SUMMARY
Many neuronal types occur as pairs that are similar in most respects but differ in a key feature. In some pairs of retinal neurons, called paramorphic, one member responds to increases and the other to decreases in luminance (ON and OFF responses). Here, we focused on one such pair, starburst amacrine cells (SACs), to explore how closely related neuronal types diversify. We find that ON and OFF SACs are transcriptionally distinct prior to their segregation, dendritic outgrowth, and synapse formation. The transcriptional repressor Fezf1 is selectively expressed by postmitotic ON SACs and promotes the ON fate and gene expression program while repressing the OFF fate and program. The atypical Rho GTPase Rnd3 is selectively expressed by OFF SACs and regulates their migration but is repressed by Fezf1 in ON SACs, enabling differential positioning of the two types. These results define a transcriptional program that controls diversification of a paramorphic pair.
Keywords: ON-OFF, cell fate, retina, transcription factor, migration, polarization
Graphical Abstract

eTOC
Two related retinal cell types called ON and OFF starburst amacrines migrate to distinct laminae and signal increased (ON) or decreased (OFF) illumination. Fezf1 acts as a fate switch between the two types by activating ON and repressing OFF genes.
INTRODUCTION
In many parts of the nervous system, one finds pairs of neuronal types that are similar in most respects, but differ in a key feature. For example, select pairs of similar mammalian geniculocortical neuron types project to the visual cortex and differ primarily in their response properties to changes in luminance (Usrey and Fitzpatrick, 1996). Many such pairs are present in the retina, some of which have been called “paramorphic,” with one member responding to increases (ON cells) and the other to decreases (OFF cells) in light intensity (Famiglietti, 2004, 2005; Rousso et al., 2016). Because inputs that endow retinal neurons with ON and OFF responses are segregated to the inner and outer portions of a synaptic region called the inner plexiform layer (IPL), the key structural difference between members of these pairs is the position of their dendrites within the IPL (Figure 1A).
Figure 1. ON and OFF Starburst Amacrine Cells are Transcriptionally Distinct Subtypes Distinguished by Fezf1. See alsoFigure S1.
(A) Development of SACs. ONBL/INBL: outer/inner neuroblastic layer; OPL/OPL: outer/inner plexiform layer; INL: inner nuclear layer; GCL: ganglion cell layer; BC: bipolar cell; ooDSGC: ON-OFF direction-selective ganglion cell.
(B) t-distributed stochastic neighbor embedding (tSNE) visualization of 136 SACs sequenced at E16.
(C) Dotplot of select genes expressed by clusters 1 and 2 of the E16 SAC dataset. Megf10, Sox2, Isl1, and Slc18a3 are known SAC marker genes (left). Genes of interest are largely selected from the P0 differentially expressed (DE) gene list. DE genes that achieve statistical significance are in bold.
(D) Violin plot of Fezf1 expression in the two E16 SAC clusters.
(E-G) Same as B-D for 2,055 SACs sequenced at P0.
(H-J) Double fluorescence in situ hybridization (FISH) using probes against Fezf1 (magenta) and Megf10 or CHAT (green) to label SACs in E15.5 and P0 mouse retina (H), P0 marmoset retina (I) and the peripheral (top) and central (bottom) regions of E8 chicken retina (J). Among Megf10- or CHAT-positive SACs, Fezf1-positive cells are ON SACs while negatives are OFF SACs. FEZF1- and CHAT-positive cells in marmoset are ON SACs.
(K) Model of Fezf1-expressing SAC soma position before and after ON and OFF SAC soma segregation.
Scale bars are 20μm. SAC somas are outlined by circles, and white dashed lines indicate the ONBL and INBL border.
In few if any cases do we know when, where, or how these closely related types acquire their distinct characteristics. To address these issues, we focus here on a paramorphic pair of retinal interneurons called starburst amacrine cells (SACs). SACs are cholinergic retinal interneurons that are critical for computing the direction of moving objects (Vaney et al., 2012). As is the case for other paramorphic pairs, ON and OFF SACs have dendrites in the ON and OFF sublaminae of the IPL, respectively; in addition, their somata are differentially positioned, with ON SACs in the ganglion cell layer (GCL) and OFF SACs in the inner nuclear layer (INL) (Figure 1A). SACs are so closely anatomically and functionally related that they have been considered mirror-symmetric cells, differing only with respect to soma and dendritic arbor placement (Famiglietti, 1983; Vaney et al., 2012).
Using single cell transcriptomic profiling, we establish that ON and OFF SACs are transcriptionally distinct cell types, not mirror symmetric versions of a single type. We show that differential gene expression patterns arise during SAC migration and identify the transcriptional regulator Fezf1 as a critical determinant of the ON phenotype. Fezf1 acts postmitotically to specify the ON and repress the OFF SAC fates, in part by regulating the atypical Rho GTPase Rnd3/RhoE (hereafter Rnd3) to promote distinct SAC migratory programs. These results identify a gene expression program that diversifies ON and OFF SACs shortly after they become postmitotic but before they complete migration, thereby determining their somatic and dendritic positions without affecting the many properties they share.
RESULTS
ON and OFF Starburst Amacrine Cells are Transcriptomically Distinct
SACs in the central mouse retina are born at the apical edge of the retinal neuroblast layer (NBL) from around embryonic day (E) 10 to E14 (Voinescu et al., 2009). They then migrate basally to reach the border of the outer and inner neuroblast layers (ONBL, INBL). After E16, about half continue basally to the INBL, where they become ON SACs; the others remain in the ONBL and become OFF SACs (Figure S1C; Knabe et al., 2007; Thangaraj et al., 2012). Over the next few days the ONBL and INBL become the INL and GCL, respectively, the border between them becomes the IPL, and SACs extend dendrites into the IPL (Ray et al., 2018; Stacy and Wong, 2003). During the first postnatal week, ON and OFF bipolar cells begin making synapses onto SACs and the SACs begin making synapses onto RGCs. SACs are fully integrated into direction-selective circuits by the time of eye opening at postnatal day (P) 14 (Figure 1A).
When we began our studies, few molecular differences between ON and OFF SACs were known. Purinergic receptor P2x (P2rx2) was detected in OFF but not ON SACs (Kaneda et al., 2010), and Glycine receptor subunit alpha 4 (Glra4) was detected in ON but not OFF SACs (Heinze et al., 2007), but these differences were apparent only in mature retina. To ask whether differences arise before SACs reach their final positions, we performed single-cell RNA-sequencing (scRNA-seq) on 136 SACs at E16 (Figures 1B–1D and S1A; see STAR Methods) and 2,055 SACs at P0, after migration is complete but before synapses form (Figures 1E–1G). All of the sequenced cells were enriched for genes expressed by ON and OFF SACs (including Megf10, Sox2, Isl1 and Slc18a3, which encodes the vesicular acetylcholine transporter [VAChT]). Markers of other retinal neuron types, including Crx (cones and rods), Nrl (rods), Vsx2 (Chx10, bipolar cells), Lhx1 (horizontal cells), Pou4f1 (retinal ganglion cells) and Hes5 (progenitor cells) were expressed at very low levels, if at all (Figure S1B).
At both ages, we identified two distinct clusters of SACs that we suspected represented ON and OFF types. To test this idea, we used fluorescence in situ hybridization (FISH) to assess differentially expressed genes. Fezf1, Gfra3, Slit2, which were enriched in Cluster 1, were all selectively expressed in ON SACs; whereas Tenm3 and Rnd3 were enriched in Cluster 2 and selectively expressed in OFF SACs (Figures 1C, 1F, 1H and S1C; and see below). Moreover, Cntn5, which we recently showed is an ON SAC marker (Peng et al., 2017), was enriched in Cluster 1. Together these results show that Clusters 1 and 2 comprise ON and OFF SACs, respectively, demonstrating that these neuronal types are transcriptionally distinct while their somata are still intermingled and before they extend dendrites or form synapses.
Fezf1 is Expressed by Postmitotic ON Starburst Amacrine Cells
Fezf1 and its homologue Fezf2 have been implicated in specification and differentiation of several central neuronal types, although roles of Fezf1 have been difficult to study owing to redundancy with Fezf2 (Eckler and Chen, 2014). We were intrigued to note that Fezf1 was selectively expressed by ON SACs and that Fezf2 was not detectably expressed by either SAC type (Figure S1B). We therefore analyzed Fezf1 expression in detail using FISH.
