Abstract
Illness caused by pathogenic strains of Vibrio bacteria incurs significant economic and health care costs in many areas around the world. In the Chesapeake Bay, the two most problematic species are V. vulnificus and V. parahaemolyticus, which cause infection both from exposure to contaminated water and consumption of contaminated seafood. We used existing Vibrio habitat models, four global climate models, and a recently developed statistical downscaling framework to project the spatiotemporal probability of occurrence of V. vulnificus and V. cholerae in the estuarine environment, and the mean concentration of V. parahaemolyticus in oysters in the Chesapeake Bay by the end of the 21st century. Results showed substantial future increases in season length and spatial habitat for V. vulnificus and V. parahaemolyticus, while projected increase in V. cholerae habitat was less marked and more spatially heterogeneous. Our findings underscore the need for spatially variable inputs into models of climate impacts on Vibrios in estuarine environments. Overall, economic costs associated with Vibrios in the Chesapeake Bay, such as incidence of illness and management measures on the shellfish industry, may increase under climate change, with implications for recreational and commercial uses of the ecosystem.
Keywords: Chesapeake Bay, climate change, Vibrio, habitat modeling
Key Points
Climate change may cause increases in season length and spatial habitat for V. vulnificus and V. parahaemolyticus in the Chesapeake Bay
Increase in favorable habitat for V. cholerae was less marked and restricted to low salinity regions of the Bay
1. Introduction
Many species of bacteria in the genus Vibrio are pathogenic to humans [Farmer et al., 2005]. Globally, the most well known is V. cholerae, the causative agent for cholera [Waldman et al., 2013]. There are estimated to be more than 1 million cases of cholera worldwide each year, with thousands of associated deaths [World Health Organization, 2016]. However, other Vibrio species also cause serious illness through consumption of contaminated seafood and environmental exposure.
In the United States, the two most problematic are V. parahaemolyticus and V. vulnificus, with other species including V. cholerae (nonepidemic strains) and V. alginolyticus also contributing to Vibrio‐associated illnesses [Ralston et al., 2011; Jones et al., 2013]. It is estimated that there are more than 34,000 cases of V. parahaemolyticus infection each year, with about $40 million in associated economic costs. V. vulnificus infections are much rarer but are more likely to lead to hospitalization and serious illness, with mortality rates of 30–40% [Ralston et al., 2011; Scallan et al., 2011; Hoffmann et al., 2015]. As a result, although there are only around 200–300 cases each year, these are associated with around $320 million in economic costs, including medical services, lost wages, and cost of premature death. V. parahaemolyticus is primarily transmitted through contaminated seafood, while V. vulnificus infections result from both foodborne sources and direct recreational exposure [e.g., from swimming with open wounds, Ralston et al., 2011].
Recent studies have suggested that the incidence of Vibrio infections may be increasing in some coastal oceans in association with warming temperatures [Andersson and Ekdahl, 2006; Martinez‐Urtaza et al., 2010; Vezzulli et al., 2010a, 2016; Le Roux et al., 2015]. Several have highlighted correlations between Vibrio outbreaks and anomalously warm conditions [Paz et al., 2007; Baker‐Austin et al., 2013, 2017]. Heat waves have been associated with the occurrence of new strains of Vibrio to a region [Martinez‐Urtaza et al., 2013] and to the outbreak of Vibrio‐related disease in areas where this was previously rare or unknown [Baker‐Austin et al., 2017]. Due to the seriousness of Vibrio‐related illnesses, the potential for increased incidence of infections in warming waters under climate change has significant economic and social implications [Lipp et al., 2002; Martinez‐Urtaza et al., 2010; Jacobs et al., 2015]. Laboratory experiments have shown optimum temperatures for several Vibrio species of 37–39°C, which is much warmer than currently observed water temperatures in the estuarine and marine environments where Vibrios are found [Kelly, 1982; Miles et al., 1997; Sedas, 2007]. In addition, current seasonal occurrences of Vibrios appear to be restricted by winter water temperatures. For example, in the southern states bordering the Gulf of Mexico, V. vulnificus has been recorded in coastal and estuarine waters year round [Lipp et al., 2001], and Vibrio‐associated illnesses are recorded in all months of the year [Altekruse et al., 2000]. In contrast, Vibrio occurrence is generally restricted to the warmest months of the year in higher latitudes of the mid‐Atlantic and New England [O'Neill et al., 1992; Jacobs et al., 2014]. Warming waters may therefore extend the length of high‐risk seasons for Vibrio‐related illnesses in regions where they are already known to occur, as well as facilitating their introduction to new areas [e.g., Baker‐Austin et al., 2013; Vezzulli et al., 2016]. However, there are relatively few studies which use projections from Intergovernmental Panel on Climate Change (IPCC) climate models to examine the risk of future Vibrio species occurrence or rates of infection.
Several Vibrio species occur naturally in the Chesapeake Bay, including V. cholerae, V. parahaemolyticus, and V. vulnificus [Colwell et al., 1977; Wright et al., 1996]. All three show positive relationships with water temperature and occur more frequently and at higher abundances over the warmer months [Kaneko and Colwell, 1973; Louis et al., 2003; Jacobs et al., 2010]. This is consistent with published relationships from other areas around the world, where Vibrios are rarely collected in waters cooler than 10–15°C and tend to increase in abundance when temperatures exceed about 20°C [O'Neill et al., 1992; Kaspar and Tamplin, 1993; Randa et al., 2004; McLaughlin et al., 2005; Vezzulli et al., 2010a, 2013; Baker‐Austin et al., 2010, 2013; Turner et al., 2014; Haley et al., 2014]. In contrast, each Vibrio species appears to be associated with a distinct, species‐specific salinity range. V. parahaemolyticus has the widest range, from approximately 5 to 30 (practical salinity unit, psu) [United States Food and Drug Administration (USFDA), 2005; Parveen et al., 2008], while V. vulnificus occurs from near‐fresh salinities to approximately 24 psu but is most common at around 8–16 [Jacobs et al., 2014]. These observations are consistent with the salinity tolerances reported elsewhere for these species [Kelly, 1982; Kaspar and Tamplin, 1993; Lipp et al., 2001; Pfeffer et al., 2003; Randa et al., 2004; Martinez‐Urtaza et al., 2010]. In contrast, V. cholerae occurs most frequently in the Chesapeake Bay where salinity is low (less than 8 psu) [Louis et al., 2003]. As these authors note, this relationship is not consistent among regions, as V. cholerae has been found from near fresh to moderately high (greater than 20 psu) salinities in other locations [Huq et al., 1984; Jiang and Fu, 2001; Louis et al., 2003; Chávez et al., 2005]. This likely reflects the strong dependence of V. cholerae on other environmental variables which are correlated with salinity, such as nutrients or concentration of zooplankton species [Louis et al., 2003; Baker‐Austin et al., 2010; Vezzulli et al., 2010b, 2016; Constantin de Magny et al., 2011]. Overall, current data suggest that climate change may lead to increasing numbers of Vibrios, and potentially of Vibrio‐related illnesses in the Chesapeake Bay, but only in those regions of the Bay where other environmental conditions remain (or become) favorable.
Jacobs et al. [2015] addressed aspects of this question by applying predictive models for V. parahaemolyticus and V. vulnificus to projections of future water temperature from downscaled global climate models. They used near‐surface air temperature as a proxy for surface water temperature, based on historical relationships from observations. Multiple sites along the United States coast were examined, including the Chesapeake Bay. These authors found that increasing temperatures would likely lead to expanded seasons of occurrence for both species. However, an important aspect of Vibrio risk assessment is the spatial location of high‐risk areas [e.g., Constantin De Magny et al., 2009; Jacobs et al., 2010; Banakar et al., 2011]. The narrower salinity range favored by V. cholerae and V. vulnificus, in particular, results in spatially localized “hot spots,” which shift according to streamflow and salinity conditions. Future precipitation patterns under climate change are uncertain for the Chesapeake Bay region [Najjar et al., 2010]. Some climate models project strong increases in freshwater inflow, which would move high‐risk areas downriver in tributaries and down bay (seawards) in the main stem. Other models project minimal change in watershed precipitation, which when added to warming watershed temperatures and increased evapotranspiration may result in increased salinity in most locations in the Bay [Muhling et al., 2017]. Under these conditions, high‐risk Vibrio areas may move upstream. Spatial projections of temperature and salinity in the Chesapeake Bay are thus required to more fully assess changes in the location of high‐risk Vibrio hot spots.
In this study, we used a recently developed statistical downscaling and spatial disaggregation modeling framework for estuarine habitats [Muhling et al., 2017] to project future spatial distribution and season length for three Vibrio species in the Chesapeake Bay, using general circulation models (GCMS) run under a high‐emission climate change scenario. We used previously published regression models to predict probability of occurrence or concentration for each species within eight regions of the Chesapeake Bay. The ability of the framework to capture habitat variability within each subregion, to the extent required to differentiate years with low versus high probability of occurrence or concentration, was assessed for each species.
2. Methodology
2.1. Habitat Models
Suitable habitat for occurrence of V. cholerae and V. vulnificus was predicted using the multivariate logistic generalized linear models (GLMs) published by Louis et al. [2003] and Jacobs et al. [2014], respectively. In addition, the concentration of V. parahaemolyticus in oysters (Crassostrea virginica) in log10 colony forming units (CFU)/gram was predicted using the relationships reported by the U.S. Food and Drug Administration [USFDA, 2005]. All models used surface temperature and surface salinity as predictor variables (Table 1). Models for Vibrio species were applied to the entire Chesapeake Bay main stem and major tributaries (Figure 1).
Table 1.
Logistic Generalized Linear Models for Three Vibrio Species (V. vulnificus, V. cholerae, and V. parahaemolyticus)a
| Species | Equation | Source |
|---|---|---|
| V. vulnificus | Logit [Vv] = 0.211*SST − 0.272*SALOPT − 4.288 ProbPres = elogit/[1 + elogit] | Jacobs et al. [2014] |
| V. cholerae | Logit [Vc] = 0.1233*SST − 0.1997*SSS − 0.0324*SST*SSS ProbPres = elogit/[1 + elogit] | Louis et al. [2003] |
| V. parahaemolyticus | log[Vp/g] = − 2.05 + 0.097*SST + 0.2*SSS − 0.0055*SSS2 | USFDA [2005] |
SST refers to surface water temperature, while SSS is surface salinity (psu). SALOPT refers to the absolute distance of measured salinity from 11.5, the optimal salinity for this species [Jacobs et al., 2014].
