Abstract
Breast cancers evolve in a multistage process that can span decades after a carcinogenic exposure. It follows that long-lived precursor breast lesions persist in a subclinical state prior to completing malignant transformation, yet widely-used breast cancer models lack an experimental framework for targeting premalignant disease. Inspired by classic multistage skin carcinogenesis protocols, we combined chemical carcinogenesis with transgenic mouse modeling to resolve mouse mammary carcinogenesis into discrete initiation and progression stages. At the initiation stage, exposure to the carcinogen 7,12-dimethylbenzanthracene (DMBA) generated “initiated mammary epithelial cells” (iMECs) by introducing a stereotyped HrasQ61L driver mutation. Whether DMBA exposure occurred during puberty or adulthood, mice efficiently acquired iMEC clones that eluded detection by conventional histology, yet were long-lived, persisting in a clinically silent state for months in the absence of a cooperating event. At the progression stage, inducible activation of oncogenic Wnt signaling drove rapid and synchronous transformation of latent iMECs into overt mammary carcinomas, while Wnt activation in neighboring normal mammary epithelium yielded only benign hyperplasia over this same time period. Although early parity (completion of a full-term pregnancy) reduces breast cancer risk in some contexts, standard parity-induced protection schemes failed to eliminate iMECs in our multistage model, suggesting Wnt-responsive iMECs are maintained by hormone-independent mechanisms. Variations on our multistage modeling strategy may help to identify and validate cellular and molecular targets for breast cancer chemoprevention.
Keywords: Mammary, Cancer, Mouse, Hras, Chemical carcinogenesis, Wnt
Introduction
Epidemiology studies provide some of the best evidence that breast cancer is a multistage disease. Exposure to carcinogens, such as ionizing radiation, increases breast cancer risk, yet the interval between exposure and clinically detectable cancer typically spans decades[1, 2]. Mechanistically, carcinogen-induced driver mutations have been proposed to create “initiated” premalignant clones, which are long-lived and predisposed to progress to frank malignancy. Alternatively, certain carcinogen exposures may not induce driver mutations directly, but instead may induce a pro-tumorigenic microenvironment (e.g., chronic inflammation) that fosters the persistence and/or outgrowth of initiated clones[3, 4]. However initiated clones originate, evolutionary modeling suggests that they frequently persist undetected for many years before finally manifesting clinically[5]. Throughout this protracted latency period, initiated clones presumably elude detection by remaining small in size and/or generating only modest changes in histomorphology. Concordantly, analysis of carcinogen-exposed human skin and esophagus specimens by high throughput, multi-region DNA sequencing has revealed normal-appearing epithelium to be a mosaic of mutant clones, many of which harbor mutations in cancer-associated genes[6, 7].
High-throughput sequencing of breast cancer genomes has generated detailed catalogs of driver mutations[8–10], but questions remain regarding the order in which these mutations accrue as well as their stage-specific roles during tumor progression. The histopathology of the direct precursor breast lesions that ultimately beget invasive breast cancers remains poorly defined, which complicates efforts aimed at identifying, monitoring, or eliminating high-risk clones. Even when breast lesion histopathology correlates with increased cancer risk, it can remain unclear whether such lesions merely serve as risk markers or whether they comprise the definitive precursors (i.e., clonal antecedents) to subsequent breast cancers[11]. Therefore, cancer models remain essential for interrogating mammary carcinogenesis across disease stages. Cell culture techniques and transplantable cell lines can be useful for modeling later stages of breast cancer progression, but autochthonous tumor models are essential for modeling earlier, subclinical disease stages.
From the very start, skin cancer modeling efforts provided an experimental framework for successful breast cancer modeling. Beginning in the 1930s, protocols for generating carcinogen-induced rodent skin cancers were adapted to generate the first transplantable mammary cancers[12]. Most notably, carcinogen-induced rat mammary cancer models became widely used and proved crucial for elucidating the biology of hormone-dependent breast cancer[13]. Later, rat mammary cancers provided the key preclinical model for developing anti-hormonal agents as therapeutics for breast cancer treatment and chemoprevention[14]. Shortly after the first transforming oncogene mutations were identified, rodent skin and mammary tumors were found to share a key driver event, namely carcinogen-induced Hras mutation, highlighting a remarkable genetic convergence between these experimental models[15, 16]. Whereas rats show a marked susceptibility to mammary tumors following carcinogen exposure, mice are relatively resistant. For example, a variety of rat strains reliably develop mammary tumors after a single carcinogen exposure administered during puberty[17–19]. By contrast, mice reliably develop mammary cancers only when carcinogen exposures are administered repeatedly or supplemented by stimulation with exogenous hormones[20, 21]. The biological explanation for this species difference remains unknown.
Transgenic mouse models reinforce the concept that mammary carcinogenesis is a multistage process. For example, whereas Wnt pathway activation drives synchronous onset of diffuse mammary hyperplasia in a matter of days, Wnt-driven mammary cancers instead arise stochastically over a period of months[22, 23]. Unlike Wnt-driven mammary hyperplasia, most Wnt-driven cancers acquire cooperating somatic mutations that create oncogenic Hras alleles[24–26], implicating Hras activation as a rate-limiting step in carcinogenesis. Supporting this model, we showed previously that iWnt transgenic mice (engineered for inducible, mammary-specific Wnt pathway activation), when subjected to Wnt induction followed by a single sub-carcinogenic DMBA exposure (i.e., a DMBA dose insufficient to generate tumors in wild-type mice), rapidly develop multiple mammary cancers, nearly all of which bear stereotyped HrasQ61L mutations attributable to DMBA[27]. Thus, the synergy between DMBA exposure and Wnt-driven mammary hyperplasia during mammary carcinogenesis closely resembles synergy between DMBA and the regenerative skin hyperplasia induced by repeated topical application of 12-O-tetradecanoyl-phorbol-13-acetate (TPA) in classic multistage skin carcinogenesis. Strikingly, in both models, chronic induction of hyperplasia culminates in the outgrowth of DMBA-initiated, HrasQ61L-mutant epithelial cell clones.
