Abstract
Schizophrenia is a highly polygenic disorder with important contributions from both common and rare risk alleles. We analysed exome-sequencing data for de novo variants (DNVs) in a new sample of 613 schizophrenia trios, and combined this with published data for a total of 3,444 trios. In our new data, loss-of-function (LoF) DNVs were significantly enriched among 3,471 LoF intolerant genes, supporting previous findings. In the full dataset, genes associated with neurodevelopmental disorders (n=159) were significantly enriched for LoF DNVs. Within these neurodevelopmental disorder genes, SLC6A1, encoding a gamma-aminobutyric acid transporter, was associated with missense-damaging DNVs. In 1,122 trios for which we had genome-wide common variant data, schizophrenia and bipolar disorder polygenic risk were significantly over-transmitted to probands. Probands carrying LoF or deletion DNVs in LoF intolerant or neurodevelopmental disorder genes had significantly less over-transmission of schizophrenia polygenic risk than non-carriers, providing robust support for these DNVs increasing liability to schizophrenia.
Introduction
Genetic liability to schizophrenia involves a combination of rare and common risk alleles distributed across the genome1. Common schizophrenia risk alleles with odds ratios < 1.3 account for at least a third of genetic liability2–4, although only a small fraction of this is captured by the 145 genome-wide significant loci that were implicated in the largest published genome-wide association study (GWAS) of the disorder5. At the other end of the frequency spectrum, rare copy number variants (CNVs) and rare coding variants, both sometimes occurring as de novo variants (DNVs), have been implicated in the disorder6–8. Although CNVs and rare coding variants are enriched in schizophrenia, not all rare variants observed in individuals with schizophrenia, including those occurring de novo, are expected to be aetiologically relevant, as there is a baseline burden of these variants in the general population.
In people with other neurodevelopmental disorders in which CNVs and rare coding variants play a role, particularly autism spectrum disorder (ASD)9,10 and developmental delay11,12, the enrichment for rare coding variants is greatest in genes classified as intolerant to loss-of-function (LoF) variants (i.e. variants that introduce premature stop codons or frameshifts in the encoded protein, or are predicted to disrupt mRNA splicing). This indicates that rare coding variants in these genes are more likely to be pathogenic for those disorders than rare coding variants occurring elsewhere in the genome. Moreover, greater enrichment is found for LoF DNVs than for missense DNVs that change an encoded amino acid, indicating the former class of mutation is particularly likely to be pathogenic. Similar observations have been made in schizophrenia, where an excess of LoF DNVs was found to be largely restricted to LoF intolerant genes7, although the degree of enrichment is lower than for ASD or developmental disorders.
In studies of ASD and developmental disorders, a significant excess of rare coding variants has been observed for 99 and 93 genes, respectively, with 33 of these genes overlapping between these disorders9,11. Only two genes, SETD1A13 and RBM1214, are currently associated with rare coding variants in schizophrenia. This is partly because of lower statistical power, as the number of trios that have been exome-sequenced in studies of schizophrenia (n=2,834) is smaller than equivalent studies of developmental disorders (n=7,580)11 and ASD (n = 6,430)9, but it also reflects the weaker enrichment in schizophrenia for this type of variant. As a set, genes disrupted by DNVs in neurodevelopmental disorders are also enriched for DNVs in schizophrenia15,16, and therefore it follows that some of the genes implicated in ASD and developmental disorders by rare coding variants are also involved in the aetiology of schizophrenia. Aiming to contribute to the schizophrenia rare variant discovery effort, we have undertaken exome-sequencing in a new sample of 613 schizophrenia trios, and combined our data with published data from 2,834 trios, which includes 617 trios previously sequenced by our group15, to provide the largest analysis of coding DNVs in schizophrenia to date. Given the anticipated modest power even of this sample, as we have successfully done before for CNV analysis17, we exploited the well documented overlap in the genetic aetiologies of schizophrenia, ASD, and developmental disorders, to undertake a hypothesis focused analysis of neurodevelopmental disorder genes in schizophrenia, which highlights SLC6A1 as a novel risk gene.
The involvement of common variant polygenic risk in schizophrenia is already established2,4,18, but few existing studies have empirically examined the relationships between different classes of rare and common variants. An early case-control exome sequencing study of schizophrenia found evidence for independent additive effects for common alleles, rare CNVs and rare coding variants when cases were compared with controls, but no within-case correlation between the burden of each type19. More recent evidence indicates a negative correlation within cases for schizophrenia-associated CNV carrier status and common risk variant burden, consistent with the hypothesis that the common and rare alleles co-act20,21. Thus, compared to controls, affected carriers of schizophrenia-associated CNVs have an increased burden of common schizophrenia risk alleles as measured by the polygenic risk score (PRS)21, but in a within case analysis, this burden is inversely proportional to the estimated effect size of the implicated CNV20. In ASD and developmental disorders, common variant polygenic risk for those disorders has been shown to be over-transmitted from parents to probands, but no difference has been reported between those that do or do not carry a disorder-associated DNV22,23. As yet, the relationship between de novo mutations and common allele risk has not been studied in schizophrenia. Here, we examine this relationship using the polygenic transmission-disequilibrium test (pTDT)23. Specifically, we show that people with schizophrenia who are carriers of DNVs in gene sets proposed to be relevant to schizophrenia have a lower common risk allele burden than people with schizophrenia who are not carriers.