Figure 1.
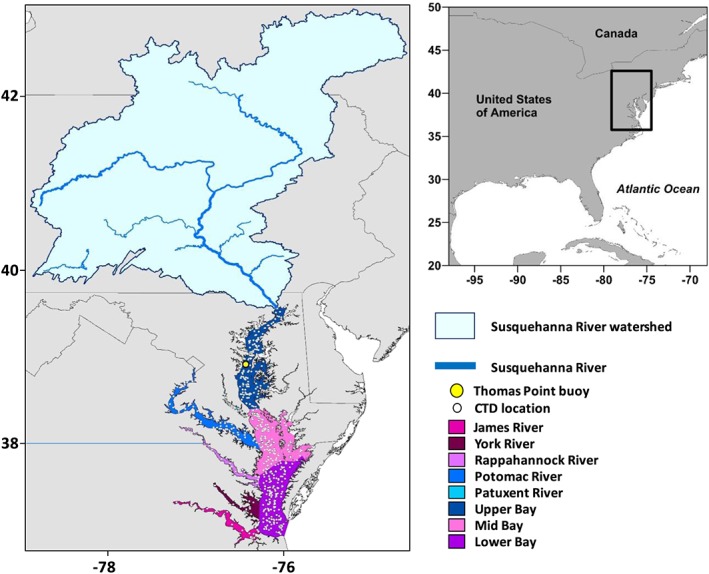
Study area showing the Susquehanna River watershed, the location of the Thomas Point buoy, and the Chesapeake Bay and major tributaries. CTD cast stations used to develop the statistical framework (Figure 3) are also shown. Colors denote the eight zones of the Bay mentioned in the text.
Sampling for V. vulnificus took place in January, April, July, and October 2007, and April, July, and October 2008–2010 [Jacobs et al., 2014]. Surface (0.5 m depth) water samples were collected from the main stem and major tributaries of the Chesapeake Bay, stored in sterile polypropylene bottles, and then frozen until sampling was completed for the month. After thawing, samples were filtered, and the filtered material was tested for V. vulnificus using quantitative Polymerase Chain Reaction (qPCR). While Jacobs et al. [2014] also determined abundance (CFU/mL), we only considered the probability of occurrence (presence/absence) for V. vulnificus in this study (Table 1). This GLM showed 86.6% agreement between modeled and observed probabilities of occurrence for V. vulnificus, with an Area Under the Receiver Operating Curve (AUC) of 0.87 [Jacobs et al., 2014].
Sampling for V. cholerae was completed at least monthly between January 1998 and February 2000 at nearshore sites in the upper Chesapeake Bay, and during summer 1999 and 2000 in the main stem [Louis et al., 2003]. Both water and plankton samples were collected, and V. cholerae was isolated using the alkaline peptone water‐enrichment procedure. Presumptive V. cholerae isolates were confirmed by PCR [Louis et al., 2003]. The GLM showed 76.9% agreement between observations and modeled probabilities of occurrence. Both temperature and salinity were statistically significant within the model (p < 0.05), as was an interaction between the two [Louis et al., 2003] (Table 1).
The United States Food and Drug Administration (USFDA) model for V. parahaemolyticus densities in oysters was developed using data from multiple studies conducted across the United States, including the Pacific, Atlantic, and Gulf of Mexico coasts. Temperature and salinity were obtained in situ where possible, and the GLM estimating V. parahaemolyticus in oysters was parameterized using the Tobit regression method [USFDA, 2005]. This study notes that water temperature explained approximately 50% of the variance in (log) V. parahaemolyticus concentrations, but no information on the overall skill of the model was given. However, both temperature and salinity were highly significant within the GLM (p < 0.0001) (Table 1).
Applying each Vibrio habitat model from Table 1 across two‐dimensional temperature/salinity space illustrates the differences in habitat preferences across species (Figure 2).
Figure 2.
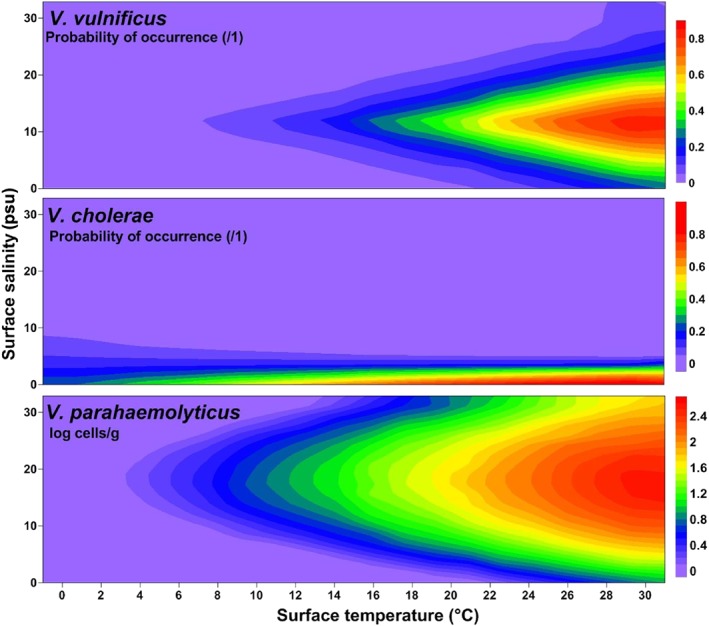
Representation of the GLMs in Table 1 in temperature‐salinity two‐dimensional space for V. vulnificus, V. cholerae, and V. parahaemolyticus.
Strong differences between V. cholerae and the other species are particularly evident. All three Vibrios had higher probability of occurrence at warmer temperatures, while V. vulnificus and V. parahaemolyticus were predicted to be most abundant at moderate salinities, and V. cholerae was associated with low salinity (less than 8 psu). The V. vulnificus and V. parahaemolyticus models suggested that these species occurred across a wider range of salinities when temperatures were very warm. Optimum salinity values stretched across a fairly broad range (8–16 psu) for V. vulnificus and an even larger range (5–30 psu) for V. parahaemolyticus.
2.2. Habitat Projection Framework
Historical estimates and future projections of surface temperature and salinity across the Chesapeake Bay were sourced from a previously published statistical downscaling framework [Muhling et al., 2017: Figure 3]. A brief summary of the framework follows, and the reader is directed to Muhling et al. [2017] for further details.
Figure 3.
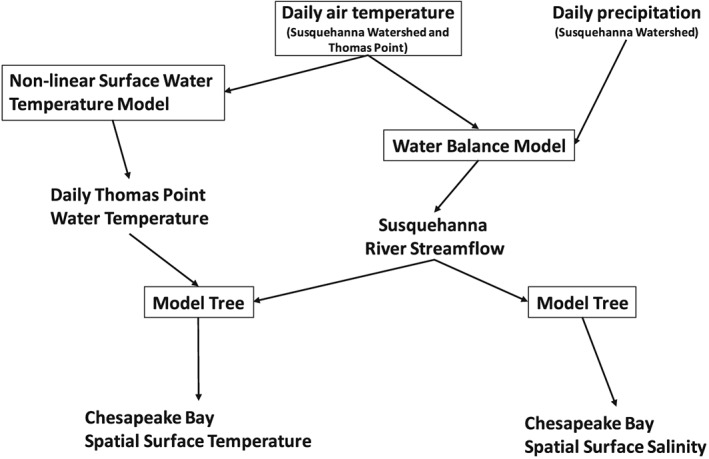
Schematic representation of the statistical framework developed by Muhling et al. [2017]. Models are boxed, and model outputs are unboxed. Adapted with permission of Springer, from Estuaries and Coasts doi: 10.1007/s12237‐017‐0280‐8, 2017.
The framework ingests air temperature at the Thomas Point buoy and air temperature/precipitation over the Susquehanna River watershed and uses a hierarchy of models to estimate spatial fields of surface temperature and salinity across the Chesapeake Bay. For future projections, the inputs to the framework are statistically downscaled air temperature and precipitation from GCMs (see section 2.3 “Future Projections” below for details). For historical estimates, inputs are observed air temperature at Thomas Point, and precipitation and air temperature in the Susquehanna River watershed from the NOAA/National Centers for Environmental Prediction GHCN (Global Historical Climatology Network) Climate Anomaly Monitoring System (CAMS) 0.5° monthly temperature data set [Fan and van den Dool, 2008], and the CPC (Climate Prediction Center) Unified Gauge‐Based 0.25° Analysis of Daily Precipitation (CPC) [Chen et al., 2008].
Streamflow is derived from downscaled air temperature and precipitation over the Susquehanna River basin using a simple water balance model [McCabe and Markstrom, 2007], calibrated for Chesapeake Bay as described in Muhling et al. [2017]. Comparison of predicted versus observed monthly Susquehanna River streamflow at Conowingo dam from 1970 to 2006 showed that the model reproduced observations with good skill (R 2 = 0.8 [Muhling et al., 2017]).
Spatial estimates of surface temperature and salinity were estimated from air temperature and streamflow anomalies using linear model trees [Quinlan, 1992] with the Cubist package in R 3.2.1 [Kuhn et al., 2015; R Core Team, 2015]. Linear model trees are similar to regression trees, but the predicted values at terminal nodes are described using multivariate linear equations rather than fixed values. This characteristic allows model trees to extrapolate beyond the range of training data, whereas many other machine‐learning techniques cannot [Quinlan, 1992, 1993]. The predictor variables are used both to split the training data into increasingly similar subsets and to parameterize the equations at the terminal nodes.
Models were trained on conductivity‐temperature‐depth (CTD) cast data from the Chesapeake Bay Program, the University of Maryland Chesapeake Biological Laboratory cruise database, and the Smithsonian Environmental Research Center database from 1986 to 2005 and then validated with out‐of‐sample data from the same programs from 2006 to 2015 [Muhling et al., 2017]. Surface temperature was predicted using 17 day moving mean air temperatures at Thomas Point, the 30 day change in this metric (to account for seasonal hysteresis [Letcher et al., 2016]), freshwater inflow at Conowingo Dam, time of day, latitude, and longitude. Surface salinity was predicted using the same variables, except that Thomas Point air temperature was not included.
