Abstract
A water-soluble self-assembled supramolecular FeII4L4 tetrahedron binds to single stranded DNA, mismatched DNA base pairs, and three-way DNA junctions. Binding of the coordination cage quenches fluorescent labels on the DNA strand, which provides an optical means to detect the interaction and allows the position of the binding site to be gauged with respect to the fluorescent label. Utilizing the quenching and binding properties of the coordination cage, we developed a simple and rapid detection method based on fluorescence quenching to detect unpaired bases in double-stranded DNA.
Interactions between DNA and synthetic molecules have enabled numerous applications in biomedicine, gene regulation, and disease diagnosis.1 For instance, many DNA-targeting anticancer drugs have been designed and developed based on their binding to double-stranded DNA (dsDNA).2 Besides the canonical B-form antiparallel DNA double helix, there are several other structural forms of DNA, including parallel duplexes, triplexes, and quadruplexes, and three- and four-way DNA junctions. These structures occurring at specific genomic locations have various biological functions in natural systems,3 but have also been used to build DNA nanostructures and devices.4
The three-way junction (3WJ) is an uncommon DNA structure, although it is implicated in many DNA metabolic processes, such as replication, transcription, recombination, and repair.5 Errors in these processes can lead to DNA mutations, which may cause a cascade of problems in gene expression.6 Hence, the development of recognition probes for 3WJs and DNA duplexes that contain a small number of unpaired bases may lead to the emergence of more efficient diagnostic tools.7
Coordination driven self-assembly has enabled the generation of useful three-dimensional molecules8 and materials9 with tunable structures and properties. Numerous applications of these metallosupramolecular complexes in nucleic acid research have been developed in recent years.10 Hannon, Coll, and co-workers have employed metallosupramolecular cylinders to induce the formation of three-way DNA and RNA junctions.11 The binding of plasmid DNA and G-quadruplexes by coordination complexes,12 including two M4L6 tetrahedral cages,12c,12e has also been reported, which demonstrates the potential for application of the metallosupramolecular cages in recognition of DNA structures. Building upon these pioneering studies, here we report the use of a simple FeII4L4 tetrahedron (1, Figure 1a)13 as a site-selective binder for 3WJs and base pair mismatches. The binding of 1 quenches the fluorescence of a proximate fluorophore, enabling an all-optical readout for the sensing of these DNA structures.
Figure 1.
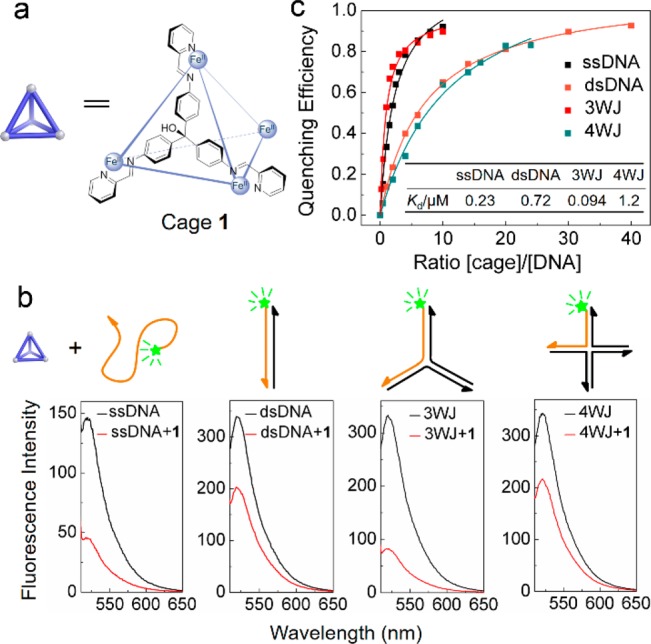
Fluorescence study of the interaction between cage 1 and different types of DNA. (a) Molecular structure of cage 1; the SO42– counterions are omitted for clarity. (b) Effects of 1 (0.2 μM) on the fluorescence intensity of different DNA structures (0.1 μM) labeled with the FAM fluorophore. (c) Quenching efficiency based on the concentration ratio of cage 1 to the whole DNA structures (0.1 μM). Inset: Kd values for different DNA structures. Further details on data fitting are given in Figure S4. All fluorescence measurements were performed in PBM buffer (10 mM phosphate buffer, 10 mM MgSO4, pH 7.5).
We selected tetrahedral cage 1 as a suitable candidate for DNA binding due to its water solubility and stability under biologically relevant conditions (Supporting Information sections S1 and S2). We also hypothesized that the trigonal three-dimensional shape of the tetrahedral cage may contribute to the recognition of 3WJ, as in the case of the 3-fold-symmetric cylinders reported by Hannon.11 Cage 1 was found by fluorescence spectroscopy to quench the fluorescent dye 6-carboxyfluorescein (FAM) attached to different DNA strands, showing a preference for single-stranded DNA (ssDNA) and 3WJ DNA. Studies of the interaction between 1 and 3WJ revealed 1 to bind preferentially at the central cavity of a 3WJ, where base-pairings are loose or even absent, according to recent reports.14 The inference that 1 bound preferentially to unpaired sites thus led to the detection of a series of base mismatches in DNA double strands.
