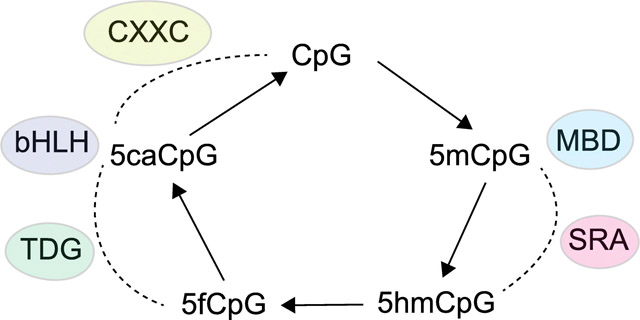
Abstract
5-methylcytosine (5mC), the major modified DNA base in mammalian cells, can be oxidized enzymatically to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by the Ten-Eleven-Translocation (TET) family of proteins. Whereas 5fC and 5caC are recognized and removed by base excision repair proteins, the 5hmC base accumulates to substantial levels in certain cell types such as brain-derived neurons and is viewed as a relatively stable DNA base. As such, the existence of ‘reader’ proteins that recognize 5hmC would be a logical assumption and various searches have been undertaken to identify proteins that specifically bind to 5hmC and the other oxidized 5mC bases. However, the existence of definitive 5hmC ‘readers’ has remained unclear and proteins interacting specifically with 5fC or 5caC are also very few. On the other hand, 5hmC is incapable of interacting with a number of proteins that recognize 5mC at CpG sequences suggesting that 5hmC is an anti-reader modification that may serve to displace 5mC readers from DNA. In this review article, we discuss candidate proteins that may positively or negatively interact with oxidized 5mC bases.
Keywords: DNA methylation, 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine
Graphical abstract
Introduction:
5-methylcytosine (5mC) was first identified in mammalian DNA in 1948 [1]. The modified cytosine occurs predominantly at CpG dinucleotides where it is produced in an enzymatic reaction carried out by DNA methyltransferases (DNMTs) [2]. In dividing cells, methylation patterns are preserved during DNA replication by a copying process catalyzed by DNMT1. According to early views, which are still mostly valid today, DNA methylation was seen as playing roles in regulating the access of DNA-binding proteins leading to changes in gene expression. DNA methylation was also thought to be critically important for X chromosome inactivation, genomic imprinting, cell differentiation and development [3-5].
Deletion of DNA methyltransferase genes in the mouse leads to arrested development and causes embryonic lethality [6, 7]. The mammalian genome is densely methylated at most CpG dinucleotide sequences. Regions that noticeably escape DNA methylation are CpG-rich sequences termed CpG islands, which are protected from methylation by a variety of specialized proteins [8, 9]. Some of these factors promote the formation of the histone modification H3K4me3, which blocks DNA methylation, and others remove any inadvertently introduced 5mC bases. At enhancer regions, CpG methylation levels are also relatively low but methylation at these regions is more dynamic and depends on cell type-dependent function of the enhancer-linked genes.
DNA methylation plays a particularly important role in negatively controlling the expression of repetitive regions of the genome including transposons and endogenous retroviral elements [10]. Methylation is also critical for maintenance of X chromosome inactivation in female cells [11], for marking one of the two alleles of imprinted genes [12], and for suppression of many germ line-expressed genes in somatic cells or tissues [13]. For these silencing pathways, which need to be maintained over the life span of an organism, the methylated state at CpG sequences is expected to be very stable during cell division or even during prolonged persistence of non-dividing cells. However, there are circumstances where methylation patterns may deteriorate (such as during aging or carcinogenesis), often involving slowly operating processes that enable a progressive loss of methylation over time [14] or that facilitate encroachment of methylation into previously unmethylated CpG islands [15].
In a more programmed manner, DNA methylation patterns may undergo drastic global changes during two stages of development when genomic methylation levels are erased and subsequently reset in primordial germ cells and then again immediately after fertilization [16]. A limited reorganization of DNA methylation patterns occurs later during developmental processes of lineage formation and cell differentiation, where methylation changes are often specified by site-specific transcription factors. These factors are capable of either excluding DNA methylation from their binding sites [17] or in certain cases, they may promote DNA demethylation in a manner dependent on 5mC oxidation [18, 19]. Connected to these models is the assumption that DNA methylation is a default state that occurs largely everywhere in the genome where it is not prevented [20] or where it is not actively removed.
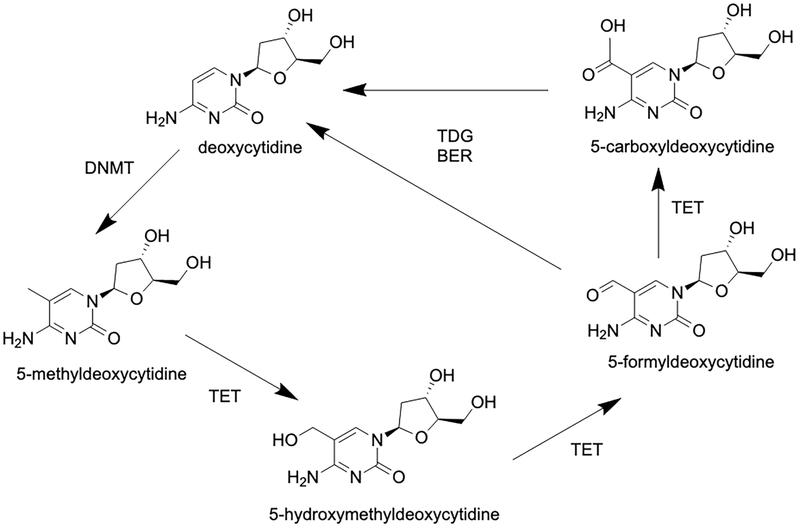
For over sixty years, 5mC was thought to be the only modified DNA base produced by endogenous enzymatic processes in mammals. Even though DNA replication-independent DNA demethylation had been observed in certain settings [21], the enzymology of demethylation remained completely obscure for decades [22]. This situation changed drastically in 2009, when substantial levels of 5hmC were found in ES cells and in certain types of neurons in the brain [23, 24]. One of these studies [24] identified an enzymatic activity responsible for the formation of 5hmC as a 5mC dioxygenase and named it Ten-Eleven Translocation 1 (TET1) after a chromosomal translocation of the TET1 gene observed earlier in leukemias [25, 26]. TET1 and its paralogues TET2 and TET3 require alpha-ketoglutarate, oxygen and Fe2+ as essential cofactors [24], and 5mC oxidation activity is enhanced in the presence of ascorbic acid [27-29]. All three TET proteins not only produce 5hmC but they can carry the oxidation reaction further in sequential enzymatic steps that lead to the formation of 5fC and ultimately 5caC (Figure 1) [30, 31]. Enzymatic 5mC oxidation was viewed as a logical pathway that may lead towards DNA demethylation. Such oxidation reactions are in principle independent of DNA replication and will allow loss of methylation by an active process as opposed to dilution of pre-existing methylation patterns in the absence of maintenance methylation during DNA replication. Interestingly, it was found that DNA molecules that contain 5hmC at CpG dinucleotides are very poor substrates for DNMT1 [32, 33] because DNMT1 forms an unproductive complex with DNA duplexes containing oxidized forms of 5mC [34]. This means that once oxidation of 5mC to 5hmC has occurred, DNA methylation patterns no longer can be maintained, even in the presence of DNMT1.
