Abstract
A large fraction of proteins naturally exist as symmetrical homooligomers or homopolymers1. The emergent structural and functional properties of such protein assemblies have inspired extensive efforts in biomolecular design2-5. As synthesized by ribosomes, proteins are inherently asymmetric. Thus, they must acquire multiple surface patches that selectively associate to generate different symmetry elements needed to form higher-order architectures1,6 – a daunting task for protein design. Here we introduce an inorganic chemical approach to address this outstanding problem, whereby multiple modes of protein-protein interactions and symmetry are simultaneously achieved by selective, “one-pot” coordination of soft and hard metal ions. We show that a monomeric protein (protomer) appropriately modified with biologically inspired hydroxamate groups and Zn-binding motifs assembles through concurrent Fe3+ and Zn2+ coordination into discrete dodecameric and hexameric cages. Closely resembling natural polyhedral protein architectures7,8 and unique among designed systems9-13, our artificial cages possess tightly packed shells devoid of large apertures, yet they can assemble and disassemble in response to diverse stimuli owing to their heterobimetallic construction on minimal interprotein-bonding footprints. With stoichiometries ranging from [2 Fe:9 Zn:6 protomer] to [8 Fe:21 Zn:12 protomer], these protein cages represent some of the compositionally most complex protein assemblies–or inorganic coordination complexes–obtained by design.
In supramolecular design, cage-like architectures have featured prominently due to their aesthetically appealing structures and their isolated interiors that enable them to encapsulate molecular cargo and to perform selective chemical transformations14-18. Inspired by naturally occurring polyhedral assemblies, protein engineers have combined principles of symmetry with proper design and arrangement of non-covalent interfaces to build diverse supramolecular architectures9-13. Yet, some of the key structural features of natural protein cages have been difficult to emulate (Fig. 1a). First, each cage is invariably composed of asymmetric protomers, which possess multiple self-associative patches to simultaneously satisfy the symmetry requirements necessary to build polyhedral assemblies (i.e., concurrent generation of at least C2 and C3 symmetries, in addition to C4 or C5 symmetries for octahedra or icosahedra)1,6. Second, these self-associative patches collectively occupy a large fraction of the surface area on each protomer, enabling the formation of tightly packed shells with small apertures to enable influx and efflux of select species8. Third, although the inter-protomer interfaces in natural protein cages are extensive to ensure stable and selective association, they are also often conformationally flexible and chemically tunable, allowing the cages to undergo cooperative motions or disassembly in response to external cues7,19.
Figure 1 ∣. Design of protein cages.
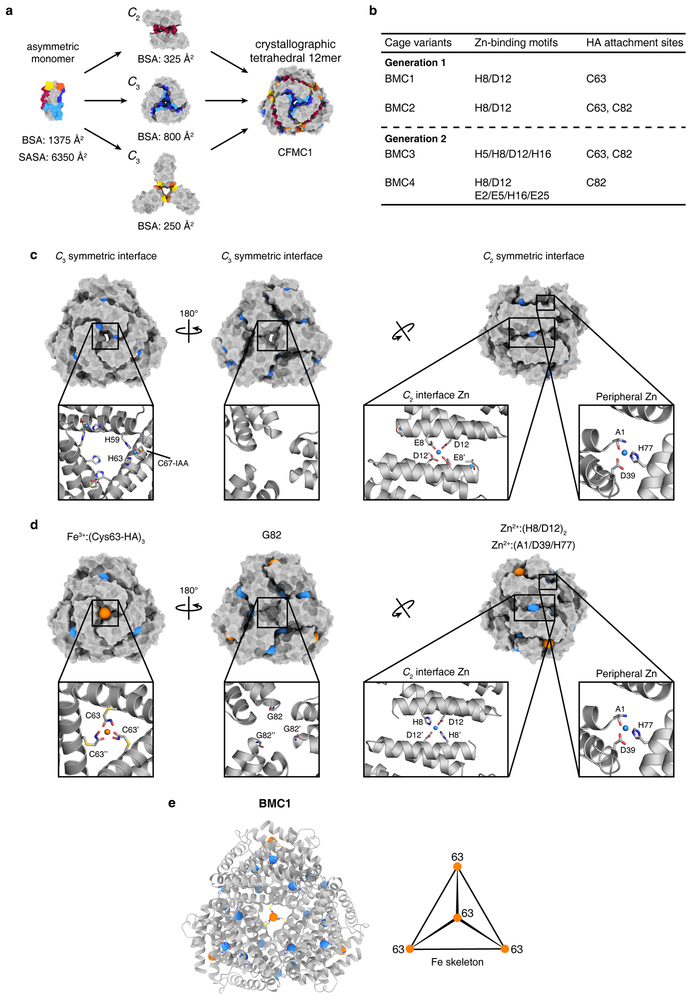
a, Representative examples of natural protein cages (DNPEP – aspartyl aminopeptidase, HuHF – human heavy-chain ferritin) and their assembly from asymmetric protomers. Per-protomer solvent-accessible surface areas (SASA) and buried surface areas (BSA) are indicated. Associative surfaces on the protomers are colored in red for homologous interactions and in orange/yellow or blue/cyan for heterologous interactions. b, C2-symmetric protein dimerization induced by tetrahedral Zn2+ coordination of native amino acid sidechains. c, C3-symmetric protein trimerization induced by octahedral Fe3+-tris-hydroxamate coordination. d, Scheme showing modification of native Cys sidechains with IHA to yield Cys-HA, which is isosteric with arginine (light grey). e, Zn-mediated solution dimerization and crystallization of CFMC1. f, Structural overview of the cytochrome cb562 scaffold. Salient structural elements are shown as sticks.
Given the difficulty of designing multiple, selectively associative surfaces on a protomer, construction of artificial cages has relied exclusively on using natively oligomeric proteins or designed peptides with Cn≥2 symmetries as building blocks and the design of a single type of binary protein-protein interactions (PPIs) through computation10, genetic fusion9,11, disulfide bond formation12 or metal coordination11,13. Although these strategies can yield polyhedral symmetries, the resulting architectures are highly porous, do not display externally controllable assembly/disassembly (with two exceptions)11,13 and cannot be easily modified to adopt alternative structures (i.e., they are not modular or flexible). Inspired by previous work on bimetallic supramolecular coordination cages20,21, we asked if these design problems could be addressed using an inorganic chemical approach, wherein a protomer is equipped with chemically orthogonal coordination motifs to self-assemble into polyhedral architectures.
Design of bimetallic protein cages
Previously, we have taken advantage of the simultaneous strength, lability and directionality of metal coordination bonds (particularly those formed by late first-row, low-valent transition metal ions) to effect the self-assembly of discrete protein complexes5 and extended 1, 2, and 3-D arrays22,23. Typically, selective nucleation sites for metal-mediated PPIs are formed by pairs of metal-binding amino acids (mainly His, Asp and Glu residues) or non-native bidentate functionalities (e.g., 2,2’-bipyridine, 1,10-phenanthroline and 8-hydroyxquinoline)5,24. However, all of these natural or synthetic coordination motifs can be considered as soft (or intermediate-soft) according to Hard-Soft Acid-Base (HSAB) classification25 and have considerable overlap in terms of their coordination preferences for soft, low-valent transition metal ions. Due to this lack of chemical discrimination, it has not been possible to design a heterometallic protein complex whose self-assembly is selectively guided by multiple metal ions that mediate different PPIs.
To achieve this goal, we turned to a bidentate chelating motif, hydroxamate (HA, the conjugate base of hydroxamic acid), a common functional group found in bacterial siderophores to enable exceptionally stable coordination of Fe3+.26,27 HA groups preferentially form octahedral Fe3+ complexes with an inherent C3 symmetry that we sought to impose on protein oligomerization (Fig. 1c). Importantly, the formation constants of Fe3+:(HA)3 complexes (>1028 M−3) are vastly higher than those of other metal-HA complexes such that they can be considered as orthogonal to the aforementioned soft metal-ligand combinations26,27. For protein derivatization, we synthesized a small reagent, iodo-hydroxamic acid (IHA), which selectively reacts with Cys residues (Fig. 1d, Extended Data Fig. 1). The resulting Cys-HA sidechain is isosteric with that of arginine and devoid of bulky aromatic moieties, furnishing a pseudo-natural amino acid functionality with the unique ability to chelate hard metal ions and induce C3 symmetric oligomerization on a single-residue footprint.