Previous validation of temperature and salinity anomalies across the regions identified in Figure 1 showed robust out‐of‐model temperature anomaly prediction skill across regions (R 2 between 0.62 and 0.77) and considerable skill for surface salinity (R 2 between 0.42 and 0.76 [Muhling et al., 2017]). The translation of these hydrographic skill metrics to accurate prediction of Vibrio habitat favorability, however, is less clear. Before making projections, we thus assessed whether out‐of‐sample estuarine Vibrio habitat estimates derived from our downscaling framework were consistent with those derived from the CTD data. That is, are Vibrio habitat projections based on statistically downscaled and spatially disaggregated habitat estimates of comparable accuracy to those based on real‐time hydrographic observations? We assessed skill for the regions defined in Figure 1 and at the subregional scale across individual CTD casts.
2.3. Future Projections
To assess the potential impacts of climate change on the occurrence of Vibrio species in the Chesapeake Bay, we used statistically downscaled projections under RCP8.5 from four GCMs with contrasting characteristics for the region, spanning the range of future warming and precipitation projections [Muhling et al., 2017]. RCP8.5 is a high‐emission scenario, which assumes that radiative forcing due to greenhouse gas emission will continue to increase strongly throughout the 21st century [Riahi et al., 2011]. The GFDL‐CM3 model (hereafter CM3 model) [Donner et al., 2011] showed strong warming of the surface air temperature (an increase of around 5.0–5.5°C) between historical (1970–1999) and end of century (2071–2100) periods under RCP8.5, and a mean annual increase in precipitation over the Susquehanna River watershed of approximately 0.75 mm/d. The MRI‐CGCM‐3 model (hereafter MRI model) [Yukimoto et al., 2012] showed a slightly lower precipitation increase (around 0.5 mm/d on average) and much weaker warming of around 2.0–2.5°C by the end of the century. The IPSL‐CM5A‐LR model (hereafter IPSL model) [Dufresne et al., 2013] warmed around 4.5–5.0°C between the historical and end century time periods but showed little change in mean precipitation over the region. The GFDL‐ESM2G model (hereafter ESM2G) [Dunne et al., 2012] showed a slight precipitation increase (around 0.2 mm/d) and weaker warming of around 3.0–3.5°C by the end of the century. The CM3 model thus represented a warmer, wetter future, the MRI model a less warm, wetter future, the IPSL model a warmer, precipitation‐neutral scenario, and the ESM2G model a less warm, precipitation‐neutral future.
Each of the four GCMs was bias corrected using four separate statistical methods—Bias‐Corrected Quantile Mapping, Change Factor Quantile Mapping, Equidistant Quantile Mapping, and the Cumulative Distribution Function Transform—and then spatially disaggregated using linear model trees. Results from Muhling et al. [2017] showed that projections using the different downscaling methods were similar except at extreme high temperatures, and that choice of downscaling method was much less influential than choice of GCM for our application. We therefore averaged projections from each GCM across all four downscaling methods and present results only among different GCMs.
The CPC daily precipitation analysis was used as the historical observations for the Susquehanna River watershed, by assigning each grid point from this 0.25 × 0.25° resolution analysis (n = 119, 1970–2005) to the closest grid point for each GCM (n = 2–6). Watershed air temperature observations for downscaling were obtained from eight weather stations (1970–2005) and then also assigned to the closest grid point for each GCM. Air temperatures at Thomas Point were obtained from only one grid point from each GCM and were downscaled using historical observed air temperatures at the Thomas Point buoy (1985–2015). Downscaled air temperature and precipitation fields were then run through the modeling framework (Figure 3) to obtain surface temperature and salinity fields in the Chesapeake Bay for the recent historical (1970–1999) and end century future (2071–2100) time periods, for each GCM [Muhling et al., 2017]. The Vibrio models in Table 1 were then applied to these fields and used to compare potential changes in probability of occurrence of V. vulnificus and V. cholerae, and the concentration of V. parahaemolyticus in oysters, between the two time periods.
3. Results
The mean habitat favorability predicted from observed temperature and salinity was similar to that predicted using out‐of‐sample temperature and salinity estimates derived from the statistical downscaling framework (Figure 4, bars). While the framework introduced some error into predictions from the Vibrio habitat models when compared to CTD observations, this was generally minimal (e.g., probabilities of occurrence by zone (Figure 1) differed by less than 5% for both V. vulnificus and V. cholerae). The R 2 values derived between CTD‐based and downscaled estuarine conditions within each zone of the Chesapeake Bay during peak seasons were above 0.6 for all species and zones, except for V. cholera within higher salinity environments where the probability of occurrence is very low (Figure 4, value above bars). In these regions, minimally varying probabilities of occurrence near zero resulted in low R 2 values.
Figure 4.
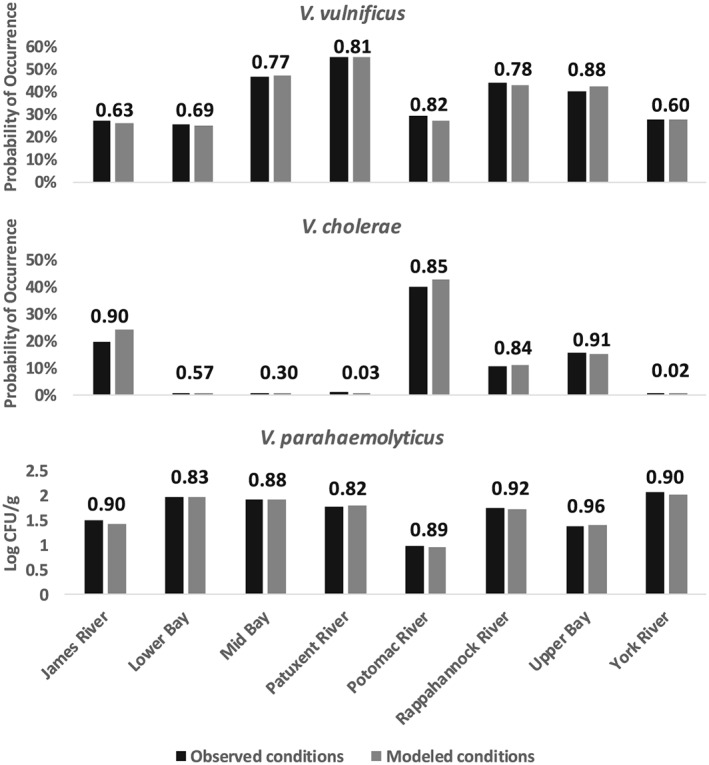
Estimated probability of occurrence (V. vulnificus and V. cholerae) and concentration of V. parahaemolyticus in oysters (log), in each region of the Chesapeake Bay, from GLMs in Table 1. Temperature and salinity values are sourced from CTD casts (black bars) and estimates from the statistical framework in Figure 3 at the same locations (gray bars), during seasons of highest detectability (May–October for V. vulnificus and V. parahaemolyticus, March–August for V. cholerae). R2 values between GLM predictions for each species from CTD casts and the statistical framework from within each region are shown. Note that only out‐of‐model test years (2006–2015) were used [see Muhling et al., 2017].
Consistent with the relationships shown in Figure 2, V. cholerae was predicted to be most prevalent in lower salinity environments (upper reaches of the James, Potomac, and Rappahannock Rivers and Upper Bay). V. vulnificus had the highest probabilities of occurrence in moderate salinity zones (Mid Bay, Patuxent and Rappahannock Rivers, and Upper Bay), while V. parahaemolyticus was abundant across most of the bay except the Potomac River (Figure 4).
Application of habitat models to future projections from downscaled GCMs resulted in higher mean monthly probability of occurrence for V. vulnificus (Figure 5, top panel) between April and October. This difference was strongest in the warmer CM3 and IPSL models and weakest in the MRI model. In contrast, probabilities of occurrence for V. cholerae (Figure 5, middle) increased substantially in winter‐spring in the warmer, wetter CM3 model, increased slightly in the MRI model, and decreased in the drier IPSL model. Probabilities of occurrence for V. cholerae between June and November were not projected to change strongly. Mean CFU/g of V. parahaemolyticus in oysters (Figure 5, bottom) was predicted to increase at all times of year, particularly in summer and fall. Increases were strongest in the warmer CM3 and IPSL models, and weaker in the MRI model.
Figure 5.
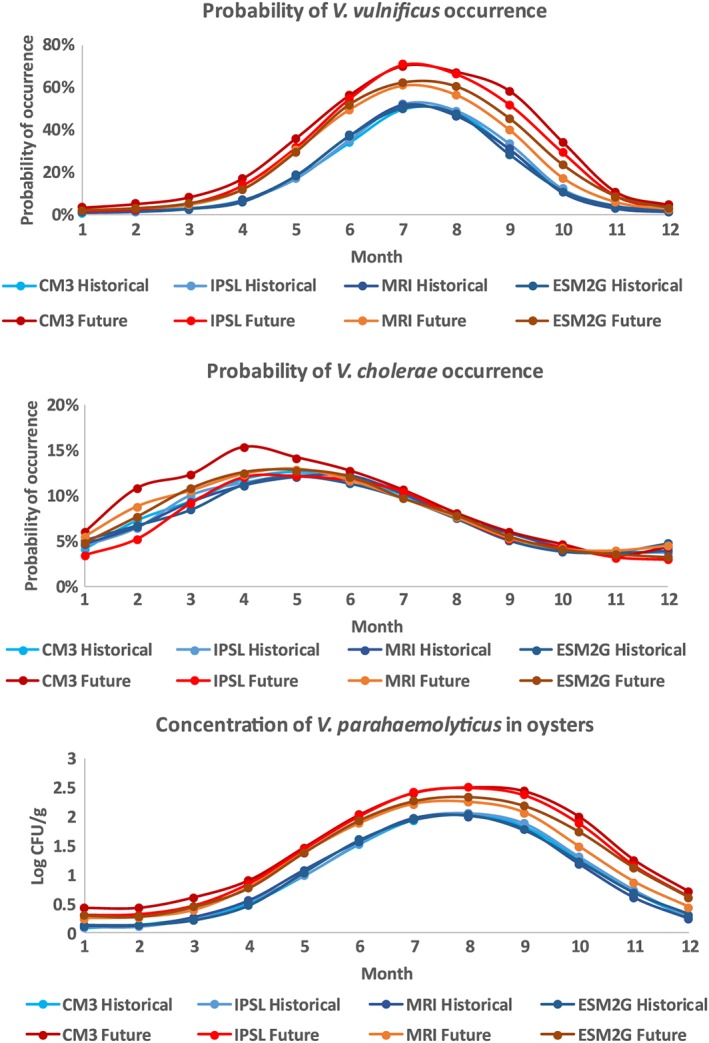
Mean monthly probability of occurrence for (top) V. vulnificus and (middle) V. cholerae, and predicted mean (bottom) V. parahaemolyticus in oysters, between a historical (1970–1999) and future (2071–2100) time period, across all regions of the Chesapeake Bay. Projections were calculated using the GLMs in Table 1, using four statistically downscaled GCMs.