Results
De novo mutation rates
After sample and variant quality control (see methods and Supplementary Figures 1-3), we observed 606 coding de novo variants (DNVs) in 613 probands (433 males, 180 females), corresponding to a rate of 0.99 (s.e = 0.041) events per proband, which is not significantly different to the rate observed in a sample of 2,831 previously published schizophrenia trios (previous de novo rate = 1.004; rate ratio (95% confidence interval (CI)) = 0.98 (0.9, 1.08); p = 0.74; Supplementary Table S1). Of the coding DNVs, 154 were synonymous, 372 were missense, 15 were inframe indels, 2 start-loss, 1 stop-lost, and 62 were LoF (19 stop-gain, 13 splice and 30 frameshift indels). The number of coding DNVs observed per-trio followed the expected Poisson distribution (Supplementary Figure S4).
De novo variant enrichment tests
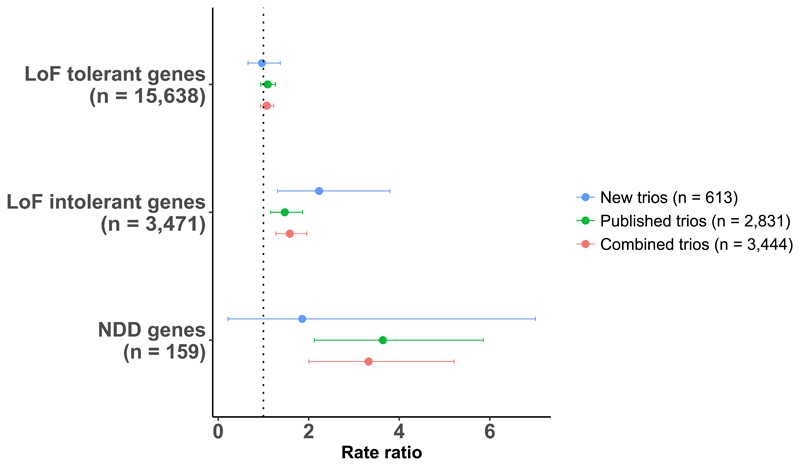
In the new data set, we observed a significant excess of LoF DNVs among LoF intolerant genes (Fig 1, rate ratio (95% CI) = 2.21 (1.3, 3.75); p = 2.3 × 10-3; Supplementary Table S2). Consistent with previous reports, we found no evidence for DNV enrichment in the following negative control gene set tests: LoF DNVs in LoF tolerant genes (Fig 1), synonymous DNVs in LoF intolerant genes; synonymous DNVs in LoF tolerant genes (Supplementary Table S3). After combining the new trio data with previously published data from 2,831 trios, LoF DNVs were enriched in LoF intolerant genes with a rate ratio (95% CI) of 1.58 (1.28, 1.96) (p = 2.5 × 10-5) (Fig 1, Supplementary Table S2). Following review, we tested alternative definitions of LoF intolerant genes based on constraint metrics generated from the gnomAD dataset24; the degree of enrichment of LoF DNVs in schizophrenia is similar regardless of the definition of LoF intolerant genes (see Supplementary Material for full results).
Figure 1.
Gene set enrichment for loss-of-function de novo variants. Loss-of-function (LoF) DNVs were tested in LoF intolerant genes and neurodevelopmental disorder genes. For LoF intolerant and neurodevelopmental disorder gene sets, rate ratios and 95% confidence intervals are relative to the baseline DNV rate, which is defined as the LoF DNV enrichment observed for all genes outside of the given set. LoF DNV enrichment for LoF tolerant genes are shown as a negative control. A breakdown of the LoF intolerant and neurodevelopmental disorder gene set results is provided in Supplementary Tables S2 and S3. NDD = neurodevelopmental disorder.
In the combined trio data, no individual gene was significantly enriched for LoF DNVs after correction for all genes tested (n=19,109). The most significant novel gene was CUL1, which had two LoF DNVs in the new trios and one additional LoF DNV in the published trios (Table 1).
Table 1.
Genes disrupted by 2 or more LoF de novo variants. Individual gene enrichment P values were generated using a one-sided Poisson test. The most significant gene, SETD1A, has been previously identified as a schizophrenia risk gene13. LoF = loss-of-function. DNV = de novo variant.
| Gene | New trios (n=613) |
Published trios (n=2,831) |
All trios (n=3,444) |
|||
|---|---|---|---|---|---|---|
| LoF DNVs | P | LoF DNVs | P | LoF DNVs | P | |
| SETD1A | 0 | 1 | 3 | 1.90E-06 | 3 | 3.00E-06 |
| CUL1 | 2 | 3.60E-05 | 1 | 0.04 | 3 | 2.00E-05 |
| TAF13 | 0 | 1 | 2 | 2.40E-05 | 2 | 3.30E-05 |
| GALNT9 | 0 | 1 | 2 | 2.90E-05 | 2 | 4.20E-05 |
| HENMT1 | 0 | 1 | 2 | 5.50E-05 | 2 | 7.90E-05 |
| PAF1 | 1 | 0.0028 | 1 | 0.013 | 2 | 0.00013 |
| SV2B | 0 | 1 | 2 | 0.00016 | 2 | 0.00023 |
| NRXN3 | 0 | 1 | 2 | 0.00022 | 2 | 0.0003 |
| HIVEP3 | 0 | 1 | 2 | 0.00026 | 2 | 0.00035 |
| RB1CC1 | 0 | 1 | 2 | 0.00046 | 2 | 0.00065 |
| SMARCC2 | 0 | 1 | 2 | 0.0005 | 2 | 0.00068 |
| MKI67 | 0 | 1 | 2 | 0.00085 | 2 | 0.0012 |
| CHD8 | 0 | 1 | 2 | 0.0009 | 2 | 0.0013 |
| TENM1 | 1 | 0.0077 | 1 | 0.046 | 2 | 0.0014 |
| TRIO | 0 | 1 | 2 | 0.0012 | 2 | 0.0016 |
| SCN2A | 1 | 0.012 | 1 | 0.057 | 2 | 0.0024 |
| DNAH9 | 0 | 1 | 2 | 0.0018 | 2 | 0.0026 |
| KMT2C | 0 | 1 | 2 | 0.0086 | 2 | 0.012 |
| KIAA1109 | 0 | 1 | 2 | 0.01 | 2 | 0.015 |
| TTN | 1 | 0.16 | 2 | 0.22 | 3 | 0.092 |
We have previously shown that rare CNVs that increase risk of schizophrenia are effectively confined to those that also influence other neurodevelopmental disorders17. Defining neurodevelopmental disorder genes as those (N=159) that are significantly enriched for rare coding variants in recent large studies of ASD9 and developmental disorders11, neurodevelopmental disorder genes were significantly enriched for LoF DNVs in the combined trio data (Fig 1; rate ratio (95% CI) = 3.3 (2.0, 5.17); p = 8.2 × 10-6; Supplementary Table S2), and this enrichment was significantly greater than for LoF intolerant genes (rate ratio (95% CI) = 2.37 (1.41, 3.8); p = 8.8 × 10-4). In the full sample of trios, we observed no enrichment of missense-damaging DNVs for sub-genic regions that have been identified as being depleted for missense variation25 (rate ratio (95% CI) = 1.004 (0.85,1.18); p = 0.9). The rate of missense-damaging DNVs in neurodevelopmental disorder genes was elevated compared with the background rate (rate ratio (95% CI) = 1.53 (0.79, 2.7)), but this is not significant (p = 0.16), possibly reflecting the small number of DNVs in neurodevelopmental disorder genes (n=13).