Fezf1 was undetectable at E13.5, but it was present in a sparse population of cells in the basal ONBL at E14.5 and E15.5 (Figures 1H and S1D). Double-labeling with the pan-SAC marker Megf10 (Kay et al., 2012) demonstrated that Fezf1-positive cells comprised a subset of SACs, presumably those destined to be ON SACs (Figure 1H). By E17.5, the Fezf1-positive SACs had migrated to the INBL (Figure S1E). Selective expression of Fezf1 by ON but not OFF SACs was maintained after birth, although levels decreased during the second postnatal week (Figures S1E and S1F).
Fezf1 Expression by ON Starburst Amacrine Cells is Evolutionarily Conserved
ON and OFF SACs are present in most, if not all, vertebrate species (Zhang et al., 2019). We asked whether Fezf1 expression is a conserved feature of ON SACs by analyzing marmoset (Callithrix jacchus) and chicken (Gallus gallus domesticus) retinas. Double FISH of FEZF1 and CHAT revealed select expression of FEZF1 in ON SACs of the neonatal marmoset retina (Figure 1I). In chicks, we analyzed retinas at E8. At that stage, owing to a central to peripheral developmental gradient, ON and OFF SACs have segregated into distinct cellular layers (INBL and ONBL) in the central retina, but remain intermingled in the ONBL of the peripheral retina. As seen in mouse, FEZF1 is selectively expressed by ON SACs in central retina at this time, but by only about half of the migratory SACs in peripheral retina (Figure 1J). Thus, Fezf1 is selectively expressed by ON SACs in multiple vertebrate species and, at least in chick and mouse, is expressed by a subset of SACs prior to the completion of their migration.
Fezf1 Determines the Position of ON Starburst Amacrine Cells
To study Fezf1 during SAC development, we used a reporter line, Fezf1GFP, in which GFP replaces the coding region of Fezf1 to generate a null allele (Eckler et al., 2011, Figure S2A). Thus, GFP marks cells that express Fezf1 in heterozygotes (Fezf1GFP/+) and cells that were fated to express Fezf1 in homozygotes (Fezf1GFP/GFP). As expected from FISH results, GFP was expressed exclusively by ON SACs in Fezf1GFP/+ retinas between E14.5 and P6 (Figures 2A and S2B). A small percentage of GFP-positive SACs remained in the INL of P0 Fezf1GFP/+ retinas (Figure 2C), suggesting that ON SAC migration is incomplete at birth. After P6, GFP was also expressed by the bipolar cell type BC1b (Figure S2C), consistent with our previous observations (Shekhar et al., 2016). The Fezf1GFP allele also allowed us to determine if Fezf1 is expressed by SAC progenitors. The lack of the progenitor marker Ki67 staining in GFP cells indicated that detectable Fezf1 expression is confined to postmitotic SACs (Figure S2D).
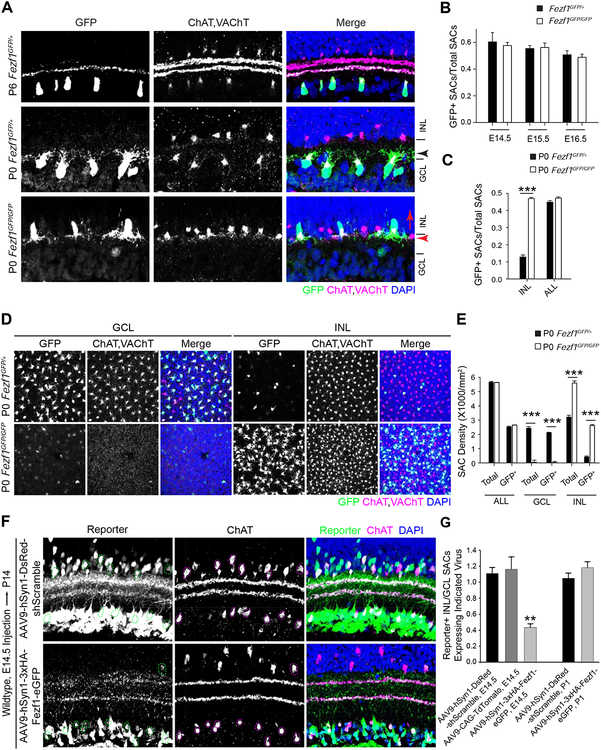
Figure 2. Fezf1 Determines the Position of ON Starburst Amacrine Cells. See alsoFigure S2.
(A) GFP-labeled ON SACs from P6 Fezf1GFP/+ (top), P0 Fezf1 GFP/+ (middle), and P0 Fezf1GFP/GFP (bottom) retinas. All SACs are labeled with antibodies against ChAT and VAChT. ON SACs are GFP-positive (green). In Fezf1GFP/+ retinas (top and middle), ON SACs are localized in the GCL while OFF SACs are localized in INL. In Fezf1GFP/GFP retinas (bottom), GFP-positive SACs are in the INL alongside OFF SACs (GFP-negative, ChAT/VAChT-positive). White and red arrows mark the position and orientation of ON SACs; arrowheads mark the stratification levels of ON SAC dendrites within the IPL.
(B) Ratios of GFP-positive SACs to total SACs at indicated embryonic time points in Fezf1 GFP/+ and Fezf1GFP/GFP retinas. Example images are in Figure S2A. Bars show mean ± SEM from 1–3 sections from each of 3–4 animals.
(C) Ratios of GFP-positive SACs to total SACs observed either in the INL or in the INL and GCL (ALL) in Fezf1GFP/+ or Fezf1GFP/GFP retinas. Bars show mean ± SEM from 8 images from each of 3–4 animals. Example images are in D.
(D) En face views of GFP-labeled SACs in the GCL (left) and INL (right) of Fezf1 GFP/+ (top) and Fezf1GFP/GFP (bottom) retinas.
(E) Cell densities of GFP-labeled SACs (GFP+) and ChAT/VAChT-labeled SACs (Total) in the GCL, INL, and both layers (ALL) in Fezf1GFP/+ and in Fezf1GFP/GFP retinas. N as in C.
(F) P14 retina sections transduced with AAVs expressing a control reporter (top) or Fezf1 overexpression construct (bottom), introduced at E14.5. In controls, AAV-expressing SACs (dashed circles) are equally likely to reside in the INL or GCL (top). Fezf1 overexpression biases infected SACs to reside in the GCL (bottom).
(G) Ratios of transduced SACs in the INL to the GCL for the indicated conditions. Bars show mean ± SEM from 4–6 images from 4 animals (Fezf1 overexpression and AAV9-hSyn1-shScramble control) or 2 animals (AAV9-CAG-TdTomato) at E14.5, and also 4–10 images from 8 animals (Fezf1 overexpression) and 5 animals (AAV9-hSyn1-shScramble control) at P1.
Scale bars are 20μm. ***, p<0.001 by Student’s t-test. **, p<0.01 by one-way ANOVA with post-hoc Tukey HSD test.
To ask if Fezf1 plays a role in ON SACs, we examined Fezf1GFP/GFP retinas. We detected no defects in overall retinal structure or development between E14.5 and ~P1–2, when Fezf1GFP/GFP mice die (Eckler et al., 2011). A normal cohort of SACs was present in mutants as early as E14.5, and the fraction that was GFP-positive was unaffected by Fezf1 deletion (Figures 2B and S2B). Strikingly, ON SACs were almost entirely depleted from the GCL in Fezf1 mutants and were instead found intermixed with OFF SACs (GFP-negative, ChAT/VAChT-positive) in the INL. This defect was apparent both in sectioned (Figure 2A) and wholemount (Figure 2D) retinas. SAC density was decreased by >90% in the GCL and correspondingly increased in the INL, with no significant change in the overall number of SACs (ChAT/VAChT-positive) or the fraction of SACs that were GFP-positive (Figures 2C and 2E). Thus, Fezf1 has no detectable effect on the generation of SACs, but it is required for their translocation into the INBL.
To test if Fezf1 is sufficient to promote SAC migration into the INBL, we generated an adeno-associated viral (AAV) vector that expresses an epitope- (3xHA-) tagged Fezf1 and GFP, driven by a human synapsin 1 promoter (hSyn1). We used AAV serotype 9, which supports strong expression in the embryonic retina within three days of infection (data not shown). We introduced the Fezf1 AAV or control AAVs at E14.5, after nearly all SACs had been born but prior to their segregation. As expected, 3xHA-Fezf1 was detected in the nuclei of infected cells (Figure S2F). SACs infected with control AAVs were equally likely to reside in either the INL or the GCL. In contrast, approximately three times as many cells infected with the Fezf1 AAV were present in the GCL compared to the INL (Figures 2F and 2G). This GCL-biased location induced by Fezf1 overexpression was specific to SACs, since other transduced amacrine cells (ChAT/VAChT-negative) and horizontal cells remained in their appropriate locations in the INL (Figure S2F). Additionally, introduction of Fezf1 at P1, after migration was complete, had no effect on the distribution of SACs (Figures 2G and S2E). Together these results demonstrate that Fezf1 is necessary and sufficient for proper SAC positioning in the GCL.