Given these deep parallels between DMBA-initiated skin and mammary cancers, we set out to develop a multistage DMBA/iWnt mammary cancer model. However, at the outset, we recognized that DMBA/iWnt mammary tumorigenesis differs from DMBA/TPA skin carcinogenesis in at least two key ways. First, in classic multistage skin cancer modeling, promotion alone is insufficient for generating tumors[28]. By contrast, strictly speaking, Wnt pathway activation can act as a “complete carcinogen” in the mammary gland, since iWnt mice monitored over many months of chronic Wnt induction eventually develop solitary mammary tumors[22, 29]. Second, whereas the first skin tumors detected during classic multistage skin cancer modeling are benign papillomas, the first mammary tumors detected in DMBA/iWnt experiments are invasive cancers, as discussed below. In light of these differences, when adapting our DMBA/iWnt protocol for multistage breast cancer modeling, we used classic terminology by referring to the DMBA “initiation” step, but adopted modified terminology by referring to a Wnt-driven “progression” (rather than “promotion”) step.
Material and Methods
Experimental animals
Mice were bred and maintained at the Pennsylvania State University College of Medicine pathogen-free rodent facility with free access to water and chow. All experimental protocols were approved by the Pennsylvania State University College of Medicine’s Institutional Animal Care and Use Committee. The MMTV-rtTA and TetO-Wnt1 transgenic mouse lines[22, 30] (Gifts from Dr. Lewis Chodosh) were generated and maintained on an inbred FVB/N background; the TetO-H2B-GFP transgenic line[31] was obtained from the Jackson Labs (Stock #005104), then backcrossed to FVB/N more than 20 generations prior to generating offspring for this study. Mice were genotyped using PCR-based assays performed on tail snip-derived genomic DNA. PCR primer pairs included the following. MMTV-rtTA transgene: 5’-ATCCGCACCCTTGATGACTCCG-3’ with 5’-GGCTATCAACCAACACACTGCCAC-3’ TetO-Wnt1 transgene: 5’-TGCGGTTCCTGATGTATTTTGC-3’ with 5’-TGCATTCCTTTGGCGAGAGG-3’. TetO-H2B-eGFP transgene: 5’-AAGTTCATCTGCACCACCG-3’ with 5’-CCTTGATGCCGTTCTTCTGCTTGTC-3’.
For Dox treatment, standard mouse chow was replaced with medicated chow containing 2 g/Kg doxycycline (Bio Serv). For chemical carcinogenesis, DMBA (Sigma-Aldrich D3254) was dissolved in sesame oil (Sigma-Aldrich N3385) at 5 mg/ml. Mice were administered a 1 mg DMBA dose by oral gavage at the indicated time points.
Tumor studies and tissue collection
Mice were examined daily for tumors by palpation. For detailed serial monitoring of tumors, caliper measurements and body photographs were obtained while mice were transiently anesthetized using inhaled isoflurane. Mice were euthanized and subjected to necropsy and tumor harvest when tumor burden reached predetermined endpoints described in the main text. Tumors were subdivided and either fixed with 4% paraformaldehyde for two hours prior to histology processing or snap-frozen on dry ice prior to storage at −80°C.
DNA preparation and Sanger sequencing
Genomic DNA was isolated from frozen tumor tissue using Promega Maxwell 16 Tissue DNA Purification Kit (Promega AS1030). A relevant Hras gene segment was PCR-amplified using the following primers: 5’GGTCAGGCATCTATTAGCCGTC with 5’GGGGAGACTCAACAGTGCGAG. PCR products were purified using either the Qiaquick PCR purification kit (28104) or Affimetrix ExoSAP-IT (75001) kit. Sanger sequencing was performed by Genewiz LLC, South Plainfield, NJ, and results were analyzed using SnapGene software.
Mammary gland imaging
For carmine staining, mammary glands were spread on glass slides, fixed in 4% PFA for 2 hours, rinsed in 70% ethanol, re-hydrated and stained with 0.2% carmine in PBS for at least 24 hours. After dehydration in graded solutions of ethanol (70%, 80%, 90% and 100%), glands were cleared with methyl salicylate, then photographed using a Nikon SMZ1000 Zoom stereomicroscope. For fluorescent imaging of mammary gland whole mounts, glands were spread on glass slides, fixed in 4% PFA for 15 minutes, rinsed with PBS, then photographed using a Leica MZ FLIII Fluorescence Stereomicroscope with a standard GFP emission filter. Image reconstructions were generated using ImageJ. Subclinical lesions and neighboring glandular tissues were identified by microscopy and were excised and collected on the stage of the dissecting microscope.
Tumor explant and ovariectomy
Fragments of DMBA/iWnt primary tumors were implanted onto the flanks of syngeneic female FVB host mice maintained on chronic Dox treatment. When explanted tumor outgrowths reached volumes of 300–500mm3, the host mice were either left untreated or subjected to ovariectomy as indicated. Successful ovariectomy was confirmed by both by histologic assessment of the extirpated tissue and by documenting atresia of the uterus at subsequent necropsy. Tumor volumes were calculated by the modified ellipsoidal formula: tumor volume = 0.5* (tumor length)* (tumor width)2.
Statistical analysis
Tumor-free survival curves were compared across cohorts using the Log-rank test. In separate analyses, time-to-first tumor and tumor multiplicity were compared across cohorts using one-way ANOVA with the Tukey correction for multiple comparisons. Data sets were not evaluated for normality. Cohort sizes were not pre-determined using a power calculation, which in principle poses an increased risk of type II error. Instead, since high tumor multiplicity rendered each mouse unusually informative, experiments were performed using cohort sizes more typical of pilot studies (4–6 mice per cohort). Most comparisons were made among contemporaneous cohorts. To minimize mouse usage, some comparisons employed a cohort from a prior experiment as an “historical control”. Comparisons involving historical controls, whenever they are employed, are indicated as such in the Results section. All graphs and statistical analyses, were generated using GraphPad Prism 8.
