Abstract
Circadian rhythms exist in nearly all organisms. In mammals, transcriptional and translational feedback loops (TTFLs) are believed to underlie the mechanism of the circadian clock. CK1δ/ε are key kinases that phosphorylate clock components such as PER proteins, determining the pace of the clock. Most previous studies of the biochemical properties of the key kinases CK1ε and CK1δ in vitro have focused upon the properties of the catalytic domains from which the auto-inhibitory C-terminus has been deleted (ΔC); those studies ignored the significance of self-inhibition by auto-phosphorylation. By comparing the properties of the catalytic domain of CK1δ/ε with the full-length kinase that can undergo auto-inhibition, we found recombinant full-length CK1 showed a sequential auto-phosphorylation process that induces conformational changes to affect the overall kinase activity. Furthermore, a direct relationship between the period change and the auto-kinase activity among CK1δ, CK1ε and CK1ε-R178C was observed. These data implicate the auto-phosphorylation activity of CK1δ & CK1ε kinases in setting the pace of mammalian circadian rhythms, and indicate that the circadian period can be modulated by tuning the auto-phosphorylation rates of CK1δ/ε.
Keywords: Circadian Clock, Casein Kinase, Kinase Activity, Auto-phosphorylation, CRISPR/Cas9
Introduction
Circadian rhythms exist universally in nearly all organisms, including bacteria, fungi, plants, and animals. Although several independent/parallel origins have been proposed for the evolution of circadian clocks [1], the clock components in animals are very similar. Current understanding of the clock mechanisms started from the discovery of the fruit fly Period mutants through chemo-mutagenetic screening [2]. Later the fly Double-time (Dbt) mutants were discovered using ethyl methane sulfonate (EMS) mutagenesis and the DOUBLETIME (DBT) protein was found to be homologous to the human casein kinases CK1δ and CK1ε [3,4]. Moreover, a spontaneous mutation Tau was identified in hamsters that dramatically shortens the period of the circadian behavioral rhythm, and ultimately the Tau mutation was located within the casein kinase 1ε gene [5,6].
The human genome includes over 500 genes that encode different kinases [7]. The CK1 kinase family has seven members, and the csnk1d and csnk1e genes that are intimately implicated in circadian rhythms were cloned by separate groups in the 1990s [8,9]. Casein kinase 1δ (CK1δ, encoded by the csnk1d gene) and casein kinase1ε (CK1ε, encoded by the csnk1e gene) are highly conserved, with 86% identity of their amino acid sequences (98% identity of the amino acid sequences within their catalytic domains [10,11]). Both kinases contain C-terminal domains (CTDs) in addition to their catalytic domains, and these CTDs have been shown to rapidly inhibit the CK1 kinase activity on substrate proteins after the CK1 is auto-phosphorylated by the catalytic domain (inhibition within 5–10 min at 37°C [12–14]). CK1ε is practically inactive when its CTD is completely auto-phosphorylated, but its activity can be restored in vitro by treatment with protein phosphatases [13].
Because both fly Dbt and hamster Tau mutants significantly affect the period of circadian clocks [3–6], CK1ε has been thought to be implicated in the circadian clockwork. In fact, both CK1δ and CK1ε phosphorylate substrates of the key mammalian clock protein family PERIOD (PER1, PER2, and PER3). These phosphorylation events regulate PER degradation and nuclear accumulation [15,16], which are considered to be a key step for determining the pace (aka period) of the circadian clock. In addition to the Tau mutant (CK1ε-R178C) with a period ~4 hours shorter than wild-type (WT) animals [5,6], other mammalian natural or transgenic mutations in the csnk1d or csnk1e genes have been reported. For example, animals bearing the T44A mutation in CK1δ have a short period [17], and humans carrying the CK1ε-S408N allele appear to display “delayed sleep phase syndrome” (DSPS) [18], which might be due to a long circadian period. Over-expression of a dominant-negative mutant form of CK1ε in mouse embryonic fibroblasts (MEFs) caused arrhythmicity [19]. Thus, both kinases contribute to setting the speed of the clock.
But which kinase (CK1δ vs. CK1ε) is the primary determinant of circadian period? Genetic-deletion and pharmacological studies have addressed which kinase principally regulates the clock speed. CK1δ-null MEFs in culture exhibited robust rhythmicity with an approximately 2 hours longer period, while the period of MEFs from CK1ε-null mice were similar to those from WT mice [20]. Circadian behavior in animals shows that CK1ε-null mice exhibited a small but significant period lengthening with the locomotor activity [21], but a comparable experiment with CK1δ-null animals is impossible because the CK1δ-null is an embryonic lethal. Recently, very specific pharmacological inhibitors towards CK1δ (PF670462, IC50 =13nM) vs.CK1ε (PF4800567, IC50 = 32 nM) have been developed [22]. In the presence of these inhibitors, PF4800567 had only a little effect on the period, but PF670462 robustly lengthened the period [22], supporting the hypothesis that CK1δ is the principal kinase that governs circadian period in mammals. On the other hand, the tau mutation of CK1ε (R178C) can dramatically shorten the circadian period of not only mutant hamsters, but also of genetically tau-site introduced mutant mice [5, 23]. Despite these clear indications that the CK1δ and CK1ε kinases play key roles in determining circadian period in mammals, at a biochemical level their action is unclear. For example, some studies have claimed that tau (R178C) in CK1ε is a gain-of-function mutation (i.e., it enhances catalytic activity [24]), while other studies claim that the R178C substitution reduces the kinase activity [6]. It thus remains unclear how CK1ε regulates the pace of circadian rhythms, and the same confusion exists for CK1δ as well.
We report here that the auto-inhibitory phosphorylation activity (rather than their catalytic activities on other substrates) of the CK1δ & CK1ε kinases is the key towards understanding their role in setting the pace of mammalian circadian rhythms. In reaching this conclusion, we apply a combination of in vitro biochemical assays with cell culture methods, including CRISPR/Cas9 technology which has recently been successfully developed for studying circadian rhythms [25]. In a circadian reporter cell line, genomic editing of the csnk1d and csnk1e loci using CRISPR/Cas9 recapitulated the phenotypes of previous reports, but the mechanism underlying the observed different period changes between these two knockout cell lines is not understood. Therefore, in order to understand the contributions of CK1δ vs.CK1ε, we studied the properties of recombinant full-length CK1δ-WT, CK1ε-WT, and CK1ε-R178C proteins in vitro. Since the initial observations of the auto-inhibitory property of the CTDs on CK1δ and CK1ε 20 years ago [12–14], there have been practically no biochemical studies of the full-length CK1δ and CK1ε kinases in vitro. We found interesting auto-phosphorylation kinetics among the CK1 proteins that revealed the auto-kinase rates of CK1 to be temperature dependent instead of the expected temperature compensation. In addition to the full-length proteins that include the CTDs, the catalytic domains of each protein (ΔC versions) were purified and their overall kinase activities measured. We found opposite trends between the activities of the catalytic domains (ΔC versions, −CTDs) and the full-length kinases (+CTDs). This result plus the close correlation between period changes and the catalytic rates on clock substrates leads us to the conclusion that the apparent strength of auto-kinase activity of CK1δ/ε may determine the period of mammalian circadian rhythms.
Materials and Methods
Reagents and Cells
Cultured cells were grown in DMEM (Gibco, #11965–092) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C (5% CO2). A mouse NIH3T3 cell line stably transfected with an mBmal1::luc reporter (made for this study, see Fig. 1C) was grown under these same conditions. Unless mentioned, the reagents used in this study were purchased from Sangon Biotech, Shanghai, China.
Figure 1. Csnk1d and Csnk1e knockout cells exhibited long circadian periods.