Bulk fluorescence measurements were used to study the interaction of cage 1 (Figure 1a) with different DNA structures (Supporting Information section S3). We found the cage to quench the fluorescence emissions of FAM dye molecules covalently bound to the 5′ end of DNA strands, but to have no effect on the fluorescence of the dye itself when free in solution (Figure S10a,b). Cage 1 was treated with four different kinds of DNA structures, each labeled with FAM (Figure 1b): ssDNA, dsDNA, 3WJ DNA, and four-way DNA junction (4WJ) DNA. The fluorescence spectrum of each sample was measured before and after mixing with cage 1 (Figure 1b). We observed cage 1 to quench the FAM fluorescence in the samples containing ssDNA and 3WJ by 69% and 75%, respectively. Thus, 1 shows higher affinities for these structures, as compared to dsDNA and 4WJ, for which only 40% and 38% of quenching were observed. The quenching efficiency (QE, Supporting Information section S3) is expected to depend on the relative concentrations of 1 and DNA. Figure 1c shows the dependence of the QE on the ratio of cage 1 to DNA for four different DNA structures. Dissociation constants (Kd) were determined for the different DNA structures (Figure 1c and Supporting Information section S3, Table S3), indicating stronger interactions of cage 1 with 3WJ and single-stranded DNA. Moreover, we found only the fully assembled cage to cause fluorescence quenching; addition of either FeII or the precursor aldehyde and triamine subcomponents of 1 to the 3WJ had a minimal effect on fluorescence intensity (Figure S10c).
To gauge further the binding between cage 1 and 3WJ, gel electrophoresis was applied to DNA samples in the presence (+) and absence (−) of cage 1. Gel results (Figure 2a, Supporting Information section S5) showed a clear change of the 3WJ band (red square in Figure 2a) following the addition of cage 1. A slight shift of the ssDNA band was also observed. In contrast, no position difference was observed for dsDNA. Although the band of the 4WJ became broadened in the presence of 1, the position of the main band did not change. The addition of increasing amounts of cage 1 into 3WJ led to a migration of the band (Figure 2b). These gel results confirm the specific binding between cage 1 and 3WJ, which increased the molecular weight of the complex, partly neutralized the DNA charge, and in consequence slowed the mobility of 3WJ. Fluorescent melting experiments were also performed to investigate the binding in Supporting Information section S4.
Figure 2.
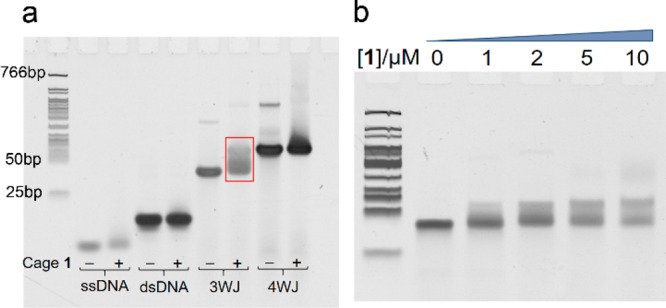
Gel results: (a) polyacrylamide gel electrophoresis of studied DNA structures (5 μM) with (+) and without (−) cage 1 (50 μM). The highlighted band has been investigated further. (b) Gel shift of 3WJ (1 μM) upon titration with increasing concentrations of cage 1. The same ladder is used in panel b).
Varying the distance between the branch point and the FAM label on different 3WJ (Figure 3a) allowed for the binding mode of 1 to be probed. Previous reports11,15 led us to the hypothesis that cage 1 binds to the 3WJ at the central branch point. Increasing the distance between the central cage binding site and the fluorescent label (FAM) is thus expected to reduce fluorescence quenching. The results of this experiment were in agreement with our hypothesis (Figure 3a); an apparent linear relationship between the normalized fluorescence intensity and the distance was observed, agreeing with the presence of cage 1 at the branch point of the 3WJ.
Figure 3.