Figure 1. Outline of the DNA demethylation pathway initiated by 5mC oxidation.
The model shows details of the TET protein-catalyzed, active DNA demethylation pathway. TET proteins oxidize 5mC leading to the formation of 5hmC, 5fC and 5caC. The oxidized bases 5fC and 5caC are recognized and excised from DNA by thymine DNA glycosylase (TDG) as part of a base excision repair (BER) process.
However, there was still a need for a demethylation pathway that could operate in the absence of DNA replication. In 2011, it was reported that such a mechanism is initiated by TET-mediated 5mC oxidation and can then be completed by removal of the 5fC and 5caC bases through base excision repair (Figure 1) [30, 35]. This DNA repair process, which recognizes 5fC and 5caC as a type of “DNA damage,” is initiated by the enzyme thymine DNA glycosylase (TDG), which effectively recognizes and removes 5fC and 5caC from DNA when these bases are paired with guanine. The TDG protein can also excise thymine from T/G mismatches as reported earlier and is thus described as a multifunctional DNA glycosylase [36-38]. Whereas 5hmC can accumulate to levels in the order of 20-30% of the levels of 5mC, specifically in mammalian neurons, 5fC and 5caC exist at almost undetectable levels in most tissues [39-41]. It is still unknown if base excision repair is the only mechanism that promotes removal of 5fC and 5caC from the genome. An alternative pathway would be C-C bond cleavage at position 5 of the pyrimidine ring, for example by decarboxylation of 5caC. However, such a decarboxylation activity has not yet been identified. Isotope labeling studies have shown that C-C bond cleavage can occur at 5fC residues resulting in their conversion to cytosine [42]. However, the mechanism and/or enzymatic activity responsible for this process have not been clarified so far.
In vitro, TET enzymes, or at least their catalytic domains, readily produce the ultimate reaction product, 5caC, although a recent study suggested that TET2-mediated 5mC oxidation has a preference for 5mC oxidation over 5hmC and 5fC oxidation [43]. These puzzling findings may be reconciled by at least two scenarios. First, the production of 5fC and 5caC by TET enzymes may be limited in vivo, perhaps due to a controlled step at the 5hmC to 5fC transition, which only occurs when true loss of methylation at specific sequences is the desired outcome. Such a step may be regulated by site-specific transcription factors that interact with a TET protein to promote active removal of 5mC. Second, the oxidized products 5fC and 5caC are very effectively removed by TDG and base excision repair. The reason for such expedited removal may be the fact that these oxidized bases are blocks to RNA polymerases [44, 45] and also block certain DNA polymerases (our unpublished data). There is currently no good evidence to favor either one or the other of the two scenarios.
5hmC is quite abundant in many cell types, for example in embryonic stem cells and in mammalian neuronal cells, suggesting that at the genome scale it rarely turns over into 5fC or 5caC and is therefore not lost subsequently due to complete demethylation. Also, stable isotope labelling suggested that 5hmC is mostly stable [46]. Its stability and abundance would be compatible with the idea that 5hmC represents a true epigenetic mark that is recognized by specific reader proteins.
Where does 5hmC occur?
Different methodologies have been used to analyze the distribution of 5hmC in the genome. The earlier studies used medium resolution level approaches such as antibody-based immunoprecipitation and sequencing (DIP-seq) [47-50] or a biotin-mediated pulldown after derivatization of the hydroxymethyl group with a modified glucose using T4 glucosyltransferase and click chemistry [51]. Single base resolution analysis of 5hmC is possible using various techniques including TET-assisted bisulfite sequencing [52], oxidative bisulfite sequencing [53], or ACE-seq, a method that uses deaminase-mediated conversion of cytosine and 5-methylcytosine, but leaves 5hmC resistant after transfer of a glucose residue [54]. Using these methods, a number of studies consistently found that 5hmC occurred preferentially in gene bodies (intragenic regions), near enhancers, and at DNA sequences immediately flanking CpG islands.
These studies initially focused on embryonic stem cells and on brain where 5hmC is relatively abundant. Several research groups showed that transcribed regions containing active genes are preferentially enriched with this modified DNA base in the brain [47, 51, 55]. Often, one can observe a direct correlation between the levels of 5hmC along gene bodies and the level of gene expression. A similar, albeit weaker correlation also exists between levels of 5mC and gene expression, although, depending on the methodology used, these studies sometimes do not distinguish between 5mC and 5hmC. Both bases are resistant to bisulfite-induced deamination and are therefore scored together as a sum of the two modified cytosines when bisulfite-based techniques are used for analysis [56]. Presence of the modified cytosines in transcribed regions may serve to prevent antisense or inappropriate transcription throughout gene bodies, which may otherwise interfere with sense transcription. This model is supported with knockout studies, in which it was shown that loss of DNMT3B, the DNA methyltransferase thought to be primarily involved in gene body methylation [57], led to an increase in transcriptional noise [58]. When extrapolating to 5hmC, this modified base may be even more effective than 5mC in preventing aberrant transcription in gene bodies. For example, it is conceivable that 5hmC could interfere with the binding of transcription initiation complexes. However, direct evidence for this model is currently not available.
