Abstract
While the fundamental importance of the white matter in supporting neuronal communication is well known, existing publications of primate brains do not feature a detailed description of its complex anatomy. The main barrier is that existing primate neuroimaging data have insufficient spatial resolution to resolve white matter pathways fully. Here, we present a resource that allows detailed descriptions of white matter structures and trajectories of fiber-pathways in the marmoset brain. The resource includes: (1) the highest resolution diffusion MRI (dMRI) data available to date, which reveal white matter features not previously described; (2) a comprehensive 3D white matter atlas depicting fiber-pathways that were either omitted or misidentified in previous atlases; and (3) comprehensive fiber-pathway maps of cortical connections combining dMRI tractography and neuronal tracing data. The resource, which can be downloaded from marmosetbrainmapping.org, will facilitate studies of brain connectivity and the development of tractography algorithms in the primate brain.
Introduction
The primate brain is evolutionarily adapted to support visual and social cognition through a large number of cortical regions1 interconnected through axonal bundles that form the white matter. Recent studies have demonstrated that the white matter is not a passive passageway of neuronal communication, but a vital component of brain plasticity and connectivity, actively affecting learning, memory, and cognitive function2. Although not traditionally emphasized in brain atlases, charting these white matter bundles is of keen interest to researchers attempting to understand the connectional principles of the primate brain and to explain symptoms in neurological diseases such as stroke and multiple sclerosis and psychiatric disorders including depression and schizophrenia2. Anterograde tracing is a traditional approach to trace discrete axonal projections3, which can be rendered in 3D to chart specific fiber pathways4, 5, but the method cannot be applied to create a complete, whole-brain white matter atlas. Recent progress in diffusion-weighted Magnetic Resonance Imaging (dMRI) has paved a new way to map fiber pathways and contributed to whole-brain white matter atlases of humans and non-human primates (NHPs)6–9. Compared with traditional tracing methods, the dMRI-based atlases have the advantage of preserving the 3D fiber orientation information across the entire brain. As digital atlases, they also provide versatile tools to facilitate fiber reconstruction and tract-based analyses10 for human and animal studies.
However, to date, dMRI atlases have had a relatively coarse spatial resolution, which limits their capacity to resolve small but essential fiber pathways known from anatomical studies. This problem stems from the fact that dMRI data acquisition on large primate brains faces inherent technical challenges that limit the signal-to-noise ratio required to acquire very small voxels. For example, larger brains require a longer scanning time, cannot fit into ultra-high-field magnets that usually have small bores, and pose difficulties for designing high-performance radiofrequency (RF) coils with whole-brain coverage. An essential resolution measure is the absolute voxel size, as opposed to the relative voxel size in proportion to overall brain size, as many white matter structures do not scale in proportion to overall brain size but remain very small even in large brains (Fig. 1). Thus, there is much to be gained by increasing the absolute dMRI resolution in a primate brain.