Results
With the goal of developing a DMBA/iWnt multistage mammary carcinogenesis model, we confirmed that no mammary tumors arose in a control cohort of iWnt mice subjected to DMBA exposure alone (“I-only” for initiation-only), indicating that the 1 mg dose administered by gavage is sub-carcinogenic. In a second control cohort, Wnt pathway activation in DMBA-naive iWnt mice (“WP-only” for Wnt Progression-only) yielded solitary, late-onset mammary tumors after a mean of 20 weeks of Dox treatment (Fig. 1A), consistent with previous studies[22, 29]. In our published DMBA/iWnt work, DMBA exposure always was administered one week after doxyxcycline (Dox)-induced Wnt pathway activation[27]. To test whether DMBA’s profound action as a co-carcinogen requires the presence of pre-existing Wnt-driven mammary hyperplasia, we reversed the experimental sequence by subjecting 5 week-old, Dox-naïve iWnt female mice to DMBA exposure up-front, then activating chronic Wnt-driven mammary hyperplasia 4 days later. This I-4d-WP protocol (Initiation-4 day interval-Wnt Progression) better recapitulates the experimental sequence in classic multistage skin carcinogenesis, where DMBA-mediated initiation precedes TPA-mediated promotion. iWnt mice subjected to the I-4d-WP protocol rapidly developed multiple palpable mammary tumors within 3 weeks of Wnt pathway activation (21 +/− 1.1 days; Fig. 1A–C). Compared to the solitary mammary tumors that arose in the WP-only cohort (0.80 +/− 0.06 mammary tumors-per-mouse), tumor multiplicity increased more than 20-fold in mice from the I-4d-WP cohort (18.6 +/− 1.8) when examined at necropsy 5–6 weeks after Wnt activation (Fig. 1D). We infer that a one-time DMBA exposure, though sub-carcinogenic on its own, reliably generated numerous iMECs in each mouse, which strictly require Wnt-driven progression to transform into malignant tumors.
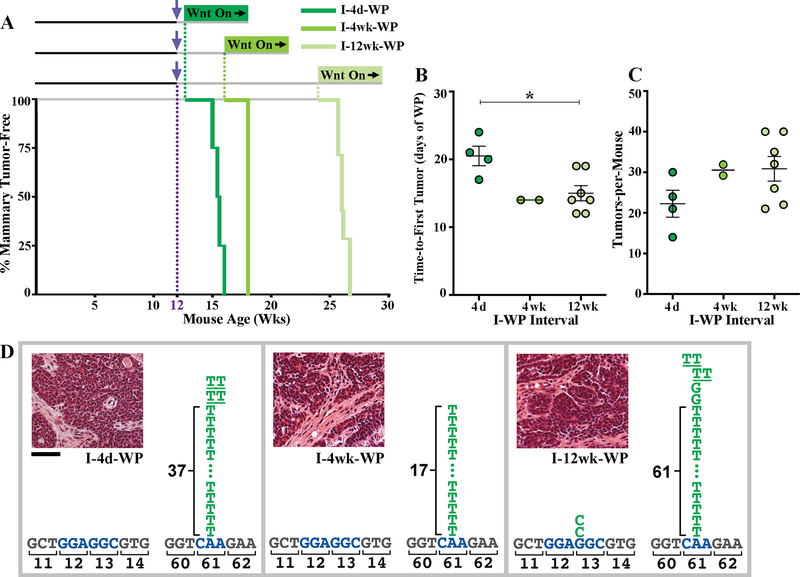
Figure 1. A DMBA/iWnt protocol yields long-lived, Hras-mutant iMECs.
(A) Tumor-free survival curves for DMBA/iWnt protocols (puberty DMBA exposure). iWnt mice, with or without DMBA exposure at 5 weeks of age (purple arrow), began chronic, Dox-induced Wnt pathway activation either 4 days, 4 weeks, or 12 weeks later (I-4d-WP, I-4w-WP and I-12w-WP, respectively). (B) Time-to-first tumor. Dot plot depicts the time interval from Dox-induced WP until the first palpable mammary tumor was detected. Each data point is derived from an individual DMBA/iWnt mouse. 4d, n=5; 4wk, n=6, 12wk, n=4. For 4d vs. 4wk, **p=0.0084; for 4d vs. 12wk, **p=0.0031; for 4wk vs. 12wk, p=0.61. (C) Representative images of a single mouse from I-4d-WP cohort at different time points after WP. (D) Tumor multiplicity. Dot plots depict discrete tumor number counts from individual mice. WP-only, n=33; 4d, n=5; 4wk, n=6, 12wk, n=4. For WP vs. 4d, WP vs. 4wk, and WP vs. 12wk, ****p<0.0001. All other comparisons were not statistically significant. For 4d vs. 4 wk, p=0.88; for 4d vs 12 wk, p=0.44; for 4wk vs. 12 wk, p=0.82. (E) Spectra of acquired somatic HRas mutations. Relevant segments of the Hras coding sequence are depicted with hot-spot codons labeled blue. Letter stacks above the coding sequence represent independent base substitutions identified in individual tumors, color-coded by treatment conditions. Tandem substitutions affecting a single mutated allele are underlined. Numbers alongside bracketed stacked sequences indicate the number of independent tumors bearing the recurrent CAA>CTA base change. Insets depict representative images of H&E-stained tissue sections. Scale bar, 50 μM. For dot plots, mean +/− SEM is shown. Cross-cohort comparisons by one-way ANOVA test with Tukey correction.
Mammary tumors generated in the context of DMBA exposure were histologically indistinguishable from DMBA-naïve iWnt tumors. Specifically, tumors from the I-4d-WP cohort were classic P-type adenocarcinomas[32], which showed mixed-lineage differentiation and well-developed stroma, frequently accompanied by small foci of squamous differentiation. Moreover, mammary tumors generated by the I-4d-WP protocol were invasive, since explanted tumor fragments readily produced tumor outgrowths when implanted onto the flanks of Dox-treated, syngeneic female host mice (Supplementary Fig. 1).