5hmC accumulates at active or poised enhancers in several cell types where this correlation has been analyzed [48, 59-61]. When enhancers are inactive, they are often embedded in a more highly methylated genomic context, although the density of CpG dinucleotides at enhancers is not as high as the CpG density at promoters. Since enhancers are cell type-dependent, they will exist in a more densely methylated configuration in one cell type but will be less methylated in another cell type in which the enhancer is active. So, why are active enhancers marked by 5hmC? During their activation, often initiated by cell type-specific transcription factors, enhancers undergo DNA demethylation. This demethylation can involve TET2-mediated 5mC oxidation and demethylation as shown in several experimental systems [59, 60, 62-66]. It is possible therefore that the accumulation of 5hmC near enhancers is a remnant of the TET-induced DNA demethylation process. This mechanism invokes that 5fC and 5caC also would accumulate at enhancers. Indeed, 5fC is enriched at poised enhancer sequences, although 5fC is best detectable only when TDG levels are reduced leading to a longer persistence of this oxidized base [67, 68]. Finally, 5hmC at enhancers may represent an activating signal by recruiting activators or by functioning as a mark that opposes the binding of repressor complexes that would normally interact with 5mC sequences.
Soon after the discovery of TET enzymes, it was proposed that one of their main functions is that of an epigenetic repair protein that maintains the unmethylated state of CpG islands [69, 70]. This model is supported by data showing that at least TET1 and TET3 are strongly targeted, to unmethylated CpG islands, likely through their CXXC zinc finger domains [71-74]. These two TET proteins are expressed as different isoforms that either contain or do not contain the CXXC domain. TET2 has mostly been implicated in enhancer demethylation and binds to enhancers [75]. The TET2 gene does not encode a CXXC domain by itself but a heterodimerization partner of TET2, named CXXC4, can provide this domain to TET2 [76]. Currently, there is insufficient information to definitively assign different functions and different genomic activities or distributions to the multiple TET isoforms.
5hmC also accumulates at sequences flanking CpG islands, for example upstream of promoters. Also at these locations, 5hmC may represent a marker of TET activity that did not get processed to the higher oxidized forms, 5fC or 5caC. The unmethylated state of CpG islands is continuously threatened by de novo DNA methylation errors. These DNMT-induced de novo methylation events will probably occur stochastically and may eventually cause gene silencing. Over evolutionary time, methylation of CpG islands will erode these genomic landmarks through DNA methylation-mediated mutagenic mechanisms [77]. De novo DNA methylation errors will likely encroach into CpG islands from their edges [78] and that is where TET activity will be most needed. The 5hmC at borders of CpG islands may simply reflect enhanced TET activity, or alternatively, the modified base may prevent binding of transcriptional repressor protein complexes that normally would interact with methylated DNA.
Several proteins function as readers of 5mC but do not bind to 5hmC
We will provide only a brief summary of the proteins that recognize 5mC at CpG sequences and will place emphasis mostly on how these proteins interact with 5hmC when such information is available.
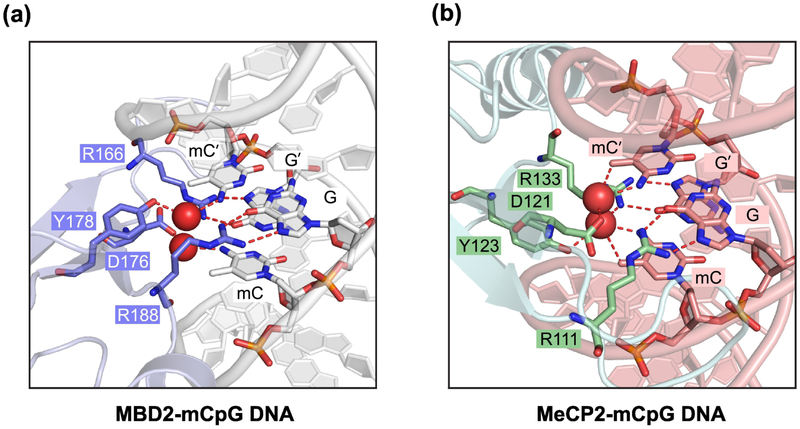
The best studied family of proteins that bind to 5mC contain a DNA binding domain referred to as the methyl-CpG binding domain (MBD). Initially, a methylated DNA binding protein complex in cell extracts was identified as MECP1 [79]. The complex contains MBD2 as a subunit that has direct DNA binding activity and a strong preference for 5mC-containing over unmethylated DNA [80]. Another protein of the MBD family is MECP2, which is mutated in the severe autism spectrum disorder Rett syndrome [81]. Structural analysis of the MBD2 – 5mC and MeCP2 – 5mC complexes indicated a similar 5mC-recognition mode between the two readers, both involving a pair of arginine residues that recognize the mCpG site through stacking and hydrogen-bonding interactions (Figure 2) [82, 83]. The protein family also includes MBD1, MBD3, and MBD4 [84]. MBD1, MBD2, MBD3 and MECP2 are subunits of larger chromatin-bound repressor complexes that also contain enzymatic activities that deacetylate histones and that remodel chromatin in an ATPase-dependent manner. Given their subunit composition, it is proposed that these complexes (for example NURD, nucleosome remodeling deacetylase) function in repression of methylated genomic regions [85].
Figure 2. Crystal structures of 5mC readers in complex with 5mC-containing DNA.
(A) Ribbon diagram of the MBD domain of MBD2 (blue-white) bound to mCpG DNA (silver) (PDB 6CNP), with the mCpG site and the interacting protein residues (slate) shown in stick representation.