Exploiting the strong enrichment among neurodevelopmental disorder genes for DNVs in schizophrenia, we undertook focused analysis of genes in this set, with the aim of identifying high probability schizophrenia risk genes. As highlighted in the study of ASD9, association to some neurodevelopmental disorder genes is driven by LoF variants alone, a combination of LoF variants and missense variants, and in some cases, primarily by missense variants. Therefore, we considered all those classes of mutation in our analysis. All LoF/missense-damaging DNVs observed in neurodevelopmental disorder genes and, where available, phenotypes observed in these carriers are presented in Supplementary Table S4.
SLC6A1 was significantly associated with missense-damaging DNVs in our new trio data after correcting for three classes of mutation (LoF, missense-damaging and LoF plus missense-damaging) and 159 neurodevelopmental disorder genes (2 damaging-missense DNVs; p = 7.46 × 10-5; pcorrected = 0.036). This finding was supported in our analysis of all trio data, where we observed one additional missense-damaging DNV (Table 2; 3 missense-damaging DNVs; p = 5.2 × 10-5, pcorrected = 0.025). It is striking that in the study of ASD9, association to SLC6A1 was also driven by missense variants (n=8) rather than LoF variants (n=1). Following the rationale outlined by the Deciphering Developmental Disorders Study26, we undertook a combined analysis of schizophrenia and ASD DNVs; the evidence for enrichment of missense-damaging DNVs (MPC ≥ 2) in SLC6A1 was more than 3 orders of magnitude stronger than for ASD alone, supporting the hypothesis that missense variants in this gene contribute to both disorders (combined p = 1.6 × 10-14; ASD alone p = 8.0 × 10-11).
Table 2.
Neurodevelopmental disorder genes with at least 1 LoF or missense-damaging de novo variant observed in schizophrenia. Enrichment P values were generated using a one-sided Poisson test from the analysis of all schizophrenia trios (n = 3,444). LoF = loss-of-function; DNV = de novo variant; Missdam = missense-damaging (MPC score ≥ 2). * indicate p values which survive correction for 159 neurodevelopmental disorder genes and three mutation classes (LoF, missense-damaging and LoF plus missense-damaging).
| Gene | Observed DNVs | P (uncorrected) | ||||
|---|---|---|---|---|---|---|
| Missdam | LoF | Missdam + LoF | Missdam | LoF | Missdam + LoF | |
| SLC6A1 | 3 | 0 | 3 | 5.20E-05* | 1 | 7.90E-05* |
| SCN2A | 1 | 2 | 3 | 0.15 | 0.0024 | 0.0019 |
| SMARCC2 | 0 | 2 | 2 | 1 | 0.00068 | 0.0019 |
| PUF60 | 1 | 1 | 2 | 0.056 | 0.022 | 0.003 |
| MED13L | 1 | 1 | 2 | 0.048 | 0.064 | 0.0062 |
| DEAF1 | 0 | 1 | 1 | 1 | 0.0082 | 0.011 |
| TRIO | 0 | 2 | 2 | 1 | 0.0016 | 0.014 |
| CHD8 | 0 | 2 | 2 | 1 | 0.0013 | 0.023 |
| CHD4 | 1 | 1 | 2 | 0.2 | 0.04 | 0.03 |
| KMT2C | 0 | 2 | 2 | 1 | 0.012 | 0.04 |
| PTEN | 1 | 0 | 1 | 0.029 | 1 | 0.044 |
| GNAO1 | 1 | 0 | 1 | 0.042 | 1 | 0.052 |
| TEK | 0 | 1 | 1 | 1 | 0.025 | 0.057 |
| AUTS2 | 0 | 1 | 1 | 1 | 0.03 | 0.057 |
| CSNK2A1 | 1 | 0 | 1 | 0.052 | 1 | 0.064 |
| POGZ | 0 | 1 | 1 | 1 | 0.048 | 0.066 |
| NACC1 | 1 | 0 | 1 | 0.08 | 1 | 0.085 |
| KDM5B | 0 | 1 | 1 | 1 | 0.084 | 0.092 |
| TLK2 | 1 | 0 | 1 | 0.075 | 1 | 0.1 |
