Abstract
Protein-coding de novo mutations (DNMs) are significant risk factors in many neurodevelopmental disorders, whereas schizophrenia (SCZ) risk associated with DNMs has thus far has been modest. We analyzed DNMs from 1,695 SCZ affected trios and 1,077 published SCZ affected trios to better understand the contribution to SCZ risk. Among 2,772 SCZ probands, exome-wide DNM burden remains modest. Gene set analyses reveal that SCZ DNMs are significantly concentrated in genes either highly brain expressed, under strong evolutionary constraint, and/or overlap with genes identified in other neurodevelopmental disorders. No single gene surpasses exome-wide significance, however sixteen genes are recurrently hit by protein-truncating DNMs, a 3.15-fold higher rate than the mutation model expectation (permuted 95% CI=1–10 genes, permuted p=3e-5). Overall, DNMs explain a small fraction of SCZ risk, and larger samples are needed to identify individual risk genes, as coding variation across many genes confer risk for SCZ in the population.
Introduction
Schizophrenia (SCZ) is a severe psychiatric disorder that affects 0.7–1% of the general population, yet its etiology and pathophysiology is only beginning to be understood. The high heritability of SCZ suggests that inherited genetic factors make up a substantial proportion of the genetic risk, and recent advances from large-scale GWAS point to a broad polygenic network of brain expressed genes contributing to its pathophysiology [1, 2]. Furthermore, genome-wide SNP-based heritability estimates suggest that ~27% of genetic variation in SCZ is effectively tagged by variants with MAF > 1% in the population [3].
Nevertheless, a large proportion of the heritability remains to be discovered and quantified. One potential source of undiscovered risk may come from rare, often deleterious, variants of more recent origin. The marked reduction in fecundity found in patients with SCZ suggests that natural selection may be removing the largest effect risk alleles from the population [4]. Indeed, the strongest individual genetic risk factors for SCZ discovered so far come from large copy number variants (CNVs) that often occur de novo in affected offspring [5–7]. With large-scale whole-exome sequencing, we can discover de novo mutations (DNMs) in the protein-coding region of the genome at base-pair resolution, facilitating the discovery of individual SCZ risk genes.
DNMs discovered from parent-offspring trios have yielded novel genetic insights for an array of severe neurodevelopmental disorders, including intellectual disability [8–10], autism spectrum disorders [11–16], epileptic encephalopathy [17, 18], and developmental delay [19, 20]. Along with an elevated burden of disruptive and damaging coding DNMs in these cohorts, specific biological pathways and individual genes have been robustly identified as de novo risk factors. For SCZ, an enrichment of damaging and disruptive DNMs has been modest by comparison. Recent published reports using either parent-child trio or case-control studies have shown enrichment in specific brain-expressed gene sets (e.g. ARC/NMDAR protein complexes, FMRP interactors) and overlap with genes implicated in the neurodevelopmental disorders listed above [21–23], however only a single gene, SETD1A, has been robustly identified as a de novo risk factor for SCZ [24, 25]. These findings suggest that coding DNMs do not explain a large fraction of SCZ diagnoses, and larger samples are needed to robustly identify genes that putatively contribute to SCZ risk.
SCZ Trio Cohort from Taiwan
Here, we report whole-exome sequencing results from 1,695 complete parent-proband trios (1,033 male and 662 female probands). Families were recruited from mental hospitals, community care centers and primary care clinics across the island of Taiwan. To our knowledge, this is the largest trio exome sequence study to date in SCZ. All trios consist of a SCZ affected proband and unaffected parents. Trios consist predominantly of sporadic case probands (91%), with the remainder reporting a history of mental illness in the family. All samples were exome-sequenced and analyzed at the Broad Institute in Cambridge, MA, USA, with a target of 20x depth in at least 80% of the exome target. All reported DNMs were either validated or showed strong confidence in the initial call (Online Methods). Exome sequencing was performed in three distinct waves, consisting of 575, 532, and 588 trios, respectively. Trio samples sequenced in the 1st wave had a non-trivial proportion of their DNA barcodes inadvertently mislabeled with a second individual sequenced in the same lane during multiplex sequencing (which we term “barcode switching”), requiring additional DNM quality control to filter out low-confidence DNMs along with validation of all DNM calls (Online Methods).
Among measured covariates that could affect our observed DNM rates, parental age was the most significant predictor of increased DNM rates (p=3.6e-7). None of the other study covariates tested significantly predicted DNM rates (supplementary section 3). The degree of barcode switching did not significantly affect post-QC DNM rates (p=0.09), although higher levels of barcode switching trended toward lower DNM rates, likely a result of the increased QC required to filter the initial DNM calls. The larger capture target used in the 3rd wave of sequencing lead to higher overall DNM rates to the other two waves (rate-ratio=1.09, p=0.07; supplementary figure 7), but attenuated after excluding DNMs outside of overlapping exome capture intervals when evaluating exome-wide DNM rate (rate-ratio=1.02, p=0.58).
The strong correlation of paternal and maternal age (r=0.49) makes it difficult to fully delineate parent-of-origin effects, however when we fit both paternal age and maternal age in the regression model, paternal age (p=3e-6) associated with increased DNM rates over and above maternal age (p=0.38). Further follow up on parental age effects revealed a modest quadratic effect of increased maternal age (p=0.01) associated with increased DNM rate, something not seen in paternal age (p=0.6). These results show a clear association of older paternal age with increasing DNM rates and suggest that older maternal age may also lead to increased DNM rates in a possibly non-linear fashion, albeit attenuated relative to paternal age.
Exome-wide rates of de novo mutation
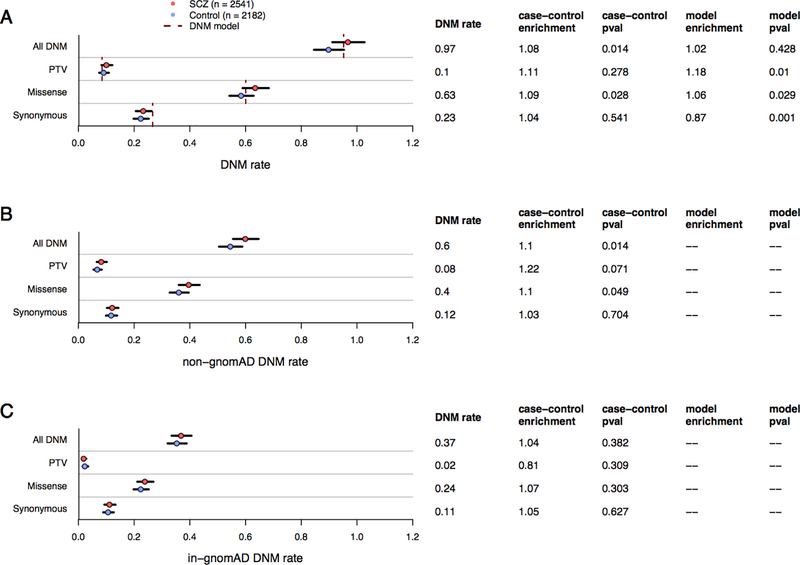
Using a Poisson distributed model specification, exome-wide DNM rates in the Taiwanese cohort were compared against 1,077 trios from previous published exome DNM studies in SCZ [21, 26–31] (see supplementary section 4 for study inclusion criteria). Of note, one cohort [31] showed a significantly lower synonymous rate relative to remaining cohorts (231 SCZ probands, 34 controls, fold-enrichment=0.47, p=7.2e-6; Supplementary Figure 13: Synonymous DNM rate by study). We omitted this cohort from whole exome DNM burden analysis, but retained it for gene set and single gene analyses. After restricting our search to shared exome capture intervals, we find no significant difference in the overall DNM rate of the three Taiwanese sequencing waves when compared to published SCZ trios (rate-ratios=1.01, 0.97, and 1.02, respectively; all p > 0.05). We combined the Taiwanese cohort with published SCZ trios (2,541 trios), and compared DNM rates against a DNM expectation model restricted to 17,925 well-covered genes [32, 33] (supplementary section 2) as well as 2,182 published unaffected siblings and control trios (collectively termed ‘controls’; Figure 1A). We find the overall DNM rate in SCZ probands only slightly above the DNM model expectation (rate-ratio=1.02, p=0.43), but significantly enriched over controls (rate-ratio=1.08, p=0.01). When we partition DNMs by coding annotation, we see a significantly lower rate of synonymous DNMs in SCZ probands relative to the DNM model (rate-ratio=0.87, p=8e-4), however the observed synonymous rate is consistent with the synonymous DNM rate observed in controls (rate-ratio=1.04, p=0.54). This discrepancy suggests that DNM calling among SCZ probands is well calibrated relative to controls, while the DNM model, despite restricting to genes with adequate sequence coverage, remains conservative in its exome-wide expectations. The discrepancy we see in the synonymous DNM rate in controls relative to expectations is not seen for either protein-truncating variants (PTV) or missense DNMs. Among PTVs, we find marginal evidence of enrichment relative to DNM model expectations (rate-ratio=1.18, p=9e-3), albeit not significantly above controls (rate-ratio=1.11, p=0.28). For missense variants, we find marginal enrichment relative to both the DNM model (rate-ratio=1.06, p=0.03) and when compared to controls (rate-ratio=1.09, p=0.03).