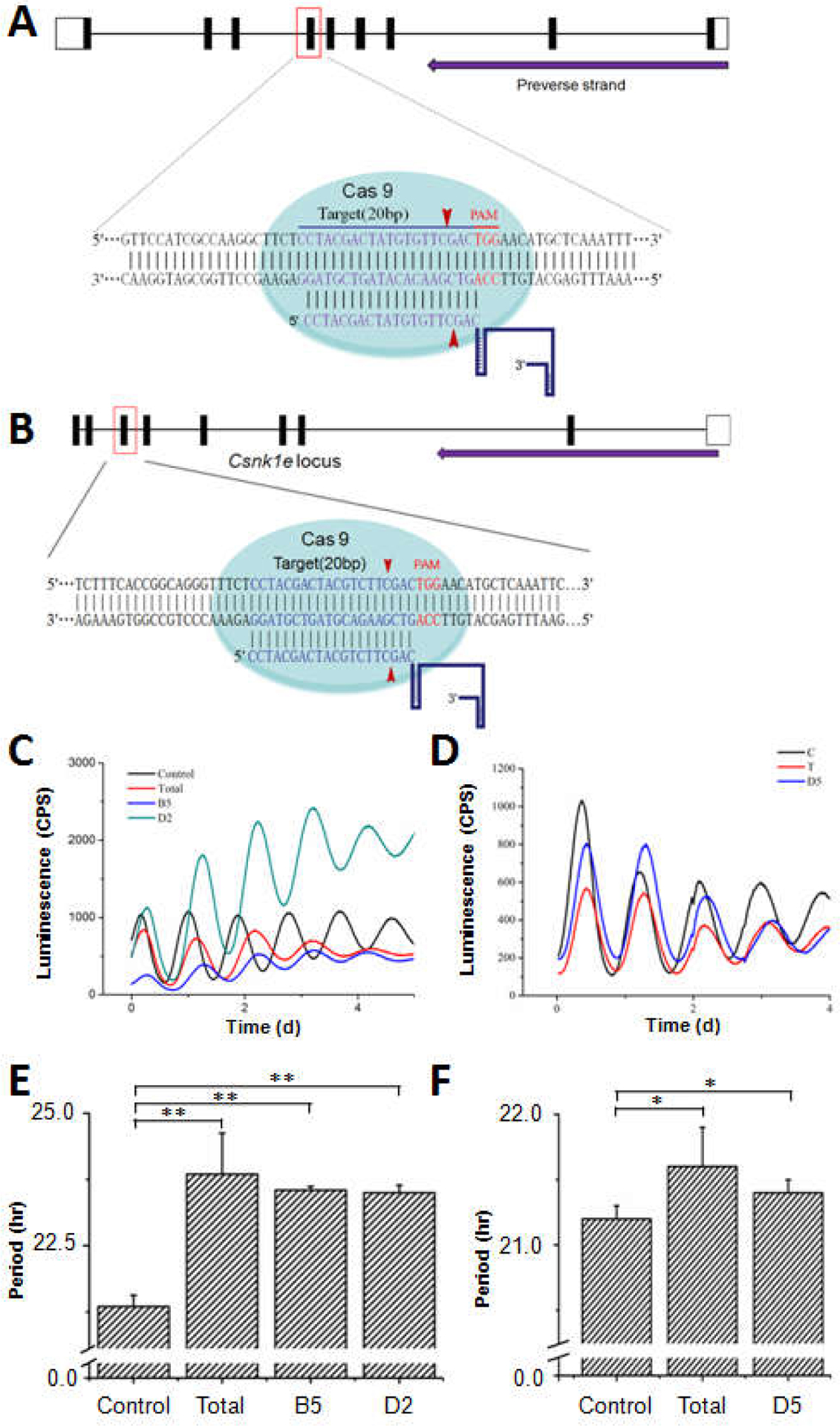
(A) Schematic representation of the CRISPR/Cas9 genome editing on the Csnk1d locus. A single guide RNA sequence (20 nt long) targeting exon 6 of mouse Csnk1d locus was selected. The PAM motif (5’-NGG) and the Cas9 cleavage positions are labeled in red. (B) Schematic representation of the CRISPR/Cas9 genome editing on the Csnk1e locus. For Csnk1e, the gRNA sequence was selected to target exon 7. (C) For Csnk1d knockout, NIH3T3 cells carrying a mBmal1 luciferase reporter were transfected with the pX459-Csnk1d construct. After screening, cell populations (named “Total”) and 2 clonal cell lines (Csnk1d-B5 and Csnk1d-D2) that survived the selection were recorded with a luminescence-monitoring apparatus. “Control” represents non-edited NIH3T3/mBmal1::luc cells.(D) ForCsnk1e knockout, circadian luminescence rhythms were recorded from the cell populations (“Total”) and a clonal Csnk1e-D5 cell line that survived the selection. (E) The circadian periods were calculated for the Csnk1d knockout cell lines depicted in panel C. Error bars are +/− standard deviation (three or more independent experiments). (F) The circadian periods were calculated for the Csnk1e knockout cell line shown in panel D (three or more independent experiments). Statistical analysis used an unpaired t-test. *p < 0.05, **p < 0.01, n≥3.
NIH3T3/mBmal1::luc cell line
The mBmal1promoter reporter pGL3-PmBmal1::luc was constructed by the following procedure. First, the mBmal1 promoter was isolated by PCR amplification from NIH3T3 genomic DNAand inserted to the pGL3-basic vector (#E1751, Promega). Then, a hygromycin resistance gene cassette (hygroR) was inserted, resulting in the reporter construct pGL3-PmBmal1::luc. Cultured NIH3T3 cells were transfected with pGL3-PmBmal1::luc, followed by hygromycin screening (300μg/mL). After a population of hygromycin-resistant cells were successfully obtained, clonal cell lines were screened by the limiting dilution method. Luminescence recording assays were performed with the cell line clones to confirm that the mBmal1 reporter was stably transfected into the NIH3T3 cells.
CRISPR/Cas9 technology to edit Csnk1d and Csnk1e genome loci
Oligonucleotides specific for the target sites of Csnk1d and Csnk1e gene loci were designed using the Optimized CRISPR Design tool (http://crispr.mit.edu/) [26]. The synthesized oligonucleotides were annealed, dephosphorylated and then ligated into the expression vector pX459 (pSpCas9{BB}−2A-Puro was a gift from Feng Zhang, Addgene plasmid # 48139) using the BbsI restriction site. NIH3T3/mBmal1::luc cells were seeded into 6-well plates 1 d before transfection at a density of 2×105 cells per well. Cells were transfected with the vector constructs pX459 targeting either the Csnk1e or the Csnk1d gene locus using Lipo6000 reagent (#C0529, Beyotime, Shanghai, China) according to the manufacturer’s instructions. 24 h after transfection, cells were screened with 1 μg/ml puromycin. Cell line clones were screened by the limiting dilution method.
T7 endonuclease assay
The screened cells were determined for gene knockout using T7 endonuclease I (T7E1) enzyme (#E001, Viewsolid, Beijing, China). A pair of primers was designed to be located about 800 bp upstream and downstream of the genomic sgRNAsite, which was used to PCR-amplify the genomic sequence. 500 ng of PCR product was denatured, reannealed and digested with T7E1, which cleaves mismatched heteroduplex DNA. The reaction was analyzed by 2% agarose gel electrophoresis.
Construction of prokaryotic expression plasmids
To generate the CKIδ/ε expression plasmid, the open reading frame of human CKIε-WT/TAU with the C-terminal truncation at position 314 (Δ314) was cloned into NdeI and Hind III /Not I sites of pET28a plasmid (#69864, Novagen). Constructs to express other proteins, such as hPP1, mPer2, and full-length CK1 kinases including CK1δ and CK1ε were described previously [27].
Protein purification
The mPER2 proteins were purified as described previously [27]. Briefly, mPER2 GST-fusion proteins were expressed in E.coli strain Rosetta2 cells (Novagen, # 71402) and purified with glutathione agarose beads (Pierce, #16100), followed by cleavage of the GST with PreScission Protease (GE healthcare, #27-0843-01). The purification procedures of hP53, hPP1 and full-length CK1 proteins were also described previously [27].
The concentration of each protein was measured with the Bradford reagent (Bio-Rad Protein Assay). The purity was determined on SDS-PAGE.
Auto-phosphorylation and kinase assays
2 μM CK1ε-FL (or CK1ε-R178C-FL) was dephosphorylated by 0.01 μM hPP1 one day before the experiment at 37 °C in a buffer system (50 mM Tris-Cl, 100 mM NaCl, 5mM DTT, 10 mM MgCl2, pH8). Then 1mM ATP was added to initiate the auto-phosphorylation assay. Samples were collected from the reactions at the time points indicated in each figure.
The kinase activity of either hCK1ε-ΔC or hCK1ε-FL was assessed by mixing 50 nM of the purified CK1 enzymes with 2 μM purified hP53 proteins (other substrates: BSA, α-Casein, or mPER2) in Kinase Reaction Buffer (20 mM Tris-Cl, 10 mM MgCl2, 5 mM DTT, pH8.0). To assess the kinase activity of hCK1ε-ΔC, UPLC was used to measure the released ADP as described in the next section. If autoradiography was used to assess the kinase activity of either the catalytic domain (ΔC) or the full-length (−FL) forms, 0.1 μCi/μl of ATP-γ−32P (PerkinElmer, #NEG002A250UC) was included as described previously [27].