Fezf1 Promotes the Polarization and Migration of ON Starburst Amacrine Cells
The loss of SACs from the GCL in Fezf1GFP/GFP mutants likely results from the effect of Fezf1 on ON SAC migration. Therefore, we next characterized the morphology of migrating wild-type and Fezf1 mutant SACs. At E14.5, GFP-positive presumptive ON SACs exhibited a similar bipolar morphology in both Fezf1GFP/+ and Fezf1GFP/GFP mice (Figure 3Ai), suggesting that the early bipolar morphology of these SACs is not affected by loss of Fezf1. In contrast, a day later at E15.5, GFP-positive SACs differed dramatically between Fezf1GFP/+ and Fezf1GFP/GFP mice (Figure 3Aii). GFP-positive SACs in Fezf1GFP/+ heterozygotes bore a leading process that penetrated the INBL, whereas GFP-positive SACs in Fezf1GFP/GFP mice maintained a prominent apical trailing process. This difference persisted as control ON SACs entered the INBL and began to elaborate dendrites in the nascent IPL (Figures 3Aiii and 3Aiv). Thus, loss of Fezf1 inverts the polarized morphology of ON SACs (Figure 3B), likely impairing their subsequent migration.
Figure 3. Fezf1 Promotes the Polarization and Migration of ON Starburst Amacrine Cells by Repressing Rnd3 Expression. See alsoFigure S3.
(A) GFP-labeled ON SACs (left, middle) and sketches of representative morphologies (right) at indicated embryonic ages in Fezf1 GFP/+ and Fezf1GFP/GFP retinas. Embryonic SACs are identified by co-expression of Sox2 (magenta) and Isl1 (blue).
(B) Polar plot of the angle (radial axes) and length (X/Y axes) of the prominent neurite of individual ON SACs in E16.5 Fezf1 GFP/+ and Fezf1GFP/GFP retinas.
(C) Percent of total SACs expressing strong or weak levels of Rnd3 transcript (defined in STAR Methods) at indicated embryonic ages (left; ON and OFF SACs are quantified separately at E16.5 based on the differential positioning of their somas across the IPL) (see Figure S3B for representative images) and in P0 Fezf1GFP/+ and Fezf1GFP/GFP retinas (right).
(D) Rnd3, Chat, and GFP expression in SACs from P0 Fezf1 GFP/+ and Fezf1GFP/GFP retinas detected by RNAscope FISH. Circles outline SACs, and mutant ‘ON’ SACs are GFP- and Chat-positive (asterisked).
(E) P14 retina sections transduced with AAVs expressing control (shScramble, top), or shRnd3 (bottom) hairpins introduced at E13.5. DsRed marks AAV-transduced cells.
(F) Ratios of transduced SACs in the INL compared to the GCL for the indicated conditions. Bars show mean ± SEM from 4–10 images from 3 animals per condition.
Scale bars are 20μm. ***, p<0.001 by Student’s t-test.
It appeared from these experiments that the morphology of Fezf1GFP/GFP mutant ON SACs resembled that of control (Fezf1GFP/+) OFF SACs. To visualize the morphology of ON and OFF SACs, we used a mouse line in which Cre recombinase was inserted into the locus encoding Megf10, which is expressed by all SACs (Kay et al., 2012; and Figures 1C and 1F). We crossed these mice to a Cre-dependent reporter (Rosa::mTmG) and visualized SAC morphology at E16.5. Wild-type ON SACs within the INBL exhibited a basally-directed leading process, similar to GFP-positive ON SACs in Fezf1GFP/+ retinas, whereas wild-type OFF SACs in the ONBL bore a prominent trailing process, similar to GFP-positive displaced ‘ON’ SACs in Fezf1GFP/GFP retinas (Figure S3E).
Fezf1 Represses Rnd3 Expression to Promote Migration into the INBL
The similarity between migrating Fezf1 mutant ‘ON’ SACs and wild-type OFF SACs raised the possibility that Fezf1 promotes an ON, or represses an OFF, SAC migratory program. We therefore sought differentially expressed, Fezf1-dependent genes that could account for the distinct polarity and migration of ON and OFF SACs. No obvious candidates were selectively expressed by ON SACs, but Rnd3, an atypical Rho family GTPase, was enriched in OFF SACs (Figures 1C, 1F and S1C). In the cerebral cortex, Rnd3 regulates interkinetic nuclear migration of neuronal progenitors and promotes neuronal migration (Pacary et al., 2011, 2013). In the embryonic retina, Rnd3 transcript is not strongly detected in retinal progenitors or radially migrating neurons in the ONBL, but it is highly expressed by differentiating neurons, including SACs, in the INBL (Figures S3A and S3B). Approximately 30% and 50% of Megf10-positive SACs express high levels of Rnd3 (>10 puncta per cell, see STAR Methods) at E14.5 and E15.5, respectively (Figures 3C and S3B). By E16.5, when ON and OFF SACs in the central retina can be distinguished by their position, 85% of OFF SACs, but only 15% of ON SACs, express high levels of Rnd3, and Rnd3 levels in ON SACs continue to decrease over time (Figures 3C, S3B, and S3D). These data suggest that Rnd3 is upregulated in OFF SACs and downregulated in ON SACs prior to ON SAC migration into the INBL. Moreover, triple FISH against Rnd3, Chat (all SACs), and GFP (ON SACs) revealed that Rnd3 was upregulated in Fezf1GFP/GFP mutant ‘ON’ SACs in the INL (Figures 3C and 3D).
These results suggest that Rnd3 could confine OFF SACs within the ONBL and, conversely, that loss of Rnd3 enables SAC migration into the INBL. To test this idea, we generated AAV9 vectors expressing DsRed and either a short hairpin RNA that potently knocked down Rnd3 expression (shRnd3) in heterologous cells (Figure S3C), or a scrambled sequence (shScramble), both driven by the hSyn1 promoter. AAVs were introduced intravitreally at E13.5, and retinas were analyzed at P14. In retinas expressing shRnd3, the ratio of DsRed-positive SACs in the INL to GCL decreased by 60% compared to retinas expressing shScramble (Figures 3E and 3F). Taken together, these data show that Fezf1 represses Rnd3 in ON SACs and that this repression enables their entry into the INBL (Figure S3F).
Fezf1 is Required for ON Starburst Amacrine Cell Fate Specification
Finally, we asked whether the role of Fezf1 in SACs is solely to control their migratory program, or if it plays a more general role in specifying SAC fate. We used two approaches, one histological and one molecular, to distinguish between these alternatives.
First, we took advantage of a feature exhibited by most retinal neurons called “mosaic spacing,” in which neurons of a single type are more evenly separated from other neurons of the same type than would be expected from a random distribution, whereas they are randomly spaced with respect to neurons of other types (Reese and Keeley, 2015; Wassle and Riemann, 1978). This spacing has been viewed as an indicator that these non-randomly spaced neurons belong to a single type (Rockhill et al., 2000; Sanes and Zipursky, 2010). Both ON and OFF SACs form mosaics in the GCL and INL, respectively (Whitney et al., 2014). Using statistical methods employed to assess mosaicism (Kay et al., 2012; Reese and Keeley, 2015; Rodieck 1991), we asked if mutant ‘ON’ SACs (ChAT/VAChT- and GFP-positive) and OFF SACs (ChAT/VAChT-positive, GFP-negative) together form a single mosaic in the INL of Fezf1GFP/GFP retinas. As expected from the increased SAC density in the Fezf1GFP/GFP mutant INL (Figure 2D), the exclusion zone radius (spacing between SACs) was reduced in-mutants, however their packing pattern and the regularity of their packing did not differ significantly between Fezf1GFP/+ and Fezf1GFP/GFP retinas (Figures S4A and S4B). Thus, by this criterion OFF SACs and GFP-positive mutant ‘ON’ SACs in the INL are members of a single type.