To show the range of possible future changes in spatial habitat, we selected the two most divergent GCMs for each Vibrio species from Figure 5 and focus on seasons of peak occurrence. For V. vulnificus these were the CM3 (most warming) and MRI (least warming) models (Figure 6). Projections suggested an overall increase in probability of occurrence across the Chesapeake Bay in summer by the end of the century. This increase was much stronger in the CM3 model, particularly in portions of the Upper Bay and parts of the Mid Bay, where mean probabilities of occurrence increased by more than 20% over late twentieth century levels. Projected increases were about half this size for the MRI model but similar in spatial character.
Figure 6.
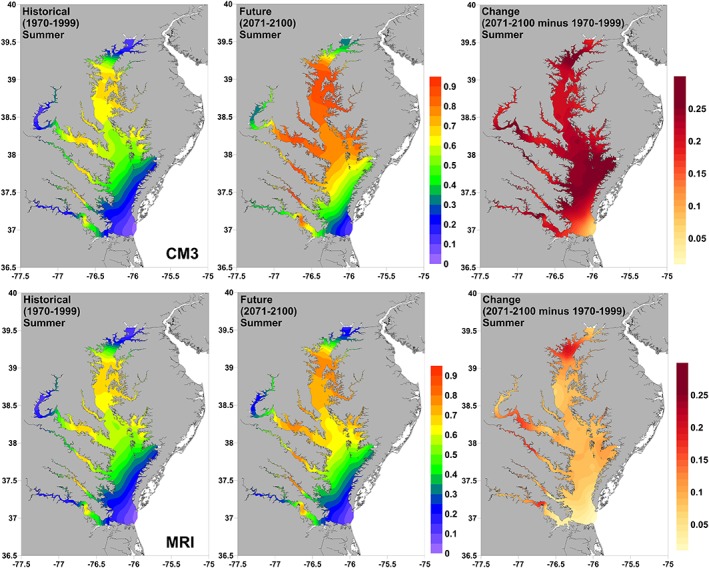
Projections of historical (1970–1999) and future (2071–2100) probability of summer (July–September) V. vulnificus occurrence in the Chesapeake Bay from two statistically downscaled GCMs (CM3 and MRI). The change between the two time periods is also shown.
In contrast, projections for the probability of occurrence of V. cholerae showed a very similar spatial distribution between the historical and future spring‐summer time periods (Figure 7).
Figure 7.
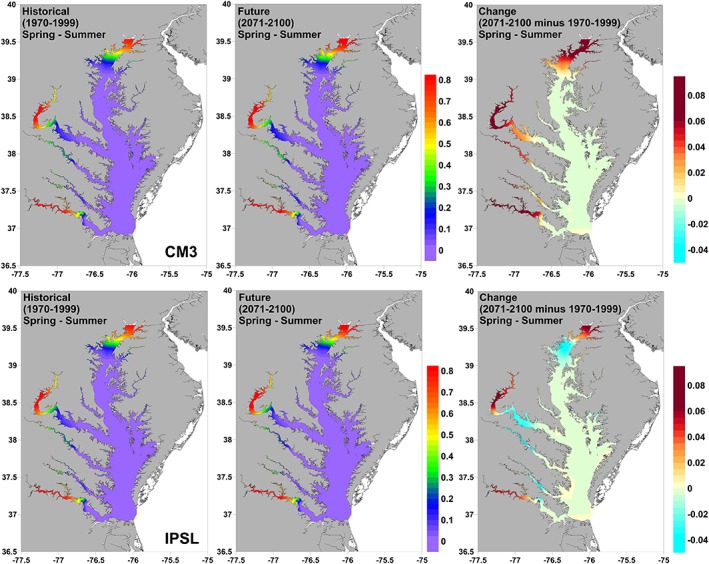
Projections of historical (1970–1999) and future (2071–2100) probability of spring‐summer (March–September) V. cholerae occurrence in the Chesapeake Bay from two statistically downscaled GCMs (CM3 and IPSL). The change between the two time periods is also shown.
While probabilities of occurrence increased within high‐risk areas, the minimal changes to summer salinity fields projected by the GCMs kept these areas restricted to the northern Upper Bay and upper reaches of the James and Potomac Rivers, where salinities remained low. Projections from the drier IPSL model also suggested an upstream contraction of high‐risk areas due to increasing salinities in currently mesohaline to oligohaline regions of the bay. This pattern was not evident in the wetter CM3 model.
The mean modeled concentration of V. parahaemolyticus CFU/(log) in oysters during summer was projected to increase between the historical and future time periods, particularly in the warmer CM3 model. In contrast to V. cholerae, high‐risk areas for V. parahaemolyticus spread across most of the Chesapeake Bay, with the exception of low salinity areas in the Upper Bay and upper reaches of rivers (Figure 8). Increases in future high‐risk areas were more spatially uniform than for V. cholerae but were slightly higher in the upper bay and the midreaches of the major rivers.
Figure 8.
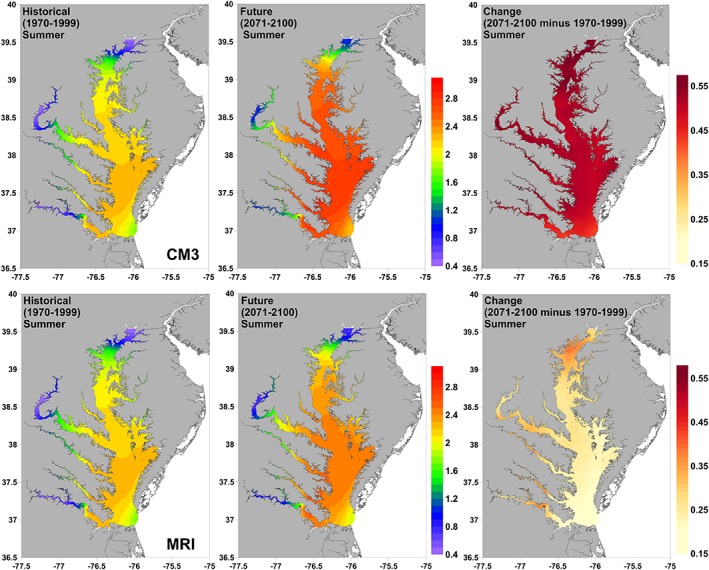
Projections of historical (1970–1999) and future (2071–2100) mean concentration of summer (July–September) V. parahaemolyticus in oysters in the Chesapeake Bay from two statistically downscaled GCMs (CM3 and MRI). The change between the two time periods is also shown.
The projected increases in probability of occurrence of V. vulnificus varied somewhat among different zones of the Chesapeake Bay (Figure 9). The Mid Bay was projected to show the greatest increase: from a mean probability of occurrence of 47.3% in 1970–1999 to a mean of 71.7% by 2071–2100 during the peak summer season. Increases in the Lower Bay and Patuxent River were weaker. The Lower Bay remained the lowest risk zone, with probability of occurrence of V. vulnificus increasing from 21.2% to 41.0% by the end of the 21st century. Conversely, the Patuxent River remained the highest risk zone: increasing from 64.6% to 84.2%.
Figure 9.
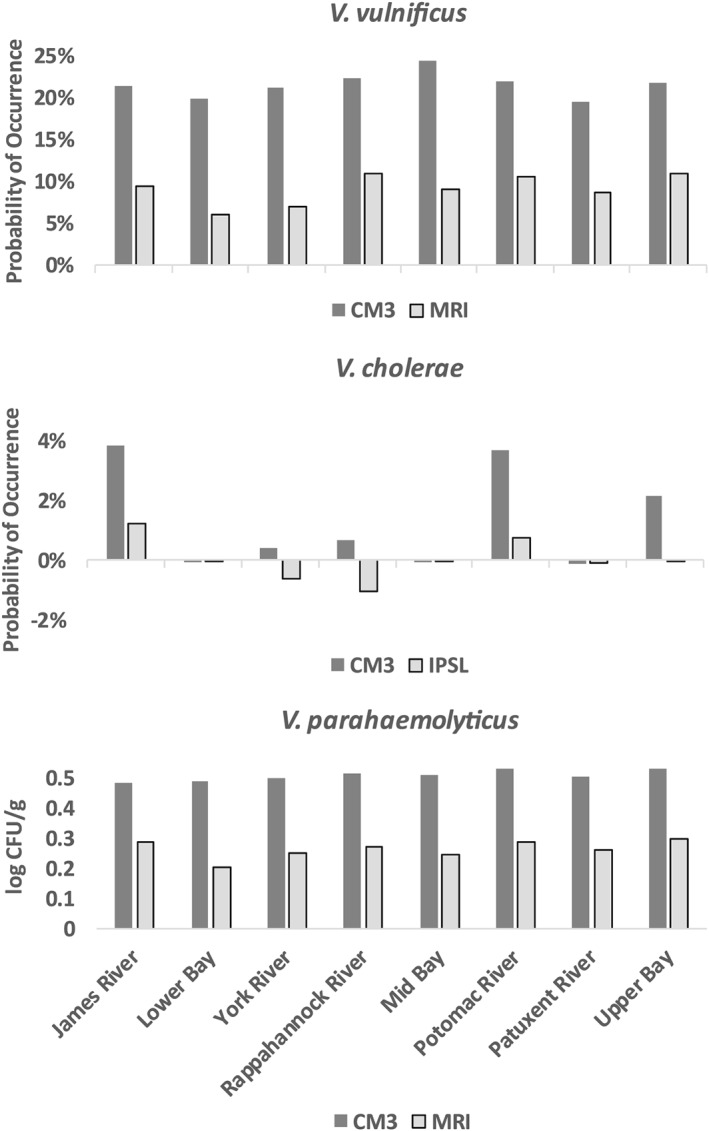
Change in mean probability of occurrence (V. vulnificus and V. cholerae) and concentration of V. parahaemolyticus in oysters (log), between historical (1970–1999) and future (2071–2100) time periods in each zone of the Chesapeake Bay. Only projections for peak seasons (summer for V. vulnificus and V. parahaemolyticus, spring‐summer for V. cholerae) are shown.