Ultra-high performance liquid chromatography (UPLC) to measure adenosine nucleotides
The kinase reactions were quenched with 3 M perchloric acid and then neutralized with 2 M potassium hydroxide and 0.5 M potassium carbonate to a pH of 5.5–6.0. The acidic homogenate was centrifuged briefly and stored at −70 °C overnight to promote precipitation of the perchlorate. Samples were diluted after centrifugation at 12,000 rpm for 25 min (4 °C). A C-18 UPLC column (#3520–39991, 5μm, 4.6 ×150 mm, Daojin, China) was used with these buffers at a rate of 0.5ml/min: (1) Buffer A contained 25 mM KH2PO4, 20 mM Triethylamine, pH 5.6, and (2) Buffer B was acetonitrile. Both buffers were vacuum filtered with a 0.22 μm filter and degassed using ultrasound.
The elution condition was 98% buffer A and 2% buffer B. 10 μl of a prepared sample or of a standards sample was auto-injected and the elution was monitored by UV absorbance (255–265nm). Peaks were identified by their retention times and compared with the standards. The concentrations of ATP and ADP in the preparation of the standard curve were 0.5 μM, 1 μM, 2 μM, 4 μM, 8 μM, and 16 μM in ultrapure water.
Circadian luminescence recording assay
1 × 105 NIH3T3/mBmal1::luc cells were inoculated into 35 mm culture plates (Costar, #9102) in triplicate. On the next day, cells in each well were synchronized with a 2-hr treatment of 100 nM dexamethasone and recorded in a LumiCycle as described previously [28]. Bioluminescence data were analyzed with the LumiCycle analysis program (Actimetrics, USA) to obtain circadian parameters such as period and amplitude.
Limited trypsin digestion assay
0.001 μg/μl trypsin was incubated with 0.1 μg/μl hyper- or hypo- phosphorylated hCK1ε-WT or TAU proteins, and the samples were collected at the indicated time points. Hypo-phosphorylated hCK1ε was prepared by mixing the purified protein with the protein phosphatase hPP1 at 200:1 molar ratio for 10 h at 37 °C. Hyper-phosphorylated hCK1ε was prepared by incubating 5 mM ATP with the hypo-phosphorylated protein for 10 h at 37 °C.
Protein extraction from cells and immunoblotting
Cultured cells were homogenized in RIPA buffer (150 mM NaCl, 5% NP-40, 50 mM Tris pH 8.0, 2mM EDTA, 10% glycerol) supplemented with protease inhibitors (1 mM PMSF and 10μg/ml leupeptin). The extracts were centrifuged to remove cell debris at 12,000 rpm for 10 min at 4 °C. Samples were heated at 95 °C for 5 min in 5X protein loading buffer, resolved on 10% SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20 and incubated with relevant antibodies. Antibodies used were: rabbit α-CK1ε (#D121893, Sangon, Shanghai, China), rabbit α-CK1δ (#D222155, Sangon, Shanghai, China), and cy3-conjugated donkey α-rabbit IgG (#D110052, Sangon, Shanghai, China) as the secondary antibody.
CK1ε structure prediction
We used a protein structure prediction tool, METATASSER, which was developed by Dr. Skolnick’s research group at the Georgia Institute of Technology. It is freely available for use on the website at http://psifr.cssb.biology.gatech.edu/ [29]. The prediction process was carried out online using the instructions on the website.
Results
CRISPR/Cas9 editing of csnk1d and csnk1e gene loci lengthen the period to different extents
Genetic loss-of-functions are informative in understanding the functions of genes and their protein products. We applied CRISPR/Cas9 for genome editing of csnk1d and csnk1e loci in NIH3T3/mBmal1::luc reporter cells. The highest score of a 20-nt guide sequence targeting exon 6 of csnk1d gene was selected (Fig. 1A). The target sites precede a 5’-NGG PAM site (TGG in this case) and the Cas9 nuclease would produce a double strand break at the −3 position (Fig. 1A). Similarly, a guide sequence targeting csnk1e gene was selected for making the pX459-Csnk1e construct (Fig. 1B). Stably transfected cells with pX459-Csnk1d or pX459-Csnk1e constructs were screened in the presence of puromycin. Three single-colony cell lines, Csnk1d-B5, Csnk1d-D2, and Csnk1e-D5 were isolated from cell populations that survived the puromycin selection. These three single-colony lines and the survived cell populations (named as Total) were synchronized with dexamethasone and the luminescence was monitored over several days (Fig. 1C&1D). Both the Total population cells and the two single-colony cell lines showed lengthened periods of ~ 2 hours (Fig. 1E). These phenotypes are similar to MEFs from CK1δ-knockout mice [20], indicating an effective Csnk1d knockout. An equivalent approach applied to the csnk1e gene yielded a single-colony line, Csnk1e-D5, in which the cells exhibited a period ~20–30 min longer than non-edited control cells (Fig. 1F).
Interestingly, two clear bands were demonstrated in our attempts to amplify the genomic region flanking the Cas9 cleavage site of csnk1d. One band was close to the WT control, while the other band was ~ 100 bp larger (Fig.S1A). To analyze if the PCR-amplified sequences contain different indels from the WT sequence, a T7E1 assay was applied to a series of annealing combinations and confirmed indels (Fig. S1B). We could not detect by immunoblotting the CK1δ protein in either of the Csnk1d-B5 or Csnk1d-D2 lines, nor could we detect CK1ε in the Csnk1e-D5 line, confirming the intended knockouts (Figs. S1C&S1D). The two PCR-amplified bands were isolated and purified separately, followed by DNA sequencing. Sequencing data showed an interesting result: one allele lacked 8 nucleotides after the Cas9 cleavage site, while the other allele had a 116-nucleotide insertion after that site (Figs. S1F&S1G.). Csnk1d-B5 and Csnk1d-D2 cells have the same genome editing results, implying that the Csnk1d-B5 and Csnk1d-D2 lines originally derived from one edited cell with these two alleles. Mysteriously, the insertion sequence was from the E.coli genome, after we blasted the sequencing data at NCBI (Fig. S1G). Our analysis of predicted open reading frames indicates that changes of both loci produce frame-shift mutations, resulting in early stop codons in CK1δ and CK1ε. But our immunoblotting results did not reveal possible truncated proteins (Fig. S1E). We PCR-amplified the targeted region from the Csnk1e-D5 genomic DNA, followed by sequencing. The results showed that both alleles in the Csnk1e-D5 cell lines were modified, with one allele losing 285 bp starting from the Cas9 cleavage site and the other allele having 1 nucleotide insertion right at that site (Fig. S1I).
CK1δ/ε auto-phosphorylation kinetics at various temperatures
Full-length recombinant casein kinase proteins (CK1δ/ε-WT) were expressed and purified by affinity chromatography. After purification, both kinases exhibited smeared bands (Fig.S2A), suggesting multiple phosphorylation forms. To confirm that the smeared bands were different phospho-forms of CK1 rather than contaminating proteins, we treated the purified proteins with human protein phosphatase 1 (hPP1) overnight and the slow-mobility bands disappeared (Fig.S2A). Addition of fresh ATP to the de-phosphorylated CK1 initiated the auto-kinase reaction. At 37 °C, CK1ε auto-phosphorylated itself rapidly so that auto-phosphorylation was complete within 30–60 min (Fig. 2A). In order to clearly observe the kinetics of CK1ε auto-phosphorylation, the autokinase assay was carried out over a range of temperatures from 4 °C to37 °C. We observed a temperature dependent change of the auto-phosphorylation kinetics (Fig. 2A). The auto-phosphorylation of purified CK1ε-R178C (the Tau mutant version of CK1ε) was also tested, and its temperature-dependent kinetics demonstrated a slower auto-kinase activity (Fig. 2B) than did the non-mutant native version of CK1ε (WT, Fig. 2A).
Figure 2. Auto-phosphorylation of CK1ε kinases is a sequential, temperature-dependent process.