Figure 1:
DNM rates (dots), 95% CI (error bars), and DNM model expectations (dotted lines) among SCZ and control probands. Figure 1A: Exome-wide DNM rates split by primary annotation, where “All DNM” encompasses protein-truncating variants (PTV), missense, and synonymous DNMs. Poisson rate p-values (all two-sided and unadjusted) and rate enrichment compared to controls are listed to the right of the DNM rate. DNM rates split by the absence (Figure 1B) or presence (Figure 1C) of an extant allele at the same position in the 104k non-psychiatric gnomAD cohort.
When we consider the exome-wide burden enrichment of DNMs among SCZ probands relative to controls, we can estimate the proportion of coding DNMs that contribute to a SCZ diagnosis. Among well covered genes, SCZ probands have 0.065 additional coding DNMs over controls (95% CI: 0.028 – 0.1 additional DNMs), which represent 6.8% of all coding DNMs. When we project this estimate as a Poisson distributed rate parameter, we estimate that around 168 of the 2772 SCZ probands carry at least one DNM that contributes to their SCZ diagnosis (95% CI: 61 – 268 SCZ probands). It is important to keep in mind that most SCZ affected trios ascertained required both parents be undiagnosed for SCZ, and the current estimate represents more of an upper bound of the contribution that DNMs have on SCZ risk.
SCZ DNMs are more likely to be annotated as deleterious
Not all protein-coding alterations lead to deleterious consequences, and population allele frequency information and predicted biological consequence can help us refine the search space towards likely pathogenic alleles contributing to SCZ risk. To see if SCZ DNMs were enriched for predicted pathogenic alleles, we first filtered out DNMs at variable sites from ~104k exomes in the non-psychiatric subset of the genome aggregation database (gnomAD release 2.1.1; [33, 34], inferring that the presence of the same allele in ostensibly healthy individuals lowers the likelihood of being under negative selection (Figure 1B and 1C). We find that 38% of DNMs found in SCZ probands are at variable sites in gnomAD, a finding consistent with DNMs in ASD probands [35]. When we filter out these alleles in both SCZ probands and controls, we see a slight increase in DNM enrichment for both PTV (from rate-ratio=1.11, p=0.28 to rate-ratio=1.22, p=0.07) and missense variants (from rate-ratio=1.09, p=0.03 to rate-ratio=1.11, p=0.05), but not in synonymous variants (from rate-ratio=1.04, p=0.54 to rate-ratio=1.03, p=0.7).
We then examined a variety of functional annotations that could further delineate the likely pathogenic alleles beyond the primary coding consequence. Previous SCZ case-control exome studies found that missense variants predicted as damaging from multiple prediction algorithms were associated with SCZ risk [22]. Eleven of the twelve missense prediction algorithms tested increased the missense DNM enrichment among SCZ probands relative to controls after filtering out predicted non-damaging missense variants, with a 1.13-fold enrichment (Polyhen2 HDIV) and 1.17-fold enrichment (CADD) in the two predictors with p < 0.05 (supplementary section 5). Outside of missense prediction algorithms, a recent analysis found that synonymous variants near exon splice junctions and within brain-derived DNAse hypersensitivity site (DHS) peaks were enriched in both published autism and SCZ DNMs [36]. When we replicate the analysis in the Taiwanese cohort (1,695 SCZ probands) and the fraction of controls not included in the initial analysis (1,485 controls), we do not find a significant enrichment in synonymous variants within 30bp of the splice site (fold-enrichment=1.14, one-tailed p=0.26) and within cerebrum-frontal cortex DHS peaks (fold-enrichment=1.26, one-tailed p=0.09). Further analysis, however, reveals that a broader definition of being near a splice site (60 bp rather than 30 bp) shows more consistency in both samples and significant enrichment in the combined SCZ cohort (fold-enrichment=1.29, p=5e-4; supplementary section 6).
DNM burden relative to other neurodevelopmental disorders
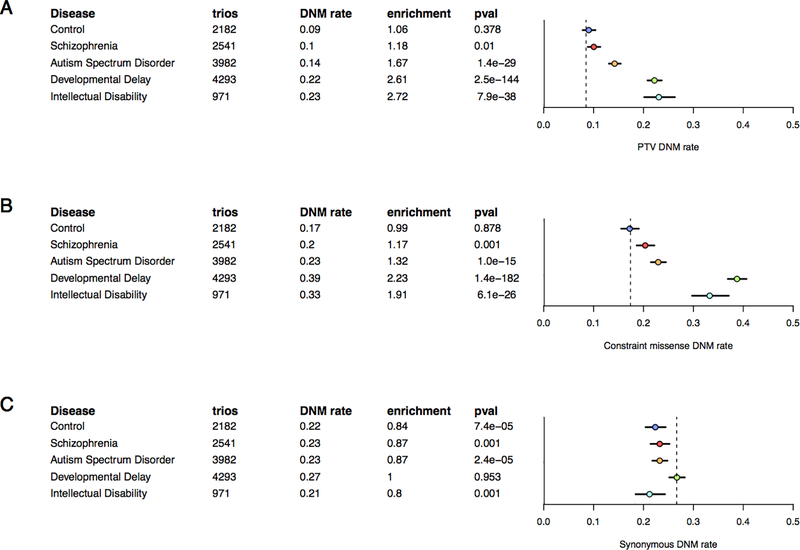
When we examine the combined cohort of SCZ probands within the larger context of mental illness, the enrichment signal is markedly reduced relative to published DNM studies of probands diagnosed with early-onset neurodevelopmental disorders among both PTVs exome-wide (Figure 2A) and missense DNMs in evolutionarily constrained genes (i.e. missense constrained [32], probability of loss-of-function intolerance (pLI) > 0.99 [33], or Residual Variation Intolerance Score (RVIS) intolerant [37] genes; Figure 2B) – two categories where there is a significant enrichment in DNM burden across all disorders analyzed. This comparison clearly indicates that the contribution of DNMs towards a SCZ diagnosis accounts for a much smaller fraction of samples than earlier onset neurodevelopmental disorders, while the fraction of synonymous DNM burden remains relatively constant across diseases (Figure 2C).
Figure 2:
DNM rates (dots) and 95% CI (error bars) compared to DNM model expectations (dotted line) for exome-sequenced trios with various mental disorders. Figure 2A: Exome-wide PTV rate and enrichment relative to DNM model expectations. Figure 2B: Missense rate among evolutionarily constrained genes (defined here as the union of high pLI, missense constraint, and RVIS intolerant gene sets) and enrichment relative to DNM model expectations. Figure 2C: Exome-wide synonymous rate and enrichment relative to DNM model expectations. All tests are Poisson rate tests and information regarding the published studies analyzed is available in Online Methods.
Enrichment in Taiwanese SCZ probands with deficits in sustained attention and executive function
One key consideration is the role that cognitive impairment plays on DNM rates within SCZ probands. For example, the elevated rate of PTV mutations in the Bulgarian SCZ trio cohort were largely confined to individuals with the lowest scholastic attainment [21], suggesting that the enrichment of deleterious coding DNMs in SCZ may arise from a subset of individuals with more severe cognitive impairment also receiving a diagnosis of SCZ. This finding has been further supported when combined with case-control SCZ exomes [23], and is more prominent in ASD probands, where the enrichment in PTV DNMs is confined to ASD probands with lower IQ [13, 32, 35, 38]. While direct measures of cognitive ability were not collected in the Taiwanese cohort, measures of sustained attention and executive function were available for most trios, providing a proxy for cognitive ability (supplementary section 3). Using a median split approach, we find a modest enrichment in missense DNMs among low scorers of sustained attention relative to high scorers (trios=1298, two-sample rate-ratio=1.16, p=0.04) and compared to the DNM model (fold-enrichment=1.11, p=0.03, supplementary figure 12). For executive function, we find a modest enrichment in PTV DNMs among low scorers relative to high scorers (trios=1319, two-sample rate-ratio=1.39, p=0.06) and compared to the DNM model (fold-enrichment=1.44, p=2e-3, supplementary figure 13). For both tests, enrichment increases when we restrict to brain expressed and constrained genes, providing further support that the elevated rate of DNMs in SCZ probands is, in part, a reflection of co-morbid intellectual impairment within these ascertained cohorts.