| KDM6B | 0 | 1 | 1 | 1 | 0.025 | 0.13 |
| GRIN2B | 1 | 0 | 1 | 0.15 | 1 | 0.17 |
| SYNGAP1 | 0 | 1 | 1 | 1 | 0.026 | 0.21 |
Polygenic transmission disequilibrium tests
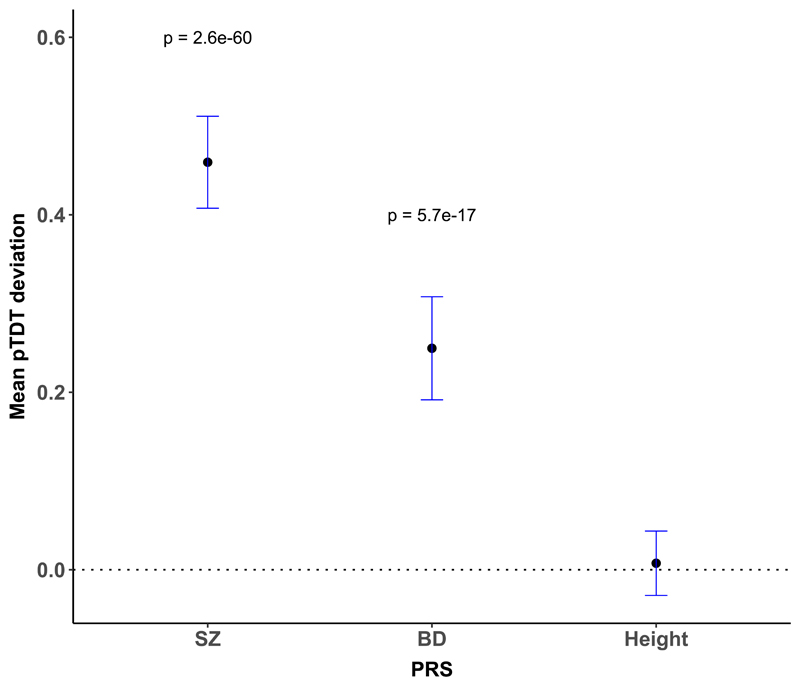
Schizophrenia and BD PRS were significantly over-transmitted from parents to probands (Fig 2, Supplementary Material Table S5). These results did not differ when the analysis was restricted to trios with European ancestry (as defined by principal component analysis; Supplementary Material Table S5).
Figure 2.
Mean pTDT deviation and 95% confidence intervals for schizophrenia, bipolar disorder (BD), and height polygenic risk scores. One-sided one-sample t tests were used to evaluate polygenic over-transmission in 1,122 schizophrenia proband-parent trios. Polygenic risk for schizophrenia and bipolar disorder is significantly over-transmitted to schizophrenia probands. PRS = polygenic risk score. pTDT = polygenic transmission disequilibrium test.
Under a liability threshold model, probands carrying DNVs of large effect size should require less transmission of polygenic risk than probands without such a variant. To test this, the mean pTDT was compared between carriers of candidate schizophrenia related DNVs and the remainder of the sample. We define candidate schizophrenia related DNVs as LoF DNVs in a LoF intolerant gene or a neurodevelopmental disorder gene. Given CNV deletions disrupting LoF intolerant genes are associated with schizophrenia7, we also included de novo CNV deletions disrupting one of these genes as candidate schizophrenia related DNVs (CNVs contributing to this analysis are presented in Supplementary Table S6. CNV calling procedure is outlined in the Supplementary Material).
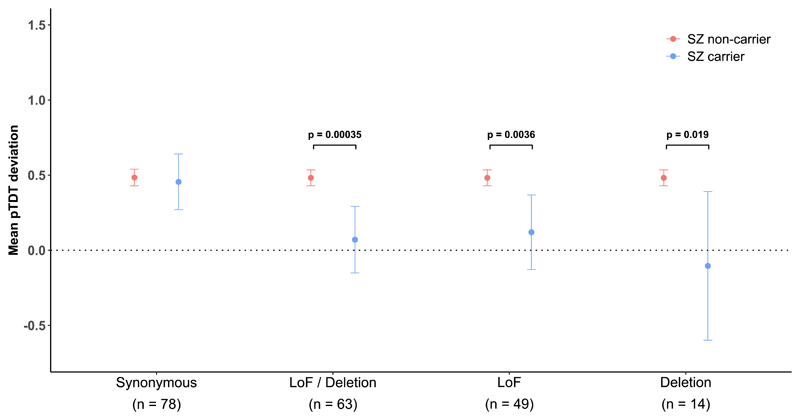
Probands carrying candidate schizophrenia related DNVs had a significantly lower mean pTDT than those who did not carry one of these DNVs (carrier mean pTDT (95% CI) = 0.07 (-0.15, 0.29); non-carrier mean pTDT (95% CI) = 0.48 (0.43, 0.54); p = 3.5 × 10-4; Fig 3). Based on mean pTDT point estimates, the over-transmission of common risk alleles from parents is about 7-fold greater to non-carriers than carriers of candidate schizophrenia related DNVs, although this estimate is imprecise given the width of the confidence intervals (Fig 3). Similar patterns were observed when LoF and deletion DNVs were tested separately (Fig 3). In a negative control test, the mean pTDT did not significantly differ between probands carrying a synonymous DNV in either a LoF intolerant or neurodevelopmental disorder gene and non-carriers (Fig 3).
Figure 3.
Mean pTDT deviation and 95% confidence intervals for schizophrenia PRS. Results are shown for probands carrying various classes of de novo variant (DNV) in a LoF intolerant gene or a neurodevelopmental disorder gene; our primary analysis defined schizophrenia carriers as probands with a LoF or deletion DNV in a LoF intolerant gene or a neurodevelopmental disorder gene (LoF/deletion label). Results are also shown separately for carriers of LoF and deletion DNVs. A one-sided two-sample t test was used to compare mean pTDT deviation scores across groups of trios (total n trios = 1,122). LoF = loss-of-function.
