 070504S2 | Sand | + | − | t002 | P, O, and E | ST231 |
| 070505S3 | Sand | + | − | t002 | P, O, and E | ST1482 |
| 070506S1 | Sand | − | − | t008 | P | ST1 |
| 070506S2 | Sand | − | − | t605 | P | ST2372 |
| 070506S3 | Sand | − | − | t14316 | P and E | ST1 |
| 070507S1 | Sand | − | + | t185 | P, C, and E | ST770 |
| 070507S2 | Sand | − | − | t2143 | P | ST45 |
| 070507S3 | Sand | − | − | t008 | P | ST8 |
| 070508S2 | Sand | − | + | t008 | P and E | ST8 |
| 070509S1 | Sand | − | − | t8337 | P | ST5 |
| 070509S2 | Sand | + | − | t008 | P, O, and E | ST8 |
| 070510S1 | Sand | − | − | t189 | P | ST188 |
| 070510S2 | Sand | − | − | t189 | P and E | ST188 |
| 070510S3 | Sand | − | − | t216 | P | ST59 |
| 070503W | Water | − | − | t021 | P and E | ST30 |
| 070505W | Water | + | − | t548 | P, O, and E | ST5 |
| 070506W | Water | − | − | t726 | P and E | ST5 |
| 070507W | Water | − | − | t2143 | P | ST45 |
| 070508W | Water | − | − | t189 | P | ST188 |
| 070509W | Water | − | − | t8337 | P | ST5 |
| 070510W | Water | − | − | t688 | P | ST5 |
| 071402S1 | Sand | − | − | t021 | P | ST30 |
| 071403S3 | Sand | − | − | t068 | P, C, and E | ST8 |
| 071404S1 | Sand | − | − | t6713 | P | ST15 |
| 071404S2 | Sand | − | − | t726 | P, E, and Cl | ST5 |
| 071404S3 | Sand | − | − | t688 | P | ST15 |
| 071402W | Water | − | − | t008 | P | ST1 |
| 071403W | Water | − | − | t189 | P | ST188 |
| 071404W | Water | − | − | t346 | P | ST130 |
| 071405W | Water | − | − | t6713 | P | ST15 |
| 071506S1 | Sand | − | − | t2143 | P | ST45 |
| 071506S2 | Sand | + | + | t216 | P and O | ST59 |
| 071506S3 | Sand | − | − | t008 | P and E | ST8 |
| 071507S1 | Sand | − | − | t346 | P | ST15 |
| 071507S2 | Sand | + | − | t688 | P and O | ST5 |
| 071507S3 | Sand | − | − | t346 | P | ST15 |
| 071510S3 | Sand | − | − | t059 | P | ST8 |
| 071507W | Water | + | − | t334 | P, O, E, and Cl | ST8 |
| 071510W | Water | − | − | t726 | P | ST5 |
| 100802S3 | Sand | − | + | t346 | P and T | ST15 |
| 100803W | Water | − | − | t084 | P | ST15 |
| 101007W | Water | − | − | t571 | P, E, and Cl | ST398 |
| 102203S3 | Sand | + | + | t008 | P, O, and E | ST8 |
| 110507S3 | Sand | − | − | t189 | P | ST188 |
| 043007S1 | Sand | − | − | t189 | P | ST188 |
| 043007S2 | Sand | + | + | t008 | P, O, and E | ST8 |
| 043008S1 | Sand | + | − | t008 | P, O, and E | ST8 |
| 043008S3 | Sand | + | − | t008 | P, O, and E | ST8 |
| 062409WCA | Water | + | − | t002 | P, O, C, L, M, E, and Cl | ST105 |
| 062410WCA | Water | + | + | t008 | P, O, C, L, and Cl | ST8 |
| 070506S1CA | Sand | + | − | t008 | P, O, E, and Cl | ST8 |
| 070506S3CA | Sand | + | − | t008 | P, O, E, and Cl | ST8 |
| 070510S1CA | Sand | − | + | t008 | P and E | ST8 |
| 070509WCA | Water | + | + | t008 | P, O, E, and Cl | ST8 |
Note. P, benzylpenicillin; O, oxacillin; E, erythromycin; Cl, clindamycin; T, tetracycline; C, ciprofloxacin; L, levofloxacin; M, minocycline; Q, quinupristin/dalfopristin; and R, rifampin. “CA” samples are those initially taken from ChromAgar plates.
The overall prevalence of PVL genes among S. aureus isolates was 21.4% (15/70). Two thirds of the PVL positive isolates were spa‐type t008/ST8 (66.7%; 10/15). More than one third of the isolates (34.3%; 24/70) were mecA positive. More than half (54.1%; 13/24) of the spa type t008 samples were positive for mecA (Table 2). spa types were grouped based on their genetic proximity to spa types associated with specific cluster complexes. BURP grouping using default parameters (“exclude parameters that are shorter than 5 repeats” and “spa types are clustered if costs are less or equal than 4”) resulted in six spa CCs, nine singletons (t065, t185, t189, t216, t267, t334, t571, t6713, and t14316), and excluded three spa types (t059, t605, and t1178).
All S. aureus isolates were subjected to MLST. A total of 18 different sequence types (STs) was detected. The most common ST type was ST8 (30%; 21/70), followed by ST5 (15.7%; 11/70), ST15 (11.4%; 8/70), ST188 (8.6%; 6/70), ST45 (7.1%; 5/70), ST1 (4.3%; 3/70), ST59 (4.3%; 3/70), ST231 (2.9%; 2/70), and ST30 (2.9%; 2/70). Remaining STs were less than 2% of the total isolates. One livestock‐associated strain, ST398 (t571), was identified (Figures 2 and 3).
Figure 3.