As a model system, we used cytochrome cb562, a monomeric four-helix-bundle protein which has proven to be a versatile building block for metal-directed protein self-assembly5. Earlier, we observed that a variant of cyt cb562 (CFMC1), designed and observed to form Zn-mediated dimers in solution, crystallized into rhombohedral lattices in which the protomers arranged into dodecameric, cage-like units via Zn-mediated crystal packing interactions (Fig. 1e)28. While Zn-mediated interactions were not sufficiently strong to maintain the tetrahedral dodecamers upon crystal dissolution, we envisioned that these lattice units could serve as a structural model to engineer the protomers such that they would form self-standing cages. Looking first to stabilize the C2 symmetric interfaces, we incorporated a bidentate His8-Asp12 motif to mediate the antiparallel association of two protomers along their Helices 1 via tetrahedral Zn2+ coordination (Fig. 1f). Given that C3 symmetric interfaces are small and heterologous (i.e., involve two different patches on each protomer) (Fig. 1e), they were unsuitable for stabilization by noncovalent interactions. Therefore, we focused on the central pores in each C3 symmetric substructure and identified positions 63 and 82 as suitable locations for installing Cys-HA functionalities, which would stabilize trimeric substructures via the formation of Fe3+:(HA)3 centers (Fig. 1f). Thus, we prepared two CFMC1 variants designated BiMetallic Cage 1 and BiMetallic Cage 2 (BMC1 and BMC2) (Fig. 1g). BMC1 and BMC2 both bear the His8-Asp12 motif on Helix 1 and the Cys63-HA along with the native peripheral Zn coordination sites (Ala1N-term, Asp39 and His77) of the parent CFMC1 structure. BMC2 additionally contains Cys82-HA (Fig. 1g, Extended Data Fig. 1).
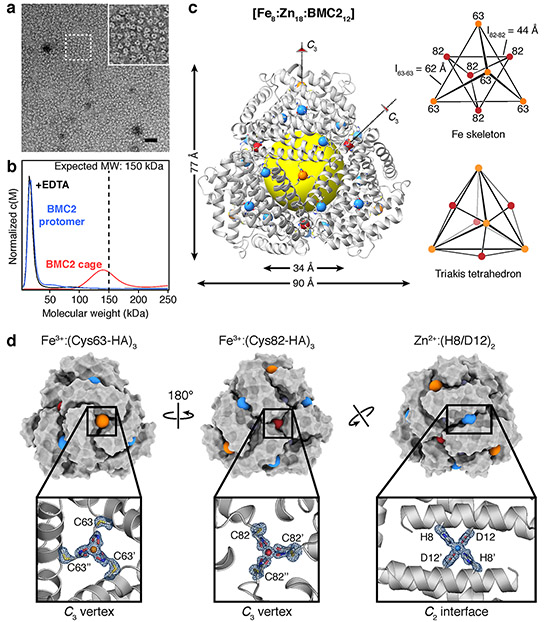
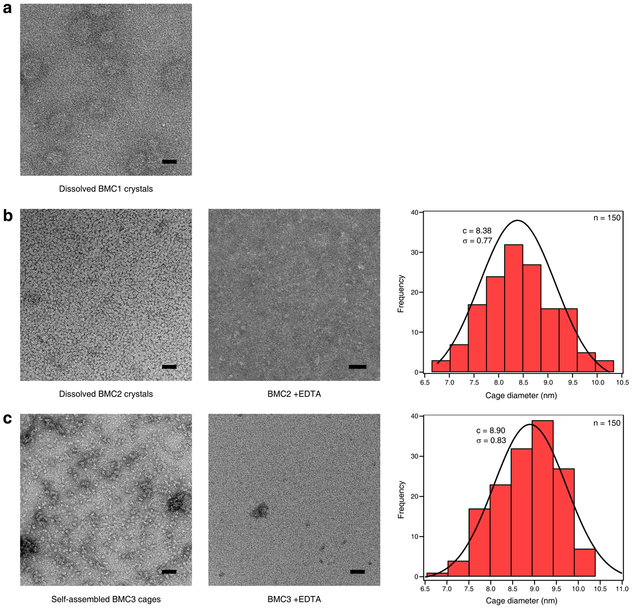
Crystals of BMC1 and BMC2 were obtained in the presence of near equimolar ZnCl2 and FeSO4. These crystals were isomorphic (R32 space group; a = b = 126±1 Å, c = 167±1 Å) with those of CFMC128, indicating that they possessed the same underlying lattice structure composed of dodecameric units (Extended Data Table 1 and Extended Data Fig. 2). Crystals were dissolved in a solution lacking the precipitating agent (PEG-400) and then analyzed by negative-stain transmission electron microscopy (ns-TEM) (Fig. 2a, Extended Data Fig. 3). The images revealed uniform particles with a diameter of 8.4 ± 0.8 nm in the case of BMC2 but not BMC1, implying that two HA coordination motifs are necessary for cage stability. Analysis of the same BMC2 solution by analytical ultracentrifugation (AUC) indicated a predominant species with a molecular weight (MWobs.) of ca. 140 kDa (Fig. 2b), approximating the calculated value (MWcalc. = 151 kDa) for a dodecamer. BMC2 particles dissociated upon treatment with ethylenediamine tetraacetic acid (EDTA), confirming their metal-dependent self-assembly (Fig. 2b and Extended Data Fig. 3).
Figure 2 ∣. Characterization of BMC2 cages.
a, ns-TEM of BMC2 cages obtained by the dissolution of 3D crystals; the inset is a close-up of the boxed region. Scale bar = 50 nm. b, AUC characterization of BMC2 protomers, BMC2 cages after crystal dissolution and after subsequent treatment with EDTA. c, Crystal structure of the BMC2 cage. Fe and Zn ions are represented as orange/red and blue spheres, respectively. The central cavity is highlighted by a yellow sphere. Two types of C3 vertices formed by Fe:(Cys63-HA)3 and Fe:(Cys82-HA)3 coordination motifs form two superimposed tetrahedra to generate a triakis tetrahedron. d, Surface representations of the BMC2 cage, with metal ions shown as colored spheres. Atomic details of each metal coordination site are shown in the insets, with the mFo-DFc electron density omit map (blue mesh) contoured at 3 σ.
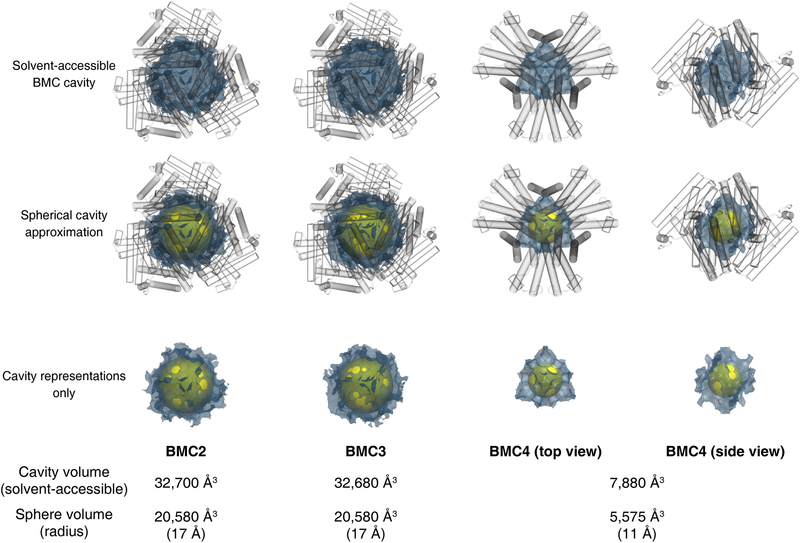
The crystal structure of the BMC2 cage was determined at 1.4 Å resolution (Extended Data Table 1), revealing a compact structure with the shape of a truncated tetrahedron, outer dimensions of 80 × 90 Å and a cavity volume of 32,700 Å3 (Fig. 2c, Extended Data Fig. 4). Like natural protein cages, the shell is tightly packed whereby the largest opening measures <4 Å across. Nearly 30% of the surface area of each protomer (1700 Å2 out of 6500 Å2) is buried in interfaces despite a design footprint of only four amino acids (His8, Asp12, Cys63 and Cys82).
The full complement of metal ions, comprising eight Fe ions (four each in the C3 symmetric pores) and eighteen Zn ions (six in C2 interfaces and twelve in peripheral sites) are clearly resolved (Fig. 2c). Anomalous X-ray diffraction data collected at and below Fe and Zn K-edges indicate that the designed Fe- and Zn-coordination sites exclusively bind their cognate ions with no evidence of crosstalk (Extended Data Fig. 5, Supplementary Tables 3-6), which establishes that the metal-dependent self-assembly of BMC2 cages occurs with absolute chemical selectivity. The Fe centers form the eight C3 vertices of a triakis tetrahedron, a Catalan solid with twelve equivalent faces (Fig. 2c). It can be viewed as the superposition of two tetrahedra: four Fe centers are coordinated by Cys63-HA motifs generate the larger of these two tetrahedra (ledge = 62 Å), and four Fe centers coordinated by Cys82-HA motifs produce the smaller one (ledge = 44 Å) (Fig. 2c). The BMC1 structure, in comparison, has a regular tetrahedral arrangement of four Fe centers as it lacks the Cys82-HA group (Extended Data Table 1, Extended Data Fig. 2).