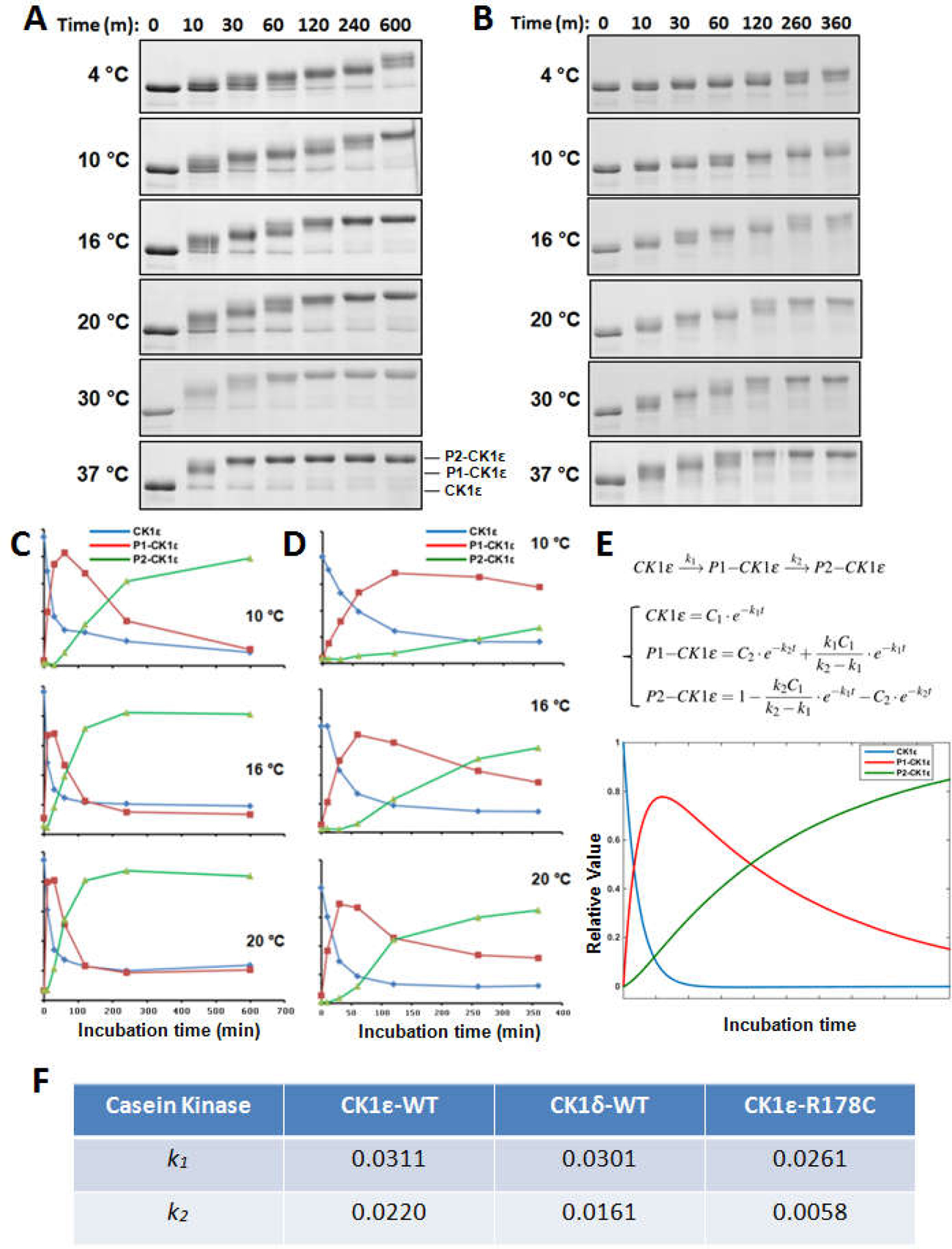
(A) The kinetics of CK1ε-WT auto-phosphorylation under a range of temperatures: 4 °C, 10 °C, 16 °C, 20 °C, 30 °C, and 37 °C. A ordered phosphorylation pattern can be observed as a progressive mobility shift. (B) Slower auto-phosphorylation kinetics was observed for CK1ε-R178C over the same temperature range. (C) The auto-phosphorylation process of CK1ε-WT at10 °C, 16 °C, and 20 °C was quantified as the time dependence of the unphosphorylated form (blue), intermediately phosphorylated P1 form (red), and fully phosphorylated P2 form (green). (D) The auto-phosphorylation processes of CK1ε-R178C at10 °C, 16 °C, and 20 °C were quantified as in panel C. Data in panels A-D are representative of two or more independent experiments. (E) A simplified two-step phosphorylation model for the sequential auto-phosphorylation of CK1ε as described in the text. Each form of CK1ε (1) un-phosphorylated, (2) P1-CK1ε, and (3) P2-CK1ε, wascalculated by solving the differential equations in the model, and then Matlab software was used to simulate the relative amount of each form over time. (F) k1 and k2 estimated for these three CK1s, indicating that CK1δ-WT has an intermediate auto-kinase rate.
Thus, both WT and R178C CK1ε showed progressive auto-phosphorylation (mobility of the proteins got slower during the incubation), indicating sequential auto-phosphorylation of CK1ε. To provide a quantifiable rate for the auto-kinase reaction that can be used to compare among CK1δ, CK1ε, and CK1ε-R178C, we simulated the auto-phosphorylation process with a simple two-step reaction model: CK1ε —> P1-CK1ε —> P2-CK1ε (Fig.2E). CK1ε is phosphorylated with reaction rate k1, and P1-CK1ε is phosphorylated with reaction rate k2. At least 8 in vivo phosphorylation sites have been identified on the CTD domain of CK1ε [30], so the P1 and P2 states are not determined by single phosphorylation events and therefore the model includes multiple phosphorylation sites that define the P1-CK1ε and P2-CK1ε forms. “P1-CK1ε” stands for the first-reached intermediate phosphorylated state, while “P2-CK1ε” represents completely auto-phosphorylated CK1ε. Each form was simulated with MATLAB using differential equations (Fig.2E), and the results of these computational analyses were consistent with the SDS-PAGE data (Figs.2C, 2D, &2E). Modeling of the CK1ε-R178C data at 10 °C vs. 20 °C suggest the phosphorylation reaction rates k1 and k2 are temperature dependent with calculated Q10 values above 2 (Fig. 2D and Fig. S3). Furthermore, the auto-phosphorylation of CK1δ was examined and similar kinetics was observed (Fig. S2). The model was used to compare the auto-kinase rates among the three enzymes and the values show that for both k1 and k2, CK1ε> CK1δ>CK1ε-R178C (Figs. 2F and S2).
Two distinct conformations revealed by computational prediction and trypsin digestion
As mentioned above, both CK1δ and CK1ε contain an N-terminal catalytic domain and a C-terminal domain (CTD) [12,13]. Although crystal structures of the N-terminal catalytic domains of CK1δ and CK1ε have been reported [31,32], the structures of the full-length CK1δ/ε including the C-terminal domains have not been determined. However, a structure-estimation program [29] applied to CK1ε predicted full-length CK1ε proteins exist in two forms. In one form, the CTD domain is unstructured and extended away from the catalytic domain (Fig. 3A), while in the other form, the CTD domain associates closely with the catalytic domain (Fig. 3B). The predicted structures suggest that CK1ε will change its conformation as it is progressively auto-phosphorylated. The catalytic domain of CK1δ/ε is positively charged and favors negatively charged substrates, such as peptides containing negative amino acids D/E or phosphorylated S/T. We therefore postulate that the compact form of CK1ε (Fig. 3B) corresponds with the structure of P2-CK1ε (Fig. 2). To confirm that unphosphorylated versus fully phosphorylated CK1ε have different 3-D conformations, limited trypsin digestion assays were performed. The results showed that unphosphorylated CK1ε degraded faster than the fully phosphorylated (P2-CK1ε) form, as indicated by the dark black arrows in Fig. 3C. Unphosphorylated CK1ε was cleaved within 10 min, while the hyper-phosphorylated form was cleaved within 30 min. These results indicate that the auto-phosphorylation events render P2-CK1ε more resistant to trypsin digestion, providing support that CK1ε undergoes a conformational change to a more compact conformation as the CTD domain gets phosphorylated. This more compact conformation may make the trypsin cleavage sites less available. Moreover, the R178C mutation did not greatly affect the digestion kinetics (Fig. 3D), indicating that even though CK1ε and CK1ε-R178C have different auto-phosphorylation rates (Fig. 2), they have similar conformations in solution at the same phosphorylation status.
Figure 3. Two distinct conformations of CK1ε revealed by computational prediction and limited trypsin digestion.