Second, and more directly, we used FACS to isolate GFP-positive SACs from Fezf1GFP/+ and Fezf1GFP/GFP retinas at E17.5 and performed RNA-seq to compare their transcriptomes. The two populations were similar but formed distinct groups (Figures 4A and S4C). Genes common to ON and OFF SACs, as judged by our initial scRNA-seq analysis (Figure 1) were present at similar levels in heterozygotes and homozygous mutants (e.g. Sox2 and Megf10; Figure S4D). However, genes selectively expressed by control ON SACs (ON genes) were downregulated in mutant compared to control GFP-positive SACs, and genes selectively expressed by OFF SACs (OFF genes) were upregulated in the mutant ‘ON’ SACs (Figure 4B). The difference was consistent across replicates, as shown by a composite index based on expression of ON and OFF genes (Figure 4C, see STAR Methods). Consistent with the RNA-seq data, ON SAC markers Gfra3 and Slit2 were absent from mutant GFP-positive ‘ON’ SACs, and their expression levels mirrored those observed in GFP-negative OFF SACs (Figures 4D, 4E, S4E, and S4F). Conversely, the mutant ‘ON’ SACs expressed high levels of the OFF SAC-specific gene Tenm3 (Figures 4F and 4G), in addition to Rnd3 (Figures 3C and 3D).
Figure 4. Fezf1 is Required for ON Starburst Amacrine Cell Fate Specification. See alsoFigure S4.
(A) PCA plot of E17.5 Fezf1 GFP/+ and Fezf1GFP/GFP ON SAC RNA-seq samples using 400 DE genes.
(B) Expression differences (lg transformation) of ON and OFF genes identified in the P0 SAC scRNA-seq dataset (Figures 1E and 1F) between Fezf1 GFP/+ and Fezf1GFP/GFP ON SACs.
(C) ON/OFF score of 13 Fezf1GFP/+ and Fezf1GFP/GFP RNA-seq replicates (see STAR Methods).
(D) RNAscope FISH using Gfra3, Chat, and GFP probes in P0 Fezf1 GFP/+ and Fezf1GFP/GFP retinas. Circles outline SACs, and mutant ‘ON’ SACs are GFP- and Chat-positive (asterisked).
(E) Percent of total SACs expressing strong or weak levels of Gfra3 in P0 Fezf1 GFP/+ and Fezf1GFP/GFP retinas.
(F) RNAscope FISH using Tenm3, Chat, and GFP probes in P0 Fezf1 GFP/+ and Fezf1GFP/GFP retinas.
(G) Percent of total SACs expressing strong or weak levels of Tenm3 in P0 Fezf1 GFP/+ and Fezf1GFP/GFP retinas.
(H) Model of ON SAC cell fate determination by Fezf1. Fezf1 inhibits OFF genes while promoting ON genes (see Discussion).
Scale bars are 20μm.
Finally, we used triple-label in situ hybridization to determine if differential expression of ON and OFF genes is established by E15.5, prior to the time when ON and OFF SACs acquire their distinct positions. We used Megf10 to label all SACs and Fezf1 to label presumptive ON SACs. Three OFF SAC enriched genes (Tenm3, Zfhx3, and Cygb) are selectively expressed in the Fezf1-negative presumptive OFF SACs at this stage (Figures S4G and S4H). These results demonstrate that multiple molecular distinctions between ON and OFF SACs are established prior to – and therefore do not depend on – their final positions. Based on these results, we conclude that Fezf1 specifies the ON SAC fate by promoting the expression of ON genes and repressing the expression of OFF genes (Figure 4H).
DISCUSSION
A promising strategy for elucidating mechanisms underlying neuronal cell type diversification is to analyze pairs of neurons that are similar in many respects but differ in a key feature. Here, we applied this strategy to a paramorphic retinal pair—ON and OFF SACs. We demonstrate that ON and OFF SACs are transcriptionally distinct cell types prior to their somatic migration, dendritic stratification, and synapse formation. We show that one differentially expressed gene, the transcriptional regulator Fezf1, is a critical ON SAC fate determinant, and another differentially expressed gene, the Rho GTPase Rnd3, is regulated by Fezf1 to control the differential migration of the SAC types.
Postmitotic Starburst Amacrine Cell Fates are Determined by Fezf1
Several lines of evidence support the conclusion that Fezf1 controls a postmitotic ON/OFF SAC fate switch. First, Fezf1 expression is detected only after SACs are born, but before they migrate to distinct locations. Second, Fezf1 is expressed in ON SACs, but not in OFF SACs or any other cell types prior to P6, at which time it appears in a subset of bipolar cells. Lastly, the function of Fezf1 promotes the expression of ON genes while suppressing OFF genes, but does not affect expression of genes shared by ON and OFF SACs. Therefore, Fezf1 joins a small number of genes shown to control a binary postmitotic cell fate choice (Jukam and Desplan, 2010).
A major outstanding question is how half of the newly postmitotic SACs “choose” to express Fezf1. We envision several possibilities. One is that the choice is stochastic, as has been observed in several neuronal and nonneural cell types. Frequently such choices result from the amplification of a small difference by a cell-intrinsic feedback mechanism (Johnston and Desplan, 2010). Another is that extrinsic factors – for example proximity to another SAC, or other cues in the extracellular environment – could affect the choice. A third possibility is that a shared progenitor undergoes an asymmetric terminal division, leading to unequal inheritance of a Fezf1 regulator by daughters (Tajbakhsh et al., 2009). Finally, we cannot rule out the possibility that ON and OFF SACs arise from separate progenitor types, one of which expresses undetectably low levels of Fezf1, or is specified to upregulate Fezf1 postmitotically. We believe this is unlikely, however, because in a prior analysis of retinal cell lineage (De la Huerta et al., 2012) we observed some clones that contained both ON and OFF SACs.
How Does Fezf1 Regulate Gene Expression in ON Starburst Amacrine Cells?
Fezf1 is a transcriptional repressor that interacts with Groucho/TLE (Transducin-Like Enhancer of Split)-type transcriptional corepressors via its EH1 (Engrailed homology1) repressor motif (Shimizu and Hibi, 2009; Shimizu et al., 2010). We found that Fezf1 not only suppresses the expression of OFF genes, but also activates the expression of ON genes. How might Fezf1 activate ON genes? One possibility is that Fezf1 derepresses an ON gene transcriptional activator, similar to derepression of neurogenin 2 by Fezf1 and Fezf2 during cortical development (Shimizu et al., 2010). Another possibility is that Fezf1 can function as an activator as well as a repressor, activating ON genes and repressing OFF genes in ON SACs, similar to the way in which Fezf2 determines the cell fate of subcerebral projection neurons by repressing Satb2 and activating Ctip2 expression (Chen et al., 2008). This could be accomplished by context-dependent association of Fezf1 with downstream coactivators and corepressors, respectively. Intriguing candidates here are the zinc finger transcription factors Zfhx3 and Zfhx4, which are selectively expressed by OFF and ON SACs, respectively. Both are predicted to have Fezf1 binding sites (GeneHancer ID: GH16J073045, GH08J076680). Zfhx3 and Zfhx4 are most closely related to each other among the four zinc finger homeobox family members, and their spatial and temporal expression patterns in the brain are consistent with cooperative function in neuronal differentiation (Kostich and Sanes, 1995; Nogami et al., 2005).
Rnd3 Regulation of Starburst Amacrine Cell Polarity and Migration
The Fezf1GFP allele allowed us to compare ON (Fezf1GFP/+) and converted OFF (Fezf1GFP/GFP) SAC migratory morphologies. Strikingly, we observed that ON and OFF SACs establish prominent neurites of opposite polarity before moving to their final destinations. Rnd3 is an attractive candidate regulator of this difference. Rnd3 lacks intrinsic GTPase activity and remains constitutively active, mediating rearrangement of the actin cytoskeleton by inhibiting the RhoA effector ROCK1 (Jie et al., 2015). In cultured hippocampal neurons, loss of Rnd3 impairs neuronal polarization and neurite outgrowth (Peris et al., 2012). Strong expression of Rnd3 by OFF but not ON SACs is consistent with Rnd3 regulating OFF SAC polarity.
Alternatively, Rnd3 may control other aspects of SAC migration. In the mouse neocortex, Rnd3 promotes neuronal radial migration through the cortical plate (Pacary et al., 2011). Given its role in radial migration, it was surprising to see high levels of Rnd3 in postmitotic neurons in the INBL rather than in radially migrating neurons in the ONBL. This raises the possibility that Rnd3 affects the local positioning of OFF SACs, rather than their radial migration, per se. It is also possible that Rnd3 promotes neurite outgrowth in OFF SACs as a prerequisite for the development of their somal-layer neurite network. This network of connected OFF SACs forms as early as E16 and depends on homotypic contacts mediated by Megf10 (Ray et al., 2018). Therefore, the somal neurite network could stabilize OFF SACs in the ONBL while Fezf1 represses Rnd3 in ON SACs, allowing for their migration into the INBL. Consistent with this idea, Rnd3 expression in ON SACs decreases over time, and knocking down Rnd3 biases SACs to reside in the GCL.