Projected increases in V. parahaemolyticus were the most spatially uniform of the three species, with little difference among zones (Figure 9). The Lower Bay showed slightly smaller increases due to the moderating influence of the continental shelf environment on water temperatures in this area, but all other zones were similar to each other. As with V. vulnificus, the variability in projections contributed by the warmer CM3 model versus the cooler MRI model was clearly evident.
In contrast to the more uniform increases projected for the other two species, projected changes in probability of occurrence of V. cholerae were much more spatially complex (Figure 9). Zones with historically low probabilities of occurrence largely remained so during the spring‐summer peak season, due to the restriction of this species to low salinity regions. In zones with historically higher concentrations of V. cholerae, probabilities of occurrence were projected to increase in the wetter CM3 model but to decrease in the drier IPSL model.
4. Discussion
4.1. Climate Change and Vibrio Risk
Several previous studies have hypothesized direct and indirect effects of climate on Vibrio presence and disease risk. The most studied species is undoubtedly V. cholerae. Strains O1 and O139 can cause epidemic disease outbreaks, and studies from different regions around the world have correlated these to air and water temperature, rainfall, river discharge, and phytoplankton/zooplankton abundance and composition [Colwell, 1996; Lobitz et al., 2000; Rodó et al., 2002; Gil et al., 2004; Koelle et al., 2005; Greer et al., 2008; Constantin de Magny et al., 2012]. Some analyses have suggested that climate change‐induced warming and eutrophication may exacerbate cholera outbreaks, with changing river flow patterns and flood events also potentially important [Martinez‐Urtaza et al., 2010; Tirado et al., 2010].
Recent work has highlighted increasing trends of Vibrio species occurrence and related infections in many areas around the world [e.g., Pascual et al., 2000; Martinez‐Urtaza et al., 2010; Tirado et al., 2010; Vezzulli et al., 2010a, 2016; Newton et al., 2012; Baker‐Austin et al., 2013]. There have also been cases of largely unprecedented occurrence of Vibrio infections in some parts of Europe, the Atlantic coast of the United States, and Alaska in recent years, associated with positive temperature anomalies and heat waves [McLaughlin et al., 2005; Paz et al., 2007; Lima and Wethey, 2012; Baker‐Austin et al., 2013; Vezzulli et al., 2016]. Several studies have shown convincing links between recent warming temperatures and associated outbreaks of Vibrio illness, including in some areas where it was previously rare or unknown [Baker‐Austin et al., 2017]. This suggests great potential for future climate change to increase habitat availability, season length, and infection rates from Vibrios. There may also be increases in associated economic costs from health care and lost wages, and increases in regulatory costs for some sectors of the seafood industry.
However, there are few studies looking at risk of future Vibrio species occurrence or infection using projections from IPCC climate models. This may be partially due to the spatial resolution required. Some brackish waters with high Vibrio risk are poorly resolved at the native resolution of most GCMs (~1–2°). The difficulties inherent in projecting future salinity fields in estuarine and nearshore environments may also have been limited. For example, Jacobs et al. [2015] projected a temperature‐driven increase in season length for V. vulnificus in the Chesapeake Bay over the next century, consistent with the present study. However, these authors used a fixed value for salinity, because no estuary‐scale projections were available. We addressed the two challenges raised above (spatial resolution and lack of salinity projections) by using a spatial modeling framework, which ingests statistically downscaled projections of air temperature and precipitation [Muhling et al., 2017]. Present‐day predictions of Vibrio probability of occurrence or concentration using modeled temperature and salinity from the statistical framework were close to those using in situ CTD casts. As a result, we can have some confidence that the framework approach is reasonable for use in developing future spatial projections of Vibrios in the Chesapeake Bay.
4.2. Future Projections
Results from future projections of Vibrio habitat in the Chesapeake Bay highlighted considerable interspecific variability. Probability of V. vulnificus occurrence increased markedly across the Bay during the peak summer season, and the overall area of high probability expanded. Increases were stronger in the warmer CM3 model than in the less warm MRI model, but the direction of change was consistent.
Similarly, the mean predicted concentration of V. parahaemolyticus in oysters increased throughout much of the Bay. Accounting for the log scale of predictions, projections from the MRI model showed V. parahaemolyticus concentrations increasing by 1.5 times, while the CM3 model suggested that concentrations would more than triple. This species had the broadest modeled environmental tolerances of all three Vibrios examined, with positive concentrations predicted at salinities from 0 to 31 psu and temperatures of greater than ~10°C, although concentrations were highest at warm temperatures (> 25°C) and moderate salinities (5–25 psu). The environmental range of V. parahaemolyticus is thus approximately the same as for the oysters it is associated with [Galtsoff, 1964; Mann and Powell, 2007].
In contrast to the other two species, the observed salinity association of V. cholerae restricted their distribution to the upper portions of the Chesapeake Bay and major rivers, with little expansion of habitat under climate change. Mean bay‐wide projected probabilities of occurrence during late winter and spring increased in the wetter CM3 model, increased slightly in the MRI and ESM2G models, and decreased slightly in the drier IPSL model. Projections for this species also showed the greatest spatial complexity. While projected probabilities of occurrence increased most strongly in the James and Potomac Rivers, they remained low in most other areas and even decreased somewhat in the Rappahannock and York Rivers in the drier IPSL model.
Projections of V. vulnificus differed somewhat among the different zones of the Chesapeake Bay, although to a lesser extent than V. cholerae. The strongest increase was in the Mid Bay, suggesting a down‐bay extension of current high‐risk areas. Projected increases in V. parahaemolyticus were the most spatially uniform of the three species. While the nature of the temperature relationships in the GLMs thus determined the general scale of future increase for all three Vibrio species, the relationships with salinity determined the spatial patterns of this increase. This result highlights the importance of salinity fields for sensitive species, if projections of future high‐risk hot spots are a priority.
4.3. Using Different Biological Habitat Models
While the use of correlative habitat models in climate change impact studies is common, the effect of model choice and parameterization on results is not often considered. However, recent work suggests that the choice of biological model can be influential [Jones et al., 2012; Payne et al., 2015]. Even where habitat models give very similar results on present‐day data, they can diverge substantially once extrapolated beyond the range of the training data. This is commonly required when projecting temperature‐based habitat models under future climate change scenarios.
The present study relied on previously existing GLMs to predict Vibrio occurrence or abundance. However, Urquhart et al. [2014] noted that a GLM for V. vulnificus trained on a subset of the data from Jacobs et al. [2010] (an earlier version of the GLM described in Jacobs et al. [2014]) gave substantially different results to a GLM trained on samples collected in the upper Chesapeake Bay over a different time period (2011–2012). In addition, a Generalized Additive Model (GAM) trained on the latter data set gave different results to both GLMs, primarily due to the fact that GAMs allow nonlinear response curves [Züur et al., 2009]. Urquhart et al. [2014] thus concluded that the impacts of climate change on V. vulnificus in the Chesapeake Bay could not be projected with any useful certainty.
As the findings of Urquhart et al. [2014] are strongly relevant to the current study, we reevaluated the robustness of our results using the full 2007–2010 data set for V. vulnificus from Jacobs et al. [2014]. The GLM in Table 1 was compared to two GAMs with logit link functions built using the mgcv package in R 3.2.1 [Wood, 2006; R Core Team, 2015]. As the number of “knots” for polynomial smoothers affects the shapes of response curves in GAMs [Keele, 2008; Züur et al., 2009] we built two models, with the maximum number of knots for both temperature and salinity set at three (GAM3) and then at five (GAM5). These values were within the range of sensible values recommended by Keele [2008], and both generated biologically plausible but slightly different response curves for both temperature and salinity.
In contrast to Urquhart et al. [2014], the three V. vulnificus habitat models (GLM, GAM3, and GAM5) gave similar seasonal predictions for the late twentieth century (Figure 10). This was likely due to the broader spatiotemporal extent of the field data available to us, compared to Urquhart et al. [2014] which emphasizes the advantages of using comprehensive data sets for habitat model training. The slightly different shapes of the modeled temperature response curves did result in diverging projections of future probability of occurrence by the late 21st century. Importantly, however, all showed a temperature‐driven increase. Laboratory experiments suggest optimum temperatures for Vibrio species of 37–39°C [Kelly, 1982; Miles et al., 1997; Sedas, 2007]: much warmer than currently observed conditions in the Chesapeake Bay. In addition, recent warming conditions have resulted in increasing rates of Vibrio‐associated illness in other parts of the world [Andersson and Ekdahl, 2006; Paz et al., 2007; Martinez‐Urtaza et al., 2010; Vezzulli et al., 2010a, 2016; Baker‐Austin et al., 2013; Le Roux et al., 2015]. If other environmental conditions remain favorable, it therefore seems reasonable to assume that warming conditions will favor Vibrios and that generally positive temperature response curves are appropriate.
Figure 10.