(B) The MBD domain of MeCP2 (light blue) bound to mCpG DNA (salmon) (PDB 3C2I), with the mCpG site and the interacting protein residues (green) shown in stick representation. In (A) and (B), the hydrogen bonds and water molecules are shown as dashed lines and red spheres, respectively.
The MBD4 protein is a somewhat unusual member of this family. MBD4 functions as a DNA glycosylase that can excise thymine from G/T mismatches that may arise through hydrolytic deamination of 5mC at methylated CpG sites [36, 77]. Interestingly, mice lacking MBD proteins (MBD1, MBD2, MBD4) have relatively mild phenotypes although knockout of MECP2 produces neurological symptoms reminiscent of Rett syndrome [86].
Given the high selectivity of proteins with an MBD domain towards CpG sequences containing 5mC, it is of interest to consider their binding to oxidized 5mC bases. As we reported in 2010, the MBD domains of MBD1, MBD2, MBD4 are in fact incapable of binding to DNA sequences containing 5hmC [56]. In other words, oxidation of the methyl group blocks the reader function of these proteins and they would not be expected to bind effectively to 5hmC-enriched regions such as gene bodies or enhancers. When the MBD complex plays a major role in repression of a methylated target, 5mC oxidation will likely alleviate this repression and make the region more permissive for gene activation.
Outside of the MBD family, there are other proteins that can preferentially interact with target sequences containing 5mC. Plant genomes encode several proteins with SET and RING finger associated (SRA) domains [87]. In mammals only two such proteins exist, UHRF1 and UHRF2. UHRF1 is well characterized as a DNMT1 regulator that ensures the maintenance of DNA methylation at hemimethylated sites after DNA replication [88]. The function of UHRF2 is less well understood.
Other DNA binding proteins with a preference for CpG-methylated DNA include certain members of the zinc finger and BTB/POZ domain (ZBTB) containing protein family, perhaps the best studied being the protein ZBTB33 (also known as KAISO) [89]. These proteins generally function as transcriptional repressors. An experimental screening system has identified additional transcription factors that preferentially bind to methylated DNA sequences [90]. Most of these factors were members of the extended homeodomain family. Not much information is available on how the different categories of 5mC-binding proteins interact with oxidized 5mC target sequences. Binding of ZBTB2 was inhibited when 5hmC was placed into its binding site [91].
In summary, one major function of 5hmC (and perhaps also 5fC and 5caC) may simply be a blocking function whereby the oxidation of 5mC prevents recognition of methylated CpG sequences by methylated DNA binding proteins [56]. If 5hmC indeed is mostly a negative mark, perhaps the term anti-reader modification may be appropriate.
5hmC-binding proteins
To identify proteins that bind to 5hmC using an unbiased approach, proteomic analysis was used to screen for proteins that bind to DNA sequences containing this modified base [92, 93]. A limited number of proteins were identified. Proteins recovered in more than one cell type included UHRF1, WDR76, THY28, and NEIL1 [93]. Some of these proteins have other known functions; for example, UHRF1 is a factor that binds to hemimethylated CpGs and NEIL1 as a DNA repair protein recognizing oxidized guanines. WDR76 has been described as a DNA damage response protein [94], but in another study it was characterized as a protein that destabilizes the RAS oncoprotein [95]. The nuclear protein THY28/THYN1 is not well characterized. The other study, also using embryonic stem cells, found only a few proteins binding to 5hmC including the ribosomal protein RPL26 and the mismatch repair protein MSH6 [92]. These proteomics results rely on incorporation of 5hmC into specific sequence contexts. However, proteins may exist that recognize the modified base in a different sequence context.
Some publications have identified putative readers of 5hmC in a candidate protein-based approach. One study suggested that the methyl-CpG binding protein MBD3 binds to 5hmC at CpG sites [96]. This result has not been confirmed in other studies [92, 93, 97]. MECP2 was initially shown to bind preferentially to 5hmC [98], but this protein seems to bind more strongly to 5mC than to 5hmC at CpG sequences [32, 93, 99, 100].
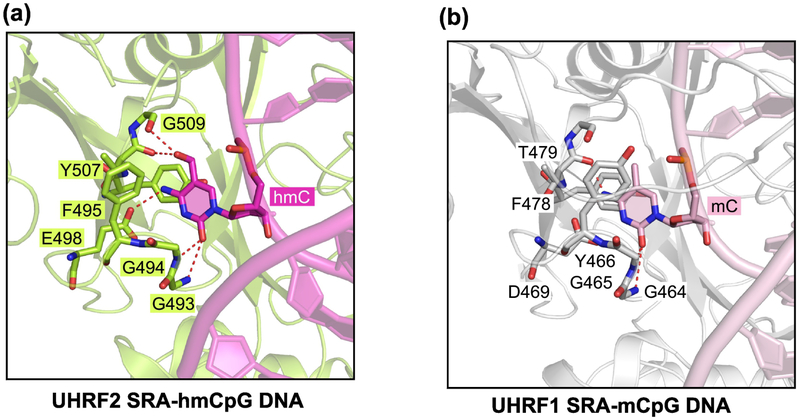
The SRA domain of UHRF1 and/or UHRF2 may be a specific reader of 5hmC [93, 101]. This idea has not remained without controversy [102]. The binding of UHRF2 to 5hmC-containing DNA has been characterized in a study using X-ray crystallography (Figure 3A) [103]. This protein can interact with symmetrically hydroxymethylated and hemi-hydroxymethylated CpG sites with moderately higher affinity than it binds to hemi-methylated DNA, that is DNA that contains 5mC on one strand only. The structural analysis of the UHRF2 SRA domain shows that the 5hmC base is flipped out of the DNA double helix and becomes inserted into the UHRF2-SRA pocket (Figure 3A), in a mechanism similar to that used by other DNA interacting baserecognizing enzymes or proteins [104], such as the interaction between the UHRF1 SRA domain and hemi-methylated CpG DNA (Figure 3B) [105-107]. It is unknown what physiological role the UHRF2-5hmC interaction may have. Binding of UHRF2 to 5hmC target sequences may be regulated by specific factors such as ZNF618 [83].
Figure 3. Structural comparison UHRF1 SRA - 5mC and UHRF2 SRA - 5hmC interactions.