From Cell Fate Determination to the Assembly of ON and OFF Circuits
Our RNA-seq analysis demonstrated that Fezf1 not only controls initial differences between ON and OFF SACs, but also instructs the early expression of adhesion molecules that later pattern the formation of ON and OFF circuits. For instance, Fezf1 promotes expression of Cntn5 in ON SACs and Cntn5 mediates the formation of ON circuits via homophilic interactions between ON SAC dendrites and ON dendrites of ooDSGCs (Peng et al., 2017). Additional intriguing candidate genes downstream of Fezf1 are Slit2 and Teneurin-3 (Tenm3), both of which are predicted to have Fezf1 binding sites (GH041020276 and GH04J182175). Slit2 is enriched in ON SACs, while Tenm3 is selectively expressed by OFF SACs. However, published images suggest that Slit2 does not affect SAC lamination (Rama et al., 2015), and our preliminary data do not support a role for Tenm3 in this process (data not shown). It remains possible that either or both might be involved, like Cntn5, in mediating interactions between SACs and their synaptic partners.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABLITY
Further information and requests for reagents may be directed and will be fulfilled by the corresponding authors Alex L. Kolodkin (kolodkin@jhmi.edu) and Joshua R. Sanes (sanesj@mcb.harvard.edu). The lead contact is Joshua R. Sanes (sanesj@mcb.harvard.edu) All unique/stable reagents generated in this study are available from the corresponding authors without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) at Harvard University and The Johns Hopkins University School of Medicine.
Mice
Mice were maintained in pathogen-free facilities at Harvard and Johns Hopkins Universities under standard housing conditions with continuous access to food and water. Timed-pregnant mice were either purchased from Charles River or set up in house. RNA sequencing experiments were carried out at embryonic age (E) 16, 17.5, and postnatal (P) 0. Histological and in situ hybridization studies used E14.5 to adult mice as indicated. Mice of both sexes were used, and no sexual dimorphisms were observed in any results reported here. None of the mice had noticeable health or immune status abnormalities and were not subject to prior procedures. The genotype of mice is described where appropriate. The following mouse lines were used:
In Fezf1GFP, eGFP was inserted into the endogenous Fezf1 locus, generating a null allele and leading to expression of eGFP in cells that normally express Fezf1 (Eckler et al., 2011).
In ChATCre mice, Cre was inserted downstream of the ChAT coding sequence, leading to expression of Cre in cholinergic neurons without disrupting endogenous ChAT expression (Rossi et al., 2011).
In Thy1-stop-YFP Line #15 transgenic mice, a lox-stop-lox eYFP cassette is expressed under control of regulatory elements from the mouse Thy1 gene, leading to cre-dependent expression of YFP in many neuronal population (Buffelli et al., 2003).
In Megf10Cre mice, Cre was inserted in the Megf10 locus without disrupting endogenous Megf10 expression. A complete description of this line will be presented elsewhere (J.N.K., in preparation). Briefly, Cre recombinase was inserted at the end of the Megf10 coding exon, with the two reading frames separated by a T2A sequence.
In ROSAmT/mG mice, a lox-membrane-localized tdTomato (mT) - membrane-localized eGFP (mG) cassette was inserted into the ROSA26 locus. Thus, mG is expressed in cells that express or previously expressed Cre recombinase, while mT is expressed in other cells (Muzumdar et al., 2007).
Timed pregnant CD1-IGS mice from Charles River were used for in situ hybridization and viral transduction experiments.
Chicken and Marmoset
Specific-Pathogen-Free (SPF) eggs were purchased from Charles River. Eyes were collected from E8 embryos. Eyes from a newborn marmoset (Callithrix jacchus) were generously provided by the McGovern Institute for Brain Research (Massachusetts Institute of Technology). We are grateful to G. Feng, Q. Zhang and C. Wu for access to this tissue. Eyes were collected and transported in ice-cold Hibernate-A (BrainBits).
METHOD DETAILS
Single Cell Isolation and Droplet Based scRNA-seq
P0 ChATCre:Thy1-STP-YFP (line#15) retinas were digested with papain (Worthington, LS003126) at 37°C for 10 mins. Retinas were then dissociated and triturated into single cell suspensions. YFP labeled SACs were isolated by FACS and loaded into 10X Chromium Single Cell A Chips at a concentration of ~1,000 cells/μL. E16 SACs were collected from an unrelated project in which RGCs were targeted, but SACs were also obtained (I. E. Whitney and J.R.S., submitted).
Single cell libraries were prepared using the Chromium 3’ v2 platform (10X Genomics, Pleasanton, CA) as previously described (Peng et al., 2019). Libraries were sequenced on the Illumina HiSeq 2500 (Paired-end reads: Read 1, 26bp, Read 2, 98bp).
RNA-seq of SACs from Fezf1 Mutants via Smart-seq2
E17.5 Fezf1GFP/+ and Fezf1GFP/GFP SACs were isolated by FACS in pools of 100 cells. 13 replicates were collected from mice of each genotype. cDNA libraries were prepared with Smart-seq2 and Nextera XT (Picelli et al., 2014). Libraries were sequenced on the Illumina NextSeq with paired-end 75bp reads.
ScRNA-seq Data Analysis
ScRNA-seq data was analyzed as previously described (Peng et al., 2019; Shekhar et al., 2016). Briefly, sequenced reads were aligned to the mouse genome mm10, and count matrix of sequenced cells was generated using the Cell Ranger software (version 2.1.0, 10X Genomics). Only cells with the expression of > 600 genes were used for further analysis. We preprocessed the count data following normalization to median UMI counts, regression out mitochondrial and ribosome genes, and extraction of all the SACs with scoring for gene signatures of SACs – Sox2, Megf10, Megf11, Chat, and Slc18a3. Lastly, we clustered SACs with graph-based clustering using significant PCs and visualized the clustering result in 2D via t-distributed stochastic neighbor embedding (t-SNE). Differential expression (DE) tests for a gene between two identified clusters (later validated as ON and OFF SAC clusters) were performed using the ‘MAST’ package with the criteria of at least expression at 20% cells within a cluster and threshold of 0.25 (Finak et al., 2015).
RNA-seq data were analyzed as previously described (Peng et al., 2017). Briefly, sequenced reads were mapped to the mouse genome (mm10) via Tophat (Trapnell et al., 2012); transcripts were counted via Cufflinks; differentially expressed genes were detected with Cuffdiff.
ON/OFF scores of all 26 replicates were calculated with a log2 transformation of the ratio of average expression levels of ON genes to that of OFF genes.
Tissue Processing and Fluorescent In Situ Hybridization
Timed-pregnant mice were anesthetized with intraperitoneal injection of euthasol (Virbac), and eyes from embryos were collected and fixed in 4% PFA for 1hr at 4°C. Retinas were then dissected and fixed with 4% PFA for additional 0.5hr at 4°C. Eyes were collected from P0–P8 mice after cervical dislocation and from adults euthanized with intraperitoneal injection of euthasol. Retinas were dissected and fixed with 4% PFA for 1.5–2hr at 4°C. E8 chicken eyes were collected and fixed with 4% PFA for 1hr at 4°C, and retinas were dissected and further fixed with 4% PFA for 0.5 hr at 4°C. Marmoset retinas were dissected in Ames from newborn marmoset eyes and fixed with 4% PFA overnight at 4°C.
After fixation, retinas were rinsed with PBS, immersed with 30% sucrose overnight at 4°C, embedded in Tissue Freezing Medium (EMS) and cryosectioned at 20μm on Superfrost Plus slides (Fisher).