A canonical feature of classic multistage skin cancer modeling is the irreversibility of the initiation step. That is, initiated skin cells persist in a clinically silent state, yet remain available for, and amenable to, TPA-mediated promotion months later[33, 34]. To determine whether transformation-competent iMECs are similarly long-lived, we tested modified DMBA/iWnt protocols in which the 4 day time interval separating initiation and progression (I-4d-WP) was increased to either 4 weeks (I-4wk-WP) or 12 weeks (I-12wk-WP). Remarkably, neither delay in Wnt-driven progression resulted in iMEC dropout. Instead, mice subjected to delayed Wnt activation remained poised for rapid outgrowth of invasive tumors following Wnt-driven progression (Fig. 1A,B). In fact, palpable mammary tumors arose even faster in the context of delayed Wnt activation (15.3 +/− 1.0 days for the I-4wk-WP cohort; 13.8 +/− 1.4 days for the I-12wk-WP cohort), suggesting iMEC clones may undergo modest subclinical expansion in the weeks preceding Wnt-driven progression. Importantly, DMBA/iWnt mice subjected to delayed Wnt activation remained comparably predisposed toward high tumor multiplicity (17.8 +/− 2.4 tumors-per mouse for the I-4wk-WP cohort; 16.5 +/− 0.29 tumors-per-mouse for the I-12wk-WP cohort; Fig. 1D), indicating that most, if not all DMBA-induced iMECs are long-lived. In addition, tumors never arose in DMBA-exposed mice during even extended intervals preceding Wnt activation, consistent with a strict requirement for Wnt activation during iMEC outgrowth.
In multistage skin carcinogenesis, initiation involves DMBA-induced HrasQ61L mutations that generate long-lived premalignant clones. To confirm that DMBA induces the identical HrasQ61L mutation in long-lived iMECs, Sanger sequencing was performed on PCR-amplified Hras alleles derived from mammary tumors that arose with and without prior DMBA exposure. Wnt-driven tumors from DMBA-naïve mice (WP-only condition) frequently harbored cooperating Hras mutations (22 of 34 tumors, 65%), which were distributed among three distinct recurring mutations (G12E, Q61R, and Q61L; Fig. 1E), consistent with previous reports[24–26]. By contrast, nearly all Wnt-driven tumors from DMBA-exposed mice harbored an activating Hras mutation (95% or more of tumors analyzed, at least 41 tumors analyzed per condition), which nearly always was the HrasQ61L mutation stereotypically induced by DMBA exposure in skin cancer modeling. This overwhelming predilection for acquiring an HrasQ61L mutation was observed regardless of whether Wnt-driven progression occurred shortly after DMBA exposure (I-4d-WP condition) or occurred weeks later (I-4wk-WP and I-12wk-WP conditions; Fig. 1E), implicating HrasQ61L mutations in the genesis of long-lived iMECs.
Next, we determined when during their genesis and outgrowth iMECs produce detectable histopathology by performing microscopic examination of carmine-stained mammary gland whole mounts and H&E-stained tissue sections. Absent activation of Wnt-driven progression, iMECs produced no overt mammary lesions, since glands harvested from mice either 4 weeks or 12 weeks after DMBA exposure were indistinguishable from those of age-matched, DMBA-naïve controls. Although iMECs generated using the I-4d-WP protocol reliably generated a dozen or more palpable mammary tumors within 3–4 weeks of Wnt progression, time course analyses of carmine-stained mammary gland whole-mounts showed that the onset of corresponding microscopic mammary ductal lesions following Wnt activation was surprisingly slow. Mammary glands from iWnt mice subjected to the I-4d-WP protocol harvested at day 6 of WP lacked any focal lesions discernible against the background Wnt-driven hyperplasia induced throughout the entire gland (0 focal lesions identified in 10 glands, 0%). Even out to day 14 of WP, most glands remained free of focal mammary ductal lesions, and the few lesions beginning to emerge were small (4 microscopic focal lesions identified in 10 glands, 40%; Fig. 2A). Nonetheless, thereafter these microscopic foci grew quickly and synchronously such that just 5 days later (d19 WP), all mammary glands harbored one or more obvious macroscopic lesions (10 of 10 glands, 100%; Fig. 2), the largest of which already were palpable in live mice prior to euthanasia and tissue harvest. Thus, iMECs comprise a cryptic reservoir of premalignant cells with an impressive capacity for rapid, Wnt-driven clonal expansion.
Figure 2. iMECs yield focal, Hras-mutant mammary lesions upon Wnt pathway activation.
(A) Detection of focal DMBA/iWnt mammary lesions. iWnt mice were subjected to the I-4d-WP protocol, including timed duration of Wnt-driven Progression (WP) for the indicated number of days. Panels depict relevant images captured during microscopic examination of carmine-stained mammary gland whole-mounts and H&E-stained sections. Arrows indicate focal, subclinical lesions. (B) Schematic depicting the transgene combinations employed for fluorescence-based detection of subclinical mammary lesions. (C) Detection and mutation analysis of focal lesions from iWnt/iGFP mammary glands. iWnt/iGFP mice were subjected to the I-4d-WP protocol, including timed duration of Wnt-driven Progression (WP) for the indicated number of days. Panels depict relevant images captured during examination of freshly-harvested mammary glands by fluorescence microscopy. Sanger sequencing panels depict relevant chromatogram segments centered on Hras codon 61. The red asterisk marks the site of the A>T base substitution, which encodes the HrasQ61L allele found recurrently within focal lesions.
If DMBA-induced HrasQ61L mutations mark the clonal link between iMECs and overt mammary cancers, then the subclinical mammary lesions that emerge during Wnt progression ought to bear HrasQ61L mutations as well. To aid in the identification of subclinical lesions within freshly harvested tissue, a reporter gene was introduced into the iWnt model via crosses with a TetO-H2B-GFP transgenic line[31, 35] (Fig. 2B). As expected, Dox-induced H2B-GFP expression in tri-transgenic offspring (MMTV-rtTA/TetO-Wnt1/TetO-H2B-GFP; hereafter iWnt/iGFP mice) permitted visualization of mammary epithelium by fluorescent microscopy (Fig. 2C). When mammary glands from iWnt/iGFP mice subjected to the I-4d-WP protocol were harvested after timed periods of Wnt-driven progression, day 11 WP glands lacked focal lesions (0 of 10 glands examined by fluorescence microscopy, 0%), and the first focal lesions emerged at day 14 WP (one focal lesion in 10 glands, 10%). By contrast, every mammary glands harvested at d18 WP harbored one or more focal mammary lesions (40 out of 40 glands, 100%; Fig. 2C), some of which were macroscopic. Subclinical focal lesions from these glands were excised on the stage of a dissecting microscope, along with separate samples of neighboring glandular tissue comprised of diffuse Wnt-driven mammary hyperplasia. Sanger sequencing performed on PCR-amplified Hras alleles showed that genomic DNA derived from focal lesions invariably carried HrasQ61L mutations, whereas neighboring epithelium lacked detectable Hras mutations (Fig. 2C). In sum, iMECs subjected to Wnt activation yielded focal mammary lesions that grew into overt mammary cancers in a quasi-synchronous manner, raising the possibility that the Wnt-driven progression from iMEC to frank malignancy may be a single-step transition. In this way, Wnt-driven progression of iMECs differs from TPA-mediated promotion of initiated skin cells, which instead yields papillomas in a stochastic manner, and only after many months.