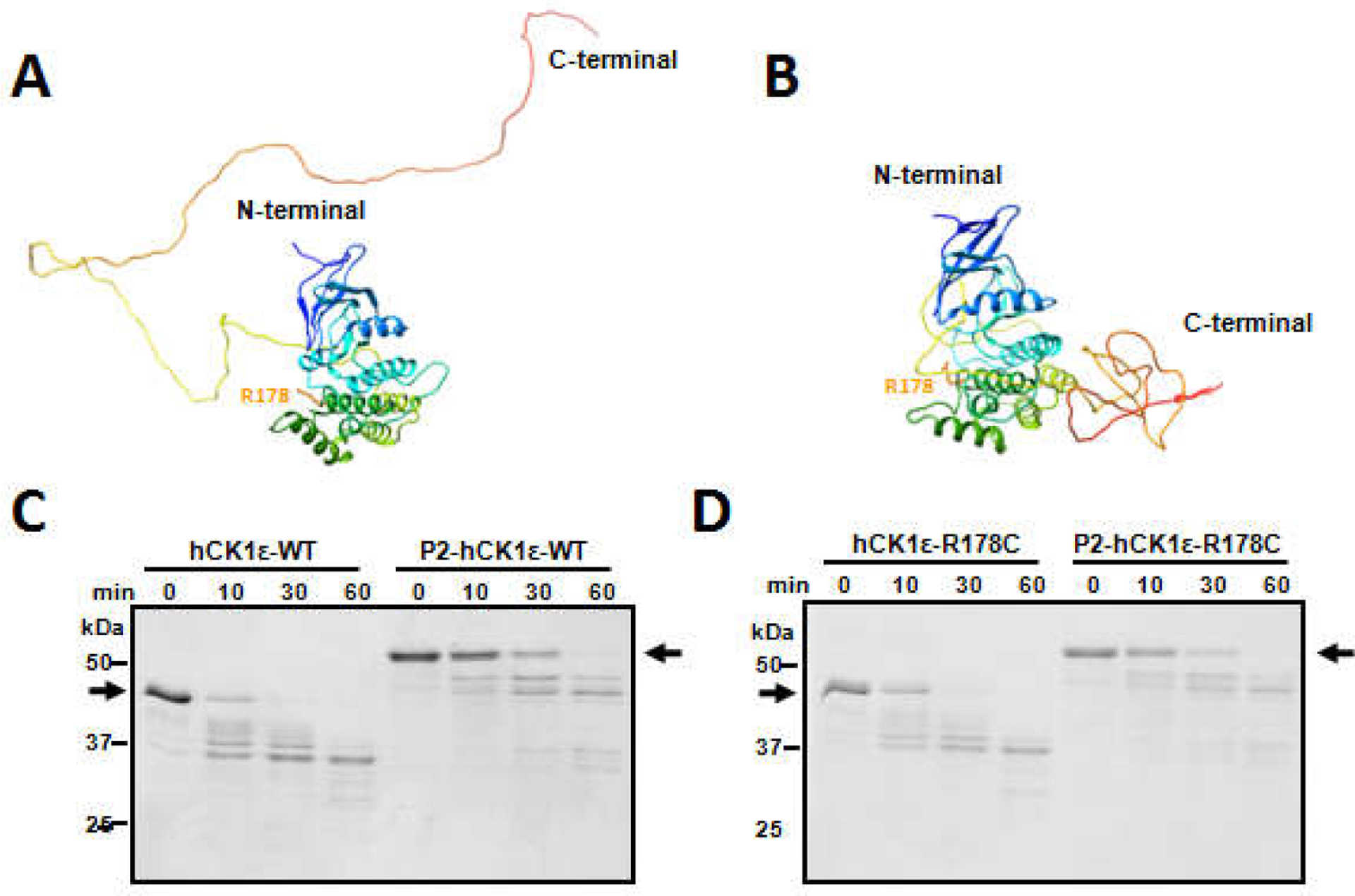
METATASSER was used to predict 3-D structures of CK1ε de novo. Two forms were extracted. In one form, the CTD domain of CK1ε is unstructured and extends away from the N-terminal catalytic domain (A). In the other form, the CTD domain is structured and present immediately adjacent to the catalytic domain (B). Limited trypsin digestion was applied to the differently phosphorylated forms of CK1ε. (C) Non-phosphorylated native CK1ε was digested faster than hyper-phosphorylated native CK1ε. Black arrows indicate the positions of the non-phosphorylatedand hyper-phosphorylatedCK1ε before trypsin digestion. (D) Similar digestion kinetics occur withCK1ε-R178C, indicating that CK1ε-WT and CK1ε-R178C have similar conformations in solution. Data are representativeof two or more independent experiments.
Circadian period changes correlate with the auto-kinase activity of casein kinases
Next, we investigated the kinase activity using a quantitative method to measure ATP conversion to ADP, ultra-high performance liquid chromatography (UPLC). Peaks of ATP and ADP were separated well by their retention times (Fig. S4A). A linear relationship existed between the detected peak areas by UPLC and the amount of ATP or ADP that we used as standards (Fig. S4B). hP53 was used as the physiological substrate for the catalytic domains of the three enzymes lacking the CTD: CK1ε-ΔC, CK1ε-ΔC-R178C, and CK1δ-ΔC. By quantifying the ratio of phosphorylated hP53 protein to total hP53 protein from SDS-PAGE images (Fig. S5A&S5B), we observed that CK1ε-ΔC exhibited the highest phosphorylation rate among these three kinases (Figs. S5B& S5C). Measured ADP production during the reaction processes confirmed that CK1ε-ΔC has the highest rate of ATP hydrolysis (synergistically with catalytic activity), while CK1ε-ΔC-R178C has the lowest activity and CK1δ-ΔC is in-between (Fig. 4A). The kinetics of the ATP hydrolysis (Fig. 4B) were different from the hP53-phosphorylation kinetics calculated from the SDS-PAGE images (Fig. S5C), which might be due (i) to continued phosphate incorporation into proteins at later time points that does not produce further mobility shifts, and/or (ii) to ongoing ATP hydrolysis by the CTD-deleted CK1s that continues after the substrate is fully phosphorylated. Nevertheless, the order of higher-to-lower rates among the three proteins is the same regardless of the method used to measure kinetics.
Figure 4. Lowest auto-phosphorylation rates correlate with the largest circadian period shortening.
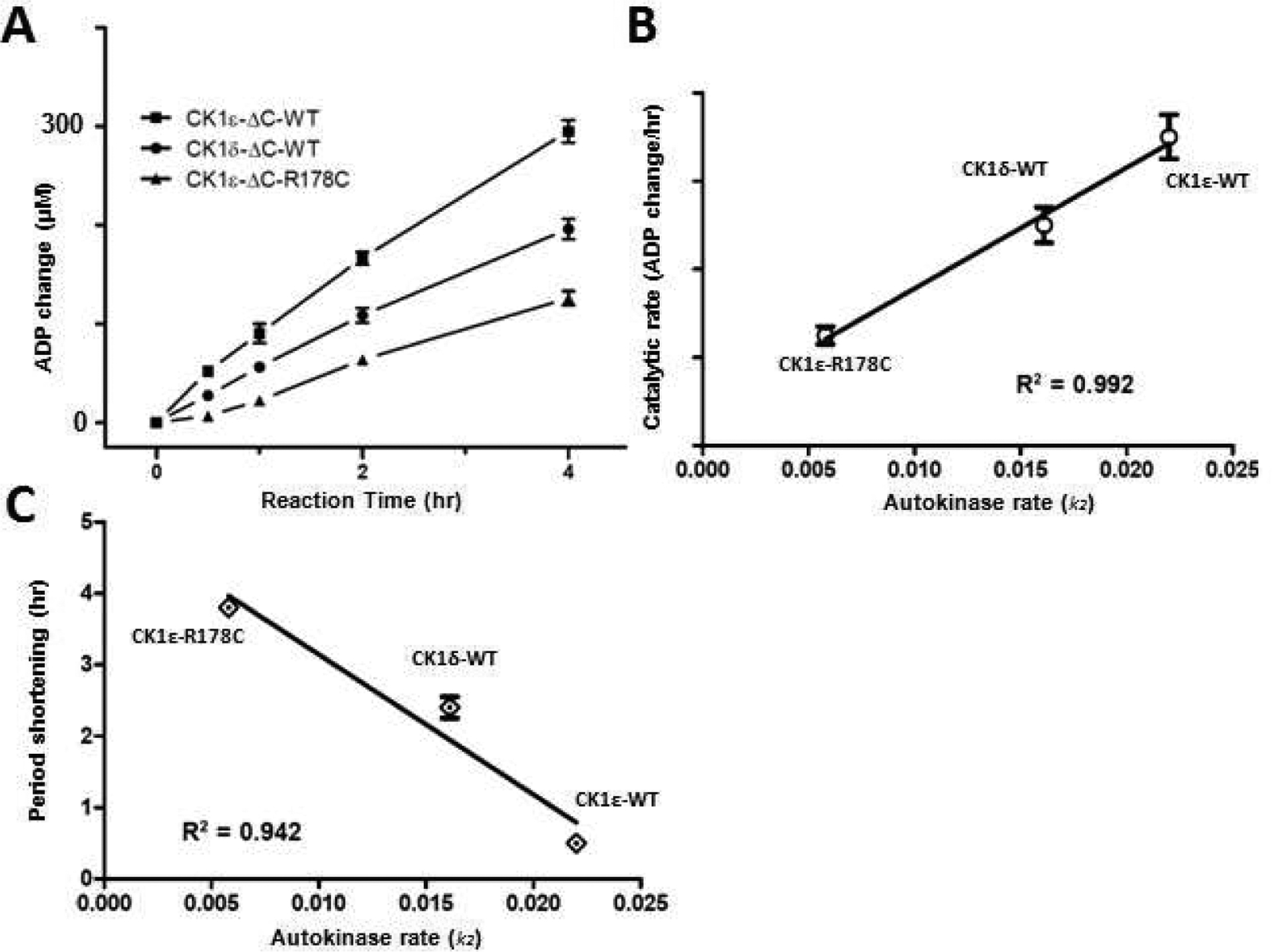
(A) Rate of conversion of ATP to ADP as measured by UPLC for CK1ε-ΔC (squares), CK1ε-ΔC-R178C (triangles), and CK1δ-ΔC(circles). (B) The catalytic rates of ΔC domains of CK1s derived from the UPLC results correlate with the autokinase rates of full-length CK1s that were derived from Figures 2, S2, and S3. (C) Correlation analysis between the circadian period shortening(see Table 1) and the auto-kinase activity of the different CK1s (Figures 2, 4C–D, S2, and S3). Error bars are +/− standard deviation (from three or more independent experiments).