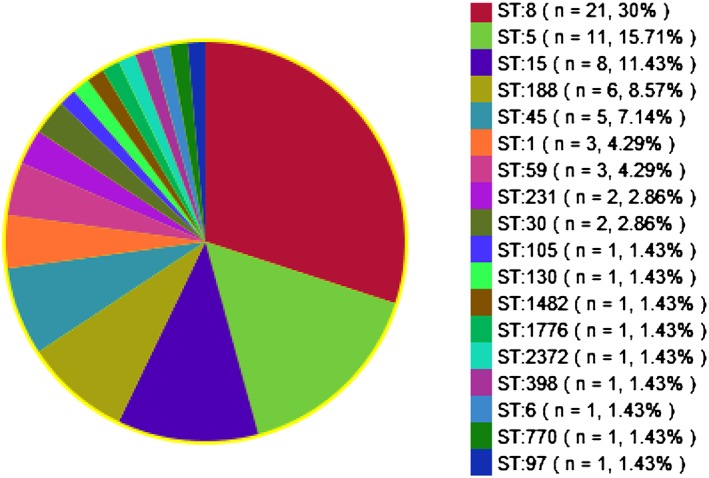
Proportion of STs (n = 70).
3.3. Antibiotic Susceptibility Testing
All isolates were tested for antibiotic susceptibility. Oxacillin resistance was observed for 34.3% (24/70) of the isolates. All isolates were resistant to penicillin. Thirty‐four isolates (48.6%) were resistant to erythromycin; 10 (14.3%) were resistant to clindamycin; 4 (5.7%) were resistant to ciprofloxacin; 3 (4.3%) were resistant to tetracycline; 2 (2.8%) were resistant to levofloxacin; and 1(1.4%) was resistant to moxifloxacin. Rifampicin and quinupristin/dalfopristin were resistant in one isolate each (1.4%). Twenty‐nine (41.4%) isolates were MDR‐S.aureus. (Figure 4).
Figure 4.
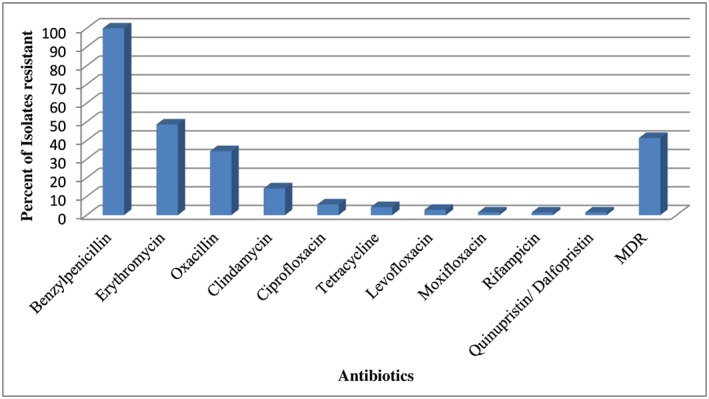
Antibiotic resistant profile of S. aureus. MDR, multidrug resistant.
3.4. Environmental Conditions
The summer 2014 sampling period was characterized by both warmer and wetter conditions than fall 2014 and spring 2015, which had very similar temperatures and average daily precipitation (Table 3). In summer 2014, there was an west‐east moisture gradient that was not present in other seasons.
Table 3.
Average Precipitation (P) and Temperature (T) for Each Sampling Period, and the Two Preceding Weeks, Based On Global Historical Climatology Network Data
| Akron‐Canton | Cleveland | Youngstown‐Warren | ||||
|---|---|---|---|---|---|---|
| P (mm/d) | T (°C) | P (mm/d) | T (°C) | P (mm/d) | T (°C) | |
| Summer 2014 | 6.5 | 22.3 | 4.9 | 22.3 | 3.6 | 21.3 |
| Fall 2014 | 2.6 | 12.2 | 2.4 | 12.1 | 2.1 | 11.5 |
| Spring 2015 | 2.4 | 10.9 | 1.8 | 10.4 | 2.1 | 10.2 |
Seven sites had WWTPs 5 km from the sampled beaches, and four rural sites potentially had septic in areas draining into the lake (Table 4). CSOs affect Lake Erie, but given the distance between CSO outfalls and sites in this study and the presence of more proximal WWTP discharges, their potential influence could not be further assessed.
Table 4.
Potential Sewage Sources at Each Site
| Site | Closest CSO outfall | Closest WWTP | Septic in surrounding area |
|---|---|---|---|
| 1 | 5 km east | 6 km west | No |
| 2 | ~25 km west | <5 km upstream on Grand River | No |
| 3 | ~25 km west | <5 km upstream on Grand River | No |
| 4 | ~50 km west | <1 km west | No |
| 5 | None | <0.6 km | No |
| 6 | None | 2 km | Possible |
| 7 | None | <0.6 km | Possible |
| 8 | None | 8 km (very small WWTP) | Possible |
| 9 | None | none | Possible |
| 10 | None | 1.2 km (two WWTPs) | No |
For the Lake Erie sites, lake current information provides additional insights into whether WWTP effluent is likely to reach the sites. At Site 1, summer currents tend to bring waters onshore from the open lake to the north of the site (Bai et al., 2013), but the GLCFS snapshots show mostly west to east currents during the summer sampling period. In the fall sampling period, all four cardinal directions of flow are represented. Based on the distance to WWTP, along with prevailing current directions, we suggest that effluent may be unlikely to reach Site 1 on a regular basis. At Sites 2–4, both summer and winter currents are generally alongshore from west to east (Bai et al., 2013). The summer sampling period, as recorded in the GLCFS snapshots, follows this general pattern, and, as at Site 1, the fall sampling period is more variable, showing both west to east and onshore from the north currents. Site 2 is located immediately (<1 km) west of the mouth of the Grand River, while Site 3 is located immediately to the east (0.5 km). Multiple WWTPs discharge into the Grand River, the closest <5 km upstream of the river mouth. Site 4 is located <1 km west of a small WWTP. Based on these distances, we suggest that WWTP treated sewage effluent may sometimes reach these beaches, even though Sites 3 and 4 are upwind relative to the dominant current directions. Imagery from mid‐June 2014, available on Google Earth, shows a sediment plume from the Grand River reaching west toward Site 2, suggesting that shortly prior to the summer sampling period, there were currents flowing in that upwind direction.