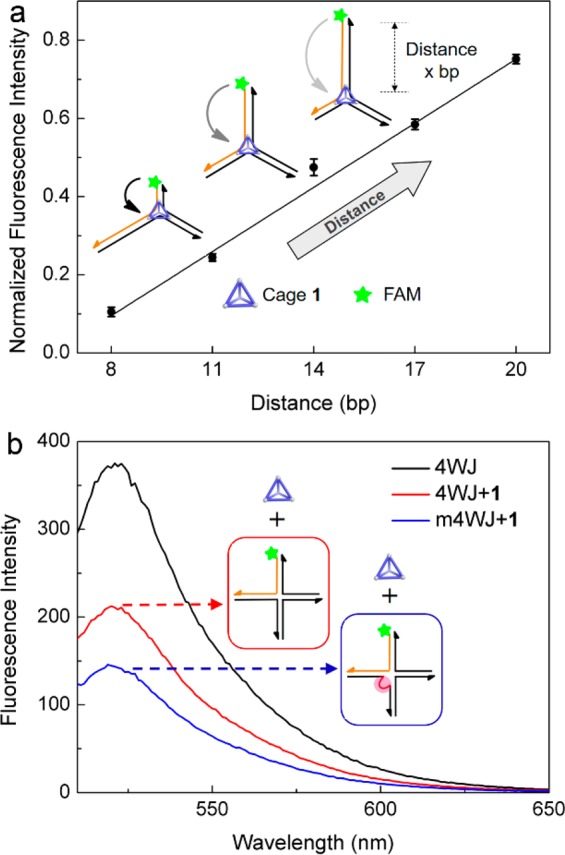
Experiments to determine the location of binding. (a) Effect of the distance (x base pairs, bp) between the label and the branch point of 3WJ on the fluorescence intensity of FAM in the presence of cage 1. Values were normalized against the mean fluorescence intensities of 3WJ-x (x = 8, 11, 14, 17, 20) in the absence of cage 1. Data were averaged over three experimental repeats. (b) Effect of mismatch (shown in red) on fluorescence quenching by cage 1 (0.2 μM) at the branchpoint of 4WJ (0.1 μM).
The complementary bases closest to the branchpoint of three-way DNA junctions are unstable and may not pair.14 This flexible configuration may provide a central trigonal cavity for the triangular shaped cage to bind, similarly to previously reported triangular cylinders.11 Cage 1 has a strong affinity toward ssDNA (Figure 1a), the looser structure of which is inferred to an optimal configuration for binding 1. In analogous fashion, we hypothesized that cage 1 binds more readily to the unpaired bases present in the central cavity of the 3WJ. To test this hypothesis, we introduced mismatched bases at the branch point of a 4WJ to destabilize the structure (Figure S7).16 As shown in Figure 1a, cage 1 minimally quenches the fluorescent dye on the 4WJ. This may be a result of the rigid branch point of the 4WJ in the presence of Mg2+ reducing the propensity of the cage to bind.17 However, when two mismatched bases were introduced at the central branch point of the 4WJ, fluorescence quenching was increased (Figure 3b). This observation indicates that the unpaired bases enable the cage to bind with higher affinity to the previously less accessible branch point of the 4WJ. It is worth noting that 3WJ is much less stable than 4WJ even if the central bases are fully paired,16b and thus its own special molecular conformation and central trigonal cavity may play more important roles in the binding in this case, which implies the cage may recognize different types of DNA structures. Further discussion of the quenching mechanism is given in Supporting Information section S7.
The DNA-cage binding that occurred specifically at unpaired bases inspired us to explore the potential of cage 1 to sense base-pair mismatches in dsDNA. We first investigated the single-base mismatch at different locations along dsDNA (M1–M3 in Figure 4). These results showed a clear dependence between the quenching efficiency and the position of the mismatch relative to the FAM label. In the case of M1, with the FAM 5 bases away from the mismatch, the quenching was minimal. However, the quenching efficiency increased in the cases of M2 and M3, in which the mismatch was closer to the label. Besides the GG mismatched M2, the cage is also sensitive to the other types of mismatches such as GA and GT at the same site (Figure S8a). We inferred that a single base mismatch was detected by the cage, most strongly for mismatches closest to the fluorescent label.
Figure 4.
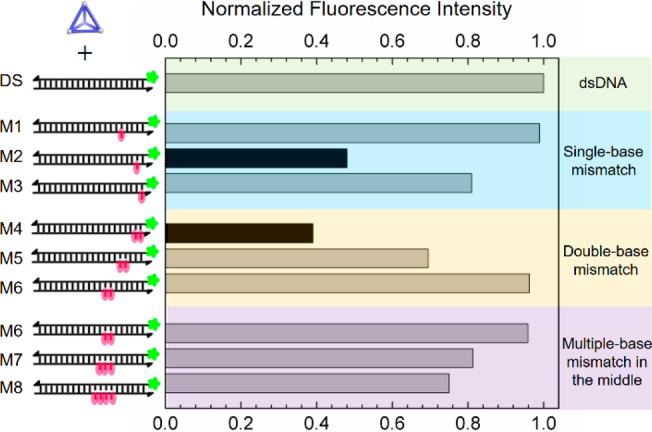
Mismatch detection in DNA duplexes (0.1 μM) using cage 1 (0.2 μM). The bar graph shows normalized fluorescence emission intensities at 520 nm of different cage 1/DNA mixtures excited at 495 nm. Mismatches in the dsDNA are marked in red.
On the basis of the distance between the mismatch and the label, we expected the quenching efficiency to be greater for M3 than M2. One possible explanation for the divergent observation is that the first A:T base pair of M2 next to the mismatch site was destabilized, rendering its behavior similar to that of a sequence containing two mismatches. Indeed, when sequences containing two mismatches (M4–M6) were investigated, we found that the quenching efficiency was increased in proportion to the distance between the mismatch point and label.