As designed, edges of the BMC2 tetrahedra are formed by six Zn ions located centrally in C2 interfaces (Fig. 2d). The Fe3+:(Cys63-HA)3 and Fe3+:(Cys82-HA)3 motifs display near-ideal octahedral geometries (Fig. 2d), with the former in Λ and the latter in Δ configuration (Extended Data Fig. 5). Interestingly, the Fe3+:(Cys82-HA)3 center also adopts an alternative conformation (20% abundance) owing to the flexibility of the Cys-HA sidechain (Extended Data Fig. 5k). All Fe-O bond distances are in the range of 1.95-2.1 Å, which are typical of Fe3+:(HA)3 complexes29. Given that a Fe2+ precursor was used to initiate self-assembly and the low reduction potentials of Fe:HA3 centers (Ered < −400 mV)26, this observation suggests that the protein self-assembly involves the initial formation of Fe2+:HA centers, followed by the thermodynamically favored oxidation of these species into Fe3+ either by the Fe3+-heme centers embedded in each protomer (see Methods) or directly by ambient O2.
Reversible assembly of dodecameric cages
Despite the stability of isolated BMC2 cages, their formation required an initial crystallization step. We reasoned that slow crystal nucleation/growth kinetics and the high attendant protein and metal concentrations likely increased the fidelity and yield of the complex self-assembly process to produce a discrete supermolecule consisting of 12 protomers and 26 metal ions of two different kinds. With the idea that strengthened Zn-mediated interactions across the C2 interface could increase the efficiency of cage self-assembly in solution, we generated two second-generation variants based on BMC2: BMC3 and BMC4 (Fig. 1g, Extended Data Fig. 1). In BMC3, the Helix 1 surface was engineered to form two Zn-coordination sites (composed of His5, His8, Asp12 and His16) across the C2 interface, whereas in BMC4 three potential Zn-coordination sites were engineered (one central site composed of two His8/Asp12 pairs as in BMC3 and two peripheral sites composed of Glu2, Glu5, His16 and Glu25). In BMC4, we also removed the Cys63-HA group with the purpose of eliminating any potential undesired assembly products that involve heteromeric Fe3+ coordination by Cys63-HA and Cys82-HA.
BMC3 indeed formed dodecameric cages in solution with high yields (>80%) as determined by ns-TEM and AUC measurements (Fig. 3a, Extended Data Fig. 3). The 1.85-Å crystal structure confirmed the eight Fe centers in the vertices and the two Zn coordination sites in each C2 interface (twelve total) as well as nine of the twelve possible peripheral Zn sites, which complete a [8 Fe:21 Zn:12 protomer] architecture (Fig. 3b). Importantly, the self-assembly of BMC3 cages in solution was dependent on the presence of both Fe and Zn ions. The absence of either metal ion or the addition of various other first-row transition metal ions instead of Fe led to smaller oligomeric forms of BMC3 or non-specific assemblies (Fig. 3a and Extended Data Fig. 6a). BMC3 cages also formed with a Fe3+ precursor, Fe(acetylacetonate)3, (Extended Data Fig. 6b). Consistent with self-assembly under thermodynamic control, dodecameric cage formation was independent of the order of the addition of Fe or Zn ions. We determined the single-particle cryo-EM structure of isolated BMC3 cages at a resolution of 2.6 Å (Fig. 3c; Extended Data Table 2). A major portion of the assembly could be resolved at ≤2.0 Å (Extended Data Fig. 7). At this resolution, nearly all side chains, Zn coordination sites and some ordered water molecules are clearly distinguished (Fig. 3d). Consistent with the crystallographically observed flexibility of Fe3+:(Cys-HA)3 coordination sites, electron densities in the C3 vertices are diffuse and some sidechains that display high temperature factors in the crystal structure are found in alternative conformations in the cryo-EM structure (Extended Data Fig. 7). These observations confirm that the solution architecture of the BMC3 cage closely reflects the solid-state structure. Due likely to lattice packing, the latter is isotropically compressed by ca. 2-3 Å compared to the former (Fig. 3e), which can be accommodated by slight changes in interfacial metal coordination.
Figure 3 ∣. Characterization of BMC3 cages.
a, AUC characterization of BMC3 self-assembly. b, Surface representation of the BMC3 cage (as derived from the crystal structure), oriented to show the incorporation of two Zn ions at the C2 symmetric interface. c, 2.6-Å density map for the BMC3 cage as determined by cryo-EM. d, Atomic details of both Zn-binding sites of the 2-fold interface overlaid with the electron density mFo-DFc omit map from the crystal structure (left) and as observed for the cryo-EM structure (right). Additional sidechains and waters are shown for the cryo-EM structure to emphasize structural robustness of the interface. e, Overlay of the BMC3 X-ray and cryo-EM structures to highlight the isotropic expansion of the cage in the absence of crystallographic packing interactions.
Next, we examined the assembly/disassembly behavior of BMC3 cages in response to different stimuli. BMC3 cages readily disassemble by treatment with EDTA (Extended Data Fig. 6c). They were stable at 50 °C but dissociated upon incubation at 70 °C (Extended Data Fig. 6d). A key feature of siderophores is that their cellular release of Fe is promoted by the destabilization and labilization of their Fe3+:(HA)3 centers through reduction to the Fe2+ form in the cytosol27. Along these lines, the treatment of BMC3 cages with a strong reductant (dithionite; Ered. < −500 mV)30 led to their disappearance and emergence of monomeric species (Extended Data Fig. 6e). In contrast, a weaker reductant (ascorbate; Ered > −100 mV at pH 7)31 with a reduction potential higher than that of Fe3+:(HA)3 had considerably less effect (Extended Data Fig. 6e), suggesting that BMC3 cage disassembly occurs through the reduction of the Fe centers. These observations establish BMC3 cages as a distinctive system among natural and artificial protein architectures whose assembly and disassembly can be controlled through multiple stimuli: chemical, thermal or redox. BMC3 cages can passively encapsulate small fluorogenic molecules either in their lumen or inter-protomer interfaces, retain them for several days and release them upon treatment with EDTA (Extended Data Fig. 8).
Formation of a hexameric cage
Surprisingly, AUC measurements indicated that the other second-generation variant, BMC4, self-assembled as a hexamer upon Fe and Zn coordination, with yields >70% (Fig. 4a). The 1.50-Å resolution crystal structure of the BMC4 assembly revealed a D3 symmetric, cage-like architecture with a composition of [2 Fe:9 Zn:6 protomer], outer dimensions of 75 Å × 50 Å and a cavity volume >7,800 Å3 (Extended Data Fig. 4). The overall shape is trigonal bipyramidal (Fig. 4b), which is the smallest polyhedral architecture with a sizeable interior cavity that can be constructed from an asymmetric building block. The apical vertex of each pyramidal half is formed by a Fe3+:(Cys82-HA)3 motif shared by three protomers (Fig. 4b). These C3 symmetric vertices are further reinforced by Zn2+ ions that link pairs of protomers through Asp21, Glu25 and His77 coordination (Fig. 4c). The pyramids are joined by three equatorial, C2 symmetric vertices mediated by Zn centers coordinated to Glu5 and His8. (Fig 4b, c). A comparison to the BMC2/3 cages indicates that this unexpected coordination motif requires a ca. 9-Å slip of each protomer along the C2 symmetric interfaces (Fig. 4d). The shift drastically reduces the C2 symmetric contact area between protomers, effectively transforming the edges in the tetrahedral BMC2/3 cages to vertices in the trigonal bipyramidal BMC4 cages. BMC4 cages exhibited similar thermal stability as BMC3 cages, with both species disassembling at <70 °C. The thermal robustness of the BMC3 and BMC4 cages appear to be limited, at least partially, by the relative instability of the individual protomers (Extended Data Fig. 6d).
Figure 4 ∣. Characterization of BMC4 cages.
a, AUC characterization of BMC4 self-assembly. b, Crystal structure of the BMC4 cage. Fe and Zn ions are represented as red and blue spheres, respectively. The central cavity is highlighted by a yellow sphere. The structural skeleton formed by Fe and Zn ions is shown below the structure. c, Surface representations of the BMC4 cage, with metal ions shown as colored spheres. Atomic details of each metal coordination site are shown in insets, with the mFo-DFc electron density omit maps (blue mesh) contoured at 3 σ. d, Comparison of the C2 symmetric protein interfaces in different BMC constructs. The residues 8 and 12, common to all constructs, are colored in purple. The slippage of the 2-fold helix interface to accommodate the hexameric architecture of BMC4 is indicated with red arrows. e, Comparison of the apical angle formed by the Fe:(Cys82-HA)3–mediated vertices in BMC3 and BMC4 cages.