For the inland lakes, four sites (5, 6, 7, and 10) had WWTPs that discharged into the lake close to the beach, while two sites (8 and 9) did not. At Sites 5 and 7, small WWTPs that serve the state parks discharge approximately <0.6 km from the beaches. At Sites 6 and 10, several small WWTPs discharge to the lakes, with the closest located 1.2–2 km from the beaches. Sites 5 and 10 are judged unlikely to have septic effluent into the lake, while Sites 6 and 7 may receive some septic effluent given their rural locations and larger watersheds. Finally, Sites 8 and 9 have WWTPs that serve lakeside areas but discharge downstream of a dam at the lake's outlet. At Site 8, one very small WWTP discharges to the lake ~7.9 km away from the beach. At both sites, there may be some septic systems in the rural areas that are tributary to the lake.
4. Discussion
This study investigated the prevalence and molecular characteristics of S. aureus and MRSA in freshwater recreational beaches sand and water samples collected from 10 beaches in Northeast Ohio. The overall prevalence of S. aureus was 22.8% (64/280) with a prevalence of 14.6% (41/280) and 8.2% (23/280) for MRSA and MSSA, respectively.
In the United States, S. aureus has been isolated from public recreational beaches in Florida (Abdelzaher et al., 2010; Elmir et al., 2007; Esiobu et al., 2004; Mohammed et al., 2012), Washington (Soge et al., 2009), Hawaii (Charoenca & Fujioka, 1993; Fowler, 2005; Seifried et al., 2007; Tice et al., 2010; Viau et al., 2011), and California (Goodwin & Pobuda, 2009; Yamahara et al., 2012), but inland freshwater beaches have not been extensively studied with the exception of a recent investigation of Great Lakes beaches (Fogarty et al., 2015) The overall MRSA prevalence in our study is similar to several of these studies (Fogarty et al., 2015; Roberts et al., 2013). However, the overall S. aureus contamination in our sample was lower than that of Goodwin and Pobuda (2009), who documented 60%–75% and 53%–72% seawater and sand, respectively, that were contaminated with S. aureus, but our study had a higher prevalence of MRSA (7% versus 11.4% in water and 0% versus 7.1% in sand) (Goodwin & Pobuda, 2009). Sampling seasons, sample processing methods, and geography may have contributed to different outcomes. For example, Goodwin and Pobuda (2009) collected samples from two California beaches from June to September and used filtration technique in water sample processing; Fogarty et al. (2015) used similar methods. We collected samples in three seasons and did not filter water.
Goodwin et al. (2012) examined S. aureus and MRSA contamination on sand and seawater at three Southern California beaches and documented that 59% (n = 328) and 53% (n = 358) of seawater and beach sand samples were contaminated with S. aureus, respectively. The MRSA contamination was 1.6% (n = 366) in seawater and 2.7% (n = 366) in beach sand (Goodwin et al., 2012). In contrast to these findings, our study found lower prevalence of S. aureus and higher prevalence of MRSA in sand and water samples. However, the summer prevalence of S. aureus in water in our study was similar to theirs (63.3% versus 59%) during the summer sampling season. Such differences in S. aureus and MRSA prevalence may be due to methodological differences between these two studies. For example, we collected sand and water samples from freshwater recreational beaches of Northeast Ohio during June and July. Their study collected coastal beach sand and sea water samples and spanned from May to September. Our sampling timeline of peak summer months for beach visitors and ideal condition for S. aureus environmental colonization (hot and humid summer days) (Lidwell & Lowbury, 1950) may have contributed for higher MRSA contamination.
Contrary to the previous study that did not find USA300 (ST 8, clonal complex 8, staphylococcal cassette chromosome mec type IV), a community‐associated S. aureus strain in recreational beach samples from the Pacific Northwest (Roberts et al., 2013), almost one third of the positive isolates (30%; 21/70) of our study were ST8. More than one third of these isolates (32.6%; 15/46) were found in sand. More than two thirds (71.4%; 15/21) of the ST8 isolates were MRSA. However, our finding of high prevalence of PVL‐positive ST8 is consistent with a study that found 18 USA300 isolates in recreational seawater samples (Seifried et al., 2007). USA300 is implicated with invasive diseases including septicemia and necrotizing fasciitis and is a major cause of community‐associated infections in North America and Europe (Diep et al., 2006). Only one MRSA isolate of the 18 USA300 isolates from recreational seawater harbored PVL gene in previous study (Seifried et al., 2007). In contrast, a previous study found nine ST133 isolates, a predominant animal strain, in both marine and fresh recreational beaches samples (Roberts et al., 2013). We did not find any ST133 but found one ST398 (t571) isolate in a water sample from a rural inland lake. ST398 is typically a livestock‐associated strain (Smith & Pearson, 2010) that has seemingly evolved from an ancestral human lineage (Price et al., 2012). This isolate was not resistant to tetracycline, suggesting that it is more likely of the human lineage.
Eleven isolates of our study were ST5 (six from sand and five from water). Of the 11 ST5 isolates, only 2 were MRSA (one from sand and one from water). One ST5 isolate was found in beach samples in a prior report (Roberts et al., 2013). ST5 (USA100) is a common human‐associated S. aureus strain that has been widely distributed (Roberts et al., 2013). Six of the 70 isolates in our study were ST188 (spa‐type t189), a double‐locus variant of ST1, which includes USA400 (Ip et al., 2014). ST188 has been documented mostly in the Asia‐Pacific region (Ip et al., 2014). Although ST188 was the first known PVL‐positive MRSA strain (Control & Prevention, 1999), none of our ST188 isolates were MRSA and none harbored PVL. Despite the previous findings that most community‐associated strains of S. aureus harbor PVL (David & Daum, 2010), only 55.5% (10/18) of our t008 (ST8) isolates were PVL‐positive.