(A) Ribbon diagram of the SRA domain of UHRF2 (limon) bound to 5hmC-containing DNA (magenta) (PDB 4PW5), with the flipped-out 5hmC nucleotide and the interacting protein residues shown in stick representation. The hydrogen bonds are shown as dashed lines.
(B) The SRA domain of UHRF1 (silver) bound to mCpG DNA (pink) (PDB 3CLZ), with the flipped-out 5mC nucleotide and the interacting protein residues (green) shown in stick representation.
It was suggested that the SOS response-associated peptidase (SRAP) domain family of proteins functions as a DNA-localized autoproteolytic switch to recruit certain DNA processing enzymes to sites of DNA damage [108]. Interestingly, C3ORF37/HMCES (hydroxymethylcytosine binding, ESC-specific), a eukaryotic member of the SRAP family, was found as a protein binding to oxidized 5mC derivatives [93]. Indeed, later it was reported that HMCES binds to oxidized forms of 5mC, including 5hmC, and catalyzes conversion of these bases to unmodified cytosine through the activity of an autopeptidase-coupled nuclease [109]. However, the HMCES protein is conserved in all domains of life including prokaryotes, in which 5hmC does not exist. In a more recent publication, HMCES was characterized as a sensor of abasic sites in single-stranded DNA, which acts at replication forks and generates a DNA-protein crosslink to shield these abasic sites from error-prone processing or breakage [110]. In that study, HMCES had no preference for modified cytosines and HMCES overexpression or deletion studies did not give evidence for altered levels of 5mC or 5hmC.
Using affinity purification, SALL1 and SALL4 were identified as having preferential affinity for 5hmC [97]. These zinc finger proteins are highly expressed in embryonic stem cells where they cooperate with NANOG in the pluripotency network. SALL4 is often overexpressed in cancer. Xiong et al reported that the longer isoform of SALL4A contains a 5hmC-binding zinc finger cluster and that both SALL1 and SALL4 preferentially occupied enhancer regions. They showed that SALL4A cooperated with TET2 to promote the further oxidation of 5hmC leading to DNA demethylation at enhancer regions. However, a direct interaction between SALL4A and TET2 could not be demonstrated [97].
In vitro studies have shown an increased binding of certain transcription factors to binding sites that contain 5hmC. 5hmC dramatically enhanced binding of the helix-loop-helix transcription factor TCF4 to E-box motif sequences ACATGTG and ACACGTG [111]. Considering the fact that there are a large number of factors that prefer 5mC in their DNA recognition sequences [90], it wouldn't be all that surprising if there were a set of sequence-specific factors that prefer motifs containing 5hmC.
To summarize current knowledge of 5hmC binders, there is structural evidence for a defined interaction of UHRF2 with 5hmC-containing DNA, although a physiological role of this interaction has not yet been determined. SALL4A and SALL1 may be specific factors that can recognize 5hmC and stimulate its conversion to 5fC and 5caC under certain circumstances. The overall evidence for other specific 5hmC readers in mammals is still incomplete, and although a small set of such interactors have been found [92, 93], there are not many follow-up studies that have characterized them in more detail. It should be kept in mind that certain cell types, such as neurons contain abundant amounts of 5hmC, and it would seem plausible that such cell types express proteins that recognize this mark. A more detailed biochemical identification and characterization of 5hmC-binding proteins in brain tissue would therefore seem necessary to make progress in this area.
5fC-binding proteins
Affinity purification and identification of bound proteins by mass spectrometry-based analysis has been used to detect candidate proteins that recognize 5fC or 5caC in DNA templates [92, 93]. Perhaps due to their increased polarity or negative charge, a greater number of putative reader proteins for 5fC and 5caC was found compared to the number of 5hmC-binding factors [92, 93]. This is in some way a counterintuitive result, because 5fC and 5caC are different from 5hmC in terms of their much lower abundance. Why would these modifications then be associated with a larger number of putative reader proteins? Regarding their genomic location, 5fC and 5hmC are most abundant near enhancer regions where they are thought to be removed by base excision repair leading to complete DNA demethylation [67, 68]. Of note, the numerous identified 5fC and 5caC interactions with candidate readers have not yet been further analyzed by biochemical methods. Recent studies have shown that 5fC can be a chemically stable DNA base in cells [39] also suggesting that there may be proteins that specifically recognize this modification.
One unique chemical property of 5fC is that the formyl group is capable of mediating DNA-protein crosslinks through the formation of a Schiff base with amino acids. This has indeed been demonstrated for 5fC interactions with lysine side chains of histones [112, 113]. The presence of 5fC in nucleosomal DNA was shown to be associated with increased nucleosome occupancy perhaps due to Schiff base formation and with increased transcription linked to such nucleosome-associated enhancers [114]. However, these 5fC-associated protein crosslinks may not always have a physiologically beneficial role but could be detrimental to the cells, for example by interfering with DNA replication [115].
5caC-binding proteins
Even though 5caC is of very low abundance in cells, several proteins have been shown to specifically recognize this modification [30, 35, 45, 74, 93, 116-118].
As first reported in 2011, the protein thymine DNA glycosylase (TDG) was found to bind and remove the 5caC base through its DNA glycosylase activity [30, 35]. TDG had earlier been characterized as an enzyme capable of excising thymine bases from T/G mispairs [36, 119]. Initial structural studies of the human TDG catalytic mutant (N140A) with a DNA duplex containing an A·caC or G·caC, respectively, showed that the 5caC base is flipped out of the DNA helix and inserts into a pocket of the enzyme [116, 118]. TDG is capable of removing 5fC and 5caC from DNA but has much lower affinity towards 5hmC or 5mC residues in DNA templates [116].