Probe templates were generated using cDNA derived from mouse, chicken, and marmoset retinas following RNA extraction and reverse transcription with AzuraQuant™ cDNA Synthesis Kit (Azura, AZ-1995). Antisense probes were generated by PCR using a reverse primer with a T7 sequence adaptor to permit in vitro transcription (see Key Resources for primer sequences). DIG rUTP (Roche, 11277073910), DNP rUTP (Perkin Elmer, NEL555001EA), and Fluorescein rUTP (Roche, 11685619910) were used for synthesizing probes for double fluorescent in situ hybridization (FISH) experiments. Details of FISH were previously described (Peng et al., 2019; Shekhar et al., 2016). Briefly, retinal sections were treated with 1.5 μg/ml of proteinase K (NEB, P8107S) and then post-fixed and treated with acetic anhydride for deacetylation. Probe detection was performed with anti-DIG HRP (1:1000) and anti-Fluorescein (1:1000) or anti-DNP HRP (1:500) followed by tyramide amplification.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Rabbit polyclonal anti-GFP | Millipore | Cat# AB3080P; RRID: AB_2630379 |
| Mouse polyclonal anti-Isl1 | DSHB | Cat# 39.4D5; RRID:AB_2314683 |
| Goat polyclonal anti-Choline Acetyltransferase | Millipore | Cat# AB144P; RRID: AB_11214092 |
| Goat polyclonal anti-Vesicular Acetylcholine Transporter | Millipore | Cat# ABN100; RRID: AB_2630394 |
| Goat polyclonal anti-Sox-2 (Y-17) | Santa Cruz | Cat# sc-17320; RRID: AB_2286684 |
| Rabbit polyclonal anti-Dsred | Living Colors | Cat# 632496; RRID: AB_10013483 |
| Chicken polyclonal anti-GFP | AVES | Cat# GFP-1020; RRID: AB_10000240 |
| Rabbit monoclonal anti-HA | Cell Signaling | Cat# 3724s; RRID: AB_1549585 |
| Mouse monoclonal anti-actin, clone C4 | Millipore | Cat# MAB1501; RRID: AB_2223041 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ames’ Medium | Sigma | Cat# A1420 |
| Papain | Worthington | Cat# LS003126 |
| Ovomucoid | Worthington | Cat# LS003087 |
| AzuraQuant cDNA synthesis kit | Azura | Cat# AZ-199 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Life Technologies | Cat# D1306; RRID: AB_2629482 |
| Lipofectamine 2000 Transfection Reagent | ThermoFisher Scientific | Cat# 11668019 |
| Critical Commercial Assays | ||
| Chromium Single Cell 3′ Library & Gel Bead Kit v2, 16 rxns | 10X Genomics | Cat# 120237 |
| Chromium Single Cell A Chip Kit, 16 rxns | 10X Genomics | Cat# 1000009 |
| Chromium i7 Multiplex Kit 96 rxns | 10X Genomics | Cat# 120262 |
| Smart-seq2 reagents | Picelli et al., 2014 | N/A |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat# FC-131–1024 |
| RNAscope Fluorescent Multiplex Kit | Advanced Cell Diagnostics | Cat# 320850 |
| RNAlater | ThermoFisher | Cat# AM7024 |
| RNeasy Mini Kit | QIAGEN | Cat# A74104 |
| RNAclean SPRI | Beckman Coulter Genomics | Cat# A63987 |
| RNAscope® Probe- eGFP | Advanced Cell Diagnostics | Cat# 400281 |
| RNAscope® Probe- GFP-C2 | Advanced Cell Diagnostics | Cat# 409011-C2 |
| RNAscope® Probe- Mm-Chat-C3 | Advanced Cell Diagnostics | Cat# 410071-C3 |
| RNAscope® Probe- Mm-Megf10-C3 | Advanced Cell Diagnostics | Cat# 406641-C3 |
| RNAscope® Probe- Mm-Rnd3-C2 | Advanced Cell Diagnostics | Cat# 424731-C2 |
| RNAscope® Probe- Mm-Gfra3-C2 | Advanced Cell Diagnostics | Cat# 467191-C2 |
| RNAscope® Probe- Mm-Slit2-C2 | Advanced Cell Diagnostics | Cat# 449691-C2 |
| RNAscope® Probe- Mm-Tenm3 | Advanced Cell Diagnostics | Cat# 411951 |
| RNAscope® 3-plex Negative Control Probe | Advanced Cell Diagnostics | Cat# 320871 |
| RNAscope® 3-plex Positive Control Probe Mm | Advanced Cell Diagnostics | Cat# 320881 |
| NEBuilder® HiFi DNA Assembly Cloning Kit | New England BioLabs | Cat# E5520S |
| Phusion® Hot Start Flex 2X Master Mix | New England BioLabs | Cat# M0536S |
| SuperScript™ III First-Strand Synthesis SuperMix | ThermoFisher | Cat# 18080400 |
| TRIzol™ Plus RNA Purification Kit | ThermoFisher | Cat# 12183555 |
| Deposited Data | ||
| Raw data files for scRNA-sequencing | This study | GEO: GSE132555 |
| Raw data files for RNA-sequencing | This study | GEO: GSE132555 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Fezf1:eGFP | Gift from B. Chen | N/A |
| Mouse: B6.Cg-Tg(Thy1-EYFP)15Jrs/J | Buffelli et al., 2003 | Stock#: 005630; RRID: IMSR_JAX:005630 |
| Mouse: CD1 | Charles River | 022 |
| Mouse: Megf10:Cre | This paper | N/A |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratory | Stock#: 007676, RRID: IMSR_JAX:007676 |
| Experimental Models: Cell Lines | ||
| Cell line: HEK293T cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Bacterial and Virus Strains | ||
| AAV9-hSyn1–3XHA-Fezf1-P2A-eGFP (4.01E+13) | Janelia Research Campus Viral Tools | N/A |
| AAV9-CAG-TdTomato (4.01E+13) | UNC Vector Core | N/A |
| AAV9-DsRed-shScrambled ((4.01E+13) | Janelia Research Campus Viral Tools | N/A |
| AAV9-hSyn1-DsRed-shRnd3 (4.8393E+13) | Janelia Research Campus Viral Tools | N/A |
| AAV9-hSyn1-DsRed-shScrambled (4.8393E+13) | Janelia Research Campus Viral Tools | N/A |
| Recombinant DNA | ||
| pCIG2 | Hand et al., 2005 | N/A |
| pCIG2-LSL-3xHA-Fezf1 | This study | N/A |
| pCIG2-LSL-3xHA-Rnd3 | This study | N/A |
| pCAG-Cre | Hand et al., 2015 | N/A |
| pPRIME-DsRed-shScramble | This study | N/A |
| pPRIME-DsRed-shRnd3 | This study | N/A |
| pAAV-hSyn1-MCS | This study | N/A |
| pAAV-hSyn1–3xHA-Fezf1-P2A-eGFP | This study | N/A |
| pAAV-hSyn1-DsRed-shScramble | This study | N/A |
| pAAV-hSyn1-DsRed-shRnd3 | This study | N/A |
| Oligonucleotides see Table S1 | ||
| Software and Algorithms | ||
| Cell Ranger v2.1.0 | 10X genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Tophat | Trapnell et al., 2012 | RRID: SCR_013035 |
| Cufflinks | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cufflinks/; RRID: SCR_014597 |
| Cuffdiff | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cufflinks/; RRID: SCR_001647 |
| R version 3.4.1 | The R project | https://www.r-project.org/ |
| ImageJ | NIH | http://rsb.info.nih.gov.ezp-prod1.hul.harvard.edu/ij/index.html; RRID:SCR_003070 |
| Pairwise stitching ImageJ plugin | NIH | http://imagej.net/Image_Stitching |
| Segmentation Simple Neurite Tracer ImageJ plugin | NIH | http://imagej.net/Simple_Neurite_Tracer |
| DRP | Rockhill et al., 2000 | http://wvad.mpimf-heidelberg.mpg.de/abteilungen/biomedizinischeOptik/software/WinDRP/index.html |
| Zen | Zeiss | RRID:SCR_013672 http://www.zeiss.com/microscopy/en_us/products/microscope-software/zen.html#introduction |
| Other | ||
| Illumina Nextseq 500 | Illumina | N/A |
| Illumina Hiseq | Illumina | N/A |
RNAscope In Situ Hybridization
Timed-pregnant mice were euthanized by continuous exposure to CO2 followed by cervical dislocation. Embryos were harvested and decapitated, and eyes were dissected and fixed overnight in RNase-free 4% PFA with rotation. Eyes were rinsed twice for 20 minutes in RNase-free PBS, followed by RNase-free 15% sucrose at room temperature for 1–2 hours, until the eyes sunk. They were then immersed in RNase-free 30% sucrose overnight, embedded in Neg-50 (Thermo Scientific) mounting medium, and cryosectioned at 14μm onto Superfrost Plus slides (Fisher). Slides were stored at −80°C until use.