The finding that the mean pTDT deviation for schizophrenia PRS was significantly different between probands carrying candidate schizophrenia related DNVs and non-carrying probands was consistent across schizophrenia PRS training p-value thresholds (Supplementary Table S7). Although the pTDT method is expected to be robust to population stratification, the efficacy of PRS as a measure of relative liability varies with the extent to which the ancestry of the sample from which risk alleles are derived (the source GWAS) matches the ancestry of those being tested (in our case the trios). Given the source GWAS is primarily of European ancestry, we tested, and confirmed, that our findings held when we restrict our analysis to trios with European ancestry (Supplementary Figure S5 and S6) despite the smaller sample size (all results for European-only trios are presented in Supplementary Tables S7 and S8).
The mean pTDT in carriers of candidate schizophrenia related DNVs was not significantly greater than the null (Fig 3). Based on the pTDT standard deviation observed for schizophrenia PRS in all trios (0.89), we only had 80% power to detect a significant (alpha = 0.05) mean pTDT of 0.4 in the 63 carriers of candidate schizophrenia related DNVs. Thus, while we can be confident that the over-transmission to candidate DNV carriers is less than to non-carriers, power limitations mean we cannot conclude that candidate DNV carriers have no contribution from common alleles.
During the review process, we performed an exploratory analysis to evaluate whether pTDT was lower in carriers of additional classes of DNV. Despite testing a wide-range of alternative variant filters (e.g. excluding DNVs observed in ExAC/gnomAD), missense annotations (e.g. MPC scores and constrained coding regions), and CNVs intersecting only LoF tolerant genes, no set of DNV carriers had a significantly greater reduction in pTDT than that observed for our primary candidate schizophrenia-related set of DNVs defined above (see Supplementary Material Table S8 for all results).
Discussion
Proband-parent trio studies have identified large numbers of genes associated with DNVs in ASD and developmental disorders9,11. Although similar studies in schizophrenia have revealed general pathophysiological insights into the disorder, such as a role for proteins involved in postsynaptic signaling complexes15,27, schizophrenia gene discovery through DNV analysis has been hindered by small samples. To add to efforts to overcome this limitation, we performed exome-sequencing of a new sample of 613 schizophrenia trios. We confirmed previous work showing schizophrenia LoF DNVs are significantly enriched among a set of 3,471 genes intolerant to this class of mutation, and identified a stronger enrichment of DNVs in a smaller set of 159 genes that are associated with rare coding variants in neurodevelopmental disorders. GWAS data suggest common risk alleles are under negative selection28 and enriched in highly conserved genes5, but are nevertheless maintained by population mechanisms related to background selection and genetic drift5,29. The findings from rare mutations, both CNVs7,30 and rare coding variants7,31, also support a role for deleterious point mutations in schizophrenia that are under more intense negative selection than alleles of weak effect. Despite this, the population burden of schizophrenia risk alleles seems to be maintained by mutation-selection balance; for CNVs, strong selection is balanced by their relatively high mutation rates30, while for exonic mutations, in the face of a low per base mutation rate, balance is likely maintained by the large size of the mutational target.
In our analysis of all schizophrenia trios, no novel gene was unequivocally associated with DNVs after correction for all genes tested. Despite conducting the largest analysis of DNVs in schizophrenia to date, it is clear that even larger samples are required to identify specific risk genes with genome-wide levels of significance. However, taking an approach based on the wealth of data showing that rare CNVs that increase risk of schizophrenia are effectively confined to those that also influence other neurodevelopmental disorders17, and exploiting the observation here for strong enrichment for DNVs in known neurodevelopmental disorder genes, we find evidence for association between SLC6A1, which encodes a sodium-dependent γ-aminobutyric acid (GABA) transporter (also known as GAT1), and missense-damaging DNVs. SLC6A1 is involved in reuptake of the inhibitory neurotransmitter GABA from the synaptic cleft; our finding therefore adds to the evidence for perturbation of GABAergic neuronal signaling in genetic risk for schizophrenia32. Congruent with our findings, the largest study of rare coding variants in ASD found SLC6A1 to be the most significant (of only four) genes where association signal was driven by missense-damaging variants (8 missense and 1 LoF DNVs)9. In myoclonic atonic epilepsy and developmental disorders, LoF variants account for 54% and 30% of the observed nonsynonymous DNVs, respectively9. Given the strong convergent evidence for this gene, and specifically for a role for missense mutations, from other neurodevelopmental disorders, SLC6A1 is highly likely also to be involved in schizophrenia. This conclusion is further supported by the result of the DNV missense meta-analysis of ASD and schizophrenia, in which the combined evidence for association is more than 3 orders of magnitude stronger than the (already strong) evidence for association to ASD alone, and surpasses genome-wide significance by 8 orders of magnitude. Given the small number of DNVs in SLC6A1, it will be important to extend our finding in other samples, and clearly, a larger number of DNVs will be required to establish that risk is conferred largely by missense rather than LoF mutations.
The role of polygenic risk in schizophrenia has been widely studied using large case-control samples. However, to our knowledge, this is the first study to investigate polygenic risk in schizophrenia using the pTDT method. The pTDT method has several advantages over case-control PRS studies as it is not confounded by ancestry or ascertainment bias or the possibility of effects arising from super-healthy controls in discovery GWAS and subsequent PRS test samples23. Our results provide strong refutation that such effects might explain the PRS effects that have been widely publicised in the literature, including that of overlap in risk between schizophrenia and BD.