The large structural transformation is accompanied by a reduction of the apical angle formed at the Fe3+:(Cys82-HA)3–mediated vertices from 101° in the BMC2/3 cages to 81° in the BMC4 cage (Fig. 4e). This observation highlights the conformational adaptability of the Fe3+:(Cys82-HA)3 coordination motif, enabling it to accommodate different polyhedral geometries. Such behavior is reminiscent of the interfacial flexibility in some icosahedral virus capsids in which the same protomer can form hexamers on capsid faces and pentamers on capsid vertices7. It is worth noting that BMC4 contains all of the Zn-coordinating residues on Helix 1 to form the C2 symmetric interfaces observed in the dodecameric BMC3 cage, indicating that the self-assembly process selects an alternative interfacial arrangement of lower free energy, enabled by the reversibility of metal coordination interactions. In terms of protein design, a caveat of interfacial flexibility is that it may lead to non-specific or unintended self-assembly products, although it can also allow error-correction during self-assembly and increase tolerance to design imperfections.
Conclusions
The self-assembly and function of biomolecular systems are predicated upon their specificity, stability and adaptiveness, which, in turn, are enabled by extensive networks of non-covalent interactions. Here, we have shown that fundamental concepts in inorganic coordination chemistry can be applied to achieve all of these attributes in protein self-assembly, and specifically, to construct complex polyhedral protein architectures from a simple, asymmetric building block. Despite their minimal design footprints, these cage-like architectures are distinguished by their structural compactness and responsiveness which are hallmarks of evolved systems like viral capsids. Key to our construction strategy was the reimagination of a biological coordination motif, hydroxamic acid, within a new structural context, i.e., as a new amino acid sidechain with the ability to chelate hard metal ions. This example expands the growing lexicon of post-translational modifications that broaden the chemical scope of proteins.
Methods
Synthesis of the IHA ligand.
O-tritylhydroxylamine was synthesized as previously described32. Chloroacetyl chloride (0.58 mL, 7.3 mmol) was dissolved in 2 mL of CH2Cl2 and added dropwise to a suspension of O-tritylhydroxylamine (2.0 g, 7.3 mmol) and N,N-Diisopropylethylamine (2.5 mL, 14.5 mmol) in 15 mL of CH2Cl2 at 0 °C. The reaction mixture was gradually warmed to room temperature and stirred at room temperature for an hour. An additional 15 mL of CH2Cl2 was added and the reaction was extracted with H2O (3 × 30 mL). The CH2Cl2 solution was collected and evaporated to dryness. A solution containing 15 mL of CH2Cl2 with 10% (v/v) trifluoroacetic acid was added and the solution was stirred for 30 min. The crude product was purified by silica gel chromatography using a gradient of 0–100% ethyl acetate in hexanes as the eluent. The product was visualized using a FeCl3 stain. Yield = 55%. Measured molecular weight (m/z): 108.37 (M − H+); Calculated: 107.99 (M − H+).
2-chloro-N-hydroxyacetamide (400 mg, 3.7 mmol) and NaI (2.7 g, 18.3 mmol) were refluxed in 30 mL of acetone for 1 h. The reaction mixture was purified by silica gel chromatography with 100% ethyl acetate as the eluent and dried in vacuo. Yield >90%. Measured molecular weight (m/z): 223.85 (M + Na+); Calculated: 223.95 (M + Na+).
Protein expression and purification.
All constructs (detailed in Supplementary Table 1) were derived from the parent pET-20b(+) plasmid containing the CFMC1 gene via site-directed mutagenesis as previously described28,32,33. The appropriate plasmids were transformed into BL21(DE3) E. Coli cells (New England Biolabs) housing a CCM (cytochrome C maturation) cassette containing a chloramphenicol resistance marker and expressed as previously described34 with minor adjustments. Multiple 2.8 L flasks containing 1.5 L of LB media were shaken at 200 RPM for 12 hours at 37 °C and then slowed down to 100 RPM for an additional period of ca.7 hours. Cells were harvested by centrifugation (5000 RPM for 10 min at 4 °C), resuspended in a buffered solution containing 5 mM sodium acetate (NaOAc) (pH 5.0) and 2 mM dithiothreitol (DTT) and lysed via sonication. The pH of the crude lysate was first raised to 10 using NaOH to precipitate cellular contaminants, then reduced to pH 4.5. After centrifugation (12000 RPM for 20 min at 4 °C), the clarified supernatant was decanted and diluted 15-fold with additional buffer. This solution was applied to a CM Sepharose gravity column (GE Healthcare) pre-equilibrated with the aforementioned buffer and subjected to multiple buffer washes prior to elution using a stepwise-gradient of NaCl (0-0.5 M). Peak elution fractions were combined and concentrated using a 400 mL Amicon Stirred Cell (Millipore) and buffer-exchanged via overnight dialysis against a buffered solution containing 10 mM phosphate (pH 8.0) at 4 °C. Next, the protein was purified via a DuoFlow workstation station fitted with a Macroprep High Q-cartridge column (BioRad) and eluted using a linear gradient over 0-0.5 M NaCl. Fractions that exhibited an RZ ratio (A421/A280)>4.4 were pooled, treated with 2 mM ethylenediamine tetraacetic acid (EDTA) for 1 hour, concentrated, and buffer-exchanged into 20 mM tris(hydroxymethyl)aminomethane (Tris) (pH 7.5) pretreated with Chelex 100 resin (BioRad), via desalting column (Econo-Pac 10DG pre-packed columns, BioRad). Demetallated and purified proteins were concentrated to ca. 2 mM and stored at 4 °C.
Protein labeling and post-labeling purification.
Purified protein solutions were treated with 100-fold excess of DTT and placed in an anaerobic Coy chamber for ca. 2 hours for slow degassing to remove dissolved oxygen. The fully reduced protein solution was buffer-exchanged into 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5) via desalting column to remove DTT, and the concentration of the resulting protein solution was determined spectroscopically (Agilent 8452 spectrophotometer) using the ε421 (red) = 162,000 M−1 cm−133. Solid iodohydroxamic acid (IHA) was dissolved in 100 μL degassed DMF to generate solutions containing a 15-fold excess IHA per protein monomer, which were then added to protein aliquots and incubated overnight. The HA-functionalized variants were removed from the Coy chamber and separated from unreacted or partially reacted protein via FPLC using a Q-column equilibrated with 10 mM N-Cyclohexyl-2-aminoethanesulfonic acid (CHES) (pH 9.3) and 2 mM DTT and eluted using a linear gradient over 0-0.5 M NaCl. Protein functionalization was verified using ESI-MS (Extended Data Fig. 1) and the resulting protein solutions were buffer-exchanged into demetallated 20 mM Tris (pH 7.5) via desalting column, concentrated to ca. 2 mM, and stored at 4 °C for further use.
Redesign of CFMC1 interfaces.
To render the CFMC1 protomer competent for the bimetallic design strategy, we first performed the following mutations to remove potential competitive interactions: C67E, H59S and H73N. A negative design strategy was then employed to disrupt a noncovalent dimerization interface found in CFMC1, leading to the mutations A34Q and A38Q. We further identified the dearth of protein-protein interactions within the core and periphery of the 3-fold axis engulfing the 82 position as a likely contributor to poor cage assembly and crystallization in general. Accordingly, as a means to facilitate cage formation, we adopted Rosetta-prescribed mutations at the following positions: A24T, Q25(T/E), N80K and E81Q.
Crystallography.
Screening and crystallization of all BMC variants were conducted via sitting drop vapor diffusion. Briefly, solutions containing 2.1-2.2 mM BMC protomer were mixed with mother liquor (1 μL + 1 μL) and equilibrated against 200 μL reservoir volumes. Supplementary Table 2 details the experimental conditions for crystal growth. Protein solutions of BMC1 and BMC4 were first incubated with FeSO4 for 1 hour prior to mixing with ZnCl2. Solutions of BMC2 and BMC3 were mixed with FeSO4 and ZnCl2 stock solutions and were immediately combined with the mother liquor (to prevent rapid aggregation of the proteins functionalized with two HA units). Crystals for all mutants typically appeared within several hours and were harvested within a week of maturation. Crystals were cryoprotected by submersion into perfluoropolyether cryo oil (Hampton Research) for a few seconds and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at 100 K at either the Advanced Light Source (ALS) beamline BL 8.3.1 (using 1.12-Å radiation for BMC3 and 1.33-Å radiation for BMC4) or at the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines 9-2 (using 0.98-Å radiation for BMC2) and 12-2 (using 0.98-Å radiation for BMC1). Data integration was performed using the XDS Program Package, truncated at CC1/2 > 0.535Datasets of the same structure recorded at different wavelengths were scaled to the highest resolution dataset with XSCALE36,37. Phaser-MR38 was employed to carry out molecular replacement with search models based on the CMFC1 monomer (PDB ID: 3M4B) containing the expected side chain mutations (generated in Pymol39) but lacking HA. Rigid-body and structure refinement was performed using multiple rounds of Phenix.refine38, interspersed with manual model rebuilding and metal/ligand placement with COOT40. Restraint files for the Cys-hydroxamic acid conjugates were generated using phenix.eLBOW to maintain the distances Cys-SG/HA-C1 (1.816 Å ± 0.02 Å) and angles Cys-CB/Cys-SG/HA-C1 as well as Cys-SG/HA-C1/HA-C2 (both 109° ± 3°) during refinement. Where necessary, the metal binding geometry of the hydroxamic acids was restrained to the distances Fe/HA-O1 (1.98 Å ± 0.05 Å) and Fe/HA-O2 (2.057 Å ± 0.05 Å) as well as through a planarity constraint for the atoms Fe/HA-O1/HA-O2/HA-C1 following data from a high-resolution structure of Fe(III)-tris-benzhydroxamate trihydrate41. Simulated annealing omit maps (metal atoms and sidechain ligands) were generated for each metal binding site and model accuracy was assessed critically against these omit maps. Electron density maps were generated using Phenix and all molecular graphics images were produced with either Pymol or the UCSF ChimeraX package from the Computer Graphics Laboratory, University of California, San Francisco42.