As shown in Table 1, the circadian period was shortened by ~ 4.0 hr when CK1ε−/− mice were compared to mice carrying the CK1εR178C/R178C alleles [21,23]. In our cell-based studies, the circadian period was shortened by ~ 2.2 hr and ~ 0.3 hr when CK1δ and CK1εWT alleles were reinserted into their respective knockout strains. The catalytic rates of different CK1 are equal to their auto-kinase activity since their C-terminal domains are substrates of these kinases. The calculated catalytic rate of ΔC enzymes (UPLC data, Fig. 4A) is highly correlated with the calculated autokinase rate (Fig. 2F) among the three CK1s (Fig. 4B). Combining the data of Table 1 and Fig. 4B, we find an obvious correlation between the autokinase rates of the different CK1 kinases and circadian period (Fig. 4C).
Table 1.
Circadian period shortening when knockouts are replaced with different CK1 alleles.
| Period of KO ctrl (hr) | Resulting Genotype | Period (hr) | Period shortening (hr) | Reference |
|---|---|---|---|---|
| 21.6 (CK1εnull)** | CK1εwt/wt | 21.3 | 0.3 | this study |
| 24.0 (CK1εnull)* | CK1εR178C/R178C | 20.0 | 4.0 | Meng et al. 2008 |
| 23.5 (CKlδnull)** | CK1δwt/wt | 21.3 | 2.2 | this study |
The periods were measured from the locomotor activities of CK1εR178C/R178C and CK1ε−/− animals.
The periods were measured from cell culture experiments (this study)
Opposite kinase activity trends between the catalytic domain and the full-length kinase
Conflicting mechanisms have been proposed for the tau mutant CK1ε-R178C: reduced activity versus gain-of-function [6, 24]. The different sequential phosphorylation kinetics of the C-terminal domain between CK1ε-WT and CK1ε-R178C suggests that their activity rates might alter as they progressively auto-phosphorylate (Figs. 2A & 2B). Since CK1ε-R178C auto-phosphorylates its CTD slower than does native CK1ε (CK1ε-WT), it therefore auto-inhibits itself more slowly. Consequently, we predict that CK1ε-R178C should exhibit enhanced kinase activity on physiologically relevant substrate proteins. To test that prediction, catalytic ΔC domains and full-length versions of CK1ε and CK1ε-R178C were tested with BSA, α-Casein, and hP53 as substrates. Compared to native CK1ε-ΔC, CK1ε-R178C-ΔC showed reduced kinase activity on all three substrates (Figs. 5A–C). Quantification and normalization of these data confirmed that the extent of phosphorylation on these substrates was less for CK1ε-R178C-ΔC than for CK1ε-WT-ΔC (Fig. S6A). When the full-length proteins were used, we observed that the R178C mutation enhances the kinase activity on physiological substrates (Fig.5 and Fig. S6B). In particular, with the physiologically relevant substrate hP53, CK1ε-R178C phosphorylates hP53 more strongly than does native CK1ε (CK1ε-WT) at the same time that CK1ε-R178C auto-phosphorylates itself slower than does CK1ε-WT (Figs. 5F & S6B). (CK1ε-WT is completely hyper-phosphorylated within 30 min in the presence of hP53, while CK1ε-R178C barely reaches the hyper-phosphorylated status at 60 min, Fig. 5F.) In the presence of its substrate, the auto-phosphorylation data of CK1ε depicted in Fig. 5F are consistent with the results shown in Figure 2 when these kinases were incubated by themselves. On the other hand, with the non-physiological substrates BSA and α-Casein, the phosphorylation level catalyzed by CK1ε-R178C was lower than that catalyzed by the CK1ε-WT control (Figs. 5D&5E). Taken together, the R178C mutation in full-length CK1ε enhances kinase activity on its physiological substrate, which supports the hypothesis that the R178C mutation reduces auto-inhibition, thereby enhancing catalytic phosphorylation of physiologically relevant substrates.
Figure 5. In vitro kinase assay comparing CK1ε-ΔC vs. R178C-ΔC and CK1ε-WT vs. CK1ε-R178C.
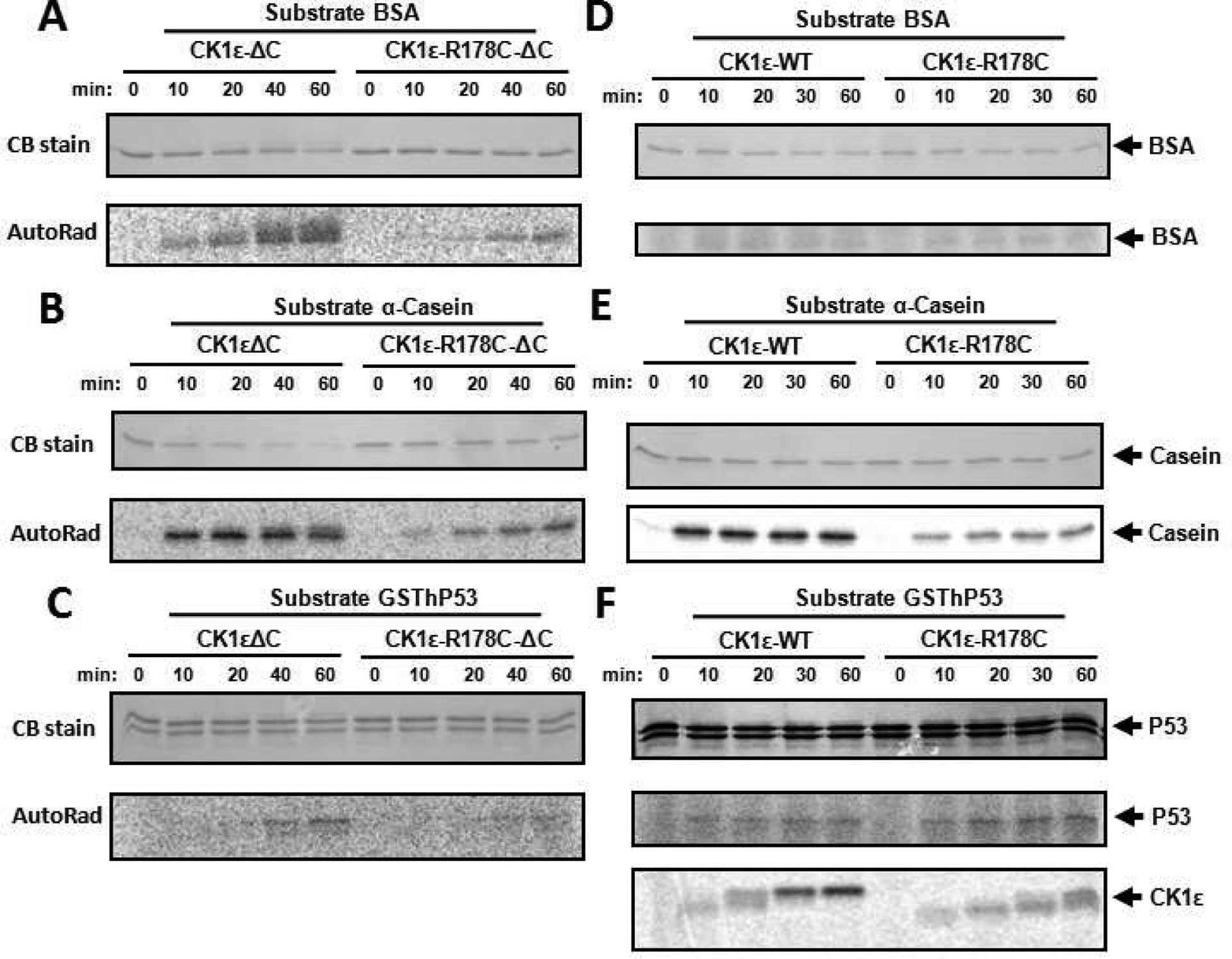
Radioactive in vitro kinase assay was performed to assess the change caused by the R178C mutation of CK1ε on different substrates. Compared to native CK1ε-ΔC, the R178C mutation reduced catalytic activity towards BSA (A), α-Casein (B), and hP53 (C). To compare with the activity of the catalytic domains (ΔC) illustrated in panels A-C, full-length CK1ε and CK1ε-R178C kinases were applied to the same substrates with the in vitro radioactive kinase assay in panels D-E: (D) BSA, (E) α-Casein, and (F) hP53. “CB stain” indicates the Coomassie Blue staining to assess the abundance of the proteins in the SDS-PAGE gels (left side). The phosphorylation of each substrate is indicated by P-32 incorporation (“AutoRad,” left side). Arrows point out each substrate (right side). The bottom of panel F adds an auto-phosphorylation time course of full-length CK1ε and CK1ε-R178C; note that native CK1ε-WT auto-phosphorylates itself faster than does CK1ε-R178C.