Gene set enrichment analyses highlights the pattern of DNM risk in SCZ probands
Along with DNM burden, we examined enrichment in specific gene sets after conditioning on overall DNM rates and examined the proportional enrichment of genes hit by DNMs in a specified gene set relative both control DNMs and the DNM model (supplementary section 8). We break down the results of our gene set enrichment analyses as follows: (1) genes implicated by multiple constraint metrics and in multiple neurodevelopmental disorders, (2) genes implicated in previous SCZ analyses, including GWAS, CNV analyses and previous SCZ trio exome studies, (3) gene sets surpassing multiple testing correction among 85 candidate gene sets tested in previous analyses, including brain expressed and biologically plausible gene sets, (4) potentially synaptic components implicated in the Swedish SCZ WES analysis [22], and (5) 911 unselected gene sets from Gene Ontology (GO) and SynaptomeDB.
Enrichment in gene sets implicated by multiple constraint metrics and in multiple neurodevelopmental disorders
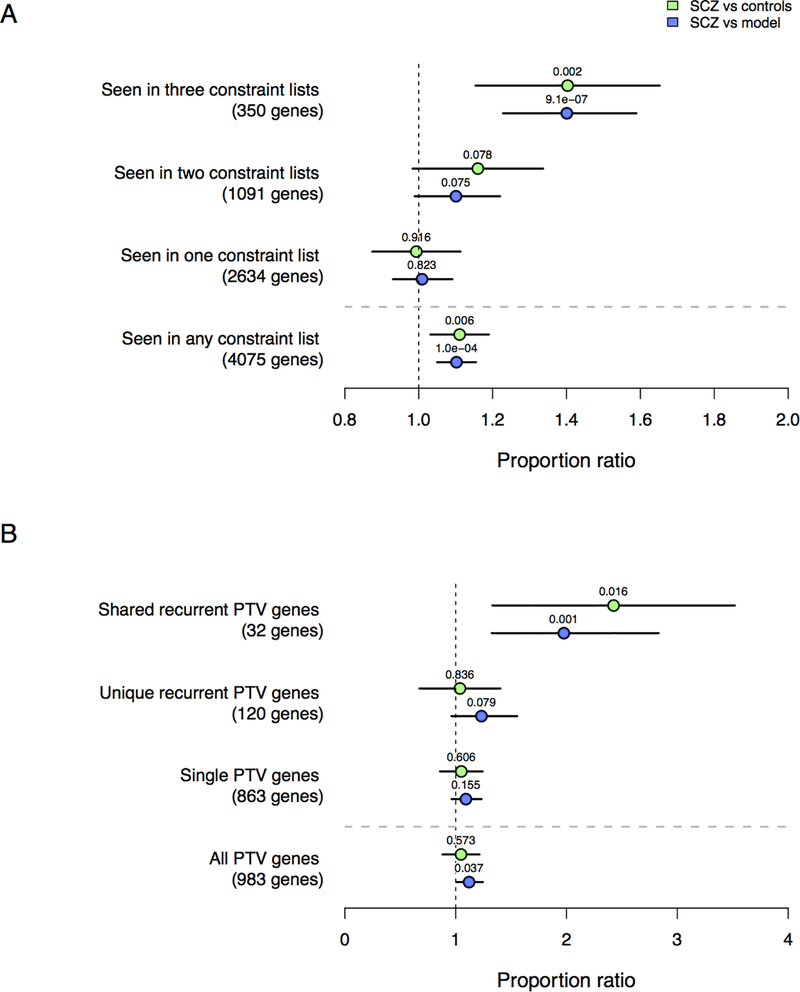
When we examine gene sets defined from evolutionarily constraint metrics (missense constraint, pLI, and RVIS), where genes show a depletion of rare coding variation relative to expectations (Figure 3A) and coding DNM studies of intellectual disability, developmental delay, and autism spectrum disorder (Figure 3B), we find DNMs in SCZ probands are significantly enriched for genes repeatedly identified across multiple metrics and diseases. Among all DNMs, enrichment is concentrated in genes implicated in all three measures of evolutionary constraint (350 genes), and among genes with recurrent PTVs identified in multiple neurodevelopmental disorders (32 genes). When we partition the enrichment results of these top gene sets by DNM annotation (Table 1), PTVs show the strongest fold-enrichment contribution, followed by missense DNMs. These results highlight the notion that despite the lower overall DNM burden in SCZ relative to other neurodevelopmental disorders, the disruption of some of the most critical genes in neurodevelopment confers risk towards a variety of mental disorders, including SCZ.
Figure 3:
Partitioning gene set enrichment among evolutionarily constrained and neurodevelopmental disorder gene sets. Proportion enrichment using a binomial test is evaluated among all DNMs in SCZ probands (n=2772) relative to controls (n=2216; green) and the DNM model (blue), with proportion enrichment (dots), 95% CI (error bars), and unadjusted two-sided p-values (above dots) displayed. Figure 3A: Gene set enrichment in three evolutionary constraint metrics (missense constraint, pLI, and RVIS), with partitioning among how often genes are present in each set. Figure 3B: Gene set enrichment for genes with observed DNM PTVs in intellectual disability, developmental delay, and ASD probands. Shared recurrent PTV genes have identified two or more PTVs in more than one disorder. Unique recurrent PTV genes have identified two or more PTVs in only one disorder. Single PTV genes are not recurrent within any disorder, although may be shared across disorders.
Table 1:
Partitioning DNM enrichment in top constraint and neurodevelopmental gene sets
| DNM annotation | SCZ DNM count | Control DNM count | SCZ-control enrichment | SCZ-control p-value | DNM model enrichment | DNM model p-value |
|---|---|---|---|---|---|---|
| Seen in all three constraint lists (350 genes) | ||||||
| All coding DNM | 220 | 118 | 1.40 | 2e-3 | 1.40 | 9e-7 |
| PTV | 38 | 7 | 3.73 | 3e-4 | 2.27 | 3e-6 |
| Missense | 135 | 77 | 1.30 | 0.05 | 1.31 | 2e-3 |
| Synonymous | 47 | 34 | 1.11 | 0.62 | 1.27 | 0.11 |
| Genes with recurrent PTVs in multiple disorders (32 genes) | ||||||
| All coding DNM | 29 | 9 | 2.42 | 0.02 | 1.98 | 8e-4 |
| PTV | 8 | 0 | NA | 0.02 | 4.61 | 4e-4 |
| Missense | 17 | 6 | 2.11 | 0.11 | 1.76 | 0.03 |
| Synonymous | 4 | 3 | 1.08 | 0.92 | 1.21 | 0.58 |
SCZ proband (n=2772) fold-enrichment and p-values (all two-sided and unadjusted) are from Poisson rate tests (Online methods).
Modest support for genes implicated in previous SCZ analyses
Among 461 genes implicated in the PGC GWAS [1] and rare CNV [5] studies of SCZ, we see a non-significant enrichment relative to the DNM model (all DNM fold-enrichment=1.2, p=0.07; PTV fold-enrichment=1.55, p=0.11) and compared to controls (all DNM fold-enrichment=1.25, p=0.16; PTV fold-enrichment=1.28, p=0.59). Moreover, no single comparison reached experiment-wide significance, with the most significant enrichment coming in genes overlapping SCZ associated CNVs (151 genes tested, case/control all DNM fold-enrichment=1.98, p=0.05). Previously published SCZ trio cohorts reported several gene sets with significant association, namely prenatally biased genes [31], chromatin modifiers [25, 29], and the ARC/NMDAR subunits [21]. None of these gene sets reached experiment-wide significance (correction set at p=8e-4, supplementary section 8) in the current analysis, with the most significant gene set coming from PTVs in proteins that interact with the ARC subunit complex (28 genes tested, PTV fold-enrichment=7.9, p = 2e-3), where an additional two genes, IQSEC1 (IQ Motif and Sec7 Domain 1) and ATP1A1 (ATPase Na+/K+ Transporting Subunit Alpha 1), are hit by PTV DNMs in the Taiwanese cohort. Notably, there is also a PTV hit in ARC in the Taiwanese cohort (which is by default not included in the ARC subunit gene set).
Enrichment in highly brain expressed and constrained genes
Many biologically plausible gene sets have been previously implicated to refine the polygenic basis of SCZ risk towards more biologically tractable components. Among 85 candidate gene sets analyzed (supplementary section 8), BrainSpan high brain expression (8928 genes tested) and GTEx brain enriched (6214 genes tested; see [39]) represented two of the four gene sets that surpassed multiple-testing correction in both the DNM model and against controls (the other two gene sets being missense constraint and RVIS intolerance). Both gene sets represent gene expression across the entirety of human brain tissues and cell types measured from post-mortem brain tissue (http://www.brainspan.org/ and https://www.gtexportal.org/home/), and reinforce the notion that the genetic risk for SCZ, while brain-centered, retains a polygenic basis even in the lowest end of allele frequency spectrum. We also see evidence that enrichment is not restricted to non-synonymous DNMs, as synonymous DNMs also show enrichment, particularly among the BrainSpan gene set (DNM model fold-enrichment=1.13, p=7e-4, control fold-enrichment=1.34, p=2.9e-6).