Slides were thawed from −80°C, dried for 20 minutes at room temperature, and then baked at 60°C for one hour prior to target retrieval. The RNAscope® assay was then conducted per the manufacturer’s protocol with the following modifications: 1) during the target retrieval step, the slides were slowly inserted into target retrieval solution at 90–95°C for two minutes and thirty seconds; 2) slides were incubated in DAPI in the final step for two minutes and thirty seconds. Protease III was used.
Images were acquired with a 40X EC Plan Apochromat NA1.3 oil immersion objective (Zeiss) at the resolution of 1024×1024 pixels and Z-step size of 0.5–0.75μm, 6–12 slices per image to encompass the entirety of the mRNA signal. To score number of mRNA puncta per cell, outlines of individual SACs were drawn on the image with the segmented line tool in ImageJ using the Megf10 (embryonic Rnd3 experiments) or Chat (P0 Fezf1GFP/GFP mutant quantifications) and DAPI channels. Cells were grouped into expression bins based on the relative numbers of puncta per gene of interest while scrolling through each optical section, rather than in a Z-projection, to enhance binning accuracy. Bins were determined for each gene separately owing to their different levels of expression. The following bins were used:
| Gene | Weak Expression (n puncta) | Strong Expression (n puncta) |
|---|---|---|
| Rnd3 (all quantifications) | ≤10 | >10 |
| Gfra3 | ≤3 | >3 |
| Tenm3 | ≤10 | >10 |
| Slit2 | ≤3 | >3 |
The quantification of embryonic Rnd3 expression was done blindly. Since the Fezf1GFP/GFP phenotype was visible, a blind analysis of this phenotype was infeasible.
Immunohistochemistry
Retinal tissues were prepared as described above. Antibodies used were as follows: chick and rabbit anti-GFP (1:500, Abcam; 1:5000, Millipore; 1:1000, AVES); goat anti-choline acetyltransferase (1:500, Millipore); goat anti-VAChT (1:1000, Millipore); goat anti-Sox2 (1:100, Santa Cruz); mouse anti-Isl1 (1:10, DSHB), rabbit anti-HA (1:500, Cell Signaling), and rabbit anti-DsRed (1:1000, Living Colors), and mouse anti-Ki67 (1:200, BD Biosciences). Nuclei were labeled with DAPI (1:1000, Invitrogen). Secondary antibodies were conjugated to Alexa Fluor 488, 568, and 647 (Invitrogen) and used at 1:1000. Fluoromount-G (SouthernBiotech) was used for mounting wholemounts. ProLong Gold Antifade was used for mounting retina section slides.
Image Acquisition and analysis
Images were acquired on Zeiss LSM 710 confocal microscopes with 405, 488–515, 568, and 647 lasers, processed using Zeiss ZEN software, and analyzed using ImageJ (NIH). Section images were acquired with 10X, 16X, 40X or 63X oil lens at the resolution of 1024×1024 pixels, a step size of 0.5–1.5μm. ImageJ (NIH) software was used to generate maximum intensity projections. Adobe Photoshop CC was used for adjustments to brightness and contrast.
Generation and use of AAV vectors
Fezf1 Overexpression.
The Fezf1 coding sequence (CDS) was amplified from a P3 retinal cDNA library (RNA was extracted using the Invitrogen™ TRIzol™ Plus RNA Purification Kit and reverse transcribed using Invitrogen™ SuperScript™ III First-Strand Synthesis SuperMix). Using NEBuilder® HiFi DNA Assembly kit, we ligated a 3x-Hemagglutinin (HA) tag to the N-terminus of the Fezf1 CDS. We then cloned the 3xHA-Fezf1 fragment into a modified pCIG2 plasmid (Hand et al., 2005), which we modified to include a Lox-STOP-Lox (Hand et al., 2015) cassette to allow Cre-dependent expression of Fezf1. Using pCAG-Cre (Hand et al., 2015), we examined expression of pCIG2-LSL-3xHA-Fezf1 in HEK293T cell, and detected specific HA signal in the nucleus with anti-HA antibody staining (data not shown). This strategy was also used to make a Cre-dependent Rnd3 overexpression construct — pCIG2-LSL-3xHA-Rnd3 (see below). We next designed an ultramer containing a furin cleavage site plus linker (RAKR-SGSG), the P2A self-cleaving peptide, and the 5’ 20 bases of the eGFP CDS to link together the Fezf1 CDS and eGFP in a HiFi reaction. The ultramer sequence was: CGCGCCAAGCGCAGTGGGAGTGGTGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGGTGAGCAAGGGCGAGGA.
The Fezf1 CDS was amplified using primers that created a 35 bp overlap between the CDS and the P2A ultramer; the P2A ultramer was amplified with complementary primers; and eGFP was amplified with primers specific to its CDS. These amplicons were then used in a HiFi reaction to generate the 3xHA-Fezf1-P2A-eGFP fragment. We then subcloned this fragment into pAAV-hSyn1-MCS to produce an AAV vector that allows for equimolar expression of 3xHA-Fezf1 and eGFP. The expression of pAAV-hSyn1–3xHA-Fezf1-P2A-eGFP was also validated in 293T cells.
shRNA Constructs.
To make pAAV-hSyn1-DsRed-shRnd3, we modified the approach taken by (Peng et al, 2017) as follows. Short hairpin (sh) oligonucleotide was designed online (http://katahdin.mssm.edu/siRNA/RNAi.cgi?type=shRNA). The following sequences were used:
shRnd3
TGCTGTTGACAGTGAGCGAACAGGCAGACTCCTGTGTCATTAGTGAAGCCACAGATGTAATGACACAGGAGTCTGCCTGTGTGCCTACTGCCTCGGA
shScramble
TGCTGTTGACAGTGAGCGGCTGTTCCGCTCGTGTAGAGTTTAGTGAAGCCACAGATGTAAACTCTACACGAGCGGAACAGCTGCCTACTGCCTCGGA
These oligos were separately amplified and cloned into pPRIME-DsRed-shCntn5 (Peng et al, 2017), replacing the shCntn5 sequence. DsRed expression was visualized in HEK293T cells for each construct. shRnd3 knockdown efficacy was determined by Western Blot as described previously (Wang et al, 2018) using lysates from HEK293T cells transfected with: 1) pCIG2-LSL-3xHA-Rnd3; 2) pCIG2-LSL-3xHA-Rnd3, pPRIME-DsRed-shRnd3, and pCAG-Cre; or 3) pCIG2-LSL-3xHA-Rnd3, pPRIME-DsRed-shScramble, and pCAG-Cre. Rabbit anti-HA (1:1000, Cell Signaling) and mouse anti-actin (1:20,000, Millipore) antibodies were used for Western Blot. The DsRed-shRnd3 or DsRed-shScramble fragments were then subcloned into pAAV-hSyn1-MCS to generate final pAAV-hSyn1-DsRed-shRnd3 and pAAV-hSyn1-DsRed-shScramble constructs for producing adeno-associated viruses at the Janelia Research Campus Viral Tools Core.
In Utero Injection of AAVs.
The procedure to introduce AAV in utero was similar to the targeted in utero electroporation surgery previously described (Hand et al., 2015). Timed pregnant E13.5 (shRNA experiments) and E14.5 (Fezf1 overexpression) CD1 females were deeply anesthetized with isoflurane and given buprenorphine at 0.1mg/kg body weight. The incision area was shaved, cleaned, and a small longitudinal incision was made to expose the embryos. The embryos were rinsed periodically with sterile dPBS. Sharp glass needles containing virus and fast green dye were pierced through the uterine wall, and into the eyes of targeted embryos. Virus was injected by mouth-pipette while slowly retracting the needle. One eye was injected per embryo, and around two-thirds of the embryos were injected per litter in order to prevent spontaneous abortion/pup loss. After injections, the embryos were gently placed back inside the abdominal wall and the incision was sutured and stapled. The dams were returned and recovered in a heated cage for several hours.
Tissue from embryonic and neonatal AAV transduction experiments were collected and processed at P14. Regions of interest for imaging and quantification were selected based on high viral transduction, defined by transduced horizontal cells in the outer retina overlying a given region of interest.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of mosaic spacing was conducted as previously described (Kay et al., 2011; Peng et al., 2017). Briefly, cell number from a 212.55 × 215.55 μm square region was counted at 8–10 locations per retina (2 retinas per genotype). X-Y cell coordinates marked manually were used to calculate DRP statistics. The distance of exclusion zone radius, packing factor and nearest-neighbor regularity indexing (data to random) were then calculated using WinDRP software.