Both epidemiology studies[1, 2] and rat models[19, 36] suggest that more mature stages of breast development may show decreased carcinogen susceptibility. In our initial DMBA/iWnt experiments, DMBA exposure was administered at mid-puberty to 5 week-old females, which are in the midst of end bud-mediated mammary ductal elongation. To test whether mature iWnt mammary glands show decreased susceptibility to DMBA-induced iMECs, we modified the I-4d-WP protocol by administering DMBA exposure to adult 12 week-old iWnt females, in whom mammary ductal elongation is complete and terminal end buds have regressed. Despite mammary gland maturation, adult females remained susceptible to DMBA-induced iMECs, which again rapidly yielded multiple palpable mammary tumor outgrowths within 3 weeks of subsequent Wnt-driven progression (Fig. 3A). Similar to the iMECs acquired during puberty, iMECs acquired via adult DMBA exposure were long-lived, since no iMEC dropout was observed upon increasing the interval separating initiation and progression to either 4 weeks (I-4wk-WP) or 12 weeks (I-12wk-WP). On the contrary, in the context of adult DMBA exposure, delayed Wnt progression trended toward producing more rapid tumor outgrowth and higher tumor multiplicity (Fig. 3B, C). As before, tumors arising from adult DMBA-exposed mice nearly always harbored the stereotyped DMBA-induced HrasQ61L mutation (Fig. 3D).
Figure 3. Adult iWnt mice are susceptible to long-lived DMBA-induced iMECs.
(A) Tumor-free survival curves for DMBA/iWnt protocols (adult DMBA exposure). iWnt mice, with or without DMBA exposure at 12 weeks of age (purple arrow), began chronic, Dox-induced Wnt pathway activation either 4 days, 4 weeks, or 12 weeks later (I-4d-WP, I-4wk-WP and I-12w-WP, respectively), as indicated. (B) Time-to-first tumor. Dot plot depicts the time interval from Dox-induced WP until the first palpable mammary tumor was detected. Each data point is derived from an individual DMBA/iWnt mouse. 4d, n=4; 4wk, n=2, 12wk, n=7. For 4d vs. 4wk, p=0.053; for 4d vs. 12wk, *p=0.025; for 4wk vs. 12wk, p=0.90. (C) Tumor multiplicity. Dot plots depict discrete tumor number counts from individual mice. 4d, n=4; 4wk, n=2, 12wk, n=7. For 4d vs. 4wk, p=0.42; for 4d vs. 12wk, p=0.19; for 4wk vs. 12wk, p=0.99. (D) Spectrum of acquired somatic HRas mutations, as depicted in Fig. 1A. Insets depict representative images of H&E-stained tissue sections. Scale bar, 50 μM. For dot plots, mean +/− SEM is shown. Cross-cohort comparisons by one-way ANOVA test with Tukey correction.
Next, we examined whether parity-induced protection schemes reduce tumor incidence in our multistage breast cancer model. While established Wnt-driven mammary tumors have been shown to grow in a hormone-independent manner[37], we reasoned that this hormone-independence might not be shared by antecedent iMEC clones, which grow slowly if at all prior to Wnt activation. First, we tested for parity-induced protection using a pre-exposure prophylaxis protocol (Fig. 4A), wherein iWnt mice began a full-term pregnancy at 5 weeks of age, nursed pups for one week up until forced weaning, then underwent mammary gland involution prior to adult DMBA exposure. Since the length of the involution period optimal for parity-induced protection is unclear, we tested separate parity schemes incorporating either a 3 week involution period (P3wk scheme) or an 8 week involution period (P8wk scheme). Neither scheme conferred parity-induced protection, since parous iWnt females remained susceptible to DMBA-induced iMECs, which rapidly yielded multiple palpable mammary tumor outgrowths within 3 weeks of Wnt-driven progression (Fig. 4B, C), nearly all of which harbored the stereotyped DMBA-induced HrasQ61L mutation (Fig. 4D).
Figure 4. Pre-exposure parity fails to prevent iMEC generation.
(A) Kaplan-Meier tumor-free survival curve with schematic depicting pre-exposure prophylaxis protocols. Female iWnt mice bred at 5 weeks of age, completed pregnancy, nursed pups for 1 week until forced weaning, then underwent either 3 or 8 weeks of involution before DMBA exposure (purple arrow). Then, 4 days after DMBA, mice began chronic Dox-induced Wnt-Progression (P3wk-I-WP and P8wk-I-WP protocols, abbreviated P3 and P8 respectively). For comparison, historical data from the adult I-4d-WP cohort presented in Figure 3 are depicted alongside the parous cohorts, providing a nulliparous control (N). (B) Time-to-first tumor. Dot plot depicts the time interval from Dox-induced WP until the first palpable mammary tumor was detected. Each data point is derived from an individual DMBA/iWnt mouse. N, n=4; P3, n=4, P8, n=6. For N vs. P3, p=0.74; for N vs. P8, p=0.99; for P3 vs. P8, p=0.78. (C) Tumor multiplicity. Dot plots depict discrete tumor number counts from individual mice. N, n=4; P3, n=4; P8, n=6. For N vs. P3, p=0.50; for N vs. P8, *p=0.048; for P3 vs. P8, p=0.35. (D) Spectrum of acquired somatic HRas mutations, as depicted in Fig. 1A. Insets depict representative images of H&E-stained tissue sections. Scale bar, 50 μM. For dot plots, mean +/− SEM is shown. Cross-cohort comparisons by one-way ANOVA test with Tukey correction.