Crystallographic metal content analysis.
Metal ions, with their relatively high-energy inner electrons, can absorb and resonate with soft X-rays leading, amongst other effects, to differences in the intensity of otherwise centro-symmetric Bragg diffraction peaks used for X-ray crystallography. Density maps calculated from these differences are routinely used to locate and identify metal ions in protein crystals. The magnitude of this anomalous X-ray diffraction varies with the X-ray energy, with stark differences around the energies of the K- and L-shell electrons of the respective element allowing one to discern between elements at a position in question, if diffraction datasets are measured at the appropriate wavelengths. For a visual analysis of the bound metals, the scaled datasets of different wavelengths were used separately as an input for a single phenix.refine run each with the final model of the highest resolution dataset. Importantly, only the B-factor or occupancy were allowed to change during refinement, resulting in anomalous difference density maps for each wavelength. Using these maps, isomorphous difference maps from data at wavelength above and below the respective element K-edges were generated (if applicable) with Phenix and were inspected manually (Extended Data Fig. 5). To gain a more quantitative understanding of the identity of the bound metals for each site, the anomalous difference signal of each dataset was used to generate ccp4 format maps with phenix.mtz2map. The generated maps were used subsequently as inputs to calculate the mean signal in a sphere of 1 Å radius centered on each metal atom with the program MAPMAN (Uppsala Software Factory). For each pair of datasets above and below a metal-absorption edge, the ratio of the anomalous signal above and below the edge for every metal atom was tabulated. The experimental ratio was compared to the theoretical ratio for both Fe and Zn (Extended Data Fig. 5) according to http://skuld.bmsc.washington.edu/scatter, which were calculated using the Cromer and Liberman approximation. Theoretical ratios were also calculated for hypothetical mixed occupancy Fe/Zn metal sites and compared to experimentally observed values (Supplementary Tables 3-6).
Protein cage sample preparation.
Self-assembled cages:
All samples were prepared in a low-[O2] atmosphere (Coy glovebox) to prevent undesired oxidation of Fe2+ ions prior to self-assembly. Protein solutions containing 20 μM BMC3 or 100 μM BMC4 in 20 mM Tris (pH 8.5) were incubated with either 20 μM FeSO4, 60 μM ZnCl2 for BMC3 or 50 μM FeSO4 and 200 μM ZnCl2 for BMC4 for 2-3 h to yield the metallated cages. We note that the addition of FeSO4 was followed by a small but observable change in the color of the solution from red to pink, attributed to a shift of the heme Soret band to longer wavelengths, which suggested reduction of the heme by the ferrous ions and generating ferric ions in close proximity to HA group(s). The final BMC3 solutions were then concentrated 7-fold prior to overnight incubation to improve the total cage yield. After self-assembly, the resulting solutions were diluted back to their original concentrations with the self-assembly buffer prior to characterization.
Dissolved crystals:
Fe:Zn:BMC1 and Fe:Zn:BMC2 crystals were dissolved using buffer containing 100 mM HEPES (pH 7.5), 200 mM MgCl2 and 800 μM ZnCl2. Mature crystals were removed from their pedestal droplet, briefly submerged in fresh buffer to remove uncrystallized protein and surface-bound precipitates and transferred into a new sitting drop crystallization well containing 8 μL of the buffer solution. The crystals were physically crushed with a small metal scalpel and vigorously pipetted until a large portion of the crystals dissolved. Undissolved crystals were removed by centrifugation (10,000 RPM for 5 min at 25 °C), yielding a light-red supernatant and dark-red precipitate.
Negative-stain transmission electron microscopy.
A 4-μL droplet of BMC cages (either self-assembled or from dissolved crystals) was deposited onto formvar/carbon-coated Cu grids (Ted Pella, Inc.) (pretreated by negative-mode glow discharge up to 15 min prior) and allowed to bind for 5 min. The grids were then washed with 50 μL of MilliQ water, blotted using Whatman filter paper and stained using 2% uranyl acetate solution in water and blotted again. Grids were imaged using a FEI Sphera transmission electron microscope operating at 200 keV, equipped with a LaB6 filament and a Gatan 4K CCD camera. Micrographs were collected using objective-lens underfocus settings ranging from 250 nm to 2 μm and analyzed using Fiji (http://fiji.sc/Fiji).
Oligomerization state determination using analytical ultracentrifugation (AUC).
Sedimentation velocity measurements were performed at 41000 RPM and 25 °C using an XL-1 analytical ultracentrifuge (Beckman Colter) equipped with an An-60 Ti rotor. Data processing was performed using Sedfit43 with the following parameters as calculated using SEDNTERP — Viscosity: 0.01000 poise, Density: 0.9988 g/mL (self-assembled samples) or Viscosity: 0.0113191 poise, Density: 1.0196 g/mL (dissolved crystals), and a partial specific volume of 0.7313 ml/g for all samples. All reported results correspond to a confidence level of 0.95.
Preparation of samples involving crystal dissolution.
Dissolved crystal samples (BMC1 and BMC2), prepared as described above at ambient conditions, were diluted to 350 μL with 10 mM HEPES (pH 7.5), 200 mM MgCl2, and 800 μM ZnCl2. The solution was clarified via brief centrifugation in order to remove crystal debris and the supernatant was placed inside the cells.
Calculation of BMC void volumes.
Structures of complete cage assemblies for BMC2, BMC3, and BMC4 were generated via the application of crystallographic symmetry operations to the fully-refined asymmetric unit of each construct. These coordinates were recentered at the origin and stripped of waters, hydrogens, alternative conformations and crystallization reagents (PEG-400). Volumetric maps and volumes for the internal cavity of each cage were calculated using VOIDOO44, and are reported as the solvent-accessible volume for a 1.4 Å rolling probe on a 0.25 Å grid spacing for all constructs. The cavity volumes using these parameters were determined to be approximately 32,700 Å3 (BMC2), 32,700 Å3 (BMC3), and 7,900 Å3 (BMC4).
Solution self-assembly, disassembly, and thermal stability of BMC3 and BMC4.
Assembled samples were prepared as described above and placed inside the AUC measurement cells anaerobically (20 μM BMC3 and 100 μM BMC4). Disassembly of the cages via metal-ion removal was performed by treating the protein cages with 2 mM EDTA for 1 h. Redox-controlled disassembly of the protein commenced by the addition of either 5 mM sodium dithionite or 5 mM sodium ascorbate to the cage solution anaerobically and subsequent incubation of the samples at ~22 °C for 16 h. Samples were then loaded into the AUC measurement cell.
For thermal stability measurements, samples were placed in a thermoregulated chamber preequilibrated at the appropriate temperature for 2 h, then subsequently removed from the chamber and equilibrated at room temperature for 30 min prior to AUC analysis. Circular dichroism (CD) spectra were measured using an Aviv 215 spectrometer. CD measurements were performed using 10 μM protein in a buffered solution containing 20 mM Tris (pH 8.5). Thermal melts were measured at 222 nm at a 1 nm slit width, scanning at 1 nm intervals with a 1 s integration time. Measurements were taken from 25 °C to 85 °C at 2-degree intervals with a 2 min equilibration at each temperature. Unfolding data were fit to a two-state model with van’t Hoff’s enthalpy using the CalFitter web server.45
Cryo-EM sample preparation.
Self-assembled BMC3 cages were removed from the anaerobic Coy chamber immediately prior to grid preparation. A 3.5-μL aliquot of self-assembled BMC3 cages was dropped onto holey carbon grids (Electron Microscopy Sciences, Quantifoil R1.2/1.3 holey carbon on 300 mesh copper) that had been freshly glow-discharged for 30 s. The initial application of the sample was side blotted manually with Whatman No. 1 filter paper immediately followed by a secondary application of a 3.5-μL aliquot, blotted for 3.5 s and plunge-frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (FEI).
Cryo-EM data acquisition and image processing.