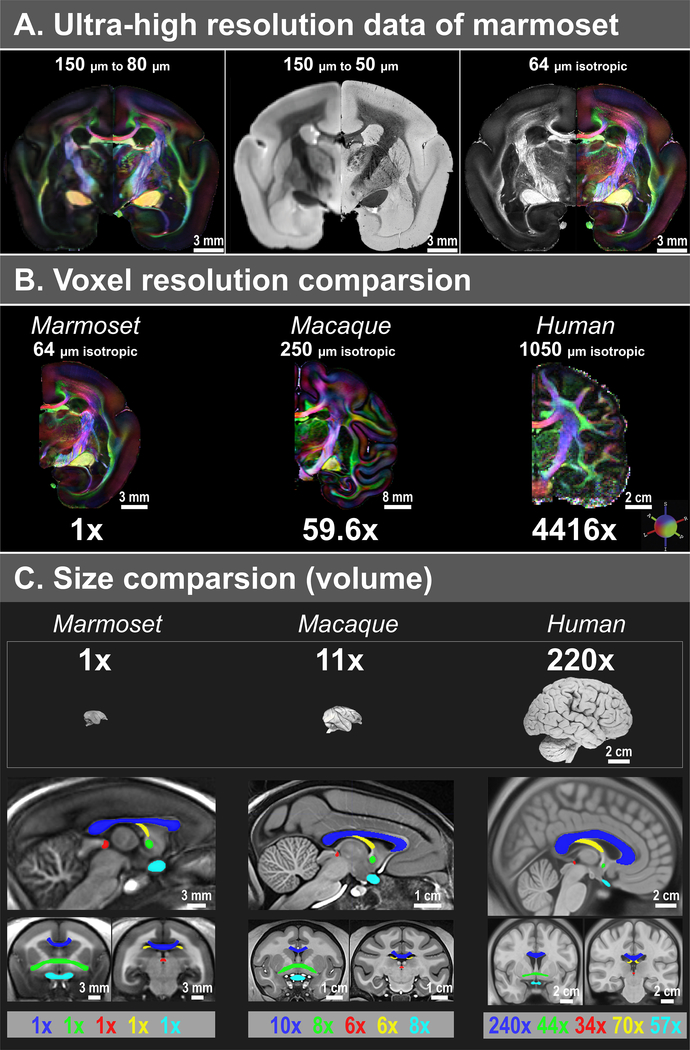
Figure 1. Comparison of isotropic data resolution, brain volume, and fiber volumes between marmosets, macaques, and humans.
(A) Compared to our previous 150 μm data16, the new dataset has a substantial improvement in spatial resolution, including 80 μm dMRI (left), 50 μm T2* (middle), and 64 μm dMRI (right). (B) Comparison of voxel resolution. The macaque ex-vivo dMRI dataset has 250 μm resolution20, and the human in-vivo dMRI dataset from the 7T human connectome project21 has 1050 μm resolution. In terms of volume (μm3), the resolution of our 64 μm data is 59.6x and 4416x higher than that of the macaque and human data, respectively. (C) Comparison of the brain and fiber volumes (μm3). Brain sample images are from brainmuseum.org. All volumes are estimated from three population-based templates, including a marmoset template from 22 adult marmosets, a NMT macaque template39, and a MNI152 adult human template40. The five white matter tracts shown are the corpus callosum (blue), the anterior commissure (green), the posterior commissure (red), the fornix (yellow), and the optic white matter (cyan; including the optic nerve, the optic chiasm, and the optic tract).
The primary aim of this study was to map white matter pathways in the primate brain with a higher level of precision than has been previously possible. For this, we turned to one of the smallest primates, the common marmoset, whose brain shares basic organizational features with larger primates11–14. The small marmoset brain allows the use of high field MRI and stronger gradients, a necessity for high-resolution dMRI15, 16. Together with improvements in RF coil selection and proper design of scanning protocols, we collected dMRI data of the marmoset brain with unprecedented spatial resolution. The data allowed us to build a fine-grained 3D white matter atlas of the marmoset brain, which depicts many fiber pathways that are either omitted or incorrectly described in previous MRI datasets or atlases of the primate brain. We also provide examples of the high similarity of white matter structures across primate species. To further extend the functionality of this atlas, we combined dMRI tractography with a neuronal tracing database (the Marmoset Brain Connectivity Atlas17, marmosetbrain.org) to provide the first and most comprehensive fiber pathway mapping of cortical connections. The white matter atlas, fiber pathways maps, and dMRI data, including both raw and processed data, are publicly available in our website (marmosetbrainmapping.org) as an open resource to facilitate mapping the white matter neuroanatomy and understanding connectional principles of the brain, as well as algorithm development and validation experiments of dMRI tractography.
Results
Ultra-high resolution dMRI datasets
We collected two sets of multi-shell 3D dMRI data (Fig. 1A). The first dataset was acquired from a whole-brain with 80 μm isotropic resolution at 7T in 15 days. We also collected a magnetization transfer ratio (MTR, a T1-like contrast) image with 80 μm isotropic resolution, and a T2* and a T2 anatomical image with 50 μm isotropic resolution. The second dMRI dataset was acquired from a half brain (right hemisphere) with 64 μm isotropic resolution at 14T in 12 days. Both dMRI datasets comprised 64 directions at b=2400 s/mm2 and 126 directions at b=4800 s/mm2.