Studies have suggested a correlation between S. aureus concentration and bather density, suggesting a possible role of human activity in S. aureus environmental contamination of beach sand and water (Goodwin & Pobuda, 2009; Plano et al., 2011; Roberts et al., 2013; Seifried et al., 2007; Soge et al., 2009). Microorganisms are shed from human skin and body cavities into water during swimming (Robinton & Mood, 1966). Previous studies have shown that bathers shed concentrations of S. aureus on the order of 1.3 × 106 (Robinton & Mood, 1966) to 7.5 × 106 (Smith & Dufour, 1993) colony‐forming unit (CFU) in freshwater and 6.1 × 106 in marine water (Elmir et al., 2007) per person in the first 15 min exposure period; slightly lower CFU were reported by Plano et al. (2011). However, S. aureus shedding was reduced by 50% in subsequent bathing cycle (Elmir et al., 2007). Human‐implicated beach sand contamination has been a public concern as beachgoers spent most of the time in contact with the sand (Heaney et al., 2009). Previous studies have implicated sand as a bacterial source in freshwater beaches in two Michigan Lakes (Alm et al., 2006; Whitman & Nevers, 2003). A prior study has noted that sand may play a protective role for the bacteria, preventing them from exposure to stressors such as “ultraviolet radiation, high salinity, high temperatures, and wave action” (Goodwin & Pobuda, 2009). A correlation between S. aureus infection rates and seawater exposure demonstrated by Charoenca and Fujioka indicated recreational water as potential reservoir for S. aureus (Charoenca & Fujioka, 1995).
Alternatively, wildlife can serve both as a reservoir and a disseminator of microorganisms into the environment (Meerburg, 2010; Woolhouse & Gowtage‐Sequeria, 2005). Studies have isolated S. aureus (including MRSA) from wild animals such as psittacine birds, turtle, bat, guinea pig, chinchilla (Walther et al., 2008), snakes (Devriese & Hajek, 1980), hamsters (Ferreira et al., 2011), Great Horned Owl, European beaver, Great Blue Heron, Rock Pigeons, fox squirrel, Screech Owl, eastern cottontail rabbits and Lesser Yellowlegs shorebird (Wardyn et al., 2012), and dolphins and walruses (Faires et al., 2009). MRSA has also been implicated with marine animals such as seals (O'Mahony et al., 2005) and bottlenose dolphins (Faires et al., 2009; Schaefer et al., 2009). We have found S. aureus and MRSA contamination in freshly voided geese fecal samples collected from the vicinity of sampling sites of this study (Thapaliya et al., 2017); however, animal samples were not collected concurrently with beach samples. While molecular epidemiology of the goose fecal samples matched in some cases (ST5 and/or ST8 were isolated in both animals and beach samples at Sites 2, 4, 5, 7, and 10), water and sand samples were far more diverse. Further, while ST398 was isolated from goose feces at Site 6, this type was not found in beach water or sand at that site, while it was found at the beach in Site 7 but not in goose feces at that location (Thapaliya et al., 2017). As such, the wild animals and migratory birds dwelling and interacting in the vicinity of lakes and state parks may have played potential role in S. aureus and MRSA contamination of beach sand and water in addition to S. aureus/MRSA shedding by beachgoers, but further research is needed to test this connection.
Location of a recreational beach on an inland lake that receives treated WWTP effluent appears to be associated with a higher occurrence of S. aureus, as found at Sites 6, 7, and 10. Site 5 is also an inland lake with WWTP effluent but did not have a significantly higher prevalence of S. aureus than other sites in the study. Further, inland lakes without WWTP effluent (Sites 8 and 9) also had S. aureus in sand and water samples. On Lake Erie, distance from WWTPs was greater for Site 1, which had no occurrence of S. aureus at any time during the study, than for Sites 2–4, where S. aureus was identified. Conversely, dominant lake currents did not appear to affect prevalence of S. aureus, as illustrated by Sites 2 and 3, and the study design did not allow us to identify the influence of CSO outfalls on S. aureus prevalence.
The strength of this study is that we conducted a longitudinal study examining S. aureus and MRSA contamination in beach sand and water in all three seasons. We collected samples from both inland lakes and Lake Erie sites. However, the use of convenience sampling techniques and inclusion of a limited number of samples from one geographic location limits the generalizability of findings. We did not measure the bacterial load or collect human samples and were unable to gather information regarding number of visitors or bathers at each park. The lack of knowledge regarding the effect of rain and other environmental and weather condition adds a further limitation to this study. The source of sand and water contamination is currently unknown. It is possible that beachgoers may have potentially contaminated beach sand and water. This hypothesis is supported from the study results that compared to fall and spring, a higher prevalence of S. aureus and MRSA was observed during the summer season when increased human activities occur in the public beaches. While further studies are necessary in order to draw conclusions, simple public health measures, such as showering at the beach after exposure, may reduce any potential health effects from beach exposure to S. aureus.
Conflict of Interest
The authors declare no conflicts of interest relevant to this study.
Acknowledgments
This study was supported by Kent State University. The data used are listed in the references, tables, and figures of this manuscript.
Thapaliya, D. , Hellwig, E. J. , Kadariya, J. , Grenier, D. , Jefferson, A. J. , Dalman, M. , … Smith, T. C. (2017). Prevalence and characterization of Staphylococcus aureus and methicillin‐resistant Staphylococcus aureus on public recreational beaches in Northeast Ohio. GeoHealth, 1, 320–332. 10.1002/2017GH000106
This article was corrected on 15 JUL 2019. The online version of this article has been modified to include a Conflict of Interest statement.