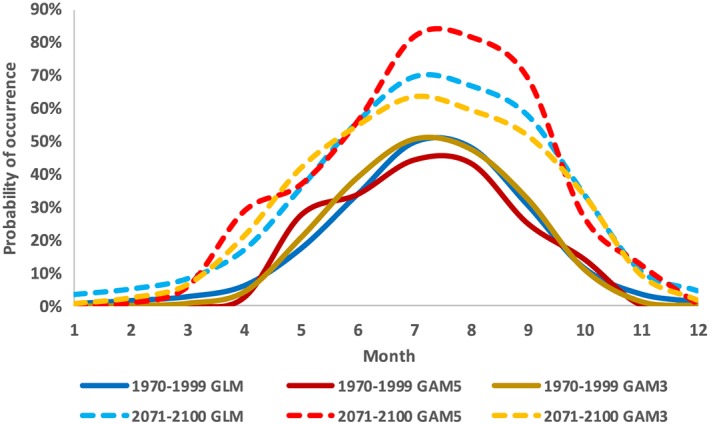
Monthly modeled probability of occurrence for V. vulnificus between a historical (1970–1999) and a future (2071–2100) time period. Projections are compared between a published GLM [Jacobs et al., 2014] (Table 1) and two GAMs and were calculated using statistically downscaled outputs from the CM3 GCM.
4.4. Management Implications
A primary management issue associated with Vibrio‐related illnesses is the economic costs of health care to affected patients. Both V. vulnificus and V. parahaemolyticus infections currently lead to millions of dollars in associated treatment costs in the United States: the former because of its severity and the latter because of its high frequency [Ralston et al., 2011; Scallan et al., 2011; Hoffmann et al., 2015]. Results from this study suggest substantially increased risk of V. vulnificus in Chesapeake Bay waters under climate change and increasing mean concentration of V. parahaemolyticus in oysters. Although the economic burden of these trends is difficult to estimate without information on virulence, future population density and recreational use of the region, and future oyster harvest rates, the potential for increasing rates of illness is a serious prospect.
In addition, best practices for the oyster harvesting industry may no longer be effective as water temperatures continue to warm. At present, Maryland and Virginia Vibrio control plans require that fishers deliver harvested oysters to dealers by certain times of day and refrigerate their catch within a certain number of hours, depending on the month [e.g., Virginia Department of Health, 2016]. These are earlier/shorter in warmer months, and later/longer in cooler months, respectively. If climate change‐induced warming leads to higher concentrations of V. parahaemolyticus in oysters, then current restrictions will likely have to be adjusted, to avoid unacceptably contaminated product and increased rates of illness.
4.5. Uncertainties and Future Work
In addition to the sources of uncertainty mentioned above, a major missing piece of the current projections is the lack of biological variables. Phytoplankton and zooplankton have been shown to be important to Vibrio ecology (especially V. cholerae), and chitinous zooplankton in particular may act as a reservoir for some species [Kaneko and Colwell, 1973, 1975; Huq et al., 1983; Rawlings et al., 2007; Gil et al., 2004; Vezzulli et al., 2010b; Turner et al., 2014; Constantin de Magny et al., 2011; Main et al., 2015].
Another consideration, which is common to many climate change impact studies, is that the environmental relationships shown by the habitat models may not represent the full extent of physiological limits. This is particularly evident for V. cholerae. While the habitat model developed by Louis et al. [2003] shows a strong association between V. cholerae and low salinity waters, this species is known to tolerate much broader salinities in other regions and in the laboratory [Singleton et al., 1982a, 1982b; Constantin De Magny et al., 2009; Banakar et al., 2011]. Louis et al. [2003] noted that other field and laboratory studies had found varying optimum salinity values for V. cholerae, and that their modeled salinity relationships for the Chesapeake Bay may have been proxies for other biologically important factors, such as turbidity or nutrient loads. While phytoplankton and zooplankton substrates are known to be important for V. cholerae, Louis et al. [2003] found no strong relationships between the occurrence of V. cholerae in the Chesapeake Bay and concentrations of chlorophyll α, or of specific zooplankton groups (e.g., copepods, rotifers, and barnacle nauplii). As a result, although the strongest predictors for V. cholerae in the Chesapeake Bay are currently temperature and salinity, the precise mechanisms controlling V. cholerae in the Chesapeake Bay remain somewhat unclear.
It is also important to note that environmental concentrations of Vibrio species do not necessarily predict disease risk, although the two are often correlated [Baker‐Austin et al., 2013; Vezzulli et al., 2016]. Some studies have recorded an increase in Vibrio infections in the past few decades [Scallan et al., 2011; Newton et al., 2012]. However, it is not yet clear if these are due to changing environmental conditions. Biogeochemical factors (e.g., nutrient concentrations, phytoplankton or zooplankton abundance, or community structure) and the presence of particular strains or virulence‐correlated genes may be more important for interannual disease risk than simple probability of occurrence. Jacobs et al. [2014] found that occurrence of a virulence‐correlated gene (Vcg) in V. vulnificus in the Chesapeake Bay was associated not only with temperature and salinity but also with nutrient and chlorophyll concentrations, which were not modeled in our study.
Projections of V. parahaemolyticus in oysters assume that suitable habitats of the Chesapeake Bay will continue to support oyster production. Although C. virginica has relatively broad environmental tolerances, populations have been strongly and negatively affected by habitat loss, eutrophication, and disease in recent decades [Rothschild et al., 1994]. It is therefore difficult to project the extent of oyster habitat, and the associated state of the oyster fishery, at the middle to end of the century with any confidence.
The use of the statistical framework to derive temperature and salinity in the Chesapeake Bay introduces additional uncertainty to the projections. These are discussed in more depth in Muhling et al. [2017], but relate primarily to the assumption of stationarity in statistically downscaled projections [e.g., Vrac et al., 2007; Gaitan and Cannon, 2013; Gaitan et al., 2014; Gaitan, 2016; Dixon et al., 2016], the simplicity of the water balance model and error from the model trees used to create the spatial fields.
Another source of uncertainty which requires further study is the interaction between future demographics and disease risk. As the United States population continues to increase and age [Ortman and Guarneri, 2009], more people may be exposed to Vibrio pathogens. An aging population, particularly with increasing prevalence of existing health conditions, may increase susceptibility, health care costs, and risk of death [Ralston et al., 2011; Weis et al., 2011].
While our projections were specific to the Chesapeake Bay, this approach may be useful in other regions of the world. Other parts of the northeast United States and northwest Europe, in particular, have been warming rapidly over the past several decades [Lima and Wethey, 2012]. Similar methods to those used in this study could be applied to assess future Vibrio prevalence in higher‐risk areas or to assess the probability of emergence of new areas of disease risk.
5. Conclusions
Overall, we found that future climate change is likely to increase the probability of occurrence of V. vulnificus in the Chesapeake Bay and increase the mean concentration of V. parahaemolyticus in oysters by the end of the 21st century. In contrast, probabilities of occurrence for V. cholerae were projected to increase only in the wetter GCMs, and high‐risk areas remained restricted to low salinity zones of the bay. The length of the high‐risk summer season for V. vulnificus and V. parahaemolyticus was also projected to increase. These findings have implications for recreational use and seafood extraction from the Chesapeake Bay, with the potential for considerable economic costs as a result.
Conflict of Interest
The authors declare no conflicts of interest relevant to this study.
Supporting information
Supporting Information S1
Acknowledgments
The manuscript was significantly improved by comments from J. Dunne and A. Leight. Assistance with the FDA model for V. parahaemolyticus was provided by J. Bowers. Primary funding and support for this study were provided by the NOAA National Ocean Service (NOS) National Centers for Coastal Ocean Science (NCCOS), with additional support from the NOAA National Marine Fisheries Service (NMFS) Office of Science and Technology, the NOAA Integrated Ecosystem Assessment (IEA) Program, and the NOAA Office of Oceanic and Atmospheric Research (OAR). Data supporting the conclusions in this study can be found in Table 1 and in the associated published manuscripts cited in the text. Climate change projections can be obtained by contacting the first author.
Muhling, B. A. , Jacobs J., Stock C. A., Gaitan C. F., and Saba V. S. (2017), Projections of the future occurrence, distribution, and seasonality of three Vibrio species in the Chesapeake Bay under a high‐emission climate change scenario, GeoHealth, 1, 278–296, doi: 10.1002/2017GH000089.
This article was corrected on 15 JUL 2019. The online version of this article has been modified to include a Conflict of Interest statement.