Three primary factors contributed to our ability to obtain the ultra-high-resolution data. The first factor was the cutting-edge gradient and RF technology applied in high-field scanners. Marmoset brains were small enough to be scanned in small-bore animal magnets with powerful gradients (450mT/m for 7T and 1500mT/m for 14T, Resonance Research Inc., Billerica, MA) and a customized 30 mm ID millipede quadrature coil (ExtendMR, LLC, Milpitas, CA) that provided 2.5x more sensitivity than the standard Bruker coil (Bruker Biospin, Ettlingen, Germany) used in our previous study16. The second central factor was an improved protocol for preparing brain samples. The small marmoset brains require less time for contrast agents to penetrate the entire tissue volume than large brains. Thus, the gadolinium contrast agent18 was effective in reducing the T1 relaxation time constants of the fixed brains, significantly improving the sensitivity per unit time of the data. The third essential factor was prolonged scanning (about two weeks) to acquire high spatial resolution data with many diffusion directions. We aimed to collect data that had not only a high spatial resolution necessary for dissecting the white matter but also a high angular resolution for dMRI tractography.
The resulting datasets provide the highest resolution dMRI data to date in any primate brain and thus reveal features never previously observed (Fig. 1 A–B). Specifically, our two datasets of dMRI data (80 μm and 64 μm isotropic) have 30.5x and 59.6x higher resolution than one of the highest ex-vivo dMRI datasets of the macaque brain (250 μm isotropic) in terms of cubic volumes (μm3)19, 20; and 2260x and 4416x higher than the 7T human connectome in-vivo dMRI data (1050 μm isotropic)21. Correspondingly, the marmoset brain volume (in μm3) is approximately 11x and 220x smaller than the macaque brain and the human brain, respectively, which is significantly less than the improvement in the spatial resolution of our data (Fig. 1C). Additionally, white matter tract sizes do not scale the same as the brain. For example, among the five tracts we estimated, only the corpus callosum follows a similar scaling factor as the brain, while the other tracts have much smaller scaling (Fig. 1C). In other words, when examining many white matter tracts, larger primates may require higher relative spatial resolution than the small marmoset after taking brain volume scaling into consideration.
Ultra-high resolution reveals previously hidden details of primate white matter
The marmoset dMRI dataset provides high spatial and angular resolutions, while fully preserving the coverage and 3D features of an intact whole brain, including the local fiber orientation. The dataset offers a unique opportunity to examine the fine-detailed 3D structures of the white matter in a primate brain. Here, we selected three examples to illustrate this point.
1). Fiber composition of the corona radiata
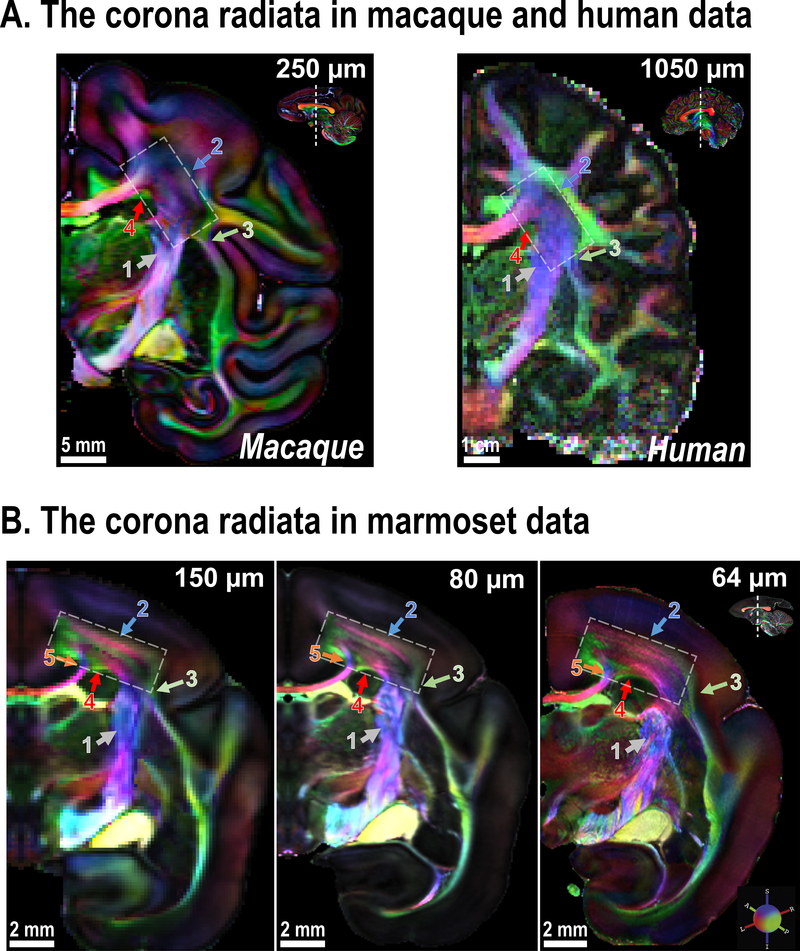