Finally, we introduced progressively more mismatched bases into the sequence at a constant distance from the FAM label (Figure 4, M6–M8). The degree of fluorescence quenching was observed to increase with the number of unpaired bases. A DNA bulge induced by more unpaired bases18 also enhanced the quenching by cage 1 (Figure S8b). We infer regions containing more unpaired bases to provide a larger cavity in dsDNA, ensuring enhanced binding to 1. Kd values for the interaction between cage 1 and mismatched and bulged DNA can be found in Figures S4, S5, and Table S4.
Overall, the above findings establish a simple and rapid new method to detect unpaired bases in dsDNA. Compared with the classic DNA mismatch-binding ligands, including metal complexes, small organic molecules, and simple metal ions, which have been reported recently,19 tetrahedral cage 1 possesses unique advantages as a mismatched DNA probe. First, although it is not a mismatch-selective binder, in the manner of other metal complexes and ions201 can sense a variety of base mismatches (Figure S8a). Second, in contrast to the luminescent metalloinsertors,21 the ability of cage 1 to give rise to fluorescence quenching allows for base mismatch detection at a lower concentration. In addition, cage 1 combined with a specifically designed fluorescently labeled probe strand would enable selective detection of target structures or sequences. Finally, the use of subcomponent self-assembly to prepare three-dimensional supramolecules enables the tuning of probe structure and selectivity. The encapsulation of guest molecules inside coordination cages could enable these guests to be released in the vicinity of specific DNA regions.
In conclusion, the interaction between FeII4L4 tetrahedron and DNA has been probed for the first time, and applied to detect mismatches in DNA base pairs. Compared with previous DNA binders, the fluorescence quenching property of cage 1 enables straightforward optical detection and hence molecular sensing applications. Our approach relying on metallosupramolecular complexes enhances the flexibility and expandability for future designs. Given the significance of DNA mismatch detection in the diagnosis of genetic diseases and the value of three-way junctions in DNA metabolic processes, cage 1 adds a promising compound for fluorescence assays especially in nanobiotechnology and biomedicine. We foresee other such metal–organic cages formed using subcomponent self-assembly to enable other new applications in medical and biological sensing.
Acknowledgments
The authors acknowledge support from the UK Engineering and Physical Sciences Research Council (EPSRC EP/M008258/1, EPSRC EP/P027067/1, and EPSRC EP/L015978/1). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 642192. We appreciate the kind suggestions on the binding mechanism study from Dr. Murray Stewart and Dr. Katherine Stott. We thank Diana Sobota for critical reading of the paper.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b03566.
DNA sequence, experimental details, supporting data, and figures (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- a Zhu R.; Regeni I.; Holstein J. J.; Dittrich B.; Simon M.; Prévost S.; Gradzielski M.; Clever G. H. Catenation and Aggregation of Multi-Cavity Coordination Cages. Angew. Chem., Int. Ed. 2018, 57 (41), 13652–13656. 10.1002/anie.201806047. [DOI] [PubMed] [Google Scholar]; b Pazos E.; Mosquera J.; Vázquez M. E.; Mascareñas J. L. DNA Recognition by Synthetic Constructs. ChemBioChem 2011, 12 (13), 1958–1973. 10.1002/cbic.201100247. [DOI] [PubMed] [Google Scholar]; c Priegue J. M.; Lostalé-Seijo I.; Crisan D.; Granja J. R.; Fernández-Trillo F.; Montenegro J. Different-Length Hydrazone Activated Polymers for Plasmid DNA Condensation and Cellular Transfection. Biomacromolecules 2018, 19 (7), 2638–2649. 10.1021/acs.biomac.8b00252. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ziach K.; Chollet C.; Parissi V.; Prabhakaran P.; Marchivie M.; Corvaglia V.; Bose P. P.; Laxmi-Reddy K.; Godde F.; Schmitter J. M.; Chaignepain S.; Pourquier P.; Huc I. Single helically folded aromatic oligoamides that mimic the charge surface of double-stranded B-DNA. Nat. Chem. 2018, 10 (5), 511–518. 