DNM enrichment in potentially synaptic components of the brain
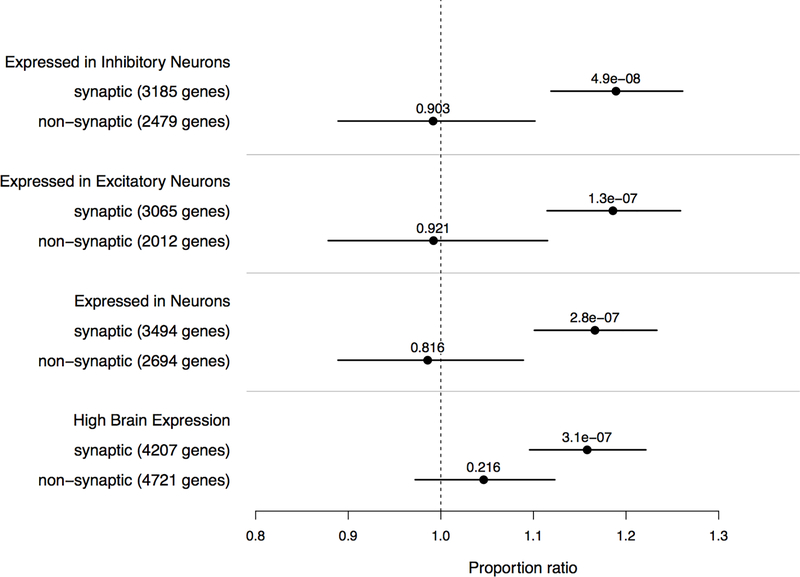
In an exome case-control study of SCZ among Swedish participants, a variety of gene sets defined as mRNA targets of regulatory proteins (FMRP, RBFOX, CELF4) highly active in the synapse were enriched for damaging rare variation in SCZ cases [22, 40]. When we compare against DNM model expectations, we see a consistent pattern of enrichment in SCZ probands, with FMRP interactors and RBFOX1/3 splicing targets surpassing multiple-testing correction (all p < 8e-4). Following the strategy of collapsing these mRNA target lists into a “potentially synaptic” gene set [22], we stratified both neuronal cell type expression and high brain expression gene sets by potentially synaptic genes (the inclusive combination of FMRP, RBFOX2, CELF4, and SynaptomeDB gene sets) identified from these mRNA targets, finding that the pattern of enrichment among neuronally expressed genes is driven primarily by synaptically localized genes (Figure 4). Of note, the signal is not significantly enriched when comparing against controls, owing in part to the statistical power differences between the model and case-control tests (supplementary section 8). These findings provide independent support for the enrichment seen among Swedish SCZ case-control exomes and indicate that these gene sets are tapping into the elements of synaptic biology that, when perturbed, increase the risk for SCZ.
Figure 4:
Partitioning of the genes expressed in neuronal cell types by their interaction with mRNAs highly active in the synapse (e.g. FMRP, RBFOX, CELF4), with interacting genes listed as “potentially synaptic”. Also included are the results from BrainSpan highly expressed genes (bottom comparison). Proportion enrichment (dots) of each gene set is tested in SCZ probands (n=2772) against the mutation model using a one-sample binomial test, with p-values (all two-sided and unadjusted) listed above each dot along with the 95% CI (error bars).
GO and SynaptomeDB databases implicate neurotransmitter secretion and chromatin organization
Outside of candidate gene sets, we ran an unbiased gene set scan using annotations from the Gene Ontology (GO) and SynaptomeDB databases. We restricted our analysis to only gene sets with at least 50 genes and analyzed a total of 911 gene sets (supplementary section 9). We ran the same permutation procedure as the candidate gene set analysis to estimate our multiple testing correction at a 5% alpha level (correction set at p=6e-5 for the DNM model test and p=2e-4 for the case/control test). No single gene set surpassed multiple testing correction in either comparison; the most significant gene set among all DNMs overlapping genes involved the neurotransmitter secretion biological process (GO:0007269, 64 genes tested, fold-enrichment=2.12, DNM model test p=4e-4). Among PTVs, the most significant enrichment overlaps genes involved in chromatin organization biological process (GO:0006325, 207 genes tested, fold-enrichment=2.81, DNM model test p=4e-4;). When comparing against controls, the most significant gene set among all DNMs overlapping genes is the biosynthetic process in Synaptome DB (73 genes tested, fold-enrichment=15.8, case-control test p = 3e-4).
Single gene association
To see if any single gene was a putative risk factor for SCZ, we tested for per-gene enrichment of DNMs in the combined SCZ cohort against the gene level mutation expectation (supplementary section 10). Exome-wide significance threshold was set at p = 8.7e-7 to correct for multiple testing, and no gene surpassed exome-wide significance, with our lowest p-value across all three tests being p=7.7e-6. Among PTVs, the most significant gene is SET Domain Containing 1A (SETD1A, p=7.7e-6), with three PTVs observed in two previously published SCZ trio cohorts [25, 27]. Notably, there were no SETD1A DNMs observed in the Taiwanese cohort. SETD1A has since been identified as an exome-wide significant gene association in combined trio and case/control exome sequencing of SCZ, whereby follow-up of patients carrying a PTV in SETD1A often presented with an associated neurodevelopmental disorder [24].
Enrichment in genes with recurrent PTVs
While no single gene association surpasses exome-wide correction, many genes are “recurrently” hit (i.e. more than one coding DNM observed in the gene) among SCZ probands. The rate of recurrently hit genes can indicate how likely such genes are SCZ risk factors. To test observed rates of gene recurrence against the mutation expectation, we used bootstrap re-sampling in the DNM model to simulate an equivalent count of observed DNMs hitting genes, with the probability of hitting a gene being its mutation expectation (supplementary section 11). We also tested control DNMs to ensure that any significant results were not the result of mis-specification in the DNM model. We observe sixteen genes with recurrent PTVs (fold-enrichment=3.15, p=3e-5), a result significantly enriched above the DNM model expectation of 5.1 genes (Table 2). When we perform the same analysis in controls, we do not find a significant enrichment in genes with recurrently hit PTVs (fold-enrichment=1.51, p=0.27). While we see modest enrichment in genes recurrently hit by missense and/or synonymous DNMs, the enrichment is similar in controls, and suggests some deviation of the DNM model to observed data through subtle effects of exome coverage variation and DNM QC parameters. Further dissection of DNM recurrence within gene sets also indicates that highly expressed brain genes, particularly those not under evolutionarily constraint, are enriched for recurrently hit genes (supplementary section 11).
Table 2:
Gene recurrence rates in SCZ probands and controls
| DNM | Observed recurrent genes | Expected recurrent genes | Fold-enrichment | Empirical p-value | |
|---|---|---|---|---|---|
| 2772 SCZ probands | |||||
| PTV | 296 | 16 | 5.1 | 3.15 | 3e-5 |
| Missense | 1763 | 150 | 136.6 | 1.10 | 0.09 |
| Synonymous | 635 | 29 | 21.2 | 1.37 | 0.05 |
| All DNM | 2694 | 314 | 286.2 | 1.10 | 0.02 |
| 2216 controls | |||||
| PTV | 212 | 4 | 2.7 | 1.51 | 0.27 |
| Missense | 1320 | 89 | 81.2 | 1.10 | 0.17 |
| Synonymous | 512 | 18 | 14.1 | 1.28 | 0.16 |
| All DNM | 2044 | 195 | 177.4 | 1.10 | 0.06 |
Fold-enrichment and empirical p-values (all one-sided and unadjusted) are from permutations using DNM model rates (Online methods).
To further assess the contribution of recurrently hit PTV genes in SCZ probands, we examined the contribution of rare transmitted variation within these genes (Table 3). While the lack of a SCZ diagnosis in the parents is expected to lower the contribution that rare inherited variants have in proband risk, the polygenic nature of SCZ risk predicts that undiagnosed individuals still carry SCZ risk alleles. Using parent-proband transmission counts in the full Taiwanese cohort (1695 trios), TDT among 15 recurrently hit PTV genes (excluding TTN) shows a 3.25-fold enriched ratio of transmitted ultra-rare PTVs to non-transmitted ultra-rare PTVs (p=0.02, supplementary section 12). The contribution of transmitted PTVs comes largely from two genes, the Trio Rho Guanine Nucleotide Exchange Factor gene (TRIO; 4 transmitted to 0 non-transmitted), and the Dynein Axonemal Heavy Chain 9 gene (DNAH9, 6 transmitted to 1 non-transmitted). While we do not observe any significant enrichment in ultra-rare damaging missense variants (defined here as ‘probably damaging’ in PolyPhen2, ‘deleterious’ in SIFT, and not seen in ExAC) among these 15 recurrently hit PTV genes (OR=1.09, p=0.64), TRIO shows a suggestive over-transmission among ultra-rare and predicted damaging missense variants (11 transmitted to 2 non-transmitted, p=0.01). Among the 20 probands in the Taiwanese cohort carrying a PTV in a gene recurrently hit by PTV DNMs, we do not see evidence of earlier SCZ symptom onset (supplementary section 11), however we are not adequately powered to make a strong inference about the impact of these specific PTVs on SCZ symptom onset.