Finally, we tested for parity-induced protection using a post-exposure prophylaxis protocol, wherein iWnt mice were subjected to DMBA exposure at 5 weeks of age, then completed either the P3wk or P8wk parity scheme prior to beginning Wnt-driven progression (Fig. 5A). Once again, neither scheme conferred parity-induced protection. DMBA-induced iMECs persisted through post-exposure parity and rapidly yielded multiple palpable mammary tumor outgrowths within 3 weeks of Wnt-driven progression (Fig. 5B,C). As before, nearly all of these tumors harbored the stereotyped DMBA-induced HrasQ61L mutation (Fig. 5D).
Figure 5. Post-exposure parity fails to eliminate established iMECs.
(A) Kaplan-Meier tumor-free survival curve with schematic summary of the post-exposure prophylaxis protocols. Dox-naïve iWnt mice were subjected to DMBA exposure at 5 weeks old, then completed either P3wk or P8wk parity scheme prior to Wnt-Progression, named as I-P3wk-WP (P3) and I-P8wk-WP (P8), respectively. For comparison, historical data from the adult I-12wk-WP cohort presented in Figure 1 are depicted alongside the parous cohorts, providing a nulliparous control (N). P=0.332. (B) Time-to-first tumor. Dot plot depicts the time interval from Dox-induced WP until the first palpable mammary tumor was detected. Each data point is derived from an individual DMBA/iWnt mouse. N, n=4; P3, n=4, P8, n=5. For N vs. P3, p=0.52; for N vs. P8, p=0.87; for P3 vs. P8, p=0.78. (C) Tumor multiplicity. Dot plots depict discrete tumor number counts from individual mice. N, n=4; P3, n=4; P8, n=5. For N vs. P3, p=0.75; for N vs. P8, p=0.86; for P3 vs. P8, p=0.42. (D) Spectrum of acquired somatic HRas mutations, as depicted in Fig. 1A. Insets depict representative images of H&E-stained tissue sections. Scale bar, 50 μM. For dot plots, mean +/− SEM is shown. Cross-cohort comparisons by one-way ANOVA test with Tukey correction.
Discussion
Our DMBA/ iWnt multistage mammary cancer model recapitulates canonical features of classic DMBA/TPA skin cancer modeling, albeit with several key differences. While DMBA is applied topically in skin models versus systemically (via gavage) in our mammary model, both modes of exposure induce stereotyped HrasQ61L mutations, thereby generating initiated epithelial cells. In both skin and mammary tissue, initiation occurs irreversibly since both TPA-mediated promotion and Wnt-driven progression yield tumors efficiently, even when started months after completing the initiation stage. Interestingly, just as most DMBA/TPA skin papillomas regress when daily TPA application is withdrawn[38], iWnt tumors consistently regress when challenged with Wnt withdrawal[22, 26, 29], including DMBA/iWnt tumors. Despite these striking similarities, the models differ starkly in how initiated cells are provoked to yield tumors. Topical TPA meets the classic definition of a tumor promoter, since TPA-induced regenerative hyperplasia never induces skin tumors in the absence of initiation. By contrast, whereas Wnt1 trangene-driven mammary hyperplasia involves diffuse, benign overgrowth of mammary epithelium in the short term, Wnt1 acts as a so-called “complete carcinogen” over the long term, in that DMBA-naïve iWnt mice stochastically develop mammary carcinomas after months of Wnt activation.
Moreover, the DMBA/TPA and DMBA/iWnt protocols differ in that only DMBA/TPA provides direct experimental access to a clinically discernible benign lesion. The DMBA/TPA protocol produces benign papillomas that arise in a stochastic manner after several months of TPA application, and only a minority of these papillomas progresses to squamous cell cancer during additional weeks of TPA[38]. By contrast, the DMBA/iWnt protocol produces prompt, quasi-synchronous outgrowth of invasive, transplantable mammary cancers within 3–4 weeks of Wnt pathway activation. Therefore, DMBA-induced iMEC clones seemingly undergo a single-step malignant conversion. In light of these key differences, we refer to the Wnt-driven “progression” of iMECs, to distinguish this process from the TPA-driven “promotion” of initiated skin cells.
Unlike some epidemiology and rat chemical carcinogenesis studies, mature mouse mammary glands were no less susceptible to DMBA-initiated mammary tumorigenesis in our multistage modeling. This discrepancy may reflect species differences, but also may reflect differences in the type and/or level of carcinogen exposure employed. Notably, our modeling produced very high tumor multiplicity, suggesting that our DMBA/iWnt protocols may overwhelm protective mechanisms, which normally might restrict tumorigenesis either by preventing iMEC generation (e.g., via DNA repair) or enabling iMEC elimination (e.g., via immune surveillance). In future studies, it will be important to test revised protocols that employ clinically relevant breast carcinogen exposures (e.g., ionizing radiation) and/or produce clinically relevant tumor multiplicity (i.e., solitary tumors).
In multistage skin carcinogenesis, initiated skin cells persist life-long, leading some to conclude that the initiating HrasQ61L mutation likely targets a stem cell. By analogy, DMBA may create iMECs by introducing mutations within a long-lived mammary stem cell compartment. In rats, the terminal end buds present during puberty have been proposed to harbor mammary cells that are uniquely susceptible to carcinogen-induced transformation[17]. In the DMBA/iWnt model, terminal end buds were dispensable for the efficient induction of iMECs. Adult iWnt mice acquired DMBA-induced iMECs, even though ductal morphogenesis had been completed and terminal end buds had regressed. That said, in a previous study[26], we found that Hras-mutant subclones from iWnt mammary tumors have basal features, which may reflect their origin from within the basal MEC compartment, where mammary stem cells are believed to reside. Alternatively, the HrasQ61L mutation, when targeted to a short-lived mammary progenitor cell, may render that cell long-lived. In lineage tracing studies performed in mouse intestinal epithelium, introducing an activating Kras mutation increased the likelihood that a cell would persist as a long-lived clone[39, 40].