Samples were imaged on a Titan Krios G3 transmission electron microscope (FEI) operating at 300 kV equipped with a K2 Summit direct electron detector (Gatan) and a GIF Quantum energy filter. The slit-width of the energy filter was set to 10 eV. Movies were collected at a magnification of 165,000x in EFTEM mode giving a physical pixel size of 0.84 Å/pixel. In total, 4,672 movie stacks (50 frames/movie) were collected using a 10 s exposure at a dose rate of 1.2 e−/Å2 per frame for a total electron dose of 60 e−/Å2 per movie. Objective-lens underfocus settings varied between 0.6 μm and 1.6 μm. Data collection was performed using software EPU (FEI). All image processing was performed in the Relion-3.0 pipeline46. Motion-correction and dose-weighting was performed using MotionCor247, and defocus values were estimated with Gctf48 using a pixel size of 0.8 Å /pixel. A total of 3,513 movie stacks were selected following motion correction and CTF estimation, and 805,156 particles were auto-picked using RELION-3.0. Particle images were extracted and binned by 2 (1.6 Å/pixel, 100 pixel box size) and subjected to two-dimensional (2D) classification. A total of 444,247 particles were selected corresponding to good 2D class averages and subjected to three-dimensional (3D) classification imposing T symmetry and using an initial model generated from a subset of the particles. A total of 129,653 particles were chosen from a 3D class showing strong secondary-structural elements and subjected to 3D auto-refinement with T symmetry. The particles were re-centered and re-extracted to their original pixel size of 0.8 Å/pixel. These particles were subjected to 3D auto-refinement with T symmetry and the yield map was then postprocessed towards 2.6 Å resolution based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion. The pixel size of the map was manually adjusted using Relion image handler to match the physical pixel size of the images. Local resolution was calculated in Relion 3.0 using ResMap49.
Model building and refinement.
The BMC3 crystal structure (PDB ID: 6OT7) stripped of hydrogens and waters was used as an initial model and manually docked into the cryo-EM density using UCSF Chimera50. The structural model was subject to real space refinement in Phenix against the cryo-EM map with geometry restraints for the Fe-binding sites and molecular coordinates for the Cys-HA ligand. The atomic model was manually improved using Coot. Tightly-bound waters were identified based on clear density in the EM density map. Whereas the structural flexibility of the hydroxamate sites manifested in poor electron density, the 2-fold interface was much more rigid and unambiguous density was observed for Zn-binding. A tryptophan at the 66 position, which had shown high temperature factors in the BMC3 crystal structure, was identified in multiple conformations in the EM density map. The final model was subjected to real space refinement using Phenix38 and evaluated using MolProbity51. All molecular graphics images were rendered in PyMoL or UCSF ChimeraX.
Encapsulation of Rhodamine in BMC3 cages.
BMC3 cages were self-assembled in a low-O2 atmosphere in the presence of rhodamine for the passive encapsulation of the dye. Solutions containing 20 μM BMC3 were incubated with 20 μM FeSO4, 60 μM ZnCl2, and 2 mM rhodamine. A control sample was prepared in the absence of added metal ions (20 μM BMC3 incubated with 2 mM rhodamine). Samples were incubated for 2-3 h and concentrated 7-fold prior to overnight incubation. Protein solutions were buffer exchanged on a PD-10 desalting column using a buffer containing 20 mM Tris (pH 8.5) (with 5 μM FeSO4 and 10 μM ZnCl2 supplemented for solutions already containing metal ions) to separate unassociated dye from protein. Cage solutions were split in two: one half was treated with 1 mM EDTA and incubated for 2 h prior to washing. All protein solutions were additionally washed 3x using a centrifugal filter to completely remove any remaining free rhodamine.
Fluorescence measurements were performed using 6 μM protein solutions after the previously mentioned wash steps. For each sample, an excitation wavelength of 555 nm with a 2 nm slit width was used and emission was measured between 560 and 650 nm with a 2 nm slit width and 0.2 s integration time. For the time-course experiments, cages encapsulating rhodamine were washed 3x after 4 d and after 7 d and diluted to 6 μM prior to fluorescence measurements. AUC measurements were performed at the λmax of the cytochrome (415 nm) and at the λmax of rhodamine (555 nm) to assess whether there was a sufficiently large rhodamine signal associated with BMC3 cages. UV-vis absorbance measurements were performed on each solution to measure the protein and rhodamine concentrations. Difference spectra were taken between each rhodamine-incubated sample and BMC3 protomer to eliminate any background signal.
Data Availability
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the corresponding authors on request.
Structural data obtained by X-ray crystallography and cryo-EM have been deposited into the RCSB and EMDB data banks under the following accession codes: 6OT4 (BMC2), 6OT7 (BMC3), 6OT8 (BMC4), 6OT9 (BMC1) and 6OVH (BMC3 Cryo-EM) or EMD-20212 at The Electron Microscopy Data Bank.
Statistics and Reproducibility
All reported samples represent technical replicates. The ns-TEM micrograph of BMC2 cages after 3D crystal dissolution (Fig. 2a) is representative of experiments repeated independently four times. AUC experiments for BMC2 (Fig. 2b) were performed in duplicate. Self-assembly of BMC3 cages and subsequent AUC characterization (Fig. 3a) were performed the following number of times: BMC3 protomer (n=2), +Fe2+ (n=4), +Zn2+ (n=4), +Fe2, +Zn2+ (n=6). Self-assembly of BMC4 cages and subsequent AUC characterization (Fig. 4a) was performed the following number of times: BMC4 protomer (n=2), +Fe2+ (n=1), +Zn2+ (n=1), +Fe2, +Zn2+ (n=5). Mass spectra (Extended Data Fig. 1c-f) were collected in duplicate for native and HA-labeled proteins; AUC experiments were performed in duplicate. TEM characterization of BMC constructs (Extended Data Fig. 3) were performed the following number of times: dissolved BMC1 crystals (n=1), dissolved BMC2 crystals (n=4), BMC2 +EDTA (n=2), self-assembled BMC3 cages (n=5), BMC3 +EDTA (n=4). AUC experiments following the incubation of BMC3 with first-row transition metals (Extended Data Fig. 6a) were performed in duplicate. Self-assembly of BMC3 in the presence of Fe(acetylacetonate)3 (Extended Data Fig. 6b) was performed in duplicate. BMC3 cage disassembly in the presence of EDTA (Extended Data Fig. 6c) was performed in triplicate. AUC characterization of BMC variants after equilibration at different temperatures (Extended Data Fig. 6d) was performed the following number of times: BMC3 at 50 °C (n=2), BMC3 at 70 °C (n=2), BMC4 at 50 °C (n=3), BMC4 at 70 °C (n=3), BMC4 at 90 °C (n=3). Thermal unfolding of BMC variants as measured by CD spectroscopy (Extended Data Fig. 6d) was performed in duplicate. Treatment of BMC3 cages with chemical reductants (Extended Data Fig. 6e) was performed in duplicate. Cryo-EM characterization of BMC3 cages was performed after collecting 4,672 movie stacks. Shown in Extended Data Fig. 7a is a representative micrograph and 3 representative 2D class averages. Fluorescence characterization of BMC3 samples incubated with rhodamine were performed (Extended Data Fig. 8a) in triplicate. AUC characterization of BMC3 cages encapsulating rhodamine (Extended Data Fig. 8b) was performed in duplicate. UV-vis characterization of BMC3 samples incubated with rhodamine (Extended Data Fig. 8c, d) was performed in triplicate. Repeated fluorescence characterization of a solution containing BMC3 cages encapsulating rhodamine (Extended Data Fig. 8e) was performed in duplicate.
Extended Data
Extended Data Figure 1 ∣. Characterization of the IHA ligand and the BMC constructs.
NMR spectra of N-hydroxy-2-iodoacetamide in DMSO-d6: a, 1H b, 13C. ESI-MS of as-isolated and HA-functionalized BMC constructs, and AUC profiles of HA-functionalized protomers for c, BMC1 d, BMC2 e, BMC3 and f, BMC4. The calculated masses for each unlabeled protein are determined by summing the mass of the polypeptide sequence and the c-type heme (618 Da) covalently linked to the cytochrome.
Extended Data Figure 2 ∣. Structural comparison of CFMC1 and BMC1 cages.
a, The symmetric substructures of the CFMC1 dodecameric unit and its per-protomer SASA and BSA values. Associative surfaces on the protomers are colored in red for homologous interactions and in red/orange or blue/cyan for heterologous interactions (right). b, Summary of engineered metal-coordination motifs for BMC constructs (see Supplementary Table 1 for all mutations). Comparison of C2 and C3 symmetric interfaces and corresponding metal binding sites for c, CFMC1 and d, BMC1. Full cages are shown as surfaces; insets show details of each interface. Fe and Zn ions are represented as orange and teal spheres, respectively. e, Cartoon representation of a full-size BMC1 cage with all metal ions shown as spheres. PDB ID: 3M4B (CFMC1), 6OT9 (BMC1).