References
- Abdelzaher, A. M. , Wright, M. E. , Ortega, C. , Solo‐Gabriele, H. M. , Miller, G. , Elmir, S. , … Bonilla, T. D. (2010). Presence of pathogens and indicator microbes at a non‐point source subtropical recreational marine beach. Applied and Environmental Microbiology, 76(3), 724–732. 10.1128/AEM.02127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm, E. W. , Burke, J. , & Hagan, E. (2006). Persistence and potential growth of the fecal indicator bacteria, Escherichia coli, in shoreline sand at Lake Huron. Journal of Great Lakes Research, 32(2), 401–405. [Google Scholar]
- Bai, X. , Wang, J. , Schwab, D. J. , Yang, Y. , Luo, L. , Leshkevich, G. A. , & Liu, S. (2013). Modeling 1993–2008 climatology of seasonal general circulation and thermal structure in the Great Lakes using FVCOM. Ocean Modelling, 65, 40–63. [Google Scholar]
- Borjesson, S. , Matussek, A. , Melin, S. , Lofgren, S. , & Lindgren, P. E. (2009). Methicillin‐resistant Staphylococcus aureus (MRSA) in municipal wastewater: An uncharted threat? Journal of Applied Microbiology, 108, 1244–51. 10.1111/j.1365-2672.2009.04515.x [DOI] [PubMed] [Google Scholar]
- Bosgelmez‐Tinaz, G. , Ulusoy, S. , Aridogan, B. , & Coskun‐Ari, F. (2006). Evaluation of different methods to detect oxacillin resistance in Staphylococcus aureus and their clinical laboratory utility. European Journal of Clinical Microbiology & Infectious Diseases, 25(6), 410–412. 10.1007/s10096-006-0153-8 [DOI] [PubMed] [Google Scholar]
- Brooks, J. P. , Adeli, A. , & McLaughlin, M. R. (2014). Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Research, 57, 96–103. 10.1016/j.watres.2014.03.017 [DOI] [PubMed] [Google Scholar]
- Charoenca, N. , & Fujioka, R. S. (1993). Assessment of Staphylococcus bacteria in Hawaii's marine recreational waters. Water Science and Technology, 27(3–4), 283–289. [Google Scholar]
- Charoenca, N. , & Fujioka, R. (1995). Association of staphylococcal skin infections and swimming. Water Science and Technology, 31(5–6), 11–17. [Google Scholar]
- Clinical and Laboratory Standards Institute (2012). Performance standards for antimicrobial susceptibility testing; Twenty‐third informational supplement.
- Control, C. F. D. , & Prevention (1999). Four pediatric deaths from community‐acquired methicillin‐resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. MMWR. Morbidity and Mortality Weekly Report, 48(32), 707. [PubMed] [Google Scholar]
- Dantes, R. , Mu, Y. , Belflower, R. , Aragon, D. , Dumyati, G. , Harrison, L. H. , … Emerging Infections Program‐Active Bacterial Core Surveillance, M. S. I (2013). National burden of invasive methicillin‐resistant Staphylococcus aureus infections, United States, 2011. JAMA Internal Medicine, 173(21), 1970–1978. 10.1001/jamainternmed.2013.10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, M. Z. , & Daum, R. S. (2010). Community‐associated methicillin‐resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clinical Microbiology Reviews, 23(3), 616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriese, L. , & Hajek, V. (1980). Identification of pathogenic staphylococci isolated from animals and foods derived from animals. Journal of Applied Bacteriology, 49(1), 1–11. [DOI] [PubMed] [Google Scholar]
- Diep, B. A. , Gill, S. R. , Chang, R. F. , Phan, T. H. , Chen, J. H. , Davidson, M. G. , … Mongodin, E. F. (2006). Complete genome sequence of USA300, an epidemic clone of community‐acquired meticillin‐resistant Staphylococcus aureus . The Lancet, 367(9512), 731–739. [DOI] [PubMed] [Google Scholar]
- Elmir, S. M. , Wright, M. E. , Abdelzaher, A. , Solo‐Gabriele, H. M. , Fleming, L. E. , Miller, G. , … Cooper, J. A. (2007). Quantitative evaluation of bacteria released by bathers in a marine water. Water Research, 41(1), 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, M. C. , Day, N. P. , Davies, C. E. , Peacock, S. J. , & Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin‐resistant and methicillin‐susceptible clones of Staphylococcus aureus . Journal of Clinical Microbiology, 38(3), 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environmental Protection Agency (2015). Combined sewer overflows. Retrieved from http://water.epa.gov/polwaste/npdes/cso/, February 16, 2015.
- Esiobu, N. , Mohammed, R. , Echeverry, A. , Green, M. , Bonilla, T. , Hartz, A. , … Rogerson, A. (2004). The application of peptide nucleic acid probes for rapid detection and enumeration of eubacteria, Staphylococcus aureus and Pseudomonas aeruginosa in recreational beaches of S. Florida. Journal of Microbiological Methods, 57(2), 157–162. [DOI] [PubMed] [Google Scholar]
- Faires, M. C. , Gehring, E. , Mergl, J. , & Weese, J. S. (2009). Methicillin‐resistant Staphylococcus aureus in marine mammals. Emerging Infectious Diseases, 15(12), 2071–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanoy, E. , Helmhout, L. C. , van der Vaart, W. L. , Weijdema, K. , van Santen‐Verheuvel, M. G. , Thijsen, S. F. , … Kingma‐Thijssen, J. L. (2009). An outbreak of non‐typeable MRSA within a residential care facility. Euro Surveillance, 14(1), 19080 10.2807/ese.14.01.19080-en [DOI] [PubMed] [Google Scholar]
- Ferreira, J. P. , Fowler, V. G. Jr. , Correa, M. T. , Lyman, R. , Ruffin, F. , & Anderson, K. L. (2011). Transmission of methicillin‐resistant Staphylococcus aureus between human and hamster. Journal of Clinical Microbiology, 49(4), 1679–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty, L. R. , Haack, S. K. , Johnson, H. E. , Brennan, A. K. , Isaacs, N. M. , & Spencer, C. (2015). Staphylococcus aureus and methicillin‐resistant S. aureus (MRSA) at ambient freshwater beaches. Journal of Water and Health, 13(3), 680–692. 10.2166/wh.2014.278 [DOI] [PubMed] [Google Scholar]
- Fowler, T. (2005). Development of methods using chromagar (TM) media to determine the prevalence of Staphylococcus aureus and methicillin‐resistant S. Aureus (MRSA) in Hawailan marine recreational waters.