More importantly in the present context, our finding that carriers of LoF DNVs in genes defined by LoF intolerance, or in a known neurodevelopmental disorder gene, have significantly lower distortion of transmission of polygenic liability from the mean parental PRS than do non-carriers provides orthogonal evidence that a substantial proportion of this class of de novo variant contribute to schizophrenia pathogenesis. This is an important observation given the possibility that previously documented gene set enrichments in cases of these variants could have been driven by errors in the calibration of the expected mutation rate, or technical issues arising from comparing cases and controls (or case and control trios) often derived opportunistically from different studies.
Our limited sample size does not allow accurate estimation of the magnitude of difference in the transmission distortion between probands carrying candidate schizophrenia related DNVs and those that do not, but the point estimate is that the distortion in non-carriers is about 7-fold of that of carriers (and almost 10-fold when restricted to those of European ancestry). This suggests that on average, the candidate DNVs contribute a substantial amount of liability in those who carry them. Indeed in the present study, carriers of candidate schizophrenia related DNVs did not significantly over-inherit a common allele burden from their parents, which is consistent with DNVs in LoF intolerant genes acting as monogenic risk factors in those who carry them. However, it is important to stress that the latter finding is also consistent with limited power (as discussed in the results) rather than no role for common variation in the carriers, and we note the point estimate for the pTDT in candidate DNV carriers is greater than 0. It will be important for future larger studies to determine whether differences in co-action between common and rare risk alleles exist between schizophrenia and neurodevelopmental disorders. Meanwhile, with respect to the genetic architecture of schizophrenia, together with previous findings from CNVs alone20,21, we interpret our data as being consistent with a polygenic liability threshold model of schizophrenia33.
In conclusion, we provide further evidence that certain classes of DNV are associated with increased risk for schizophrenia. We highlight strong evidence that mutations in SLC6A1, a known ASD, developmental disorders and epilepsy gene, confer high risk of schizophrenia. Through combining exome-sequencing and GWAS data, we show that carriers of candidate schizophrenia related DNVs inherit significantly fewer common risk alleles than non-carrying cases, providing strong orthogonal evidence that these DNVs contribute to schizophrenia liability.
Methods
Sample overview
674 schizophrenia proband-parent trios, consisting of 2,000 individuals, were exome-sequenced on Illumina HiSeq 4000 platforms. The proband-parent trios were composed of 653 trios, 9 quads (two affected children) and one family with 3 affected children. None of these samples have been previously exome-sequenced. The families were recruited by six independent groups (Supplementary Table S9), and were ascertained from general psychiatric wards or outpatient clinics. Proband-parent trios were ascertained blind to any genome analysis. Randomization of experimental groups was not applicable to this study (see Life Sciences Reporting Summary for more detail). All probands had received a DSM-IV or ICD-10 diagnosis of schizophrenia or schizoaffective disorder. Individuals with a known diagnosis of intellectual disability or other neurodevelopmental disorder were not included. For probands passing quality control (quality control procedure described below), information on family history of schizophrenia/psychosis was available for 552 trios; 66% of probands were recorded as family history negative. Further details on the recruitment and diagnostic criteria for each cohort are provided in the Sample Description section of the Supplementary Material.
Exome sequence generation
Exome sequence was generated using the Nextera DNA Exome capture kit and HiSeq 3000/4000 PE Cluster Kit and HiSeq 3000/4000 SBS Kit. Raw sequencing reads were processed according to GATK best practice guidelines34,35. Reads were aligned to the human reference genome (GRCh37) using bwa version 0.7.1536. Variants were called using GATK haplotype caller (v3.4) and filtered using the GATK Variant Quality Score Recalibration (VQSR) tool. For all samples passing quality control (criteria outlined below), we generated sequence data for a median of 83% of the exome target at ≥ 10X coverage. We discuss sequencing coverage further in the Supplementary Material. For future users of our new dataset, we provide in Supplementary Table S10 the median proportion each gene is covered at ≥ 10X coverage.
Sample quality control
Trios (n=27) were excluded for low sequencing coverage if less than 70% of the exome target achieved ≥10X coverage in the proband or either parent (Supplementary Figure S1). An additional 27 trios were excluded for excess heterozygosity (heterozygote:homozygote ratio > 1.9) or evidence of cross sample contamination (as measured by the FREEMIX sequence only estimate of contamination37) (Supplementary Figure S2). The last two metrics are highly correlated. Identity-by-descent (IBD) analysis (plink v1.9) to ensure expected proband-parent relationships resulted in exclusion of 3 trios. Four additional trios were excluded as outliers for the number of DNVs (Supplementary Figure S3). Following implementation of all the above sample quality control steps, 613 proband-parent trios were retained for DNV analysis.
Variant quality control
In each of our newly sequenced samples, we excluded genotypes if they did not meet the following criteria: depth ≥ 10X; genotype quality score ≥ 30; allele balance ≤ 0.1 and ≥ 0.9 for homozygous genotypes for the reference and alternative allele, respectively; allele balance between 0.2 and 0.8 for heterozygous genotypes. For samples and variants that passed quality control, we observed no difference in the number of heterozygous variants transmitted or non-transmitted from parents to probands (transmission disequilibrium test p = 0.53), indicating high data quality.
De novo variant calling
Putative DNVs in the new trios were identified as sites that were heterozygous in the proband and homozygous for the reference allele in both parents. All trio members were required to pass genotype quality control described above. We considered as putative DNVs 1) those where there were no reads for the mutant allele in either parent, and the mutant allele was not called in any other sample of the new trios (parent or proband) and 2) those where the mutant allele met all of the following; an allele count ≤3 in all newly sequenced samples, no mutant allele variant reads in either parent, and at least 5 reads of the mutant allele in the proband. Read alignments for all putative DNVs were manually inspected using IGV (http://software.broadinstitute.org/software/igv/) and variants were reassigned as high or low confidence if there was, respectively, no evidence or evidence for, read misalignment.