10.1038/s41557-018-0018-7. [DOI] [PubMed] [Google Scholar]; e Yu G.; Yu S.; Saha M. L.; Zhou J.; Cook T. R.; Yung B. C.; Chen J.; Mao Z.; Zhang F.; Zhou Z.; Liu Y.; Shao L.; Wang S.; Gao C.; Huang F.; Stang P. J.; Chen X. A discrete organoplatinum(II) metallacage as a multimodality theranostic platform for cancer photochemotherapy. Nat. Commun. 2018, 9 (1), 4335. 10.1038/s41467-018-06574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Zhu J.; Yan Z.; Zhou W.; Liu C.; Wang J.; Wang E. Lighting Up the Thioflavin T by Parallel-Stranded TG(GA)n DNA Homoduplexes. ACS Sensors 2018, 3 (6), 1118–1125. 10.1021/acssensors.8b00141. [DOI] [PubMed] [Google Scholar]
- a Hurley L. H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 188. 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]; b Erkkila K. E.; Odom D. T.; Barton J. K. Recognition and Reaction of Metallointercalators with DNA. Chem. Rev. 1999, 99 (9), 2777–2796. 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]; c Howson S. E.; Bolhuis A.; Brabec V.; Clarkson G. J.; Malina J.; Rodger A.; Scott P. Optically pure, water-stable metallo-helical ‘flexicate’ assemblies with antibiotic activity. Nat. Chem. 2012, 4, 31. 10.1038/nchem.1206. [DOI] [PubMed] [Google Scholar]; d Kaner R. A.; Allison S. J.; Faulkner A. D.; Phillips R. M.; Roper D. I.; Shepherd S. L.; Simpson D. H.; Waterfield N. R.; Scott P. Anticancer metallohelices: nanomolar potency and high selectivity. Chem. Sci. 2016, 7 (2), 951–958. 10.1039/C5SC03677A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhao J.; Bacolla A.; Wang G.; Vasquez K. M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010, 67 (1), 43–62. 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kaushik M.; Singh A.; Kumar M.; Chaudhary S.; Ahmed S.; Kukreti S. Structure-Specific Ligand Recognition of Multistranded DNA Structures. Curr. Top. Med. Chem. 2017, 17 (2), 138–147. 10.2174/1568026616666160530154239. [DOI] [PubMed] [Google Scholar]; c Bacolla A.; Wells R. D. Non-B DNA Conformations, Genomic Rearrangements, and Human Disease. J. Biol. Chem. 2004, 279 (46), 47411–47414. 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- a Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Seeman N. C.; Sleiman H. F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]; c Zhang T.; Hartl C.; Frank K.; Heuer-Jungemann A.; Fischer S.; Nickels P. C.; Nickel B.; Liedl T. 3D DNA Origami Crystals. Adv. Mater. 2018, 30 (28), 1800273. 10.1002/adma.201800273. [DOI] [PubMed] [Google Scholar]; d Yatsunyk L. A.; Mendoza O.; Mergny J. L. Nano-oddities”: Unusual Nucleic Acid Assemblies for DNA-Based Nanostructures and Nanodevices. Acc. Chem. Res. 2014, 47 (6), 1836–1844. 10.1021/ar500063x. [DOI] [PubMed] [Google Scholar]
- a Singleton M. R.; Scaife S.; Wigley D. B. Structural Analysis of DNA Replication Fork Reversal by RecG. Cell 2001, 107 (1), 79–89. 10.1016/S0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]; b Slean M. M.; Reddy K.; Wu B.; Nichol Edamura K.; Kekis M.; Nelissen F. H. T.; Aspers R. L. E. G.; Tessari M.; Schärer O. D.; Wijmenga S. S.; Pearson C. E. Interconverting Conformations of Slipped-DNA Junctions Formed by Trinucleotide Repeats Affect Repair Outcome. Biochemistry 2013, 52 (5), 773–785. 10.1021/bi301369b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kunkel T. A.; Erie D. A. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu. Rev. Genet. 2015, 49 (1), 291–313. 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Widłak W.DNA Replication, Mutations, and Repair. In Molecular Biology: Not Only for Bioinformaticians; Widłak W., Ed.; Springer: Berlin, Heidelberg, 2013; pp 49–69. [Google Scholar]