Table 3:
Genes recurrently hit by PTV DNMs
| Gene symbol | Coding bases (canonical) | pLI | PTV DNM Poisson p-value | PTV DNM | Missense DNM | PTV transmitted | PTV non-transmitted | Damaging Missense transmitted | Damaging Missense non-transmitted |
|---|---|---|---|---|---|---|---|---|---|
| SETD1A | 5124 | 1 | 7.77E-06 | 3 | 0 | 0 | 0 | 1 | 4 |
| GALNT9 | 714 | 0.56 | 1.27E-05 | 2 | 0 | 0 | 0 | 2 | 0 |
| TAF13 | 375 | 0.08 | 2.43E-05 | 2 | 0 | 0 | 0 | 0 | 0 |
| HENMT1 | 1182 | 0 | 3.61E-05 | 2 | 0 | 1 | 0 | 0 | 0 |
| SV2B | 2052 | 0.34 | 1.50E-04 | 2 | 0 | 1 | 0 | 1 | 2 |
| NRXN3 | 3186 | 1 | 2.12E-04 | 2 | 0 | 0 | 0 | 6 | 6 |
| SMARCC2 | 3645 | 1 | 6.03E-04 | 2 | 0 | 0 | 0 | 0 | 1 |
| RB1CC1 | 4785 | 1 | 8.32E-04 | 2 | 0 | 0 | 0 | 3 | 3 |
| HIVEP3 | 7221 | 0.84 | 1.04E-03 | 2 | 0 | 0 | 0 | 2 | 4 |
| MKI67 | 9771 | 0 | 1.55E-03 | 2 | 1 | 0 | 3 | 4 | 3 |
| CHD8 | 7746 | 1 | 3.04E-03 | 2 | 1 | 0 | 0 | 1 | 3 |
| TRIO | 9294 | 1 | 3.11E-03 | 2 | 0 | 4 | 0 | 11 | 2 |
| DNAH9 | 13461 | 0 | 5.07E-03 | 2 | 0 | 6 | 1 | 8 | 7 |
| KIAA1109 | 15018 | 0.72 | 7.70E-03 | 2 | 1 | 1 | 0 | 14 | 12 |
| KMT2C | 14736 | 1 | 8.13E-03 | 2 | 2 | 0 | 0 | 6 | 7 |
| TTN | 107976 | 0 | 1.83E-01 | 2 | 3 | 14 | 8 | 59 | 58 |
Single gene p-values (all one-sided and unadjusted) are from a Poisson rate test of PTV counts against the DNM model (Online methods).
Finally, we examined if recurrently hit PTV genes are also enriched for ultra-rare PTVs in an independent sample of case-control exomes. Using 5,140 SCZ cases and 17,175 controls from the published UK10K and Swedish SCZ case-control cohorts [23], we ran a logistic regression model predicting case status from ultra-rare variants in recurrently hit PTV genes (excluding TTN) and controlling for exome-wide ultra-rare variant count, the 1st five PCs, and participant sex. When using all 15 genes, ultra-rare PTVs are significantly enriched in SCZ cases over controls (OR=2.89, 95% CI=1.61–5.18, p=3.6e-4), and increases further when we remove the three genes with pLI=0 (removing HENMT1, MKI67, and DNAH9; OR=5.53, 95% CI=2.72–11.23, p=2.3e-6). We see no significant enrichment for ultra-rare synonymous variants (OR=1.12, 95% CI=0.79–1.58, p=0.53), suggesting that the PTV enrichment signal is unlikely to be driven by population stratification or technical bias.
Discussion
By combining the Taiwanese and published SCZ trio cohorts, we report on 2772 trios, the largest exome-wide SCZ study on coding DNMs to date. SCZ trios show only a modest elevation of PTV and missense DNMs over both DNM model expectations and published control trios. Restricting to in silico predictors of DNM deleteriousness, such as absence in exome sequence reference panels, missense damaging predictors, and near splice-site synonymous changes further elevate the DNM burden among SCZ cases above controls. Gene set analysis reveals that DNM enrichment in SCZ probands is spread across highly brain-expressed and evolutionarily constrained genes, and further enriched within putatively synaptic pathways and among genes identified as DNM risk factors in other neurodevelopmental disorders. In general, however, the signal is not confined to a specific cell type or curated biological pathway. We find a significant elevation of genes recurrently hit by DNM PTVs, and these genes are significantly enriched for ultra-rare PTVs in independent SCZ case-control cohorts. Despite this robust candidate list, no single gene association surpasses exome-wide significance. Power analyses informed by the recurrence rate seen in PTVs suggest that we are currently somewhere between 62–73% power to detect individual gene associations, and between 3,200 and 4,000 trios will be necessary to achieve 80% detection power (supplementary section 14). While these findings further support the polygenic nature of SCZ risk, larger samples will detect individual gene associations at strict exome-wide significance.
Exome sequence of SCZ case-control samples offers a natural comparison of the efficacy of different study designs and ascertainment criteria. The exome analysis of a large Swedish SCZ cohort [22] examined 4.9k SCZ cases and 6.2k controls. In parsing the contribution of rare coding variation to individual SCZ diagnoses, the Swedish SCZ cohort estimated that ~10% of damaging and disruptive ultra-rare variants (dURVs) analyzed are DNMs, leaving 90% as inherited variants. One question that arises is how much DNMs are driving the enrichment seen among dURVs. Among the SCZ trio cohorts, we find a per-trio rate of 0.38 damaging and disruptive DNMs among SCZ probands compared to 0.3 per-trio in controls (fold enrichment=1.25), whereas case-control exomes find 0.25 additional dURVs in cases over controls (fold enrichment=1.07). Thus, our current estimate is that approximately 1/3 of the enrichment (or 0.08 of the 0.25 dURVs) observed in the case-control exomes is driven by DNMs. While this difference supports the notion that damaging and disruptive DNMs are more penetrant than inherited variants in terms of their influence on an SCZ diagnosis, the general pattern is that both de novo and inherited rare coding variants confer a small, but significant impact on SCZ risk. When we consider the pattern of enrichment among various gene sets analyzed, most show a consistent direction and effect size in both trio DNM and case-control dURVs in brain expressed and constrained gene sets (supplementary section 13), indicating a similar pattern of polygenic burden emerges across study designs. For rare variant analysis in SCZ, the results suggest that both case-control and trio-based studies are likely to reveal a similar genetic signature of rare variation conferring risk for SCZ.
Outside of sequence data, large structural variation analyzed from genotype array studies provides another source of rare genetic variation in SCZ. A mega-analysis of rare copy number variation (CNV) in SCZ case-control data from the Psychiatric Genetics Consortium (PGC; [5]) has refined the established association of rare CNV with SCZ. While a handful of previously SCZ -associated CNV (most co-morbid with severe neurodevelopmental features) are confirmed in the mega-analysis, there persists a modest genome-wide enrichment of ultra-rare CNV (ultra-rare being defined as MAF < 0.1%, OR=1.11, p=1.3e-7) overlapping genes outside of known disease-relevant CNV hotspots. Provided these ultra-rare CNVs are presumably a mixture of de novo and inherited events, the enrichment observed is consistent with coding DNMs described here, and further supports the notion that rare coding variants confer a small, but significant impact on SCZ risk.
Overall, the rates and patterns of coding DNMs observed in SCZ probands do not implicate individual loci explaining a large fraction of SCZ risk, but rather a modest elevation of risk for DNM carriers, and one that is largely polygenic in its genetic architecture. The overlap in relative risk and gene set enrichment between trio and case-control designs so far strongly suggests that either approach will identify individual risk genes in larger samples.
Methods
Taiwanese trio sample ascertainment
3,093 patients with schizophrenia and their unaffected parents from 3,008 families were recruited from mental hospitals, community care centers, and primary care clinics across the Island country of Taiwan. The average age of patients at recruitment was 35.7 years (SD=8.3 years), and had 2.1 siblings on average (SD=1.3). Of these 3,093 trios, 1,732 were recruited for the current project. The Department of Psychiatry, National Taiwan University Hospital and College of Medicine, National Taiwan University, Taipei, Taiwan served as the headquarters for data collection and research diagnosis.