- Francisco, A. P. , Bugalho, M. , Ramirez, M. , & Carriço, J. A. (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics, 10(1), 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco, A. P. , Vaz, C. , Monteiro, P. T. , Melo‐Cristino, J. , Ramirez, M. , & Carriço, J. A. (2012). PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics, 13(1), 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, S. A. , Epplin, E. K. , Garbutt, J. , & Storch, G. A. (2009). Skin infection in children colonized with community‐associated methicillin‐resistant Staphylococcus aureus . The Journal of Infection, 59, 394–401. 10.1016/j.jinf.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, K. , & Pobuda, M. (2009). Performance of CHROMagar™ Staph aureus and CHROMagar™ MRSA for detection of Staphylococcus aureus in seawater and beach sand—Comparison of culture, agglutination, and molecular analyses. Water Research, 43(19), 4802–4811. [DOI] [PubMed] [Google Scholar]
- Goodwin, K. D. , McNay, M. , Cao, Y. , Ebentier, D. , Madison, M. , & Griffith, J. F. (2012). A multi‐beach study of Staphylococcus aureus, MRSA, and enterococci in seawater and beach sand. Water Research, 46(13), 4195–4207. [DOI] [PubMed] [Google Scholar]
- Graham, P. L. 3rd , Lin, S. X. , & Larson, E. L. (2006). A U.S. population‐based survey of Staphylococcus aureus colonization. Annals of Internal Medicine, 144(5), 318–325. doi: 144/5/318 [pii] [DOI] [PubMed] [Google Scholar]
- Heaney, C. D. , Sams, E. , Wing, S. , Marshall, S. , Brenner, K. , Dufour, A. P. , & Wade, T. J. (2009). Contact with beach sand among beachgoers and risk of illness. American Journal of Epidemiology, 170(2), 164–172. [DOI] [PubMed] [Google Scholar]
- Ip, M. , Wang, Z. , Lam, W. Y. , Zhou, H. , & Tsui, S. (2014). Draft genome sequence of methicillin‐resistant Staphylococcus aureus CUHK_188 (ST188), a health care‐associated bacteremic isolate from Hong Kong. Genome Announcements, 2(2), e00255–e00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, T. C. , McCollister, B. D. , Sharma, R. , McFann, K. K. , Madinger, N. E. , Barron, M. , … Burman, W. J. (2009). Epidemiology of healthcare‐associated bloodstream infection caused by USA300 strains of methicillin‐resistant Staphylococcus aureus in 3 affiliated hospitals. Infection Control and Hospital Epidemiology, 30(3), 233–241. 10.1086/595963 [DOI] [PubMed] [Google Scholar]
- Kadariya, J. , Smith, T. C. , & Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food‐borne disease: An ongoing challenge in public health. BioMed Research International, 2014, 827965 10.1155/2014/827965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens, R. M. , Morrison, M. A. , Nadle, J. , Petit, S. , Gershman, K. , Ray, S. , … Fridkin, S. K. (2007). Invasive methicillin‐resistant Staphylococcus aureus infections in the United States. JAMA, 298(15), 1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Koreen, L. , Ramaswamy, S. V. , Graviss, E. A. , Naidich, S. , Musser, J. M. , & Kreiswirth, B. N. (2004). spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro‐and macrovariation. Journal of Clinical Microbiology, 42(2), 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourbatova, E. V. , Halvosa, J. S. , King, M. D. , Ray, S. M. , White, N. , & Blumberg, H. M. (2005). Emergence of community‐associated methicillin‐resistant Staphylococcus aureus USA300 clone as a cause of health care‐associated infections among patients with prosthetic joint infections. American Journal of Infection Control, 33(7), 385–391. 10.1016/j.ajic.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Lidwell, O. , & Lowbury, E. (1950). The survival of bacteria in dust. II. The effect of atmospheric humidity on the survival of bacteria in dust. Epidemiology and Infection, 48(1), 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina, G. , Piemont, Y. , Godail‐Gamot, F. , Bes, M. , Peter, M. O. , Gauduchon, V. , … Etienne, J. (1999). Involvement of Panton‐Valentine leukocidin‐producing Staphylococcus aureus in primary skin infections and pneumonia. Clinical Infectious Diseases, 29(5), 1128–1132. doi: CID990251 [pii] [DOI] [PubMed] [Google Scholar]
- Magiorakos, A. P. , Srinivasan, A. , Carey, R. , Carmeli, Y. , Falagas, M. , Giske, C. , … Olsson‐Liljequist, B. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281. [DOI] [PubMed] [Google Scholar]
- Meerburg, B. (2010). Rodents are a risk factor for the spreading of pathogens on farms. Veterinary Microbiology, 142(3), 464–465. [DOI] [PubMed] [Google Scholar]
- Mellmann, A. , Weniger, T. , Berssenbrügge, C. , Rothgänger, J. , Sammeth, M. , Stoye, J. , & Harmsen, D. (2007). Based Upon Repeat Pattern (BURP): An algorithm to characterize the long‐term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiology, 7(1), 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne, M. , Durre, I. , Korzeniewski, B. , McNeal, S. , Thomas, K. , Yin, X. , … Gleason, B. (2012a). Global historical climatology network‐daily (GHCN‐Daily), Version 3. NOAA National Climatic Data Center. 2012. [Google Scholar]