- a Nano A.; Boynton A. N.; Barton J. K. A Rhodium-Cyanine Fluorescent Probe: Detection and Signaling of Mismatches in DNA. J. Am. Chem. Soc. 2017, 139 (48), 17301–17304. 10.1021/jacs.7b10639. [DOI] [PMC free article] [PubMed] [Google Scholar]; b He H.; Xia J.; Chang G.; Peng X.; Lou Z.; Nakatani K.; Zhou X.; Wang S. Selective recognition of G–G mismatch using the double functional probe with electrochemical activeferrocenyl. Biosens. Bioelectron. 2013, 42, 36–40. 10.1016/j.bios.2012.10.042. [DOI] [PubMed] [Google Scholar]; c Kong J.; Zhu J.; Keyser U. F. Single molecule based SNP detection using designed DNA carriers and solid-state nanopores. Chem. Commun. 2017, 53 (2), 436–439. 10.1039/C6CC08621G. [DOI] [PubMed] [Google Scholar]
- a Kim Y.; Koo J.; Hwang I. C.; Mukhopadhyay R. D.; Hong S.; Yoo J.; Dar A. A.; Kim I.; Moon D.; Shin T. J.; Ko Y. H.; Kim K. Rational Design and Construction of Hierarchical Superstructures Using Shape-Persistent Organic Cages: Porphyrin Box-Based Metallosupramolecular Assemblies. J. Am. Chem. Soc. 2018, 140 (44), 14547–14551. 10.1021/jacs.8b08030. [DOI] [PubMed] [Google Scholar]; b Preston D.; Sutton J. J.; Gordon K. C.; Crowley J. D. A Nona-nuclear Heterometallic Pd3Pt6 “Donut”-Shaped Cage: Molecular Recognition and Photocatalysis. Angew. Chem., Int. Ed. 2018, 57 (28), 8659–8663. 10.1002/anie.201804745. [DOI] [PubMed] [Google Scholar]; c Handke M.; Adachi T.; Hu C.; Ward M. D. Encapsulation of Isolated Luminophores within Supramolecular Cages. Angew. Chem., Int. Ed. 2017, 56 (45), 14003–14006. 10.1002/anie.201707097. [DOI] [PubMed] [Google Scholar]; d Burke B. P.; Grantham W.; Burke M. J.; Nichol G. S.; Roberts D.; Renard I.; Hargreaves R.; Cawthorne C.; Archibald S. J.; Lusby P. J. Visualizing Kinetically Robust CoIII4L6 Assemblies in Vivo: SPECT Imaging of the Encapsulated [99mTc]TcO4– Anion. J. Am. Chem. Soc. 2018, 140 (49), 16877–16881. 10.1021/jacs.8b09582. [DOI] [PubMed] [Google Scholar]; e Samanta D.; Galaktionova D.; Gemen J.; Shimon L. J. W.; Diskin-Posner Y.; Avram L.; Král P.; Klajn R. Reversible chromism of spiropyran in the cavity of a flexible coordination cage. Nat. Commun. 2018, 9 (1), 641. 10.1038/s41467-017-02715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Zhou J.; Zhang Y.; Yu G.; Crawley M. R.; Fulong C. R. P.; Friedman A. E.; Sengupta S.; Sun J.; Li Q.; Huang F.; Cook T. R. Highly Emissive Self-Assembled BODIPY-Platinum Supramolecular Triangles. J. Am. Chem. Soc. 2018, 140 (24), 7730–7736. 10.1021/jacs.8b04929. [DOI] [PubMed] [Google Scholar]
- a Wong Y. S.; Leung F. C. M.; Ng M.; Cheng H. K.; Yam V. W. W. Platinum(II)-Based Supramolecular Scaffold-Templated Side-by-Side Assembly of Gold Nanorods through Pt···Pt and π–π Interactions. Angew. Chem., Int. Ed. 2018, 57 (48), 15797–15801. 10.1002/anie.201810302. [DOI] [PubMed] [Google Scholar]; b Sinn S.; Yang L.; Biedermann F.; Wang D.; Kübel C.; Cornelissen J. J. L. M.; De Cola L. Templated Formation of Luminescent Virus-like Particles by Tailor-Made Pt(II) Amphiphiles. J. Am. Chem. Soc. 2018, 140 (6), 2355–2362. 10.1021/jacs.7b12447. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Learte-Aymamí S.; Curado N.; Rodríguez J.; Vázquez M. E.; Mascareñas J. L. Metal-Dependent DNA Recognition and Cell Internalization of Designed, Basic Peptides. J. Am. Chem. Soc. 2017, 139 (45), 16188–16193. 10.1021/jacs.7b07422. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Calabrese C. M.; Merkel T. J.; Briley W. E.; Randeria P. S.; Narayan S. P.; Rouge J. L.; Walker D. A.; Scott A. W.; Mirkin C. A. Biocompatible Infinite-Coordination-Polymer Nanoparticle–Nucleic-Acid Conjugates for Antisense Gene Regulation. Angew. Chem., Int. Ed. 2015, 54 (2), 476–480. 10.1002/anie.201407946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Singh K.; Kumari S.; Jana A.; Bhowmick S.; Das P.; Das N. Self-assembled neutral [2 + 2] platinacycles showing minimal DNA interactions. Polyhedron 2019, 157, 267–275. 10.1016/j.poly.2018.09.075. [DOI] [Google Scholar]; b Hotze A. C. G.; Kariuki B. M.; Hannon M. J. Dinuclear Double-Stranded Metallosupramolecular Ruthenium Complexes: Potential Anticancer Drugs. Angew. Chem., Int. Ed. 2006, 45 (29), 4839–4842. 10.1002/anie.200601351. [DOI] [PubMed] [Google Scholar]