Overall demographic information regarding the ascertainment of samples is listed below. The island country of Taiwan is situated in the Pacific Ocean about 160 km from the southern coast of the Chinese mainland. As of September 2014, the population size of Taiwan is 23,271,643. Recruitment catchment areas included metropolitan Taipei (population 7,030,620), Northern Taiwan (population 3,588,318), Middle Taiwan (population 4,520,155), Southern Taiwan (population 3,387.035), KaoPing (population 3,728,269) and Eastern Taiwan (population 1,017,246). In these six recruitment areas, 76, 28, 45, 31, 36, and 24 clinical settings participated in recruitment, respectively. Successfully recruited patients for family ascertainment was 704, 558, 569, 576, 497 and 104, respectively. In turn, the recruitment ratio of patients per 10,000 in the population is 1.0, 1.5, 1.26, 1.70, 1.33, and 1.02, respectively.
No statistical methods were used to pre-determine the sample size, but our sample size is larger to those reported in previous publications [21,25–31].
Clinical ascertainment and diagnostic assessment
The clinical ascertainment of schizophrenia patients and parents was comprised of three stages: (1) Obtaining IRB approval and informed consent, (2) Recruitment of patients diagnosed with schizophrenia and their parents, and (3) Collection of clinical data and diagnostic assessment.
Stage 1
We obtained approval from all IRBs of the hospitals participating in this study for patients and parent recruitment, data and sample collection, and informed consent approval for both patient and parents. The major content of the informed consent form composed of the following elements: goal of the study, inclusion/exclusion criteria of study subjects, methods of the study (diagnostic interview, blood sample collections), management of any remaining samples (e.g. DNA samples), potential adverse effects of study activities and its management, the expected results, requested activities of the recruited participants of this study, confidentiality, compensation and insurance of the participants for any adverse effect encountered in this study, rights of the recruited participants, withdrawal and termination for participating this study, and de-linking the basic identifiable personal data from the sample. Of the 813 IRB approvals, almost all are written in Chinese, and can be provided upon request.
Stage 2
Recruitment of patients and parents spanned a 5-year period, starting in June 2009 and concluding in March 2014. Recruitment included clinical screening and obtaining informed consent. Clinical screening, using a clinical screening sheet, was employed to (a) exclude those potential subjects with an ancestor of aboriginal origin; (b) include those potential subjects fulfilling the DSM-IV criteria of schizophrenia for patient recruitment based on clinical observation and interview by the attending psychiatrist providing the psychiatric services.
After identifying the potential patient, parents were informed about the details of this study, and initial oral consent was obtained. The patient and parents were then given the IRB-approved consent forms and the details of this study were explained. We then obtained the signed informed consent documents, and the patient entered the study.
Stage 3
Collection of clinical data and diagnostic assessment consisted of four main steps. 1) A DIGS (Diagnostic Interview for Genetic Studies) interview of each patient was performed by trained research assistants with backgrounds in nursing, psychology, or social work. Following the interview, a review of clinical chart records in the hospital or in the clinical service settings was made prior to completing the clinical summary. 2) Blood samples were drawn for DNA analysis. 3) Two board-certified psychiatrists independently completed an initial research diagnostic assessment based on integrated clinical information in the DIGS interview data and summary notes of clinical course, symptom manifestations, and social functioning derived from the records of medical charts. If both research diagnostic assessments reached a consensus diagnosis of schizophrenia, the research diagnosis was finalized. If there was a discrepancy in the diagnostic assessment, then the case was subject to the last step of research diagnosis assessment. 4) The last research diagnostic assessment was done by senior research psychiatrist, Professor Hai-Gwo Hwu, based on the information in the clinical screening sheet, the DIGS interview data, and the clinical summary note. If necessary, the research psychiatrist would call up the field-attending psychiatrists for clarification of clinical information crucial for the diagnostic assessment. Twenty-seven subjects were excluded from this study due to the diagnostic deviation from schizophrenia. After exclusion, 3,093 patients with schizophrenia and their unaffected parents from 3,008 families in total were recruited for research. Among 3,093 patients, 2,923 did not have apparent family history upon brief clinical screening (i.e. sporadic cases).
Parents of patients were not subject the interview stage (stage 3) of diagnostic assessment, and designation of a schizophrenia diagnosis in parents was based on family history information. From this information, 1,732 trios were identified with unaffected parents, and recruited for exome sequencing.
Exome sequence generation
After sample collection, blood DNA samples were shipped to Rutgers University Cell and DNA Repository. Samples identified for project inclusion were sent to the Broad Institute for exome sequencing.
Exome targeting of DNA for the first two waves of data used hybrid selection capture from the Agilent SureSelect Human All Exon v2, targeting 33Mb of exon sequence, or 99% of human exons as defined by NCBI September 2009 consensus. Illumina HISeq 2000 sequencers were used to generate sequence reads, producing paired-end reads spanning 76 bases on average, with coverage goals of 20X sequencing depth or greater for at least 80% of the exome target. Sequencing was performed in two separate waves, consisting of 582 and 558 trios, respectively. Trios that did not meet coverage goals in initial sequencing passes were attempted in subsequent passes. Exome targeting of DNA for the third wave of data used hybrid selection capture using Illumina’s Nextera Rapid Capture Exome v1 (ICE), targeting 37.7Mb of exon sequence. ICE capture covers the same exome target as Agilent SureSelect, with additional capture designed to include added content from the consensus coding sequence (CCDS) and RefSeq in the March 2012 UCSC genome database, as well as Gencode V11 database. Illumina X10 sequencers were used to generate sequence reads producing paired-end reads spanning 76 bases on average, with coverage goals of 20X sequencing depth or greater for at least 80% of the exome target. Sequencing was performed on 598 trios.
We performed additional quality control (QC) to confirm relatedness among trios, ensure DNA was whole blood, and sequence coverage was met. In all, 43 trios were removed in these QC steps, leaving 1695 trios for downstream analysis. On average, samples had 86% of the exome target meeting 20X sequencing depth (87% for Agilent captured samples and 84% for ICE captured samples), and 93% of the exome target meeting 10X sequencing depth (93% for both Agilent and ICE captured samples). Sample information for the 1695 trios passing QC are available in Supplementary Data File 2: Taiwanese cohort sample list and Supplementary Data File 3: Taiwanese cohort trio list, with column level descriptions available in Supplementary Data File 1: Data file Descriptions.
Variant calling
The Burrows-Wheeler Algorithm (BWA: [41]) was used to align unmapped sequence reads to the human reference (hg19), and the Picard pipeline (https://github.com/broadinstitute/picard) performed additional sequence QC checks and metrics to create a final BAM (Binary Sequence Alignment/Map format) file for each sample. GATK version 3.4 [42] was used to call single-nucleotide (SNV) and small insertion/deletion (indel) variants from individual BAM files, and recalibrated variant quality scores (VQSR) were generated to determine the overall variant quality in the VCF (variant calling format) file.
DNM calling and filtering
De novo single nucleotide variants and short indels were identified in the VCF calls, whereby the proband offspring genotype was heterozygous while the parental genotypes were both homozygous reference that the genotype call. For all three cohort waves, genotype calls were required to have a minimum PHRED-scaled likelihood (PL) > 20, analogous to at most a 1% probability of being a false genotype call. Allelic depth, or the proportion of non-reference genotype reads, was required to be at least 20% in offspring, and no more than 5% in either parent. We also removed sites where the offspring had < 10% read depth than the combined parental read depth, suggesting poor sequence capture or copy number deletion in the offspring. Cohort waves 1 and 2: DNM calling did not use the conditional probability model or ExAC allele counts as filters (supplementary section 1), and validation was pursued for all putatively called DNMs. Validated calls, even if not passing all filters in the final combined VCF, were retained in the final set. Cohort wave 3: DNM calling used the conditional probability model and the ExAC allele counts as filters. Validation was pursed only for protein truncating DNMs.
A small fraction of DNM calls from a single individual were observed in the same gene, often adjacent to one another. Most of these calls were confirmed in validation, suggesting a more “complex” DNM event is occurring at the site. Similar events have been reported and confirmed in previous studies [21]. Following previous studies, we selected the single DNM with the most severe consequence in our final call set. In all, we observed 27 DNMs from 12 individuals considered “complex” DNMs, and selected 12 DNMs that had the most severe consequence (any ties were broken by selecting the higher depth call).
DNA barcode switching
During the first wave of sequence generation, it was discovered that DNA barcodes inadvertently mislabeled individuals sequenced in the same lane during multiplex sequencing. During the process of adding DNA barcode identifiers to everyone’s sheared DNA prior to sequencing, the active enzyme carrying the individual-specific barcode did not fully denature after bringing to higher temperature. Thus, when two samples were mixed (duplexed) together during sequence generation, there was still active enzyme labeling additional DNA fragments among the mixed DNA. This led to a non-trivial proportion of DNA barcodes of one individual attaching to the DNA of another individual, which we term “DNA barcode switching”.