Several features of our multistage breast cancer model complicate a straightforward translation of our findings to clinical scenarios. While activating Hras mutations are a recurring genetic event in breast cancer, they occur only rarely (<1% of cases)[8], whereas such mutations are relatively common in skin cancer (~20% of cases)[41]. That said, Hras mutations are key driver events in rodent breast cancer models with proven clinical utility. Although chemical carcinogen-induced rat mammary cancers carry highly recurrent Hras mutations, they nonetheless provided an indispensible preclinical model for the development of key anti-hormonal agents, including Tamoxifen and aromatase inhibitors[14]. Likewise, mutation-based activation of the Wnt pathway seems to be uncommon in breast cancer. With respect to modeling of parity-induced protection, some epidemiology data suggests that early parity primarily reduces the risk of hormone-dependent breast cancer[42]. Consistent with this idea, established Wnt-driven mammary cancers have been shown to grow in a hormone-independent manner[37], which may help to explain why Wnt-responsive iMECs are refractory to parity protection schemes. Conversely, preventive anti-hormonal treatment with Tamoxifen delays onset of mammary tumors in the classic MMTV-Wnt1 model, suggesting that different disease stages may show distinct hormonal requirements[43].
Thus far, iMECs remain abstract entities, whose existence is inferred from the requirement that DMBA/iWnt mammary cancers arise from clonal antecedents. Our early efforts to visualize latent iMEC clones in situ have proved unsuccessful thus far, leaving key features of iMEC biology uncharacterized. Outstanding questions include, whether latent iMEC clones are maintained as single cells or multicellular structures, and whether such cells remain quiescent or undergo dynamic turnover. Addressing these questions, among others, will require new experimental strategies. In the case of multistage skin carcinogenesis, numerous insights have emerged by superimposing lineage tracing methodology on the classic DMBA/TPA protocol[44]. Similarly, lineage tracing performed in the context of DMBA/iWNT experiments might prove informative. In the long term, identifying the culprit iMECs that bridge carcinogen exposure and tumor progression should inform efforts to design rational breast cancer chemoprevention strategies. In the short term, ongoing work in our group is testing pharmacologic strategies aimed at eradicating iMECs using FDA-approved drugs.
Supplementary Material
Acknowledgements
We thank members of the Gunther laboratory for critical review of the manuscript. This work was supported by grants from the National Cancer Institute to E. Gunther (R01CA152222 and R01CA212584) and funding received from the benefactors of the Jake Gittlen Laboratories for Cancer Research. Animal housing was provided through a facility constructed with support from a Research Facilities Improvement Grant (C06 RR-15428-01) from the National Center for Research Resources.
Footnotes
This manuscript has been reviewed and approved by all three authors. The authors have no conflicts of interest to declare.
References
- [1].Land CE, “Studies of cancer and radiation dose among atomic bomb survivors. The example of breast cancer,” JAMA, vol. 274, pp. 402–7, August 2 1995. [PubMed] [Google Scholar]
- [2].Land CE, Tokunaga M, Koyama K, Soda M, Preston DL, Nishimori I, et al. , “Incidence of female breast cancer among atomic bomb survivors, Hiroshima and Nagasaki, 1950–1990,” Radiat Res, vol. 160, pp. 707–17, December 2003. [DOI] [PubMed] [Google Scholar]
- [3].Nguyen DH, Oketch-Rabah HA, Illa-Bochaca I, Geyer FC, Reis-Filho JS, Mao JH, et al. , “Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type,” Cancer Cell, vol. 19, pp. 640–51, May 17 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barcellos-Hoff MH, “New biological insights on the link between radiation exposure and breast cancer risk,” J Mammary Gland Biol Neoplasia, vol. 18, pp. 3–13, March 2013. [DOI] [PubMed] [Google Scholar]
- [5].Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. , “The life history of 21 breast cancers,” Cell, vol. 149, pp. 994–1007, May 25 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. , “Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin,” Science, vol. 348, pp. 880–6, May 22 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, et al. , “Somatic mutant clones colonize the human esophagus with age,” Science, vol. 362, pp. 911–917, November 23 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. , “Landscape of somatic mutations in 560 breast cancer whole-genome sequences,” Nature, vol. 534, pp. 47–54, June 2 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. , “The genomic landscapes of human breast and colorectal cancers,” Science, vol. 318, pp. 1108–13, November 16 2007. [DOI] [PubMed] [Google Scholar]
- [10].Cancer Genome Atlas N, “Comprehensive molecular portraits of human breast tumours,” Nature, vol. 490, pp. 61–70, October 4 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Morrow M, Schnitt SJ, and Norton L, “Current management of lesions associated with an increased risk of breast cancer,” Nat Rev Clin Oncol, vol. 12, pp. 227–38, April 2015. [DOI] [PubMed] [Google Scholar]
- [12].Huggins C, Grand LC, and Brillantes FP, “Critical Significance of Breast Structure in the Induction of Mammary Cancer in the Rat,” Proc Natl Acad Sci U S A, vol. 45, pp. 1294–300, August 1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huggins C and Yang NC, “Induction and extinction of mammary cancer. A striking effect of hydrocarbons permits analysis of mechanisms of causes and cure of breast cancer,” Science, vol. 137, pp. 257–62, July 27 1962. [DOI] [PubMed] [Google Scholar]
- [14].Jordan VC, “Proven value of translational research with appropriate animal models to advance breast cancer treatment and save lives: the tamoxifen tale,” Br J Clin Pharmacol, vol. 79, pp. 254–67, February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Balmain A and Pragnell IB, “Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene,” Nature, vol. 303, pp. 72–4, May 5–11 1983. [DOI] [PubMed] [Google Scholar]