Although TDG effectively recognizes and removes 5caC, this reaction may not represent a real epigenetic reader function. However, a few other studies have now shown that 5caC may have a more direct role in regulating transcription and DNA methylation turnover. For example, Wilms tumor protein 1 (WT1), a transcription factor with a recognition sequence 5’-GCG(T/G)GGGCG-3’, can interact with its recognition site when 5caC is present, as determined by a crystal structure [117]. The MAX protein, a binding partner of the MYC transcription factor, has a preference for a recognition sequence (5’-CACGTG-3’) when the central CpG site contains 5caC or C, but a much lower affinity when there is 5mC, 5hmC, or 5fC [120]. Similarly, cytosine carboxylation has a strong positive impact on TCF4 binding to DNA [121]. This data suggests that 5caC can have a different epigenetic meaning relative to the other cytosine modifications.
Other structural work has shown that 5caC, when located within transcribed gene regions, can result in diminished RNA polymerase II-mediated transcription elongation due to arrest of the polymerase by the 5caC modification [45]. Certain DNA polymerases may similarly be impeded by this base. These negative effects of 5caC on polymerase progression in transcription or DNA replication may explain its rapid removal from cells by DNA repair pathways.
The biochemical regulation of TET protein-mediated 5mC oxidation is not completely understood. When the enzymatic reaction proceeds beyond 5hmC, and 5fC or 5caC are formed, DNA repair may commence relatively quickly. However, it is not known how the individual steps are controlled or coordinated. In many cases, DNA demethylation of a specific genomic region requires removal of multiple 5mC bases. We found that TET3 contains a domain that acts as a specific reader module for 5caC [74]. Interestingly, this domain, which consists of CXXC-type zinc fingers, is present in only one of the two major isoforms of TET3. CXXC domains are present in only about a dozen mammalian proteins. Almost all of them have a known epigenetic function and they have commonly been characterized by their ability to bind to unmethylated CpG-rich DNA sequences [9]. Indeed, the CXXC domain of mouse and Xenopus TET3 can bind to unmodified CpG sequences (Figure 4A) [74, 122]. Surprisingly, we found that the mouse TET3 CXXC domain binds most strongly to 5’CcaCG-containing DNA and has a very low affinity to 5mC, 5hmC and 5fC in the same sequence context. Binding to 5caC-containing DNA is about 3-times stronger than its binding to unmethylated DNA. The crystal structure of the mouse TET3-CXXC domain with a 12-mer DNA sequence containing a central CcaCG site was determined at 1.3 Å resolution (Figure 4B) [74]. Supporting the in vitro binding studies, we observed that the 5-carboxyl group of 5caC forms direct and water-mediated hydrogen bonds with the side chain amino group of TET3 at Lys88 and forms additional backbone contacts. The CcaCG-interacting residues of TET3 are conserved within the TET protein-associated CXXC domains, but not, for example, in the DNMT1-associated CXXC domains. To develop a model for the biological function of 5caC binding by TET3, one can view this mechanism as a property of TET3 to get anchored to its reaction product (probably transiently), which provides opportunity to TET3 for oxidizing additional 5mC bases in its immediate vicinity through its catalytic domain. A number of epigenetic modifier proteins, both “writers” and “erasers,” are characterized by the dual presence of a catalytic domain and an additional reader domain (often near the N-terminus or C-terminus of the proteins) that recognizes the reaction product. This dual domain structure has been described for histone acetyltransferases [123], histone methyltransferases [124-126] and histone demethylases [127-129]. The double domain organization has been proposed as a means to promote genomic specificity and processivity of the epigenetic modifiers at their target sequences in vivo.
Figure 4. Structural comparison of TET3 CXXC - CG and TET3 CXXC - CcaCG interactions.
(A) The CXXC domain of Xenopus Tet3 (aquamarine) bound to unmodified CpG DNA (orange) (PDB 4HP3), with the interacting nucleotides and protein residues (green) shown in stick representation (B) Ribbon diagram of the CXXC domain of mouse TET3 (slate) bound to CcaCG-containing DNA (wheat) (PDB 5EXH), with the interacting nucleotides and protein residues (marine) shown in stick representation. The hydrogen bonds are shown as dashed lines. The zinc ions and water molecules are shown as purple and red spheres, respectively.
Conclusions and Perspectives:
Since its discovery about a decade ago, the mechanistic details and biological function of 5mC oxidation are still only partially understood. These oxidized bases, in particular 5hmC, can reach substantial levels in certain tissues or cell types. Therefore, it seems plausible that proteins would specifically recognize and interact with DNA sequences that contain oxidized 5mC. However, it has been difficult to substantiate reader proteins for 5hmC, 5fC or 5caC. The latter two modified bases are clearly recognized by the DNA base excision repair protein TDG, which is well supported by biochemical and structural studies. The few proteins identified as readers of 5hmC include the SRA domain-containing protein UHRF2, but the biological meaning of this binding needs further investigation. SALL4A recently has emerged as a protein interacting with 5hmC but no structural study has yet shown the details of this interaction. Proteomics and affinity purification approaches have been used to identify a larger set of proteins that may bind to 5fC and 5caC, but for the most part there is not yet much further information about the details or biological meaning of these interactions. A few specific protein domains that bind to oxidized 5mC bases have emerged (e.g. the SRA domain for 5hmC and the CXXC domain for 5caC) but additional oxidized 5mC-recognizing proteins or domains may exist and may function only in specific sequence contexts or in specific cell types.
Oxidized 5mC bases may primarily be intermediate products of TET protein-initiated DNA demethylation pathways. It is likely that in many settings the oxidized bases are able to promote replication-dependent inhibition of DNA methyltransferase function, which will lead to loss of DNA methylation. One should also consider the possibility that 5hmC is a remnant of TET protein activity. For example, 5hmC accumulates at enhancers in the context of enhancer activation, a process that involves active DNA demethylation [66]. A well-supported view has been that oxidized 5mC is primarily a ‘negative’ epigenetic mark that prevents the binding of proteins that would otherwise strongly interact with 5mC, such as the methyl-CpG binding protein (MBD) family, and that the purpose of 5mC oxidation is to alleviate the repression imposed by MBDs and their associated repressor complexes. Further in vivo studies should be performed to test this model.
Highlights:
In this review, we discuss the potential function of 5-methylcytosine oxidation in the context of DNA-protein interactions.
We examine putative readers of oxidized bases in terms of available structural and mechanistic insights.