References
- Altekruse, S. F. , Bishop R. D., Baldy L. M., Thompson S. G., Wilson S. A., Ray B. J., and Griffin P. M. (2000), Vibrio gastroenteritis in the US Gulf of Mexico region: The role of raw oysters, Epidemiol. Infect., 124, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, Y. , and Ekdahl K. (2006), Wound infections due to Vibrio cholerae in Sweden after swimming in the Baltic Sea, summer 2006, Eurosurveillance, 11, E060803. [DOI] [PubMed] [Google Scholar]
- Baker‐Austin, C. , Stockley L., Rangdale R., and Martinez‐Urtaza J. (2010), Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective, Environ. Microbiol. Rep., 2, 7–18. [DOI] [PubMed] [Google Scholar]
- Baker‐Austin, C. , Trinanes J. A., Taylor N. G., Hartnell R., Siitonen A., and Martinez‐Urtaza J. (2013), Emerging Vibrio risk at high latitudes in response to ocean warming, Nat. Clim. Change, 3, 73–77. [Google Scholar]
- Baker‐Austin, C. , Trinanes J., Gonzalez‐Escalona N., and Martinez‐Urtaza J. (2017), Non‐cholera Vibrios: The microbial barometer of climate change, Trends Microbiol., 25, 76–84. [DOI] [PubMed] [Google Scholar]
- Banakar, V. , Constantin De Magny G., Jacobs J., Murtugudde R., Huq A., Wood R. J., and Colwell R. R. (2011), Temporal and spatial variability in the distribution of Vibrio vulnificus in the Chesapeake Bay: A hindcast study, EcoHealth, 8, 456–467. [DOI] [PubMed] [Google Scholar]
- Chávez, M. D. R. C. , Sedas V. P., Borunda E. O., and Reynoso F. L. (2005), Influence of water temperature and salinity on seasonal occurrences of Vibrio cholerae and enteric bacteria in oyster‐producing areas of Veracruz, México, Mar. Pollut. Bull., 50, 1641–1648. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Shi W., Xie P., Silva V., Kousky V. E., Higgins R. W., and Janowiak J. E. (2008), Assessing objective techniques for gauge‐based analyses of global daily precipitation, J. Geophys. Res., 113, D04110, doi: 10.1029/2007JD009132. [DOI] [Google Scholar]
- Colwell, R. R. (1996), Global climate and infectious disease: The cholera paradigm, Science, 274, 2025–2031. [DOI] [PubMed] [Google Scholar]
- Colwell, R. R. , Kaper J., and Joseph S. W. (1977), Vibrio cholerae, Vibrio parahaemolyticus, and other Vibrios: Occurrence and distribution in Chesapeake Bay, Science, 198, 394–396. [PubMed] [Google Scholar]
- Constantin de Magny, G. , Long W., Brown C. W., Hood R. R., Huq A., Murtugudde R., and Colwell R. R. (2009), Predicting the distribution of Vibrio spp. in the Chesapeake Bay: A Vibrio cholerae case study, EcoHealth, 6, 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin de Magny, G. , Mozumder P. K., Grim C. J., Hasan N. A., Naser M. N., Alam M., Sack R. B., Huq A., and Colwell R. R. (2011), Role of zooplankton diversity in Vibrio cholerae population dynamics and in the incidence of cholera in the Bangladesh Sundarbans, Appl. Environ. Microbiol., 77, 6125–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin de Magny, G. , Thiaw W., Kumar V., Manga N. M., Diop B. M., Gueye L., Kamara M., Roche B., Murtugudde R., and Colwell R. R. (2012), Cholera outbreak in Senegal in 2005: Was climate a factor?, PLoS One, 7, e44577, doi: 10.1371/journal.pone.0044577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, K. W. , Lanzante J. R., Nath M. J., Hayhoe K., Stoner A., Radhakrishnan A., Balaji V., and Gaitán C. F. (2016), Evaluating the stationarity assumption in statistically downscaled climate projections: Is past performance an indicator of future results?, Clim. Change, 135, 395–408. [Google Scholar]
- Donner, L. J. , et al. (2011), The dynamical core, physical parameterizations, and basic simulation characteristics of the atmospheric component AM3 of the GFDL global coupled model CM3, J. Clim., 24, 3484–3519. [Google Scholar]
- Dufresne, J.‐L. , et al. (2013), Climate change projections using the IPSL‐CM5 Earth System Model: From CMIP3 to CMIP5, Clim. Dyn., 40, 2123–2165. [Google Scholar]
- Dunne, J. P. , et al. (2012), GFDL's ESM2 global coupled climate‐carbon Earth system models. Part I: Physical formulation and baseline simulation characteristics, J. Clim., 2, 6646–6665. [Google Scholar]
- Fan, Y. , and van den Dool H. (2008), A global monthly land surface air temperature analysis for 1948‐present, J. Geophys. Res., 113, D01103, doi: 10.1029/2007JD008470. [DOI] [Google Scholar]
- Farmer, J. J., III , Janda J. M., Brenner F. W., Cameron D. N., and Birkhead K. M. (2005), Genus I. Vibrio Pacini 1854, 411AL, Bergey's Manual Syst. Bacteriol., 2B, 494–546. [Google Scholar]
- Gaitan, C. F. (2016), Effects of variance adjustment techniques and time‐invariant transfer functions on heat wave duration indices and other metrics derived from downscaled time‐series. Study case: Montreal, Canada, Nat. Hazards, 83, 1661–1681. [Google Scholar]
- Gaitan, C. F. , and Cannon A. J. (2013), Validation of historical and future statistically downscaled pseudo‐observed surface wind speeds in terms of annual climate indices and daily variability, Renew. Energy, 51, 489–496. [Google Scholar]
- Gaitan, C. F. , Hsieh W. W., and Cannon A. J. (2014), Comparison of statistically downscaled precipitation in terms of future climate indices and daily variability for southern Ontario and Quebec, Canada, Clim. Dyn., 43, 3201–3217. [Google Scholar]
- Galtsoff, P. S. (1964), The American oyster Crassostrea virginica Gmelin, Fish. Bull., 64, 1–480. [Google Scholar]
- Gil, A. I. , et al. (2004), Occurrence and distribution of Vibrio cholerae in the coastal environment of Peru, Environ. Microbiol., 6, 699–706. [DOI] [PubMed] [Google Scholar]
- Greer, A. , Ng V., and Fisman D. (2008), Climate change and infectious diseases in North America: The road ahead, Can. Med. Assoc. J., 178, 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley, B. J. , et al. (2014), Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea, Front. Microbiol., 5, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, S. , Maculloch B., and Batz M. (2015), Economic burden of major foodborne illnesses acquired in the United States, United States Department of Agriculture Economic Research Service Economic Information Bulletin, 140, May 2015, 59p.
- Huq, A. , Small E. B., West P. A., Huq M. I., Rahman R., and Colwell R. R. (1983), Ecological relationships between Vibrio cholerae and planktonic crustacean copepods, Appl. Environ. Microbiol., 45, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, A. , West P. A., Small E. B., Huq M. I., and Colwell R. R. (1984), Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar 01 associated with live copepods in laboratory microcosms, Appl. Environ. Microbiol., 48, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J. M. , Rhodes M., Brown C. W., Hood R. R., Leight A., Long W., and Wood R. (2010), Predicting the distribution of Vibrio vulnificus in Chesapeake Bay, NOAA Technical Memorandum NOA NCCOS 112, 24p. [DOI] [PubMed]
- Jacobs, J. M. , Rhodes M., Brown C. W., Hood R. R., Leight A., Long W., and Wood R. (2014), Modeling and forecasting the distribution of Vibrio vulnificus in Chesapeake Bay, J. Appl. Microbiol., 117, 1312–1327. [DOI] [PubMed] [Google Scholar]
- Jacobs, J. , Moore S. K., Kunkel K. E., and Sun L. (2015), A framework for examining climate‐driven changes to the seasonality and geographical range of coastal pathogens and harmful algae, Clim. Risk Manag., 8, 16–27. [Google Scholar]
- Jiang, S. C. , and Fu W. (2001), Seasonal abundance and distribution of Vibrio cholerae in coastal waters quantified by a 16S‐23S intergenic spacer probe, Microb. Ecol., 42, 540–548. [DOI] [PubMed] [Google Scholar]
- Jones, E. H. , Feldman K. A., Palmer A., Butler E., Blythe D., and Mitchell C. S. (2013), Vibrio infections and surveillance in Maryland, 2002–2008, Public Health Rep., 128, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. C. , Dye S. R., Pinnegar J. K., Warren R., and Cheung W. W. (2012), Modelling commercial fish distributions: Prediction and assessment using different approaches, Ecol. Model., 225, 133–145. [Google Scholar]
- Kaneko, T. , and Colwell R. R. (1973), Ecology of Vibrio parahaemolyticus in Chesapeake Bay, J. Bacteriol., 113, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, T. , and Colwell R. R. (1975), Adsorption of Vibrio parahaemolyticus onto chitin and copepods, J. Appl. Microbiol., 29, 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar, C. W. , and Tamplin M. L. (1993), Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish, Appl. Environ. Microbiol., 59, 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele, L. J. (2008), Semiparametric Regression for the Social Sciences, John Wiley, Chichester, U. K. [Google Scholar]
- Kelly, M. T. (1982), Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment, Appl. Environ. Microbiol., 44, 820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle, K. , Rodó X., Pascual M., Yunus M., and Mostafa G. (2005), Refractory periods and climate forcing in cholera dynamics, Nature, 436, 696–700. [DOI] [PubMed] [Google Scholar]
- Kuhn, M. S. , Weston S., Keefer C., and Coulter N (2015), Cubist: Rule‐ and instance‐based regression modeling, R package version 0.0.18. [Available at http://CRAN.R‐project.org/package=Cubist.]
- Le Roux, F. L. , et al. (2015), The emergence of Vibrio pathogens in Europe: Ecology, evolution, and pathogenesis (Paris, 11–12th March 2015), Front. Microbiol., 6, 830, doi: 10.3389/fmicb.2015.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher, B. H. , Hocking D. J., O'Neil K., Whiteley A. R., Nislow K. H., and O'Donnell M. J. (2016), A hierarchical model of daily stream temperature using air‐water temperature synchronization, autocorrelation, and time lags, Peer J, 4, e1727, doi: 10.7717/peerj.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, F. P. , and Wethey D. S. (2012), Three decades of high‐resolution coastal sea surface temperatures reveal more than warming, Nat. Commun., 3, 704. [DOI] [PubMed] [Google Scholar]
- Lipp, E. K. , Rodriguez‐Palacios C., and Rose J. B. (2001), Occurrence and distribution of the human pathogen Vibrio vulnificus in a subtropical Gulf of Mexico estuary, in The Ecology and Etiology of Newly Emerging Marine Diseases, edited by Porter J. W., pp. 165–173, Springer, Dordrecht, Netherlands. [Google Scholar]
- Lipp, E. K. , Huq A., and Colwell R. R. (2002), Effects of global climate on infectious disease: The cholera model, Clin. Microbiol. Rev., 15, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobitz, B. , Beck L., Huq A., Wood B., Fuchs G., Faruque A. S. G., and Colwell R. (2000), Climate and infectious disease: Use of remote sensing for detection of Vibrio cholerae by indirect measurement, Proc. Natl. Acad. Sci. U.S.A., 97, 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, V. R. , Russek‐Cohen E., Choopun N., Rivera I. N., Gangle B., Jiang S. C., Rubin A., Patz J. A., Huq A., and Colwell R. R. (2003), Predictability of Vibrio cholerae in Chesapeake Bay, Appl. Environ. Microbiol., 69, 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main, C. R. , Salvitti L. R., Whereat E. B., and Coyne K. J. (2015), Community‐level and species‐specific associations between phytoplankton and particle‐associated Vibrio species in Delaware's inland bays, Appl. Environ. Microbiol., 81, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, R. , and Powell E. N. (2007), Why oyster restoration goals in the Chesapeake Bay are not and probably cannot be achieved, J. Shellfish Res., 26, 905–917. [Google Scholar]
- Martinez‐Urtaza, J. , Bowers J. C., Trinanes J., and DePaola A. (2010), Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illness, Food Res. Int., 43, 1780–1790. [Google Scholar]
- Martinez‐Urtaza, J. , Baker‐Austin C., Jones J. L., Newton A. E., Gonzalez‐Aviles G. D., and DePaola A. (2013), Spread of Pacific Northwest Vibrio parahaemolyticus strain, N. Engl. J. Med., 369, 1573–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe, G. J. , and Markstrom S. I. (2007), A monthly water‐balance model driven by a graphical user interface, U.S. Geol. Surv. Open File Rep., 2007‐1088, p. 6.