- a Phongtongpasuk S.; Paulus S.; Schnabl J.; Sigel R. K. O.; Spingler B.; Hannon M. J.; Freisinger E. Binding of a Designed Anti-Cancer Drug to the Central Cavity of an RNA Three-Way Junction. Angew. Chem., Int. Ed. 2013, 52 (44), 11513–11516. 10.1002/anie.201305079. [DOI] [PubMed] [Google Scholar]; b Boer D. R.; Kerckhoffs J. M. C. A.; Parajo Y.; Pascu M.; Usón I.; Lincoln P.; Hannon M. J.; Coll M. Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders. Angew. Chem., Int. Ed. 2010, 49 (13), 2336–2339. 10.1002/anie.200906742. [DOI] [PubMed] [Google Scholar]; c Malina J.; Hannon M. J.; Brabec V. Recognition of DNA Three-Way Junctions by Metallosupramolecular Cylinders: Gel Electrophoresis Studies. Chem. - Eur. J. 2007, 13 (14), 3871–3877. 10.1002/chem.200700159. [DOI] [PubMed] [Google Scholar]; d Oleksi A.; Blanco A. G.; Boer R.; Usón I.; Aymamí J.; Rodger A.; Hannon M. J.; Coll M. Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate. Angew. Chem., Int. Ed. 2006, 45 (8), 1227–1231. 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]; e Cerasino L.; Hannon M. J.; Sletten E. DNA Three-Way Junction with a Dinuclear Iron(II) Supramolecular Helicate at the Center: A NMR Structural Study. Inorg. Chem. 2007, 46 (16), 6245–6251. 10.1021/ic062415c. [DOI] [PubMed] [Google Scholar]
- a Zhao C.; Song H.; Scott P.; Zhao A.; Tateishi-Karimata H.; Sugimoto N.; Ren J.; Qu X. Mirror-Image Dependence: Targeting Enantiomeric G-Quadruplex DNA Using Triplex Metallohelices. Angew. Chem., Int. Ed. 2018, 57 (48), 15723–15727. 10.1002/anie.201809207. [DOI] [PubMed] [Google Scholar]; b Drożdż W.; Walczak A.; Bessin Y.; Gervais V.; Cao X. Y.; Lehn J. M.; Ulrich S.; Stefankiewicz A. R. Multivalent Metallosupramolecular Assemblies as Effective DNA Binding Agents. Chem. - Eur. J. 2018, 24 (42), 10802–10811. 10.1002/chem.201801552. [DOI] [PubMed] [Google Scholar]; c Xi S. F.; Bao L. Y.; Xu Z. L.; Wang Y. X.; Ding Z. D.; Gu Z. G. Enhanced Stabilization of G-Quadruplex DNA by [Ni4L6]8+ Cages with Large Rigid Aromatic Ligands. Eur. J. Inorg. Chem. 2017, 2017 (29), 3533–3541. 10.1002/ejic.201700409. [DOI] [Google Scholar]; d Garci A.; Castor K. J.; Fakhoury J.; Do J. L.; Di Trani J.; Chidchob P.; Stein R. S.; Mittermaier A. K.; Friščić T.; Sleiman H. Efficient and Rapid Mechanochemical Assembly of Platinum(II) Squares for Guanine Quadruplex Targeting. J. Am. Chem. Soc. 2017, 139 (46), 16913–16922. 10.1021/jacs.7b09819. [DOI] [PubMed] [Google Scholar]; e Xi S. F.; Bao L. Y.; Lin J. G.; Liu Q. Z.; Qiu L.; Zhang F. L.; Wang Y. X.; Ding Z. D.; Li K.; Gu Z. G. Enantiomers of tetrahedral metal–organic cages: a new class of highly efficient G-quadruplex ligands with potential anticancer activities. Chem. Commun. 2016, 52 (67), 10261–10264. 10.1039/C6CC05743H. [DOI] [PubMed] [Google Scholar]; f Rakers V.; Cadinu P.; Edel J. B.; Vilar R. Development of microfluidic platforms for the synthesis of metal complexes and evaluation of their DNA affinity using online FRET melting assays. Chem. Sci. 2018, 9 (14), 3459–3469. 10.1039/C8SC00528A. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Engelhard D. M.; Nowack J.; Clever G. H. Copper-Induced Topology Switching and Thrombin Inhibition with Telomeric DNA G-Quadruplexes. Angew. Chem., Int. Ed. 2017, 56 (38), 11640–11644. 10.1002/anie.201705724. [DOI] [PubMed] [Google Scholar]; h Engelhard D. M.; Meyer A.; Berndhäuser A.; Schiemann O.; Clever G. H. Di-copper(ii) DNA G-quadruplexes as EPR distance rulers. Chem. Commun. 2018, 54 (54), 7455–7458. 10.1039/C8CC04053B. [DOI] [PubMed] [Google Scholar]
- Bilbeisi R. A.; Clegg J. K.; Elgrishi N.; Hatten X. d.; Devillard M.; Breiner B.; Mal P.; Nitschke J. R. Subcomponent Self-Assembly and Guest-Binding Properties of Face-Capped Fe4L48+ Capsules. J. Am. Chem. Soc. 2012, 134 (11), 5110–5119. 10.1021/ja2092272. [DOI] [PubMed] [Google Scholar]
- a Sabir T.; Toulmin A.; Ma L.; Jones A. C.; McGlynn P.; Schröder G. F.; Magennis S. W. Branchpoint Expansion in a Fully Complementary Three-Way DNA Junction. J. Am. Chem. Soc. 2012, 134 (14), 6280–6285. 10.1021/ja211802z. [DOI] [PubMed] [Google Scholar]; b Toulmin A.; Baltierra-Jasso L. E.; Morten M. J.; Sabir T.; McGlynn P.; Schröder G. F.; Smith B. O.; Magennis S. W. Conformational Heterogeneity in a Fully Complementary DNA Three-Way Junction with a GC-Rich Branchpoint. Biochemistry 2017, 56 (37), 4985–4991. 10.1021/acs.biochem.7b00677. [DOI] [PubMed] [Google Scholar]