Quantifying the level of the DNA barcode switching was determined by examining the correlation coefficient between a) shared alleles of duplexed samples and b) 1K Genomes allele frequency. Once the enzyme protocol was fixed, evidence of DNA barcode switching disappeared. For the current analysis, DNA barcode switching presented a unique challenge to the discovery of de novo variation, as overall Mendelian error rates were increased across the board. Given that DNA barcode switching was restricted to sample pairs mixed in multiplex sequence, we could run direct comparisons of variant quality for DNMs between samples.
To identify DNMs that were the result of barcode switching, we examined the variant quality of everyone’s putative DNM call in their barcode switching partner (BSP). In general, variants present in an individual due barcode switching will be of lower sequence quality than the true variant. To start, we removed any DNM call where the BSP was called as homozygous alternate. We then compared the phred-likelihood (PL) score in both individuals, and removed DNM calls that had greater than 1.2 times higher PL score in the BSP. While many variants were removed using this approach, we still observed a significantly higher rate than expectation. In turn, we pursued an aggressive validation approach to confirm the presence of true DNMs in the first wave of sequencing.
DNM validation
For the first two waves of sequencing, we performed molecular validation of 1688 putative DNMs using an Illumina targeted amplicon sequencing framework. With the presence of DNA barcode switching in most of wave 1, we expected a substantial fraction of putative DNM calls in this cohort to be rejected in the validation process (See Online Methods Table 1 for validation counts).
First, target region specific primers were designed using Primer3 [43] and checked for specificity using the in-silico PCR tool from the UCSC Genome Browser. Oligo tails were added to the designed primers for subsequent hybridization to Illumina specific adapter sequence. Target regions were amplified in a first-round PCR for each father, mother, and child corresponding to a given DNM call. These PCR products were pooled by individual of origin, purified using the Agencourt AMPure XP system, then put into a second-round PCR in which Illumina specific adapter sequence and barcodes were added via hybridization to the complementary oligo tail sequence. After purification of the round 2 PCR products, completed libraries were pooled, with quantification and quality control being done by QuBit fluorometric quantitation, Agilent BioAnalyzer high sensitivity DNA kits, and Kapa Biosystems Illumina library quantification kits. Amplicon libraries were loaded on a MiSeq or HiSeq 2500 with a 5% PhiX spike in for sequencing.
Demultiplexed fastq files from the validation sequencing runs were aligned with BWA-MEM [41], reads with greater than 3 soft clipped bases in the beginning of the alignment were removed to control for sequencing artifacts. Allele pile-ups downsampled to 1000 were counted using the GATK UnifiedGenotyper [42] in a targeted fashion after filtering for mapping quality, base quality, and edit distance. Only sites with at least 30 reads overlapping the variant site were considered. Genotype calls were made based on allelic fraction of reads at each locus, with a non-reference allele fraction within 30–70% being considered heterozygous. All target sites were visually inspected in IGV [44] for accuracy, with a selected subset of calls confirmed by Sanger sequencing.
For the third wave of trios, validation was pursued specifically on protein-truncating variants, and use more stringent filtering criteria as a general method to keep the false positive call rate low. Primers were designed for 68 PTV variants, with validation performed through an external contract with Sequenom. The 9 variants that didn’t return a confident call either way were followed up using Sanger sequencing. In all, we validated 59 PTVs (86.8% validation rate) from the putative DNM list (Online Methods Table 1). Final DNM calls for the full Taiwanese trio cohort are listed in Supplementary Data File 4: Taiwanese cohort DNM list.
Online Methods Table 1:
DNM validation counts
| Taiwanese cohort | Trios | Putative called DNMs | DNMs submitted for validation | Validated DNMs | Final DNM count |
|---|---|---|---|---|---|
| Agilent wave 1 | 575 | 1060 | 1060 | 586 (55.3%) | 586 |
| Agilent wave 2 | 532 | 628 | 628 | 517 (82.3%) | 517 |
| Nextera wave 3 | 588 | 653 | 68 | 59 (86.8%) | 644 |
DNM annotation
Putative DNMs were primarily annotated using the Variant Effect Predictor (VEP) version 81, which uses GENCODE v19 mapped to the GRCh37 genome build. All non-coding variants were removed from further analysis. Remaining variants comprise three broad categories – synonymous, missense, and protein-truncating variants. Synonymous annotation includes synonymous amino acid changes and stop-retained changes. Missense annotation includes non-synonymous single amino acid changes, inframe insertions and deletions, and stop-lost changes. Protein-truncating variant (PTV) annotation includes stop-gain, start-lost, frameshift, essential splice site changes. To maintain consistency, annotation was updated for published DNMs, resulting in a few minor changes to published annotations.
Within missense mutations, we used annotations curated through the dbNSFP v2.9 database [45, 46]. We assessed PolyPhen2 HDIV and HVAR, SIFT, LRT, MutationTaster, MutationAssessor, FATHMM, MetaSVM, MetaLR, Provean, and CADD v1.2 predictions and scores to measure predicted pathogenicity. Secondary analyses of published synonymous DNMs reported a significant enrichment in near-splice site synonymous changes among ASD probands, as well as enrichment in DNAse Hypersensitivity sites (DHS) drawn from the cerebrum, cerebellum, and frontal cortex region of post-mortem tissues in SCZ probands [36]. Following their protocol, we annotated the distance to the nearest splice site using SeattleSeq Annotation 137 [47], exon-splicing enhancers (ESE) / silencers (ESS) from hexamer motifs [48–50], and candidate DHS regions from ENCODE Experiment Matrix (https://genome.ucsc.edu/ENCODE/dataMatrix/encodeDataMatrixHuman.html).
We leveraged variant sites from the genome aggregation database (gnomAD, release 2.1.1) to further interpret the potential for pathogenicity of DNM calls. While the Taiwanese cohort were included in the full 141k gnomAD call set, we used allele counts from the non-psychiatric subset (104k samples), which did not include either probands or parents from the Taiwanese cohort. For exome-wide burden and gene-set analyses, we annotated DNM calls by their presence as a variable site in gnomAD, predicting that DNM sites not seen in gnomAD are more likely to be pathogenic and therefore enriched in SCZ cases relative to controls.
Incorporation of published DNM
Along with our mutational model, we also wanted to assess how the patterns and rates of observed DNMs compare with published control trios and unaffected siblings in the published literature, as well as incorporate published DNMs in SCZ to further refine their impact of on SCZ risk. We collected results from independent exome-sequenced SCZ trios, their unaffected siblings and control trios, as well as unaffected siblings collected from other neuropsychiatric disorders. Specific publications and descriptive data are listed in Supplementary Data File 5: DNM studies. In total, we assessed 1077 published SCZ trios and 2216 control trios and unaffected siblings of ASD probands.
To assess overall DNM rates, we performed cross-study comparisons and checked against the mutational model expectation. Datasets with significantly different synonymous or insertion/deletion (indel) rates when compared with similar studies and against the mutation model were flagged and not incorporated in exome-wide DNM rate calculations, but were retained for single gene and gene-set analyses (Supplementary Figure 1: SCZ vs control synonymous DNM rate and Supplementary Figure 2: SCZ vs control indel DNM rate). Within the Taiwanese trio cohort, the wave 3 cohort had a significantly higher synonymous rate due to the larger exome target in Nextera capture (37.7 Mb vs 33 Mb in previous waves). When we restricted to only DNMs within the Agilent capture targets, the synonymous rate was comparable to previous waves (Supplementary Figure 3: Taiwanese vs published SCZ synonymous DNM rate). We therefore restricted to only DNMs within the Agilent capture targets among all cohorts when analyzing exome-wide burden. The full list of DNMs analyzed are available in Supplementary Data File 6: Combined cohorts DNM list.
Along with published SCZ and control trios, we also incorporated published DNMs from autism (3982 probands [11–16], intellectual disability (971 probands, [8–10], and developmental delay (4293 probands, [20]) for comparison purposes in exome-wide burden and as candidate gene sets in gene set enrichment analyses. Given the overlapping nature of data freezes for these studies, we included only the most recent DNM list among overlapping waves, and collapsed multiple DNMs from the same gene/same individual (when proband ID was available) into a single DNM with the most severe consequence.
Statistical tests used to analyze DNM rates and patterns
All statistical tests were performed using R statistical software (www.r-project.org). To examine overall rates, recurrence, and single gene enrichment of DNMs in SCZ probands, we fit the data to Poisson distributed models. To examine the role of covariates on DNM rates within the Taiwanese trio cohorts, we use a Poisson-distributed multiple regression model. For comparing against mutation model expectations, we used a one-sample exact Poisson test, with the mutation expectation as our lambda parameter. For comparing against control DNMs, we used a two-sample exact Poisson test. To test gene recurrence against the mutation model, we used a bootstrap re-sampling method to sample recurrent genes using per-gene probabilities and used the empirical distribution to test for significance.