We consider the genomic positions of oxidized 5-methylcytosine residues and how this knowledge may be used to deduce a potential role of the oxidation process.
Acknowledgements:
Research of the authors has been supported by NIH grant CA160965 (to G.P.P.).
Footnotes
Declarations of interest: none
Credit author statement:
All authors participated in writing, editing and reviewing the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. The Journal of biological chemistry. 1948;175:315–32. [PubMed] [Google Scholar]
- [2].Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–32. [PubMed] [Google Scholar]
- [4].Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. [DOI] [PubMed] [Google Scholar]
- [5].Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. [DOI] [PubMed] [Google Scholar]
- [6].Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. [DOI] [PubMed] [Google Scholar]
- [7].Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. [DOI] [PubMed] [Google Scholar]
- [8].Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochemical Society transactions. 2013;41:727–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. [DOI] [PubMed] [Google Scholar]
- [11].Riggs AD, Pfeifer GP. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–74. [DOI] [PubMed] [Google Scholar]
- [12].Mann JR, Szabo PE, Reed MR, Singer-Sam J. Methylated DNA sequences in genomic imprinting. Crit Rev Eukaryot Gene Expr. 2000;10:241–57. [DOI] [PubMed] [Google Scholar]
- [13].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- [14].Zhou W, Dinh HQ, Ramjan Z, Weisenberger DJ, Nicolet CM, Shen H, et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Skvortsova K, Masle-Farquhar E, Luu PL, Song JZ, Qu W, Zotenko E, et al. DNA Hypermethylation Encroachment at CpG Island Borders in Cancer Is Predisposed by H3K4 Monomethylation Patterns. Cancer Cell. 2019;35:297–314 e8. [DOI] [PubMed] [Google Scholar]
- [16].Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol. 2013;25:281–8. [DOI] [PubMed] [Google Scholar]
- [17].Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. [DOI] [PubMed] [Google Scholar]
- [18].Thakur A, Wong JCH, Wang EY, Lotto J, Kim D, Cheng JC, et al. HNF4A is essential for the active epigenetic state at enhancers in mouse liver. Hepatology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hahn MA, Jin SG, Li AX, Liu J, Huang Z, Wu X, et al. Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Science Advances. 2019;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Singh P, Li AX, Tran DA, Oates N, Kang ER, Wu X, et al. De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell reports. 2013;4:205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wilks A, Seldran M, Jost JP. An estrogen-dependent demethylation at the 5' end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984;12:1163–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–8. [DOI] [PubMed] [Google Scholar]
- [23].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17:637–41. [DOI] [PubMed] [Google Scholar]
- [26].Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res. 2002;62:4075–80. [PubMed] [Google Scholar]
- [27].Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. The Journal of biological chemistry. 2013;288:13669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–403. [DOI] [PubMed] [Google Scholar]
- [30].He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.Science. 2011;333:1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–50. [DOI] [PubMed] [Google Scholar]
- [34].Seiler CL, Fernandez J, Koerperich Z, Andersen MP, Kotandeniya D, Nguyen ME, et al. Maintenance DNA Methyltransferase Activity in the Presence of Oxidized Forms of 5-Methylcytosine: Structural Basis for Ten Eleven Translocation-Mediated DNA Demethylation. Biochemistry. 2018;57:6061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. The Journal of biological chemistry. 2011;286:35334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bellacosa A, Drohat AC. Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites. DNA repair. 2015;32:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schuermann D, Weber AR, Schar P. Active DNA demethylation by DNA repair: Facts and uncertainties. DNA repair. 2016. [DOI] [PubMed] [Google Scholar]
- [39].Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nature chemical biology. 2015;11:555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS one. 2010;5:e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhu C, Gao Y, Guo H, Xia B, Song J, Wu X, et al. Single-Cell 5-Formylcytosine Landscapes of Mammalian Early Embryos and ESCs at Single-Base Resolution. Cell Stem Cell. 2017;20:720–31 e5. [DOI] [PubMed] [Google Scholar]
- [42].Iwan K, Rahimoff R, Kirchner A, Spada F, Schroder AS, Kosmatchev O, et al. 5-Formylcytosine to cytosine conversion by C-C bond cleavage in vivo. Nature chemical biology. 2018;14:72–8. [DOI] [PubMed] [Google Scholar]
- [43].Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, et al. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527:118–22. [DOI] [PubMed] [Google Scholar]
- [44].Kellinger MW, Song CX, Chong J, Lu XY, He C, Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523:621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nature chemistry. 2014;6:1049–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome biology. 2011;12:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. [DOI] [PubMed] [Google Scholar]
- [51].Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–7. [DOI] [PubMed] [Google Scholar]
- [54].Schutsky EK, DeNizio JE, Hu P, Liu MY, Nabel CS, Fabyanic EB, et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat Biotechnol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell reports. 2013;3:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–7. [DOI] [PubMed] [Google Scholar]
- [58].Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–7. [DOI] [PubMed] [Google Scholar]
- [59].Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Molecular cell. 2014;56:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28:2103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic acids research. 2012;40:8255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sardina JL, Collombet S, Tian TV, Gomez A, Di Stefano B, Berenguer C, et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell. 2018;23:727–41 e9. [DOI] [PubMed] [Google Scholar]
- [64].Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, et al. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv. 2018;4:eaau6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, et al. TET2 Mutations Affect Non-CpG Island DNA Methylation at Enhancers and Transcription Factor Binding Sites in Chronic Myelomonocytic Leukemia. Cancer Res. 2015;75:2833–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hahn MA, Jin SG, Li AX, Liu J, Huang Z, Wu X, et al. Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation. Science Advances. 2019;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pfeifer GP, Rauch TA, Tommasi S, Besaratinia A, Hahn MA, Iqbal K, et al. Profiling of modified cytosines in normal and malignant cells. Princess Takamatsu Symp. 2011;41:69–72. [Google Scholar]