To define the prominent process, processes from a cell were traced and measured using simple neurite tracing (ImageJ). The longest process, longer than 20 μm, was defined as a prominent process. The angle ROI function (ImageJ) was then used to measure the angle of the prominent process with 0° corresponding to a line perpendicular to the apical surface of the retina. A polar plot of showing lengths and angles was generated using SigmaPlot (Sigma) software.
All data are shown as Mean ± SEM with n representing the sample number from at least two mice or independent experimental replicates. Two-tailed Student’s t tests were used for two group comparisons. One-way ANOVA with post-hoc Tukey HSD was used for multi-group comparisons. Statistical details can be found in Figures and Figure Legends.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw and processed scRNA-seq and RNA-seq data reported in this paper is GEO: (GSE132555).
Supplementary Material
Highlights.
ON and OFF SACs are transcriptionally distinct prior to migration
Fezf1 is selectively expressed by newly postmitotic ON SACs Fezf1 functions as a fate switch by activating ON and repressing OFF SAC genes
Rnd3 acts downstream of Fezf1 to enable distinct positioning of ON and OFF SACs
ACKNOWLEDGMENTS
This work was supported by NIH grant R37NS029169-29 (J.R.S.); funding from the Howard Hughes Medical Institute (A.L.K.); and VSTP 5T32EY007143-20 and F32EY025114 (R.E.J.). We are grateful to Dr. Bin Chen (University of California Santa Cruz) for generously providing Fezf1GFP mutant mice. We thank Drs. Loyal A. Goff and Karthik Shekhar for advice on scRNA-seq analysis strategies. We thank Mallory Laboulaye at Harvard, and Nicole Kropowski, Randal Hand, Natalie Hamilton, and Tim Al-Khindi at Johns Hopkins, for technical assistance. We also thank members of the J.R.S. and A.L.K. laboratories for helpful discussions and comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests
REFERENCES
- Antinucci P, Suleyman O, Monfries C, and Hindges R (2016). Neural Mechanisms Generating Orientation Selectivity in the Retina. Curr Biol 26, 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, and Sanes JR (2003). Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature 424, 430–434. [DOI] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, and McConnell SK (2008). The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A 105, 11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Huerta I, Kim IJ, Voinescu PE, and Sanes JR (2012). Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci U S A 109, 17663–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckler MJ, and Chen B (2014). Fez family transcription factors: controlling neurogenesis and cell fate in the developing mammalian nervous system. Bioessays 36, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckler MJ, McKenna WL, Taghvaei S, McConnell SK, and Chen B (2011). Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. J Comp Neurol 519, 1829–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiglietti EV (2004). Class I and class II ganglion cells of rabbit retina: a structural basis for X and Y (brisk) cells. J Comp Neurol 478, 323–346. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV (2005). “Small-tufted” ganglion cells and two visual systems for the detection of object motion in rabbit retina. Vis Neurosci 22, 509–534. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV Jr. (1983). ‘Starburst’ amacrine cells and cholinergic neurons: mirror-symmetric on and off amacrine cells of rabbit retina. Brain Res 261, 138–144. [DOI] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. (2005). Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48, 45–62. [DOI] [PubMed] [Google Scholar]
- Heinze L, Harvey RJ, Haverkamp S, and Wassle H (2007). Diversity of glycine receptors in the mouse retina: localization of the alpha4 subunit. J Comp Neurol 500, 693–707. [DOI] [PubMed] [Google Scholar]
- Jie W, Andrade KC, Lin X, Yang X, Yue X, and Chang J (2015). Pathophysiological Functions of Rnd3/RhoE. Compr Physiol 6, 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ Jr., and Desplan C (2010). Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu Rev Cell Dev Biol 26, 689–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, and Desplan C (2010). Binary fate decisions in differentiating neurons. Curr Opin Neurobiol 20, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Ito K, Shigematsu Y, and Shimoda Y (2010). The OFF-pathway dominance of P2X(2)-purinoceptors is formed without visual experience. Neurosci Res 66, 86–91. [DOI] [PubMed] [Google Scholar]
- Kay JN, Chu MW, and Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knabe W, Washausen S, Happel N, and Kuhn HJ (2007). Development of starburst cholinergic amacrine cells in the retina of Tupaia belangeri. J Comp Neurol 502, 584–597. [DOI] [PubMed] [Google Scholar]
- Kostich WA, and Sanes JR (1995). Expression of zfh-4, a new member of the zinc finger-homeodomain family, in developing brain and muscle. Dev Dyn 202, 145–152. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nogami S, Ishii Y, Kawaguchi M, Sakata N, Oya T, Takagawa K, Kanamori M, Sabit H, Obata T, Kimura T, et al. (2005). ZFH4 protein is expressed in many neurons of developing rat brain. J Comp Neurol 482, 33–49. [DOI] [PubMed] [Google Scholar]
- Pacary E, Azzarelli R, and Guillemot F (2013). Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nat Commun 4, 1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E, Heng J, Azzarelli R, Riou P, Castro D, Lebel-Potter M, Parras C, Bell DM, Ridley AJ, Parsons M, et al. (2011). Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron 69, 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237 e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron 95, 869–883 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris B, Gonzalez-Granero S, Ballester-Lurbe B, Garcia-Verdugo JM, Perez-Roger I, Guerri C, Terrado J, and Guasch RM (2012). Neuronal polarization is impaired in mice lacking RhoE expression. J Neurochem 121, 903–914. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Rama N, Dubrac A, Mathivet T, Ni Charthaigh RA, Genet G, Cristofaro B, Pibouin-Fragner L, Ma L, Eichmann A, and Chedotal A (2015). Slit2 signaling through Robo1 and Robo2 is required for retinal neovascularization. Nat Med 21, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Roy S, Kozlowski C, Wang J, Cafaro J, Hulbert SW, Wright CV, Field GD, and Kay JN (2018). Formation of retinal direction-selective circuitry initiated by starburst amacrine cell homotypic contact. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese BE, and Keeley PW (2015). Design principles and developmental mechanisms underlying retinal mosaics. Biol Rev Camb Philos Soc 90, 854–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, and Masland RH (2000). Spatial order within but not between types of retinal neurons. Proc Natl Acad Sci U S A 97, 2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW (1991). The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci 6, 95–111. [DOI] [PubMed] [Google Scholar]
- Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, and Elmquist JK (2011). Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab 13, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Qiao M, Kagan RD, Yamagata M, Palmiter RD, Sanes JR.(2016) Two Pairs of ON and OFF Retinal Ganglion Cells Are Defined by Intersectional Patterns of Transcription Factor Expression. Cell Rep. 15, 1930–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Zipursky SL (2010). Design principles of insect and vertebrate visual systems. Neuron 66, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, and Hibi M (2009). Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev Growth Differ 51, 221–231. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Nakazawa M, Kani S, Bae YK, Shimizu T, Kageyama R, and Hibi M (2010). Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development 137, 1875–1885. [DOI] [PubMed] [Google Scholar]
- Stacy RC, and Wong RO (2003). Developmental relationship between cholinergic amacrine cell processes and ganglion cell dendrites of the mouse retina. J Comp Neurol 456, 154–166. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocheteau P, and Le Roux I (2009). Asymmetric cell divisions and asymmetric cell fates. Annu Rev Cell Dev Biol 25, 671–699. [DOI] [PubMed] [Google Scholar]
- Thangaraj G, Greif A, Bachmann G, and Layer PG (2012). Intricate paths of cells and networks becoming “Cholinergic” in the embryonic chicken retina. J Comp Neurol 520, 3181–3193. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usrey WM, and Fitzpatrick D (1996). Specificity in the axonal connections of layer VI neurons in tree shrew striate cortex: evidence for distinct granular and supragranular systems. J Neurosci 16, 1203–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney DI, Sivyer B, and Taylor WR (2012). Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat Rev Neurosci 13, 194–208. [DOI] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, and Sanes JR (2009). Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol 517, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassle H, and Riemann HJ (1978). The mosaic of nerve cells in the mammalian retina. Proc R Soc Lond B Biol Sci 200, 441–461. [DOI] [PubMed] [Google Scholar]
- Whitney IE, Keeley PW, St John AJ, Kautzman AG, Kay JN, and Reese BE (2014). Sox2 regulates cholinergic amacrine cell positioning and dendritic stratification in the retina. J Neurosci 34, 10109–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yu WQ, Hoshino A, Huang J, Rieke F, Reh TA, and Wong ROL (2019). Development of ON and OFF cholinergic amacrine cells in the human fetal retina. J Comp Neurol 527, 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the raw and processed scRNA-seq and RNA-seq data reported in this paper is GEO: (GSE132555).