Lower temperature accelerates the clock
Since we found that lower rates of the auto-kinase activity (Fig. 2F) and catalytic domains (Fig. 4A) correlate with a higher activity for the full-length kinase on the physiological substrate hP53 (Fig. 5), we sought another way to manipulate the activity of the catalytic center. When we decreased the temperature, a lower kinase activity of the catalytic domain (ΔC) was observed as expected (Fig. S7A–C). We reasoned that for the full-length kinase, lowering the temperature would reduce the auto-phosphorylation of the inhibitory CTD domain, therefore slowing the rate of auto-inhibition and thereby allowing a higher rate of phosphorylation of a physiological substrate. We tested this prediction with full-length CK1ε. When full-length CK1ε was mixed with its physiological clock substrate mPER2 in the presence of radioactive γ-P32-ATP, the auto-phosphorylation rate of CK1ε itself was slower at the lower temperature of 25 °C (Fig. S7D). In contrast, the phosphorylation of mPER2 was faster at 25 °C (Fig. S7E), which supports the prediction that full-length CK1ε exhibits a higher kinase activity at lower temperature due to less auto-inhibition. In addition, we executed a luminescence reporter assay with our NIH3T3/mBmal1::luc cells under two different physiological temperatures, 32 °C and 37 °C. The pace of the clock was significantly accelerated at the lower temperature with amuch shorter period: 19.7±0.43 hr at 32 °C versus 21.3±0.36 hr at 37 °C (Fig. 6A), consistent with the inference that the overall apparent kinase activity may govern the speed of the clock.
Figure 6. Lower temperature in the physiological range accelerates the clock.

(A) Comparing two different temperatures, the circadian clock of NIH3T3/mBmal1::luc cells oscillated faster at the lower temperature, whereas native CK1ε exhibits a higher kinase activity on the clock substrate PER2 at lower temperature (Fig. S6). At 32 °C, the period is 19.7±0.43 hr, while at 37 °C the period is 21.3±0.36 hr. Error bars are +/− standard deviation (n≥6 independent repeats). Statistical analysis used an unpaired t-test. ****p < 0.0001. (B) A schematic model illustrates the relationships among (1) the kinase activities of CK1δ/ε, (2) the structure and auto-phosphorylation of CK1δ/ε, and (3) the period of mammalian circadian rhythms. On physiological substrates, the activities of full-length CK1δ/ε kinases are inverted relative to their catalytic domain activities by the regulating influence of the auto-phosphorylation of their CTD domains.
Discussion
Most previous studies of the circadian properties of the key kinases CK1ε and CK1δ have focused upon the properties of the catalytic (ΔC) domains and ignored the significance of the auto-inhibition by auto-phosphorylation. We found that the kinetics of CK1ε auto-phosphorylation revealed a sequential process; the catalytic domain first auto-phosphorylates the CTD domain at initial sites, which leads to an ordered phosphorylation of later Ser/Thr sites. In our simplified model, the activity of each form of CK1ε (CK1ε, P1-CK1ε, and P2-CK1ε) can be simulated by differential equations (Fig.2E). The fitted data indicate that the auto-phosphorylation rates are temperature dependent. The auto-kinase reaction rate for the mutant CK1ε-R178C is much slower than that of native CK1ε-WT because the mutation site at residue 178 is in the crucial R178-G215-K224 triad that is predicted to recognize the substrate [6]. When this key Arg178 residue is replaced with Cysteine, the affinity between the catalytic domain and the CTD domain is likely to decrease, leading to a lower auto-kinase rate. We further showed that the auto-phosphorylation rate for CK1δ is intermediate between those of CK1ε andCK1ε-R178C (Figs. S2&S3).
We predicted that the rates of the three catalytic domains (ΔC versions) would exhibit the same trend pattern as their auto-phosphorylation rates. Indeed, when we measured the rate of phosphorylation of the substrate hP53 for CK1ε-ΔC, CK1ε-R178C-ΔC and CK1δ-ΔC using UPLC, the results supported the prediction (Fig. 4). Substrate phosphorylation of both BSA and hP53 also exhibited a higher catalytic activity by CK1ε-ΔC than by CK1δ-ΔC (Figs. 5&S6). Measurements of the overall activity of full-length CK1ε using a radioactive in vitro assay supports the model prediction that reduction of auto-inhibition by the R178C mutation enhances the rate of phosphorylation of physiological substrates (Fig. 5 & Fig. S6B).
We were unsuccessful to introduce the CK1ε-R178C mutation in our reporter cell lines by CRISPR, and therefore our comparisons in Fig. 4B/C include data from animal studies. Table 1 summarizes CK1-null animal studies and cell-based analyses (including our data) to show ~ 2 h longer period in CK1δ−/− animals/cells, and ~ 20–30 min longer periods in CK1ε−/− animals/cells, while Tau mutant mice exhibited ~ 4 h shorter periods for homozygous animals than that of CK1ε−/− animals [21, 23]. Even though the actual period value of the CK1ε−/− genotype is different between animals and cell cultures, there is a comparable trend in the period change between animals and cell cultures upon re-introduction of CK1 alleles. In particular, when these data are compared as the period shortening caused by the re-introduction of the WT or mutant allele to the relevant null, there is a clear correlation between the period change and the auto-phosphorylation rates of the three kinases; the lowest auto-kinase rate (CK1ε-R178C) corresponds with the largest degree of period shortening(Table 1, Fig. 4C). Thus, we propose a model that the higher initial catalytic activity of CK1ε would auto-phosphorylate its inhibitory CTD domain faster, thereby retarding substrate phosphorylation more effectively with the final result of a slower circadian clock. On the other hand, CK1ε-R178C with the lowest auto-phosphorylation rate would suppress its substrate phosphorylation less rapidly; the resulting enhanced/accelerated rate of substrate phosphorylation generates a faster circadian clock (Fig. 6B).
Manipulating the catalytic activity in another way would further support our model. Though lowering temperature would be expected to reduce the kinase activity towards substrates (and this was indeed true for CK1ε-ΔC, Fig. S7A–C), we found that PER2 was phosphorylated to a stronger extent by full-length CK1ε at lower physiological temperature (Fig. S7). This apparently counter-intuitive result is consistent with our hypothesis; namely, that the cooler temperature slowed auto-inhibition of substrate phosphorylation in full-length CK1ε but this temperature compensating reaction is defective in CK1ε-ΔC. This is a very interesting observation in the context of the canonical property of circadian rhythms, temperature compensation of period, which is present even in isolated cells of endothermic mammals (e.g. Rat-1/Pper1-luc cells, NIH3T3 cells, skin fibroblast cells, and Rat-1/Pbmal1-luc cells [33–36]).