- [70].Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO reports. 2012;13:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, et al. Tet3 Reads 5-Carboxylcytosine through Its CXXC Domain and Is a Potential Guardian against Neurodegeneration. Cell reports. 2016;14:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Rasmussen KD, Berest I, Kebetaler S, Nishimura K, Simon-Carrasco L, Vassiliou GS, et al. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res. 2019;29:564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Pfeifer GP. Mutagenesis at methylated CpG sequences. Current topics in microbiology and immunology. 2006;301:259–81. [DOI] [PubMed] [Google Scholar]
- [78].Manzo M, Wirz J, Ambrosi C, Villasenor R, Roschitzki B, Baubec T. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 2017;36:3421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. [DOI] [PubMed] [Google Scholar]
- [80].Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Molecular cell. 2008;29:525–31. [DOI] [PubMed] [Google Scholar]
- [83].Liu Y, Zhang B, Kuang H, Korakavi G, Lu LY, Yu X. Zinc Finger Protein 618 Regulates the Function of UHRF2 (Ubiquitin-like with PHD and Ring Finger Domains 2) as a Specific 5-Hydroxymethylcytosine Reader. The Journal of biological chemistry. 2016;291:13679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–73. [DOI] [PubMed] [Google Scholar]
- [86].Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–6. [DOI] [PubMed] [Google Scholar]
- [87].Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Buck-Koehntop BA, Defossez PA. On how mammalian transcription factors recognize methylated DNA. Epigenetics. 2013;8:131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lafaye C, Barbier E, Miscioscia A, Saint-Pierre C, Kraut A, Coute Y, et al. DNA binding of the p21 repressor ZBTB2 is inhibited by cytosine hydroxymethylation. Biochem Biophys Res Commun. 2014;446:341–6. [DOI] [PubMed] [Google Scholar]
- [92].Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome biology. 2013;14:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. [DOI] [PubMed] [Google Scholar]
- [94].Gallina I, Colding C, Henriksen P, Beli P, Nakamura K, Offman J, et al. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat Commun. 2015;6:6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jeong WJ, Park JC, Kim WS, Ro EJ, Jeon SH, Lee SK, et al. WDR76 is a RAS binding protein that functions as a tumor suppressor via RAS degradation. Nat Commun. 2019;10:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, et al. Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Molecular cell. 2016;64:913–25. [DOI] [PubMed] [Google Scholar]
- [98].Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004;32:4100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mellen M, Ayata P, Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci U S A. 2017;114:E7812–E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PloS one. 2011;6:e21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Otani J, Kimura H, Sharif J, Endo TA, Mishima Y, Kawakami T, et al. Cell cycle-dependent turnover of 5-hydroxymethyl cytosine in mouse embryonic stem cells. PloS one. 2013;8:e82961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B, et al. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Molecular cell. 2014;54:879–86. [DOI] [PubMed] [Google Scholar]
- [104].Hong S, Cheng X. DNA Base Flipping: A General Mechanism for Writing, Reading, and Erasing DNA Modifications. Adv Exp Med Biol. 2016;945:321–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–21. [DOI] [PubMed] [Google Scholar]
- [106].Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–5. [DOI] [PubMed] [Google Scholar]
- [107].Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Aravind L, Anand S, Iyer LM. Novel autoproteolytic and DNA-damage sensing components in the bacterial SOS response and oxidized methylcytosine-induced eukaryotic DNA demethylation systems. Biol Direct. 2013;8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kweon SM, Zhu B, Chen Y, Aravind L, Xu SY, Feldman DE. Erasure of Tet-Oxidized 5-Methylcytosine by a SRAP Nuclease. Cell reports. 2017;21:482–94. [DOI] [PubMed] [Google Scholar]
- [110].Mohni KN, Wessel SR, Zhao R, Wojciechowski AC, Luzwick JW, Layden H, et al. HMCES Maintains Genome Integrity by Shielding Abasic Sites in Single-Strand DNA. Cell. 2019;176:144–53 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Khund-Sayeed S, He X, Holzberg T, Wang J, Rajagopal D, Upadhyay S, et al. 5-Hydroxymethylcytosine in E-box motifs ACAT∣GTG and ACAC∣GTG increases DNA-binding of the B-HLH transcription factor TCF4. Integr Biol (Camb). 2016;8:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ji S, Shao H, Han Q, Seiler CL, Tretyakova NY. Reversible DNA-Protein Cross-Linking at Epigenetic DNA Marks. Angew Chem Int Ed Engl. 2017;56:14130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Li F, Zhang Y, Bai J, Greenberg MM, Xi Z, Zhou C. 5-Formylcytosine Yields DNA-Protein Cross-Links in Nucleosome Core Particles. J Am Chem Soc. 2017;139:10617–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Raiber EA, Portella G, Martinez Cuesta S, Hardisty R, Murat P, Li Z, et al. 5-Formylcytosine organizes nucleosomes and forms Schiff base interactions with histones in mouse embryonic stem cells. Nat Chem. 2018;10:1258–66. [DOI] [PubMed] [Google Scholar]
- [115].Ji S, Fu I, Naldiga S, Shao H, Basu AK, Broyde S, et al. 5-Formylcytosine mediated DNA-protein cross-links block DNA replication and induce mutations in human cells. Nucleic Acids Res. 2018;46:6455–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nature chemical biology. 2012;8:328–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014;28:2304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hashimoto H, Zhang X, Cheng X. Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH. DNA repair. 2013;12:535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. The Journal of biological chemistry. 2000;275:33449–56. [DOI] [PubMed] [Google Scholar]
- [120].Wang D, Hashimoto H, Zhang X, Barwick BG, Lonial S, Boise LH, et al. MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res. 2017;45:2396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Yang J, Horton JR, Li J, Huang Y, Zhang X, Blumenthal RM, et al. Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine. Nucleic Acids Res. 2019;doi: 10.1093/nar/gkz381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, et al. Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development. Cell. 2012;151:1200–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–79. [DOI] [PubMed] [Google Scholar]
- [124].Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nature cell biology. 2008;10:1291–300. [DOI] [PubMed] [Google Scholar]
- [125].Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Dumesic PA, Homer CM, Moresco JJ, Pack LR, Shanle EK, Coyle SM, et al. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell. 2015;160:204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 2014;6:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nature communications. 2015;6:6204. [DOI] [PMC free article] [PubMed] [Google Scholar]