Extended Data Figure 3 ∣. ns-TEM characterization of BMC constructs.
Dissolved a, Fe:Zn:BMC1 and b, Fe:Zn:BMC2 crystals in a buffer containing 100 mM HEPES (pH 7.5), 200 mM MgCl2 and 800 μM ZnCl2. c, Self-assembled Fe:Zn:BMC3 cages in a buffer containing 20 mM Tris (pH 8.5), 20 μM FeSO4 and 60 μM ZnCl2. Histograms in b and c reflect the size distributions of Fe:Zn:BMC2 and Fe:Zn:BMC3 cage diameters as measured from ns-TEM images. Gaussian fits to both distributions are drawn as solid lines along with their centers and standard deviations reported. Scale bars are 50 nm.
Extended Data Figure 4 ∣. Cavity volumes of BMC cages.
Solvent-accessible cavity volumes within BMC cages as calculated by a 1.4-Å rolling probe are shown visually as blue meshes and reported numerically below. Spherical cavities, shown as yellow spheres in Fig. 2 and Fig. 4, are reproduced for comparison to the calculated volumes. BMC proteins are represented as transparent cylinders.
Extended Data Figure 5 ∣. Anomalous densities of engineered metal binding sites and conformational flexibility of Cys82-HA site.
Cartoon and stick representations of the a, b, BMC1, c-e, BMC2, f-h, BMC3, and i-j, BMC4 symmetric interfaces showing the engineered metal binding sites with the C63-HA ligands (a, c, f), C82-HA ligands (d, g, i) and the Zn binding sites (b,e,h,j). To discern between bound Zn or Fe, the difference of the anomalous signal between pairs of datasets above and below the K-shell energy of Zn and Fe respectively, are depicted as blue or orange meshes. A strong signal illustrates strong change in anomalous signal across the respective edge, in turn suggesting the presence of the respective metal. The upper right corner of each panel indicates the energies of the datasets used for the map of the respective color. All anomalous difference maps were contoured at 3 σ. As datasets around the Fe-edge were not available for BMC1 and BMC3 (necessitating calculations using anomalous difference density of singular datasets), the calculated f’’ for Zn at 7.3 and 9.3 keV are 0.82 and 0.52 (i.e. non-zero) and thus some residual anomalous signal of the lower energy maps around the Zn atoms is expected to result even from strictly selective Zn loading. For a more quantitative analysis of the nature of the bound metal, ratios of the anomalous signal to the expected values (lower left corner of each panel) were calculated as described in the Methods. k, Stick representation of the BMC2 Cys82-HA binding site in both alternative conformations with the anomalous difference density over the “Fe-edge” shown as orange mesh and a simulated annealing omit map (omitting all C82-HA atoms and Fe) of the normal electron density as light blue mesh contoured at 2 σ. For all Cys-HA binding sites, arrows indicate the handedness of the binding site as Δ (right handed) or Λ (left handed). The reversion of handedness in k with the respective view angle is indicated by arrows. Color code for atoms in all panels: Fe in orange, Zn in blue, S in yellow, O in red and N in dark blue.
Extended Data Figure 6 ∣. Solution characterization of self-assembled BMC3 and BMC4 cages.
The oligomerization state of BMC3 cages as monitored by AUC measurements following: a, incubation with various first-row transition metal ions b, incubation with Zn2+ and Fe3+ (Fe(acetylacetonate)3) and c, Disassembly via sequestration of metal ions by EDTA. d, AUC profiles of BMC variants after equilibration for two hours at the indicated temperatures (top). Thermal unfolding of BMC variants as measured by circular dichroism spectroscopy at 222 nm (bottom). e, Treatment with chemical reductants of different reduction potentials. ns-TEM micrographs are shown for cage samples incubated with chemical reductants.
Extended Data Figure 7 ∣. Cryo-EM analysis of BMC3 cages.
a, Representative cryo-EM micrograph and 2D class averages. b, Flowchart detailing image processing from collected movie stacks to final map. Additional details can be found in the Methods. c, FSC curves calculated between the half-maps (black line), atomic model to the unmasked full map (purple line) and atomic model to the masked full map (blue line). Resolution values are indicated at the gold-standard FSC 0.143 criterion. d, Local resolution estimates of the final reconstruction calculated using ResMap. e, Electron density shown at BMC3 C3 interfaces highlighting poorly-resolved density (reflecting high flexibility) at hydroxamate sites and multiple conformations of W66.
Extended Data Figure 8 ∣. Encapsulation of rhodamine inside BMC3 cages.
a, Fluorescence characterization of BMC3 samples incubated with rhodamine. Cages encapsulating rhodamine were treated with EDTA and washed prior to measuring fluorescence intensity. b, AUC profiles of cages encapsulating rhodamine monitored at the heme Soret absorption maximum (λmax = 415 nm) and rhodamine absorption maximum (λmax = 555 nm). c, UV-vis characterization of BMC3 samples incubated with rhodamine. d, Difference spectra of BMC3 samples and BMC3 protomer shown in c. Free rhodamine dissolved in buffer is shown as dark-red dashes. e, Repeated fluorescence characterization of a solution containing BMC3 cages encapsulating rhodamine over several days. The sample was washed 3x prior to each fluorescence measurement.
Extended Data Table 1 ∣. X-ray data collection, processing and refinement statistics.
Numbers in parentheses correspond to the highest resolution shell.
| BMC1 | BMC2 | BMC3 | BMC4 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | R 3 2 | R 3 2 | R 3 2 | P 63 2 2 |
| Cell dimensions | ||||
| a, b, c (Å) | 125.6, 125.6, 166.4 | 126.1, 126.1, 168.2 | 126.7, 126.7, 167.8 | 87, 87, 63.3 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 39.92 – 2.40 (2.46 – 2.40) |
39.25 – 1.40 (1.44 – 1.40) |
91.83 – 1.85 (1.90 – 1.85) |
48.47 – 1.50 (1.54 – 1.50) |
| No. Reflections Observed | 393414 (28956) | 1863896 (88779) | 885564 (66924) | 734764 (18399) |
| No. Reflections Unique | 38245 (2875) | 195424 (14465) | 85783 (6342) | 37652 (1334) |
| Rmerge | 0.185 (2.986) | 0.056 (1.830) | 0.096 (2.377) | 0.053 (1.936) |
| I / σI | 8.1 (0.96) | 20.3 (0.84) | 13.7 (1.04) | 26.6 (1.23) |
| CC1/2 | 0.998 (0.501) | 1.000 (0.410) | 0.999 (0.468) | 1.000 (0.480) |
| Completeness (%) | 99.81 (100.00) | 99.37 (98.9) | 100.00 (100.00) | 87.66 (42.2) |
| Redundancy | 10.29 (10.01) | 9.54 (6.138) | 10.32 (10.54) | 19.51 (13.94) |
| Wilson B (Å)2 | 52 | 19 | 36 | 29 |
| Refinement | ||||
| Resolution (Å) | 36.37 – 2.40 (2.43 – 2.40) |
31.54 - 1.40 (1.42 - 1.40) |
91.83 - 1.85 (1.88 - 1.85) |
48.47 - 1.50 (1.53 - 1.50) |
| No. reflections | 38194 (1286) | 195270 (8377) | 85755 (3974) | 37642 (810) |
| Rwork/Rfree | 0.2174/0.2718 | 0.1659/0.1909 | 0.1826/0.2106 | 0.1900/0.2181 |
| No. atoms | ||||
| Protein | 3292 | 3502 | 3459 | 925 |
| Ligand/ion | 200 | 244 | 240 | 58 |
| Water | 19 | 786 | 342 | 124 |
| B-factors (Å)2 | ||||
| Protein | 70 | 24 | 41 | 42 |
| Ligand/ion | 68 | 22 | 43 | 35 |
| Water | 63 | 37 | 46 | 49 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.010 | 0.011 | 0.009 | 0.013 |
| Bond angles (°) | 1.29 | 1.33 | 1.15 | 1.33 |
| Clashscore | 7 | 5 | 7 | 10 |
| Ramachandran favored (%) | 100 | 100 | 100 | 97 |
| Ramachandran allowed (%) | 0 | 0 | 0 | 3 |
| Ramachandran outliers (%) | 0 | 0 | 0 | 0 |
| Rotamer outliers (%) | 1 | 2 | 2 | 1 |
Extended Data Table 2 ∣. Cryo-EM data collection, processing, and refinement statistics.