- a Barros S. A.; Chenoweth D. M. Recognition of Nucleic Acid Junctions Using Triptycene-Based Molecules. Angew. Chem., Int. Ed. 2014, 53 (50), 13746–13750. 10.1002/anie.201407061. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Duprey J. L. H. A.; Takezawa Y.; Shionoya M. Metal-Locked DNA Three-Way Junction. Angew. Chem., Int. Ed. 2013, 52 (4), 1212–1216. 10.1002/anie.201207338. [DOI] [PubMed] [Google Scholar]; c Barros S. A.; Yoon I.; Chenoweth D. M. Modulation of the E. coli rpoH Temperature Sensor with Triptycene-Based Small Molecules. Angew. Chem., Int. Ed. 2016, 55 (29), 8258–8261. 10.1002/anie.201601626. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Takezawa Y.; Yoneda S.; Duprey J. L. H. A.; Nakama T.; Shionoya M. Metal-responsive structural transformation between artificial DNA duplexes and three-way junctions. Chem. Sci. 2016, 7 (5), 3006–3010. 10.1039/C6SC00383D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Duckett D. R.; Lilley D. M. J. Effects of base mismatches on the structure of the four-way DNA junction. J. Mol. Biol. 1991, 221 (1), 147–161. 10.1016/0022-2836(91)80211-C. [DOI] [PubMed] [Google Scholar]; b Kadrmas J. L.; Ravin A. J.; Leontis N. B. Relative stabilities of DNA three-way, four-way and five-way junctions (multi-helix junction loops): unpaired nucleotides can be stabilizing or destabilizing. Nucleic Acids Res. 1995, 23 (12), 2212–2222. 10.1093/nar/23.12.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. M.; Murchie A. I.; Lilley D. M. The solution structure of the four-way DNA junction at low-salt conditions: a fluorescence resonance energy transfer analysis. Biophys. J. 1994, 66 (1), 99–109. 10.1016/S0006-3495(94)80765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cardo L.; Nawroth I.; Cail P. J.; McKeating J. A.; Hannon M. J. Metallo supramolecular cylinders inhibit HIV-1 TAR-TAT complex formation and viral replication in cellulo. Sci. Rep. 2018, 8 (1), 13342. 10.1038/s41598-018-31513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Malina J.; Hannon M. J.; Brabec V. Recognition of DNA bulges by dinuclear iron(II) metallosupramolecular helicates. FEBS J. 2014, 281 (4), 987–997. 10.1111/febs.12696. [DOI] [PubMed] [Google Scholar]
- Granzhan A.; Kotera N.; Teulade-Fichou M. P. Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014, 43 (10), 3630–3665. 10.1039/c3cs60455a. [DOI] [PubMed] [Google Scholar]
- a Jackson B. A.; Barton J. K. Recognition of DNA Base Mismatches by a Rhodium Intercalator. J. Am. Chem. Soc. 1997, 119 (52), 12986–12987. 10.1021/ja972489a. [DOI] [Google Scholar]; b Ono A.; Togashi H. Highly Selective Oligonucleotide-Based Sensor for Mercury(II) in Aqueous Solutions. Angew. Chem., Int. Ed. 2004, 43 (33), 4300–4302. 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]; c Ono A.; Cao S.; Togashi H.; Tashiro M.; Fujimoto T.; Machinami T.; Oda S.; Miyake Y.; Okamoto I.; Tanaka Y. Specific interactions between silver(i) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 2008, 39, 4825–4827. 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- a Gabr M. T.; Pigge F. C. Platinum(II) Complexes with Sterically Expansive Tetraarylethylene Ligands as Probes for Mismatched DNA. Inorg. Chem. 2018, 57 (20), 12641–12649. 10.1021/acs.inorgchem.8b01782. [DOI] [PubMed] [Google Scholar]; b Fung S. K.; Zou T.; Cao B.; Chen T.; To W. P.; Yang C.; Lok C. N.; Che C. M. Luminescent platinum(II) complexes with functionalized N-heterocyclic carbene or diphosphine selectively probe mismatched and abasic DNA. Nat. Commun. 2016, 7, 10655. 10.1038/ncomms10655. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Boynton A. N.; Marcélis L.; McConnell A. J.; Barton J. K. A Ruthenium(II) Complex as a Luminescent Probe for DNA Mismatches and Abasic Sites. Inorg. Chem. 2017, 56 (14), 8381–8389. 10.1021/acs.inorgchem.7b01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