To test for gene set enrichment, we fit the data to a binomial model. This model is conditional on the overall rate, and examines the relative proportions of enrichment rather than the observed rate. For example, if SCZ probands had twice the rate of PTV DNMs than controls, but 50% fell in brain-expressed genes in each sample, then we would have two-fold enrichment in the PTV rate, but no enrichment for brain-expressed genes among PTVs. For comparing against mutation model expectations, we used a one-sample exact binomial test, with the mutation expectation as our null probability of gene set overlap. For comparing against control DNMs, we used a two-sample exact binomial test. Gene set enrichment analyses were divided into two sets: ‘candidate’ gene sets and ‘discovery’ gene sets. Candidate gene sets represent previously reported gene sets associated with schizophrenia and other neurodevelopmental disorders. Discovery gene sets are derived from Gene Ontology (GO) annotations (http://www.geneontology.org/GO.downloads.ontology.shtml) and SynaptomeDB (http://metamoodics.org/SynaptomeDB/index.php). Candidate and discovery gene sets were corrected for multiple testing independently of each other.
Additional information about the tests used for each analysis are provided in their appropriate section.
Supplementary Material
Acknowledgements
This study was supported by grants from The National Human Genome Research Institute (U54 HG003067, R01 HG006855), The Stanley Center for Psychiatric Research, and the National Institute of Mental Health (R01 MH077139, R01 MH085521, and RC2 MH089905).
Footnotes
Competing Interests
B.M.N. is on the Scientific Advisory Board at Deep Genomics and Camp4 Therapeutics Corporation, and on the Biogen Genomics Advisory Panel. M.F. is an employee of Verily Life Sciences.
Accession codes
Data included in this manuscript have been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs001196.v1.
Data collection and analysis were not performed blind to the conditions of the experiments.
Data availability
Data included in this manuscript have been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs001196.v1.
Data collection and analysis were not performed blind to the conditions of the experiments.
Code availability
Code used to identify coding DNMs and assess enrichment are publicly available at: https://github.com/howrigan/trio_sequence_analysis
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics, C., Biological insights from 108 schizophrenia-associated genetic loci. Nature, 2014. 511(7510): p. 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekar A, et al. , Schizophrenia risk from complex variation of complement component 4. Nature, 2016. 530(7589): p. 177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, et al. , Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet, 2012. 44(3): p. 247–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Power RA, et al. , Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry, 2013. 70(1): p. 22–30. [DOI] [PubMed] [Google Scholar]
- 5.Marshall CR, et al. , Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet, 2017. 49(1): p. 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malhotra D and Sebat J, CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell, 2012. 148(6): p. 1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees E, et al. , Analysis of copy number variations at 15 schizophrenia associated loci. Br J Psychiatry, 2014. 204(2): p. 108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ligt J, et al. , Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med, 2012. 367(20): p. 1921–9. [DOI] [PubMed] [Google Scholar]
- 9.Rauch A, et al. , Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet, 2012. 380(9854): p. 1674–82. [DOI] [PubMed] [Google Scholar]
- 10.Lelieveld SH, et al. , Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci, 2016. 19(9): p. 1194–6. [DOI] [PubMed] [Google Scholar]
- 11.De Rubeis S, et al. , Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 2014. 515(7526): p. 209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iossifov I, et al. , De novo gene disruptions in children on the autistic spectrum. Neuron, 2012. 74(2): p. 285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iossifov I, et al. , The contribution of de novo coding mutations to autism spectrum disorder. Nature, 2014. 515(7526): p. 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neale BM, et al. , Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature, 2012. 485(7397): p. 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Roak BJ, et al. , Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature, 2012. 485(7397): p. 246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders SJ, et al. , De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature, 2012. 485(7397): p. 237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epi KC, et al. , De novo mutations in epileptic encephalopathies. Nature, 2013. 501(7466): p. 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epi KC, De Novo Mutations in SLC1A2 and CACNA1A Are Important Causes of Epileptic Encephalopathies. Am J Hum Genet, 2016. 99(2): p. 287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deciphering Developmental Disorders S, Large-scale discovery of novel genetic causes of developmental disorders. Nature, 2015. 519(7542): p. 223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deciphering Developmental Disorders, S., Prevalence and architecture of de novo mutations in developmental disorders. Nature, 2017. 542(7642): p. 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromer M, et al. , De novo mutations in schizophrenia implicate synaptic networks. Nature, 2014. 506(7487): p. 179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese G, et al. , Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci, 2016. 19(11): p. 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh T, et al. , The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet, 2017. 49(8): p. 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh T, et al. , Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci, 2016. 19(4): p. 571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takata A, et al. , Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron, 2014. 82(4): p. 773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard SL, et al. , Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet, 2011. 43(9): p. 860–3. [DOI] [PubMed] [Google Scholar]
- 27.Guipponi M, et al. , Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One, 2014. 9(11): p. e112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulsuner S, et al. , Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell, 2013. 154(3): p. 518–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy SE, et al. , De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry, 2014. 19(6): p. 652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu B, et al. , Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet, 2011. 43(9): p. 864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B, et al. , De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet, 2012. 44(12): p. 1365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samocha KE, et al. , A framework for the interpretation of de novo mutation in human disease. Nat Genet, 2014. 46(9): p. 944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lek M, et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature, 2016. 536(7616): p. 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski KJFLCTG, Cummings BB; Alföldi J; Wang Q; Collins RL, Laricchia KM; Ganna A; Birnbaum DP; Gauthier Laura D, Brand Harrison, Solomonson Matthew, Watts Nicholas A, Rhodes Daniel, Singer-Berk Moriel, Seaby Eleanor G, Kosmicki Jack A, Walters Raymond K, Tashman Katherine, Farjoun Yossi, Banks Eric, Poterba Timothy, Wang Arcturus, Seed Cotton, Whiffin Nicola, Chong Jessica X, Samocha Kaitlin E, Pierce-Hoffman Emma, Zappala Zachary, O’Donnell-Luria Anne H, Minikel Eric Vallabh, Weisburd Ben, Lek Monkol, Ware James S, Vittal Christopher, Armean Irina M, Bergelson Louis, Cibulskis Kristian, Connolly Kristen M, Covarrubias Miguel, Donnelly Stacey, Ferriera Steven, Gabriel Stacey, Gentry Jeff, Gupta Namrata, Jeandet Thibault, Kaplan Diane, Llanwarne Christopher, Munshi Ruchi, Novod Sam, Petrillo Nikelle, Roazen David, Ruano-Rubio Valentin, Saltzman Andrea, Schleicher Molly, Soto Jose, Tibbetts Kathleen, Tolonen Charlotte, Wade Gordon, Talkowski Michael E, The Genome Aggregation Database Consortium, Neale Benjamin M, Daly Mark J, MacArthur Daniel G, Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv, 2019. [Google Scholar]
- 35.Kosmicki JA, et al. , Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet, 2017. 49(4): p. 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takata A, et al. , De Novo Synonymous Mutations in Regulatory Elements Contribute to the Genetic Etiology of Autism and Schizophrenia. Neuron, 2016. 89(5): p. 940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrovski S, et al. , Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet, 2013. 9(8): p. e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson EB, et al. , Autism spectrum disorder severity reflects the average contribution of de novo and familial influences. Proc Natl Acad Sci U S A, 2014. 111(42): p. 15161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganna A, et al. , Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. Nat Neurosci, 2016. 19(12): p. 1563–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell SM, et al. , A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 2014. 506(7487): p. 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 41.Li H and Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009. 25(14): p. 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, et al. , The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res, 2010. 20(9): p. 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Untergasser A, et al. , Primer3--new capabilities and interfaces. Nucleic Acids Res, 2012. 40(15): p. e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson JT, et al. , Integrative genomics viewer. Nat Biotechnol, 2011. 29(1): p. 24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Jian X, and Boerwinkle E, dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat, 2011. 32(8): p. 894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Jian X, and Boerwinkle E, dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat, 2013. 34(9): p. E2393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng SB, et al. , Targeted capture and massively parallel sequencing of 12 human exomes. Nature, 2009. 461(7261): p. 272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairbrother WG, et al. , Predictive identification of exonic splicing enhancers in human genes. Science, 2002. 297(5583): p. 1007–13. [DOI] [PubMed] [Google Scholar]
- 49.Ke S, et al. , Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res, 2011. 21(8): p. 1360–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, et al. , Systematic identification and analysis of exonic splicing silencers. Cell, 2004. 119(6): p. 831–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.