- Menne, M. J. , Durre, I. , Vose, R. S. , Gleason, B. E. , & Houston, T. G. (2012b). An overview of the global historical climatology network‐daily database. Journal of Atmospheric and Oceanic Technology, 29(7), 897–910. [Google Scholar]
- Mohammed, R. , Echeverry, A. , Stinson, C. , Green, M. , Bonilla, T. , Hartz, A. , … Esiobu, N. (2012). Survival trends of Staphylococcus aureus, Pseudomonas aeruginosa, and Clostridium perfringens in a sandy South Florida beach. Marine Pollution Bulletin, 64(6), 1201–1209. [DOI] [PubMed] [Google Scholar]
- O'Mahony, R. , Abbott, Y. , Leonard, F. , Markey, B. , Quinn, P. , Pollock, P. , … Rossney, A. (2005). Methicillin‐resistant Staphylococcus aureus (MRSA) isolated from animals and veterinary personnel in Ireland. Veterinary Microbiology, 109(3), 285–296. [DOI] [PubMed] [Google Scholar]
- Ohio Department of Health (2013). Household sewage treatment failures in Ohio. Retrieved February 16, 2015, from http://www.odh.ohio.gov/~/media/ODH/ASSETS/Files/eh/STS/2012HSTSSystemsandFailures.ashx
- Plano, L. R. , Garza, A. C. , Shibata, T. , Elmir, S. M. , Kish, J. , Sinigalliano, C. D. , … Solo‐Gabriele, H. M. (2011). Shedding of Staphylococcus aureus and methicillin‐resistant Staphylococcus aureus from adult and pediatric bathers in marine waters. BMC Microbiology, 11(1), 5 10.1186/1471-2180-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrero, M. C. , Valverde, A. , Fernandez‐Llario, P. , Diez‐Guerrier, A. , Mateos, A. , Lavin, S. , … Dominguez, L. (2014). Staphylococcus aureus carrying mecC gene in animals and urban wastewater, Spain. Emerging Infectious Diseases, 20(5), 899–901. 10.3201/eid2005.130426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, L. B. , Stegger, M. , Hasman, H. , Aziz, M. , Larsen, J. , Andersen, P. S. , … Aarestrup, F. M. (2012). Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. MBio, 3(1), e00305‐11 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijen, M. M. , Bosch, T. , Heck, M. E. , & Kluytmans, J. A. (2009). Meticillin‐resistant Staphylococcus aureus epidemiology and transmission in a Dutch hospital. The Journal of Hospital Infection, 72(4), 299–306. 10.1016/j.jhin.2009.05.006 [DOI] [PubMed] [Google Scholar]
- van Rijen, M. M. , Van Keulen, P. H. , & Kluytmans, J. A. (2008). Increase in a Dutch hospital of methicillin‐resistant Staphylococcus aureus related to animal farming. Clinical Infectious Diseases, 46(2), 261–263. 10.1086/524672 [DOI] [PubMed] [Google Scholar]
- Roberts, M. C. , Soge, O. O. , & No, D. (2013). Comparison of multi‐drug resistant environmental methicillin‐resistant Staphylococcus aureus isolated from recreational beaches and high touch surfaces in built environments. Frontiers in Microbiology, 4, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton, E. D. , & Mood, E. W. (1966). A quantitative and qualitative appraisal of microbial pollution of water by swimmers: A preliminary report. Journal of Hygiene, 64(04), 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg Goldstein, R. E. , Micallef, S. A. , Gibbs, S. G. , Davis, J. A. , He, X. , George, A. , … Sapkota, A. R. (2012). Methicillin‐resistant Staphylococcus aureus (MRSA) detected at four U.S. wastewater treatment plants. Environmental Health Perspectives, 120(11), 1551–1558. 10.1289/ehp.1205436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, A. M. , Goldstein, J. D. , Reif, J. S. , Fair, P. A. , & Bossart, G. D. (2009). Antibiotic‐resistant organisms cultured from Atlantic bottlenose dolphins (Tursiops truncatus) inhabiting estuarine waters of Charleston, SC and Indian River Lagoon, FL. EcoHealth, 6(1), 33–41. [DOI] [PubMed] [Google Scholar]
- Seifried, S. E. , Tice, A. D. , Eischen, M. , Diep, B. A. , Sensabaugh, G. F. , & Perdreau‐Remington, F. (2007). Diversity of community‐associated strains of methicillin‐resistant Staphylococcus aureus in Hawaii [with reply]. The Journal of Infectious Diseases, 195(2), 305–307. [DOI] [PubMed] [Google Scholar]
- Seybold, U. , Kourbatova, E. V. , Johnson, J. G. , Halvosa, S. J. , Wang, Y. F. , King, M. D. , … Blumberg, H. M. (2006). Emergence of community‐associated methicillin‐resistant Staphylococcus aureus USA300 genotype as a major cause of health care‐associated blood stream infections. Clinical Infectious Diseases, 42(5), 647–656. 10.1086/499815 [DOI] [PubMed] [Google Scholar]
- Shopsin, B. , Gomez, M. , Montgomery, S. O. , Smith, D. H. , Waddington, M. , Dodge, D. E. , … Kreiswirth, B. N. (1999). Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. Journal of Clinical Microbiology, 37(11), 3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, B. , & Dufour, A. (1993). Effects of the microbiological quality of recreational waters: A simulation study. Paper presented at the American Society for Microbiology 93rd General Meeting: May 16–20 1993; Atlanta, GA.