We used Sanger sequencing to perform a validation experiment, where DNA was available and primers could be designed, on all high confidence LoF DNVs, as well as additional putative DNVs. In total, primers were successfully designed for 205 putative DNVs. We observed high validation rates for high confidence DNVs (95.5%) and low rates (3.4%) for low confidence DNVs (Supplementary Table S11). Following these results, in our new trios we included in the downstream analyses all high confidence DNVs (N = 606 coding DNVs, Supplementary Table S12).
Adding published de novo data
To increase the power of our analysis, we included previously published DNVs from 2,831 schizophrenia trios. When combined with our new trios, this resulted in a sample size of 3,444 schizophrenia trios. No statistical methods were used to pre-determine sample sizes but our trio sample is the largest reported to date and consists of all publicly available data from exome-sequencing studies of de novo variants in schizophrenia15,16. We note that no DNV from our new trios was also observed among the previously published schizophrenia de novo data, thus confirming the independence of our new trio dataset. A summary of the published data can be found in Supplementary Table S13.
Statistics
De novo variant analysis
We tested whether DNVs were enriched in single genes or sets of genes using the statistical framework described in Samocha et al 201438. Here, for a given set of genes we estimated the number of DNVs expected in our new sample using per-gene mutation rates39, adjusted for sequence coverage. When estimating the number of expected DNVs in previously published trios, we did not adjust per-gene mutation rates for coverage as coverage metrics were not available for all samples; the use of unadjusted per-gene mutation would over-estimate the expected number of DNVs in these trios, producing more conservative enrichment results. For our gene-set analysis, we define LoF intolerant genes as genes with a pLi score ≥ 0.9, using pLi metrics generated from the non-psychiatric component of ExAC31 (available from http://exac.broadinstitute.org/downloads). For single genes, we tested whether the overall burden of DNVs was significantly greater than that expected using a one-sided Poisson test (implemented in R). For our primary de novo gene set analysis, we controlled for background de novo rates by using a two-sample Poisson rate ratio test, which compared the DNV enrichment observed for genes in the set to that in genes outside the set.
DNVs from both the new trios and previously published de novo data were annotated using Ensemble Variant Effect Predictor (version 96)40. We define LoF variants as stop-gain, splice-acceptor, splice-donor and frameshift mutations. Although we observed a small number of start-loss and stop-loss DNVs, we did not include them in our LoF annotation as mutation rates are not available for these variants. We classify missense-damaging variants as missense variants with an MPC score ≥ 2, as this metric has proven effective at identifying variants associated with ASD9,25. Missense-damaging mutation rates for individual genes were calculated by summing tri-nucleotide mutation probabilities for all sites with an MPC score ≥ 2. Following previous work by us and others15,16, if an individual carried multiple de novo variants in the same gene, we conservatively considered these to be the result of a single mutation event, and retained for analysis only the variant predicted to be most deleterious.
Polygenic risk scores
Where available (n=1,122 trios), we used SNP genotype data to generate polygenic risk scores. We confirmed that genotype and exome-sequence data belonged to the same individual through IBD analysis (plink v1.9). A summary of the data sets for which we had both exome-sequencing and SNP genotype data can be found in Supplementary Table S14. To derive PRS for schizophrenia, bipolar disorder (BD) and height, we used the largest available GWAS summary statistics that were independent from our trio test data. Given some samples overlapped between our Bulgarian trios and PGC2, we computed schizophrenia PRS in the Bulgarian trios using custom PGC2 GWAS summary statistics that omitted the Bulgarian samples. We used BD PRS as previous studies have shown that common variant liability to schizophrenia and BD is substantially shared41. Height PRS was used as a negative control. A summary of the training data used to generate PRS can be found in Supplementary Table S14.
For quality control purposes, SNP genotype data were first harmonised to the Haplotype Reference Consortium panel using the Genotype Harmonizer package42 and then subjected to standard quality control, which included exclusion of samples with a call rate < 95%, SNPs with a MAF < 0.1, SNPs with > 1% missingness, or SNPs with a Hardy-Weinberg equilibrium exact test p value < 1 × 10-6. PRS were generated using PRSice 2 software43, where SNPs were clumped based on a window of 250 kb and a maximum r2 of 0.2. We generated PRS across a range of training data P-value thresholds (P < 0.5, 0.1, 0.05, 0.001).
pTDT deviation
To test for a significant over-transmission of polygenic risk, we used the polygenic transmission disequilibrium test (pTDT) as described in Weiner et al (2017)23. Here, pTDT deviation scores were generated for each trio by subtracting the mean-parental PRS from the child PRS (Equation 1). pTDT deviation scores were standardised by dividing them by the cohort-specific mean-parental PRS standard deviation.
| Equation 1 |
We tested whether the mean pTDT deviation was significantly greater than 0, representing an over-transmission of polygenic risk, by using a one-sided one-sample t test. A one-sided two-sample t test was used to compare mean pTDT deviation scores across groups of trios.
The primary pTDT results were produced using PRS generated with a P-threshold of 0.05, as this threshold explained the most case-control variance in the 2014 schizophrenia PGC analysis4. However, we also present in the Supplementary Material Table S5 pTDT results obtained for PRS generated across different P-value thresholds.