| BMC3 (EMDB ID: EMD-20212) (PDB ID: 6OVH) |
|
|---|---|
| Data collection and processing | |
| Magnification | 165,000x |
| Voltage (kV) | 300 |
| Electron dose (e−/Å2) | 60 |
| Exposure rate (e−/ Å2/s) | 6 |
| Defocus range (μm) | 0.84 |
| Pixel size (Å) | 0.3 – 2.7 |
| Symmetry imposed | T |
| Total extracted particles (no.) | 805,156 |
| Final refined particles (no.) | 25,391 |
| Map resolution (Å) | 2.57 |
| FSC 0.143 (unmasked/masked) | 2.62/2.57 |
| Map resolution range (Å) | ∞– 2.57 |
| Applied B-factor (Å2) | −79 |
| Refinement | |
| Initial model used (PDB code) | 6OT7 |
| Model resolution (Å) | 2.57 |
| FSC 0.5 (unmasked/masked) | 2.74/2.72 |
| FSC 0.143 (unmasked/masked) | 2.62/2.57 |
| Model resolution range (Å) | ∞– 2.57 |
| Map sharpening B factor (Å2) | −79 |
| Model composition | |
| Non-hydrogen atoms | 10895 |
| Protein residues | 1272 |
| Ligand/ion | 68 |
| Water | 171 |
| B-factors (Å)2 | |
| Protein | 51 |
| Ligand/ion | 59 |
| Water | 48 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.876 |
| Validation | |
| MolProbity score | 1.33 |
| Clashscore | 1.15 |
| Rotamer outliers (%) | 4.55 |
| Ramachandran plot | |
| Favored (%) | 99.04 |
| Allowed (%) | 0.96 |
| Disallowed (%) | 0.00 |
Supplementary Material
Acknowledgements
This work was supported by the U.S. Department of Energy (Division of Materials Sciences, Office of Basic Energy Sciences; DE-SC0003844; for the design strategy, EM imaging and analysis, and biochemical analysis) and by the National Science Foundation (Division of Materials Research; DMR-1602537; for crystallographic analysis). E.G. acknowledges support by an EMBO Long-Term Postdoctoral Fellowship (ALTF 1336-2015). J.E. acknowledges support by a DFG Research Fellowship (DFG 393131496). R.H.S. was supported by the National Institute of Health Chemical Biology Interfaces Training Grant UC San Diego (T32GM112584). We acknowledge the use of the UCSD Cryo-EM Facility which is supported by NIH grants to T. S. B. and a gift from the Agouron Institute to UCSD. Crystallographic data were collected either at Stanford Synchrotron Radiation Lightsource (SSRL) or at the Lawrence Berkeley Natural Laboratory on behalf of the Department of Energy.
Footnotes
Competing financial interests
The authors declare no competing interests.
References
- 1.Marsh JA & Teichmann SA Structure, Dynamics, Assembly, and Evolution of Protein Complexes. Annu. Rev. Biochem 84, 551–575 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Padilla JE, Colovos C & Yeates TO Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. USA 98, 2217–2221 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, Luo Q & Liu J Protein self-assembly via supramolecular strategies. Chem. Soc. Rev 45, 2756–2767 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hamley IW Protein Assemblies: Nature-Inspired and Designed Nanostructures. Biomacromolecules (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churchfield LA & Tezcan FA Design and Construction of Functional Supramolecular Metalloprotein Assemblies. Acc. Chem. Res 52, 345–355 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Yeates TO Geometric Principles for Designing Highly Symmetric Self-Assembling Protein Nanomaterials. Annu. Rev. Biophys 46, 23–42 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Johnson JE & Speir JA Quasi-equivalent viruses: a paradigm for protein assemblies. J. Mol. Biol 269, 665–675 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Lawson DM et al. Solving the Structure of Human H-Ferritin by Genetically Engineering Intermolecular Crystal Contacts. Nature 349, 541–544 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Lai Y-T, Cascio D & Yeates TO Structure of a 16-nm Cage Designed by Using Protein Oligomers. Science 336, 1129 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Bale JB et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristie David A & Marsh ENG Metal-dependent assembly of a protein nano-cage. Protein Sci. 28, 1620–1629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher JM et al. Self-Assembling Cages from Coiled-Coil Peptide Modules. Science 340, 595–599 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malay AD et al. An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature 569, 438–442 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Pluth MD, Bergman RG & Raymond KN Acid catalysis in basic solution: A supramolecular host promotes orthoformate hydrolysis. Science 316, 85–88 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Chakrabarty R, Mukherjee PS & Stang PJ Supramolecular Coordination: Self-Assembly of Finite Two- and Three-Dimensional Ensembles. Chem. Rev 111, 6810–6918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshizawa M, Klosterman JK & Fujita M Functional Molecular Flasks: New Properties and Reactions within Discrete, Self-Assembled Hosts. Angew. Chem. Int. Ed 48, 3418–3438 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Mal P, Breiner B, Rissanen K & Nitschke JR White Phosphorus Is Air-Stable Within a Self-Assembled Tetrahedral Capsule. Science 324, 1697–1699 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Hu C, Comotti A & Ward MD Supramolecular Archimedean Cages Assembled with 72 Hydrogen Bonds. Science 333, 436–440 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Mateu MG Assembly, stability and dynamics of virus capsids. Arch. Biochem. Biophys 531, 65–79 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Johnson DW, Caulder DL, Raymond KN & Wong EH Rational Design and Assembly of M2M’3L6 Supramolecular Clusters with C3h Symmetry by Exploiting Incommensurate Symmetry Numbers. J. Am. Chem. Soc 123, 2752–2763 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Smulders MMJ, Jiménez A & Nitschke JR Integrative Self-Sorting Synthesis of a Fe8Pt6L24 Cubic Cage. Angew. Chem. Int. Ed 51, 6681–6685 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Brodin JD et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem 4, 375–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radford RJ, Nguyen PC & Tezcan FA Modular and Versatile Hybrid Coordination Motifs on Alpha-Helical Protein Surfaces. Inorg. Chem 2010, 7106–7115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson RG Hard and Soft Acids and Bases. J. Am. Chem. Soc 85, 3533–3539 (1963). [Google Scholar]
- 26.Wong GB, Kappel MJ, Raymond KN, Matzanke B & Winkelmann G Coordination chemistry of microbial iron transport compounds. 24. Characterization of coprogen and ferricrocin, two ferric hydroxamate siderophores. J. Am. Chem. Soc 105, 810–815 (1983). [Google Scholar]
- 27.Crumbliss AL Iron bioavailability and the coordination chemistry of hydroxamic acids. Coord. Chem. Rev 105, 155–179 (1990). [Google Scholar]
- 28.Ni TW & Tezcan FA Structural Characterization of a Microperoxidase Inside a Metal-Directed Protein Cage. Angew. Chem. Int. Ed 49, 7014–7018 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Failes TW & Hambley TW Crystal Structures of Tris(hydroxamato) Complexes of Iron(III). Aust. J. Chem 53, 879–881 (2001). [Google Scholar]
- 30.Mayhew SG The Redox Potential of Dithionite and SO2− from Equilibrium Reactions with Flavodoxins, Methyl Viologen and Hydrogen plus Hydrogenase. Eur. J. Biochem 85, 535–547 (1978). [DOI] [PubMed] [Google Scholar]
- 31.Borsook H & Keighley G Oxidation-Reduction Potential of Ascorbic Acid (Vitamin C). Proc. Natl. Acad. Sci. USA 19, 875–878 (1933). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalak K, Wicha J & Wójcik J Studies towards dynamic kinetic resolution of 4-hydroxy-2-methylcyclopent-2-en-1-one and its E-O-trityloxime. 72, 4813–4820 (2016). [Google Scholar]
- 33.Liu H & Naismith JH An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faraone-Mennella J, Tezcan FA, Gray HB & Winkler JR Stability and folding kinetics of structurally characterized cytochrome cb562. Biochemistry 45, 10504–10511 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Bailey JB, Subramanian RH, Churchfield LA & Tezcan FA in Methods in Enzymology Vol. 580 (ed Pecoraro Vincent L.) 223–250 (Academic Press, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karplus PA & Diederichs K Linking Crystallographic Model and Data Quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC et al. Phenix - a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr, Medium: ED; (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3. (2010).
- 41.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindner HJ & Gottlicher S Die Kristall- und Molekulstruktur des Eisen(III)-benzhydroxamat-Trihydrates. Acta Crystallogr. B Struct. Cryst. Cryst. Chem 25, 832–842 (1969). [Google Scholar]
- 43.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck P A model for sedimentation in inhomogeneous media. I. Dynamic density gradients from sedimenting co-solutes. Biophys. Chem 108, 187–200 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Kleywegt GJ & Jones TA Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D 50, 178–185 (1994). [DOI] [PubMed] [Google Scholar]
- 46.Mazurenko S et al. CalFitter: a web server for analysis of protein thermal denaturation data. 46, W344–W349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The principal data supporting the findings of this work are available within the figures and the Supplementary Information. Additional data that support the findings of this study are available from the corresponding authors on request.
Structural data obtained by X-ray crystallography and cryo-EM have been deposited into the RCSB and EMDB data banks under the following accession codes: 6OT4 (BMC2), 6OT7 (BMC3), 6OT8 (BMC4), 6OT9 (BMC1) and 6OVH (BMC3 Cryo-EM) or EMD-20212 at The Electron Microscopy Data Bank.