- McLaughlin, J. B. , DePaola A., Bopp C. A., Martinek K. A., Napolilli N. P., Allison C. G., Murray S. L., Thompson E. C., Bird M. M., and Middaugh J. P. (2005), Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters, N. Engl. J. Med., 353, 1463–1470. [DOI] [PubMed] [Google Scholar]
- Miles, D. W. , Ross T., Olley J., and McMeekin T. A. (1997), Development and evaluation of a predictive model for the effect of temperature and water activity on the growth rate of Vibrio parahaemolyticus , Int. J. Food Microbiol., 38, 133–142. [DOI] [PubMed] [Google Scholar]
- Muhling, B. A. , Gaitan C. F., Stock C. A., Saba V. S., Tommasi D., and Dixon K. W. (2017), Potential salinity and temperature futures for the Chesapeake Bay using a statistical downscaling spatial disaggregation framework, Estuar. Coasts, doi: 10.1007/s12237-017-0280-8. [DOI] [Google Scholar]
- Najjar, R. G. , et al. (2010), Potential climate‐change impacts on the Chesapeake Bay, Estuar. Coast. Shelf Sci., 86, 1–20. [Google Scholar]
- Newton, A. , Kendall M., Vugia D. J., Henao O. L., and Mahon B. E. (2012), Increasing rates of vibriosis in the United States, 1996‐2010: Review of surveillance data from 2 systems, Clin. Infect. Dis., 54, S391–S395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, K. R. , Jones S. H., and Grimes D. J. (1992), Seasonal incidence of Vibrio vulnificus in the Great Bay estuary of New Hampshire and Maine, Appl. Environ. Microbiol., 58, 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortman, J. M. , and Guarneri C. E. (2009), United States population projections: 2000 to 2050, U. S. Census Bur., 1–19. [Available at http://www.census.gov/population/www/projections/analytical-document09.pdf.] [Google Scholar]
- Parveen, S. , Hettiarachchi K. A., Bowers J. C., Jones J. L., Tamplin M. L., McKay R., Beatty W., Brohawn K., DaSilva L. V., and DePaola A. (2008), Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters, Int. J. Food Microbiol., 128, 354–361. [DOI] [PubMed] [Google Scholar]
- Pascual, M. , Rodó X., Ellner S. P., Colwell R., and Bouma M. J. (2000), Cholera dynamics and El Niño‐southern oscillation, Science, 289, 1766–1769. [DOI] [PubMed] [Google Scholar]
- Payne, M. R. , et al. (2015), Uncertainties in projecting climate‐change impacts in marine ecosystems, ICES J. Mar. Sci., 73, 1272–1282. [Google Scholar]
- Paz, S. , Bisharat N., Paz E., Kidar O., and Cohen D. (2007), Climate change and the emergence of Vibrio vulnificus disease in Israel, Environ. Res., 103, 390–396. [DOI] [PubMed] [Google Scholar]
- Pfeffer, C. S. , Hite M. F., and Oliver J. D. (2003), Ecology of Vibrio vulnificus in estuarine waters of eastern North Carolina, Appl. Environ. Microbiol., 69, 3526–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, J. R. (1992), Learning with continuous classes, 5th Australian Joint Conference on Artificial Intelligence, 92, 343–348.
- Quinlan, J. R. (1993), Combining instance‐based and model‐based learning, in Proceedings of the Tenth International Conference on Machine Learning, pp. 236–243. [Google Scholar]
- R Core Team (2015), R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing URL, Vienna: [Available at http://www.R‐project.org/.] [Google Scholar]
- Ralston, E. P. , Kite‐Powell H., and Beet A. (2011), An estimate of the cost of acute health effects from food‐ and water‐borne marine pathogens and toxins in the USA, J. Water Health, 9, 680–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randa, M. A. , Polz M. F., and Lim E. (2004), Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR, Appl. Environ. Microbiol., 70, 5469–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, T. K. , Ruiz G. M., and Colwell R. R. (2007), Association of Vibrio cholerae O1 El Tor and O139 Bengal with the copepods Acartia tonsa and Eurytemora affinis , Appl. Environ. Microbiol., 73, 7926–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi, K. , Rao S., Krey V., Cho C. H., Chirkov V., Fischer G., Kindermann G., Nakicenovic N., and Rafaj P. (2011), RCP 8.5—A scenario of comparatively high greenhouse gas emissions, Clim. Change, 109, 33–57. [Google Scholar]
- Rodó, X. , Pascual M., Fuchs G., and Faruque A. S. G. (2002), ENSO and cholera: A nonstationary link related to climate change, Proc. Natl. Acad. Sci. U.S.A., 99, 12901–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild, B. J. , Ault J. S., Goulletquer P., and Heral M. (1994), Decline of the Chesapeake Bay oyster population: A century of habitat destruction and overfishing, Mar. Ecol. Prog. Ser., 111, 29–39. [Google Scholar]
- Scallan, E. R. , Hoekstra M., Angulo F. J., Tauxe R. V., Widdowson M.‐A., Roy S. L., Jones J. L., and Griffin P. M. (2011), Foodborne illness acquired in the United States—Major pathogens, Emerg. Infect. Dis., 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedas, V. T. P. (2007), Influence of environmental factors on the presence of Vibrio cholerae in the marine environment: A climate link, J. Infect. Dev. Ctries., 1, 224–241. [PubMed] [Google Scholar]
- Singleton, F. L. , Attwell R., Jangi S., and Colwell R. R. (1982a), Effects of temperature and salinity on Vibrio cholerae growth, Appl. Environ. Microbiol., 44, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, F. L. , Attwell R. W., Jangi M. S., and Colwell R. R. (1982b), Influence of salinity and organic nutrient concentration on survival and growth of Vibrio cholerae in aquatic microcosms, Appl. Environ. Microbiol., 43, 1080–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirado, M. C. , Clarke R., Jaykus L. A., McQuatters‐Gollop A., and Frank J. M. (2010), Climate change and food safety: A review, Food Res. Int., 43, 1745–1765. [Google Scholar]
- Turner, J. W. , Malayil L., Guadagnoli D., Cole D., and Lipp E. K. (2014), Detection of Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae with respect to seasonal fluctuations in temperature and plankton abundance, Environ. Microbiol., 16, 1019–1028. [DOI] [PubMed] [Google Scholar]
- Urquhart, E. A. , Zaitchik B. F., Waugh D. W., Guikema S. D., and Del Castillo C. E. (2014), Uncertainty in model predictions of Vibrio vulnificus response to climate variability and change: A Chesapeake Bay case study, PLoS One, 9, e98256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Food and Drug Administration (2005), Quantitative risk assessment on the public health impact of pathogenic Vibrio parahaemolyticus in raw oysters. Center for Food Safety and Applied Nutrition. [Available at http://www.fda.gov/Food/FoodScienceResearch/RiskSafetyAssessment/ucm050421.htm.]
- Virginia Department of Health (2016), Vibrio control plan, 11 pp.
- Vezzulli, L. , Previati M., Pruzzo C., Marchese A., Bourne D. G., and Cerrano C. (2010a), Vibrio infections triggering mass mortality events in a warming Mediterranean Sea, Environ. Microbiol., 12, 2007–2019. [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , Pruzzo C., Huq A., and Colwell R. R. (2010b), Environmental reservoirs of Vibrio cholerae and their role in cholera, Environ. Microbiol. Rep., 2, 27–33. [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , Colwell R. R., and Pruzzo C. (2013), Ocean warming and spread of pathogenic Vibrios in the aquatic environment, Microb. Ecol., 65, 817–825. [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , Grande C., Reid P. C., Hélaouët P., Edwards M., Höfle M. G., Brettar M. G., Colwell R. R., and Pruzzo C. (2016), Climate influence on Vibrio and associated human diseases during the past half‐century in the coastal North Atlantic, Proc. Natl. Acad. Sci. U.S.A., 113, E5062–E5071, doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrac, M. , Stein M. L., Hayhoe K., and Liang X.‐Z. (2007), A general method for validating statistical downscaling methods under future climate change, Geophys. Res. Lett., 34, L18701, doi: 10.1029/2007GL030295. [DOI] [Google Scholar]
- Waldman, R. J. , Mintz E. D., and Papowitz H. E. (2013), The cure for cholera—Improving access to safe water and sanitation, N. Engl. J. Med., 368, 592–594. [DOI] [PubMed] [Google Scholar]
- Weis, K. E. , Hammond R. M., Hutchinson R., and Blackmore C. G. M. (2011), Vibrio illness in Florida, 1998–2007, Epidemiol. Infect., 139, 591–598. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2016), Cholera: Fact sheet. [Available at http://www.who.int/mediacentre/factsheets/fs107/en, Accessed November 11th, 2016.]
- Wood, S. (2006), Generalized Additive Models: An Introduction With R, CRC Press, Boca Raton, Fla. [Google Scholar]
- Wright, A. C. , Hill R. T., Johnson J. A., Roghman M. C., Colwell R. R., and Morris J. G. (1996), Distribution of Vibrio vulnificus in the Chesapeake Bay, Appl. Environ. Microbiol., 62, 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukimoto, S. , et al. (2012), A new global climate model of the Meteorological Research Institute: MRI‐CGCM3—Model description and basic performance, J. Meteor. Soc. Jpn., 90A, 23–64. [Google Scholar]
- Züur, A. , Ieno E., Walker N., Saveliev A., and Smith G. (2009), Mixed Effects Models and Extensions in Ecology With R, Statistics for Biology and Health, 574 pp., Springer New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1