Supplementary Material
Acknowledgments
The work at Cardiff University was supported by Medical Research Council Centre Grant No. MR/L010305/1 (to MJO) and Program Grant No. G0800509 (to MJO, MCOD, JTRW, VE-P, PH, AJP), European Community Seventh Framework Programme Grant No. HEALTH-F2-2010-241909 (Project EU-GEI to MCOD), and European Union Seventh Framework Programme for research, technological development, and demonstration Grant No. 279227 (CRESTAR Consortium, to MCOD, JTRW). We acknowledge L Bates and L Hopkins, at Cardiff University, for laboratory sample management. We acknowledge M Einon, at Cardiff University, for support with the use and setup of computational infrastructures.
Footnotes
Data availability
De novo variants discovered from the new trios are published in Supplementary Table S12. The data that support the findings of this study are available from the corresponding author upon request.
Code availability
A description of the R functions used for statistical analysis can be found in the Life Sciences Reporting Summary.
Author contributions
MCOD, MJO, JTRW, PH and ER conceived and designed the research. ER analysed the data. JH, JM and NC performed and managed the sequencing experiments. JH and MD performed the Sanger sequencing validation experiment. VEP, AJP, LH, SEL, AFP and ALR contributed to the interpretation of the results. TL, NK, VK, VG, MP, JGP, CA, GI, MG, JR, GROUP Investigators, GK, JTRW, MCOD and MJO led the acquisition of the clinical samples. ER, MCOD, MJO wrote the manuscript, which was read, edited and approved by all authors.
Competing interests
MOCD, MJO, PH, JTRW, and AJP are supported by a collaborative research grant from Takeda. Takeda played no part in the conception, design, implementation, funding or interpretation of this study. All other authors declare no competing interests.
Genetic Risk and Outcome of Psychosis (GROUP) Investigators are:
Behrooz Z. Alizadeh6,7, Therese van Amelsvoort8, Richard Bruggeman6,9, Wiepke Cahn10,11, Lieuwe de Haan12,13, Jurjen J. Luykx10,14, Bart P.F. Rutten8, Jim van Os10,15, Ruud van Winkel8,16
1. University of Groningen, University Medical Center Groningen, University Center for Psychiatry, Rob Giel Research center, Groningen, The Netherlands;
2. University Medical Center Groningen, Department of Epidemiology, Groningen, The Netherlands
3. Maastricht University Medical Center, Department of Psychiatry and Neuropsychology, School for Mental Health and Neuroscience, Maastricht, The Netherlands;
4. University of Groningen, Department of Clinical and Developmental Neuropsychology, Groningen, The Netherlands
5. University Medical Center Utrecht, Department of Psychiatry, Brain Centre Rudolf Magnus, Utrecht University, Utrecht, The Netherlands;
6. Altrecht, General Menthal Health Care, Utrecht, The Netherlands
7. Amsterdam UMC, University of Amsterdam, Department of Psychiatry, Amsterdam, The Netherlands;
8. Arkin, Institute for Mental Health, Amsterdam, The Netherlands
9. University Medical Center Utrecht, Department of Translational Neuroscience, Brain Center Rudolf Magnus, Utrecht, The Netherlands
10. King’s College London, King’s Health Partners, Department of Psychosis Studies, Institute of Psychiatry, London, United Kingdom
11. KU Leuven, Department of Neuroscience, Research Group Psychiatry, Leuven, Belgium;
References
- 1.Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ripke S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardiñas AF, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees E, O’Donovan MC, Owen MJ. Genetics of schizophrenia. Curr Opin Behav Sci. 2015;2:8–14. [Google Scholar]
- 7.Singh T, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genovese G, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19 doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterstrom FK, et al. Novel genes for autism implicate both excitatory and inhibitory cell lineages in risk. bioRxiv. 2018:484113. doi: 10.1101/484113. [DOI] [Google Scholar]
- 10.Sanders Stephan J, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosmicki JA, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh T, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg S, et al. Truncating mutations in RBM12 are associated with psychosis. Nat Genet. 2017;49:1251–1254. doi: 10.1038/ng.3894. [DOI] [PubMed] [Google Scholar]
- 15.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howrigan D, et al. Schizophrenia risk conferred by protein-coding de novo mutations. bioRxiv. 2018:495036. doi: 10.1101/495036. [DOI] [Google Scholar]
- 17.Rees E, et al. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry. 2016;73:963–969. doi: 10.1001/jamapsychiatry.2016.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Schizophrenia Consortium et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergen SE, et al. Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. Am J Psychiatry. 2018;176:29–35. doi: 10.1176/appi.ajp.2018.17040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tansey KE, et al. Common alleles contribute to schizophrenia in CNV carriers. Mol Psychiatry. 2015;21:1085–1089. doi: 10.1038/mp.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemi MEK, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiner DJ, et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49:978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karczewski K, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 25.Samocha KE, et al. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017:148353. doi: 10.1101/148353. [DOI] [Google Scholar]
- 26.The Deciphering Developmental Disorders, S. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gazal S, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller MC. Evolutionary Perspectives on Genetic and Environmental Risk Factors for Psychiatric Disorders. Annu Rev Clin Psychol. 2018;14:471–493. doi: 10.1146/annurev-clinpsy-050817-084854. [DOI] [PubMed] [Google Scholar]
- 30.Rees E, Moskvina V, Owen MJ, O'Donovan MC, Kirov G. De novo rates and selection of schizophrenia-associated copy number variants. Biol Psychiatry. 2011;70:1109–1114. doi: 10.1016/j.biopsych.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pocklington Andrew J, et al. Novel Findings from CNVs Implicate Inhibitory and Excitatory Signaling Complexes in Schizophrenia. Neuron. 2015;86:1203–1214. doi: 10.1016/j.neuron.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci U S A. 1967;58 doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7.25.21–27.2515. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anttila V, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360 doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deelen P, et al. Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Research Notes. 2014;7:901. doi: 10.1186/1756-0500-7-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Euesden J, Lewis CM, O'Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.