Many small molecules have been developed to modulate the activity of CK1ε and CK1δ, such as CKI-7, PF670462 andPF4800567 [37]. However, these inhibitors mainly lengthen circadian period. In general, people with longer period go to sleep and wake up later so that they are often considered as “owls.” If drugs can be developed that manipulate the period in either an accelerating or decelerating directions, people could adjust their clocks optimally to their environment. For example, such pharmacological agents could be applied as a treatment of Delayed Sleep Phase Syndrome (DSPS). The model we discuss in this paper may suggest a novel pathway; to develop molecules that interfere with the interface between the catalytic domain and the CTD domain, thereby suppressing the auto-inhibition. In this way, the overall activity of CK1 can be enhanced on substrates such as PER1/2 so that the mammalian clock may run faster, consequently correcting the DSPS phenotype.
Supplementary Material
Acknowledgments
We would like to thank Dr. Masaaki Ikeda for providing us with the original mouse Bmal1 reporter construct Bp/527-dluc. This work was financially supported by grants from the National Natural Science Foundation of China (31571208 and 31771302 to XQ), the Anhui Provincial Natural Science Foundation (1608085MH212 to XQ), and the USA National Institutes of Health (NIH/NIGMS grant # GM 067152 to CHJ). Ximing Qin acknowledges the start-up fund provided by Anhui University. We are also grateful to the staff for providing technical support with using the facility of Institute of Physical Science and Information Technology.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Dunlap JC and Loros JJ (2018) Just-So Stories and Origin Myths: Phosphorylation and Structural Disorder in Circadian Clock Proteins. Mol Cell. 69(2):165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konopka RJ, Benzer S. (1971) Clock mutants of Drosophila melanogaster. ProcNatlAcadSci U S A. 68(9):2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 94(1): 83–95. [DOI] [PubMed] [Google Scholar]
- 4.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 94(1): 97–107. [DOI] [PubMed] [Google Scholar]
- 5.Ralph MR and Menaker M. (1988) A mutation of the circadian system in golden hamsters. Science. 241: 1225–1227. [DOI] [PubMed] [Google Scholar]
- 6.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 288(5465):483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. (2002) The protein kinasecomplement of the human genome. Science. 298: 1912–1934. [DOI] [PubMed] [Google Scholar]
- 8.Graves PR, Haas DW, Hagedorn CH, DePaoli-Roach AA, Roach PJ. (1993) Molecular cloning, expression, and characterization of a 49-kilodalton casein kinase I isoform from rat testis. J Biol Chem. 268(9): 6394–6401. [PubMed] [Google Scholar]
- 9.Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. (1995) Isolation and characterization of human casein kinase I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem. 270(25):14875–14883. [DOI] [PubMed] [Google Scholar]
- 10.Cheong JK, Virshup DM. (2011) Casein kinase 1: complexity in the family, Int. J. Biochem. Cell Biol 43: 465–469. [DOI] [PubMed] [Google Scholar]
- 11.Knippschild U, Krüger M, Richter J, Xu P, García-Reyes B, Peifer C, Halekotte J, Bakulev V, Bischof J. (2014) The CK1 family: contribution to cellular stress response and its role in carcinogenesis. Front Oncol. 4: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves PR and Roach PJ. (1995) Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem 270: 21689–21694. [DOI] [PubMed] [Google Scholar]
- 13.Cegielska A, Gietzen KF, Rivers A, Virshup DM (1998) Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J. Biol. Chem 273:1357–1364. [DOI] [PubMed] [Google Scholar]
- 14.Rivers A, Gietzen KF, Vielhaber E, Virshup DM. (1998) Regulation of casein kinase I epsilon and casein kinase I delta by an in vivo futile phosphorylation cycle. J Biol Chem. 273(26): 15980–15984. [DOI] [PubMed] [Google Scholar]
- 15.Camacho F, Cilio M, Guo Y, Virshup DM, Patel K, Khorkova O, Styren S, Morse B, Yao Z, Keesler GA. (2001) Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 489(2–3):159–165. [DOI] [PubMed] [Google Scholar]
- 16.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. (2002) Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells, Mol. Cell. Biol 22:1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptácek LJ, and Fu YH. (2005) Functional consequences of a CKIdelta mutation causing familialadvanced sleep phase syndrome. Nature. 434: 15240–15245. [DOI] [PubMed] [Google Scholar]
- 18.Takano A, Uchiyama M, Kajimura N, Mishima K, Inoue Y, Kamei Y, Kitajima T, Shibui K, Katoh M, Watanabe T, Hashimotodani Y, Nakajima T, Ozeki Y, Hori T, Yamada N, Toyoshima R, Ozaki N, Okawa M, Nagai K, Takahashi K, Isojima Y, Yamauchi T, Ebisawa T. (2004) A missense variation in human casein kinase I epsilon gene that induces functional alteration and shows an inverse association with circadian rhythm sleep disorders. Neuropsychopharmacology. 29(10):1901–1909. [DOI] [PubMed] [Google Scholar]
- 19.Lee H, Chen R, Lee Y, Yoo S, Lee C. (2009) Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. ProcNatlAcadSci U S A. 106(50):21359–21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, Weaver DR. (2009) Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 29(14):3853–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loudon AS, Meng QJ, Maywood ES, Bechtold DA, Boot-Handford RP, Hastings MH. (2007) The biology of the circadian Ck1epsilon tau mutation in mice and Syrian hamsters: a tale of two species. Cold Spring HarbSymp Quant Biol. 72: 261–71. [DOI] [PubMed] [Google Scholar]
- 22.Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sládek M, Adams J, Bass M, Chandrasekaran R, Butler T, Griffor M, Rajamohan F, Serpa M, Chen Y, Claffey M, Hastings M, Loudon A, Maywood E, Ohren J, Doran A, Wager TT. (2009) Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period.JPharmacolExpTher. 330(2):430–439. [DOI] [PubMed] [Google Scholar]
- 23.Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sládek M, Semikhodskii AS, Glossop NRJ, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon ASI. (2008) Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 58(1): 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. (2006) An opposite role for tau in circadian rhythms revealed by mathematical modeling. ProcNatlAcadSci U S A. 103(28):10618–10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korge S, Grudziecki A, Kramer A. (2015) Highly Efficient Genome Editing via CRISPR/Cas9 to Create Clock Gene Knockout Cells. J Biol Rhythms. 30(5):389–395. [DOI] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin X, Mori T, Zhang Y, Johnson CH. (2015) PER2 Differentially Regulates Clock Phosphorylation versus Transcription by Reciprocal Switching of CK1ε Activity. J Biol Rhythms. 30(3): 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki S and Takahashi JS. (2005) Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 393: 288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandit SB, Brylinski M, Zhou H, Gao M, Arakaki AK, Skolnick J. (2010) PSiFR: an integrated resource for prediction of protein structure and function. Bioinformatics. 26(5): 687–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gietzen KF and Virshup DM. (1999) Identification of inhibitory autophosphorylation sites in casein kinase I epsilon, J. Biol. Chem 274: 32063–32070. [DOI] [PubMed] [Google Scholar]
- 31.Long A, Zhao H, Huang X. (2012) Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor. J Med Chem. 55: 956–960. [DOI] [PubMed] [Google Scholar]
- 32.Long AM, Zhao H, Huang X. (2012) Structural basis for the potent and selective inhibition of casein kinase 1 epsilon. J Med Chem. 55: 10307–10311. [DOI] [PubMed] [Google Scholar]
- 33.Izumo M, Johnson CH, Yamazaki S. (2003) Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: temperature compensation and damping. Proc Natl AcadSci U S A. 100(26): 16089–16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchiya Y, Akashi M, Nishida E. (2003) Temperature compensation and temperature resetting of circadian rhythms in mammalian cultured fibroblasts. Genes Cells. 8(8): 713–720. [DOI] [PubMed] [Google Scholar]
- 35.Dibner C, Sage D, Unser M, Bauer C, d’Eysmond T, Naef F, Schibler U. (2009) Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 28(2): 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. (2010) Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl AcadSci U S A. 107(21): 9665–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Xu T, Zhang Y, Qin X. (2017) Molecular basis for the regulation of the circadian clock kinases CK1δ and CK1ε. Cell Signal. 31: 58–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