- [16].Sukumar S, Notario V, Martin-Zanca D, and Barbacid M, “Induction of mammary carcinomas in rats by nitroso-methylurea involves malignant activation of H-ras-1 locus by single point mutations,” Nature, vol. 306, pp. 658–61, December 15–21 1983. [DOI] [PubMed] [Google Scholar]
- [17].Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR, and van Zwieten MJ, “Comparative study of human and rat mammary tumorigenesis,” Lab Invest, vol. 62, pp. 244–78, March 1990. [PubMed] [Google Scholar]
- [18].Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, et al. , “Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy,” Cancer Res, vol. 66, pp. 6421–31, June 15 2006. [DOI] [PubMed] [Google Scholar]
- [19].Huggins C, Grand LC, and Brillantes FP, “Mammary cancer induced by a single feeding of polymucular hydrocarbons, and its suppression,” Nature, vol. 189, pp. 204–7, January 21 1961. [DOI] [PubMed] [Google Scholar]
- [20].Medina D, Butel JS, Socher SH, and Miller FL, “Mammary tumorigenesis in 7,12-dimethybenzanthracene-treated C57BL x DBA/2f F1 mice,” Cancer Res, vol. 40, pp. 368–73, February 1980. [PubMed] [Google Scholar]
- [21].Aldaz CM, Liao QY, LaBate M, and Johnston DA, “Medroxyprogesterone acetate accelerates the development and increases the incidence of mouse mammary tumors induced by dimethylbenzanthracene,” Carcinogenesis, vol. 17, pp. 2069–72, September 1996. [DOI] [PubMed] [Google Scholar]
- [22].Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, et al. , “Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis,” Genes Dev, vol. 17, pp. 488–501, February 15 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, and Varmus HE, “Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice,” Cell, vol. 55, pp. 619–25, November 18 1988. [DOI] [PubMed] [Google Scholar]
- [24].Podsypanina K, Li Y, and Varmus HE, “Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype,” BMC Med, vol. 2, p. 24, June 15 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jang JW, Boxer RB, and Chodosh LA, “Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis,” Mol Cell Biol, vol. 26, pp. 8109–21, November 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cleary AS, Leonard TL, Gestl SA, and Gunther EJ, “Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers,” Nature, vol. 508, pp. 113–7, April 3 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Keller RR, Gestl SA, Lu AQ, Hoke A, Feith DJ, and Gunther EJ, “Carcinogen-specific mutations in preferred Ras-Raf pathway oncogenes directed by strand bias,” Carcinogenesis, vol. 37, pp. 810–6, August 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Balmain A and Yuspa SH, “Milestones in skin carcinogenesis: the biology of multistage carcinogenesis,” J Invest Dermatol, vol. 134, pp. E2–7, October 10 2014. [DOI] [PubMed] [Google Scholar]
- [29].Debies MT, Gestl SA, Mathers JL, Mikse OR, Leonard TL, Moody SE, et al. , “Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss,” J Clin Invest, vol. 118, pp. 51–63, January 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, et al. , “A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology,” FASEB J, vol. 16, pp. 283–92, March 2002. [DOI] [PubMed] [Google Scholar]
- [31].Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. , “Defining the epithelial stem cell niche in skin,” Science, vol. 303, pp. 359–63, January 16 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, et al. , “Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors,” Am J Pathol, vol. 161, pp. 1087–97, September 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Loehrke H, Schweizer J, Dederer E, Hesse B, Rosenkranz G, and Goerttler K, “On the persistence of tumor initiation in two-stage carcinogenesis on mouse skin,” Carcinogenesis, vol. 4, pp. 771–5, 1983. [DOI] [PubMed] [Google Scholar]
- [34].Berenblum I and Shubik P, “The persistence of latent tumour cells induced in the mouse’s skin by a single application of 9:10-dimethyl-1:2-benzanthracene,” Br J Cancer, vol. 3, pp. 384–6, September 1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Plichta KA, Mathers JL, Gestl SA, Glick AB, and Gunther EJ, “Basal but not luminal mammary epithelial cells require PI3K/mTOR signaling for Ras-driven overgrowth,” Cancer Res, vol. 72, pp. 5856–66, November 15 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thompson TA, Haag JD, and Gould MN, “ras gene mutations are absent in NMU-induced mammary carcinomas from aging rats,” Carcinogenesis, vol. 21, pp. 1917–22, October 2000. [DOI] [PubMed] [Google Scholar]
- [37].Zhang X, Podsypanina K, Huang S, Mohsin SK, Chamness GC, Hatsell S, et al. , “Estrogen receptor positivity in mammary tumors of Wnt-1 transgenic mice is influenced by collaborating oncogenic mutations,” Oncogene, vol. 24, pp. 4220–31, June 16 2005. [DOI] [PubMed] [Google Scholar]
- [38].Kemp CJ, “Multistep skin cancer in mice as a model to study the evolution of cancer cells,” Semin Cancer Biol, vol. 15, pp. 460–73, December 2005. [DOI] [PubMed] [Google Scholar]
- [39].Snippert HJ, Schepers AG, van Es JH, Simons BD, and Clevers H, “Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion,” EMBO Rep, vol. 15, pp. 62–9, January 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vermeulen L, Morrissey E, van der Heijden M, Nicholson AM, Sottoriva A, Buczacki S, et al. , “Defining stem cell dynamics in models of intestinal tumor initiation,” Science, vol. 342, pp. 995–8, November 22 2013. [DOI] [PubMed] [Google Scholar]
- [41].Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, et al. , “Mutational landscape of aggressive cutaneous squamous cell carcinoma,” Clin Cancer Res, vol. 20, pp. 6582–92, December 15 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Colditz GA, Rosner BA, Chen WY, Holmes MD, and Hankinson SE, “Risk factors for breast cancer according to estrogen and progesterone receptor status,” J Natl Cancer Inst, vol. 96, pp. 218–28, February 4 2004. [DOI] [PubMed] [Google Scholar]
- [43].Fuchs-Young R, Shirley SH, Lambertz I, Colby JK, Tian J, Johnston D, et al. , “P53 genotype as a determinant of ER expression and tamoxifen response in the MMTV-Wnt-1 model of mammary carcinogenesis,” Breast Cancer Res Treat, vol. 130, pp. 399–408, November 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Reeves MQ, Kandyba E, Harris S, Del Rosario R, and Balmain A, “Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis,” Nat Cell Biol, vol. 20, pp. 699–709, June 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.