- Smith, T. C. , & Pearson, N. (2010). The emergence of Staphylococcus aureus ST398. Vector Borne and Zoonotic Diseases, 11, 327–39. 10.1089/vbz.2010.0072 [DOI] [PubMed] [Google Scholar]
- Soge, O. O. , Meschke, J. S. , No, D. B. , & Roberts, M. C. (2009). Characterization of methicillin‐resistant Staphylococcus aureus and methicillin‐resistant coagulase‐negative Staphylococcus spp. isolated from US West Coast public marine beaches. The Journal of Antimicrobial Chemotherapy, 64(6), 1148–1155. 10.1093/jac/dkp368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattevin, P. , Diep, B. A. , Jula, M. , & Perdreau‐Remington, F. (2009). Methicillin‐resistant Staphylococcus aureus USA300 clone in long‐term care facility. Emerging Infectious Diseases, 15(6), 953–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchobanoglous, G. , Feldman, B. , & Stensel, H. D. (2003). Wastewater engineering: Treatment and reuse. McGraw‐Hill. [Google Scholar]
- Thapaliya, D. , Dalman, M. , Kadariya, J. , Little, K. , Mansell, V. , Taha, M. Y. , … Smith, T. C. (2017). Characterization of Staphylococcus aureus in Goose Feces from State Parks in Northeast Ohio. EcoHealth, 14(2), 303–309. 10.1007/s10393-017-1227-z [DOI] [PubMed] [Google Scholar]
- Thompson, J. M. , Gundogdu, A. , Stratton, H. M. , & Katouli, M. (2013). Antibiotic resistant Staphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin‐resistant Staphylococcus aureus (MRSA). Journal of Applied Microbiology, 114(1), 44–54. 10.1111/jam.12037 [DOI] [PubMed] [Google Scholar]
- Tice, A. D. , Pombo, D. , Hui, J. , Kurano, M. , Bankowski, M. J. , & Seifried, S. E. (2010). Quantitation of Staphylococcus aureus in seawater using CHROMagar SA. Hawaii Medical Journal, 69(1), 8. [PMC free article] [PubMed] [Google Scholar]
- Tolba, O. , Loughrey, A. , Goldsmith, C. E. , Millar, B. C. , Rooney, P. J. , & Moore, J. E. (2008). Survival of epidemic strains of healthcare (HA‐MRSA) and community‐associated (CA‐MRSA) meticillin‐resistant Staphylococcus aureus (MRSA) in river‐, sea‐ and swimming pool water. International Journal of Hygiene and Environmental Health, 211(3–4), 398–402. 10.1016/j.ijheh.2007.06.003 [DOI] [PubMed] [Google Scholar]
- Viau, E. J. , Goodwin, K. D. , Yamahara, K. M. , Layton, B. A. , Sassoubre, L. M. , Burns, S. L. , … Boehm, A. B. (2011). Bacterial pathogens in Hawaiian coastal streams—Associations with fecal indicators, land cover, and water quality. Water Research, 45(11), 3279–3290. [DOI] [PubMed] [Google Scholar]
- Wagenvoort, J. H. , Sluijsmans, W. , & Penders, R. J. (2000). Better environmental survival of outbreak vs. sporadic MRSA isolates. The Journal of Hospital Infection, 45(3), 231–234. 10.1053/jhin.2000.0757 [DOI] [PubMed] [Google Scholar]
- Walther, B. , Wieler, L. H. , Friedrich, A. W. , Hanssen, A.‐M. , Kohn, B. , Brunnberg, L. , & Lübke‐Becker, A. (2008). Methicillin‐resistant Staphylococcus aureus (MRSA) isolated from small and exotic animals at a university hospital during routine microbiological examinations. Veterinary Microbiology, 127(1), 171–178. [DOI] [PubMed] [Google Scholar]
- Wan, M. T. , & Chou, C. C. (2014). Spreading of beta‐lactam resistance gene (mecA) and methicillin‐resistant Staphylococcus aureus through municipal and swine slaughterhouse wastewaters. Water Research, 64, 288–295. 10.1016/j.watres.2014.07.014 [DOI] [PubMed] [Google Scholar]
- Wardyn, S. E. , Kauffman, L. K. , & Smith, T. C. (2012). Methicillin‐resistant Staphylococcus aureus in central Iowa wildlife. Journal of Wildlife Diseases, 48(4), 1069–1073. [DOI] [PubMed] [Google Scholar]
- Wertheim, H. F. , Melles, D. C. , Vos, M. C. , van Leeuwen, W. , van Belkum, A. , Verbrugh, H. A. , & Nouwen, J. L. (2005). The role of nasal carriage in Staphylococcus aureus infections. The Lancet Infectious Diseases, 5(12), 751–762. 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- Whitman, R. L. , & Nevers, M. B. (2003). Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Applied and Environmental Microbiology, 69(9), 5555–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse, M. E. J. , & Gowtage‐Sequeria, S. (2005). Host range and emerging and reemerging pathogens. Emerging Infectious Diseases, 11(12), 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf, M. , & Voss, A. (2008). MRSA in livestock animals—An epidemic waiting to happen? Clinical Microbiology and Infection, 14(6), 519–521. 10.1111/j.1469-0691.2008.01970.x [DOI] [PubMed] [Google Scholar]
- Wulf, M. W. , Markestein, A. , van der Linden, F. T. , Voss, A. , Klaassen, C. , & Verduin, C. M. (2008). First outbreak of methicillin‐resistant Staphylococcus aureus ST398 in a Dutch hospital, June 2007. Euro Surveillance, 13(9), 3–4. [PubMed] [Google Scholar]
- Yamahara, K. M. , Sassoubre, L. M. , Goodwin, K. D. , & Boehm, A. B. (2012). Occurrence and persistence of bacterial pathogens and indicator organisms in beach sand along the California coast. Applied and Environmental Microbiology, 78(6), 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]