The corona radiata is one of the most complex white matter zones in the primate brain. Multiple fiber pathways with different directions pass through the corona radiata, including, but not limited to, projection fibers from/to the internal capsule, commissure fibers from/to the corpus callosum, striatal fibers from/to the external capsule, association fibers from/to the cingulum (Cg) and the superior longitudinal fasciculus (SLF). Due to the complex fiber-crossing and partial volume effects in MRI data, standard directionally-encoded color (DEC) maps22 from dMRI cannot reveal the exact fiber structures in the corona radiata. This limitation is corroborated by anterograde neuronal tracing data3, which reveals many fibers from the superior longitudinal fasciculus that run in an anterior-posterior direction through the coronal plane at the level of the central corona radiata. Neither macaque nor human dMRI data are currently able to identify this portion of the superior longitudinal fasciculus (DEC-green) inside the central corona radiata, which is dominated by fibers running in an inferior-superior direction (DEC-purple) (Fig. 2A; white rectangle).
Figure 2. The corona radiata imaged at different spatial resolutions.
(A) Coronal slices of the corona radiata (white rectangle) of an ex-vivo macaque brain (at 250 μm) and an in-vivo human brain (at 1050 μm). Note that the corona radiata is mostly dominated by projection fibers from the internal capsule (ic; gray arrow-1). (B) Coronal slices that show the corona radiata of marmoset brain acquired at increasing spatial resolution. Colored arrows highlight areas that benefit from the use of higher spatial resolution (see text for details; the zoomed-in view is provided in Supplementary Fig. 1).
In the marmoset data, although all resolutions (150 μm, 80 μm, and 64 μm) are high enough to reveal the superior longitudinal fasciculus, which divides the DEC-purple zone of the corona radiata, different resolutions present varying patterns (Fig. 2B; white rectangle). At 150 μm, the corona radiata appears to have two layers of DEC-purple fibers that are separated by the DEC-green fasciculus. At 80 μm, one more layer (DEC-purple) fades in near the gray-white-matter boundary (Fig. 2B, blue arrow-2). At 64 μm, the corona radiata presents many DEC-purple fibers interspaced by thin DEC-green fibers, in clear demonstration that its real fiber organization is significantly more complex than the previously acquired lower-resolution data could depict.
The higher resolution also provides finer details for each fiber pathway (Fig. 2 and Supplementary Fig. 1 for a zoomed-in view). Fibers from/to the corpus callosum are split into two different branches in the corona radiata (Fig. 2B, orange arrow-5). Both 80 μm and 64 μm datasets show this, but not in the 150 μm one. Fibers from/to the external capsule contribute a small portion of the “DEC-purple” fibers in the corona radiata, but the connecting fibers (Fig. 2B, green arrow-3) are only clearly visible at 64 μm. In the 80 μm and 150 μm dataset, the connecting fibers appear as a “dark” crossing-fiber zone due to the partial volume effect. Finally, near the bottom of the corona radiata, the 80 μm and 64 μm datasets show a branch of the corpus callosum that separates two distinct DEC-green fibers (Fig. 2B, red arrow-4). This separation is not visible at 150 μm. These examples demonstrate the advantages offered by ultra-high-resolution scans in the fine-grained identification of white matter pathways.
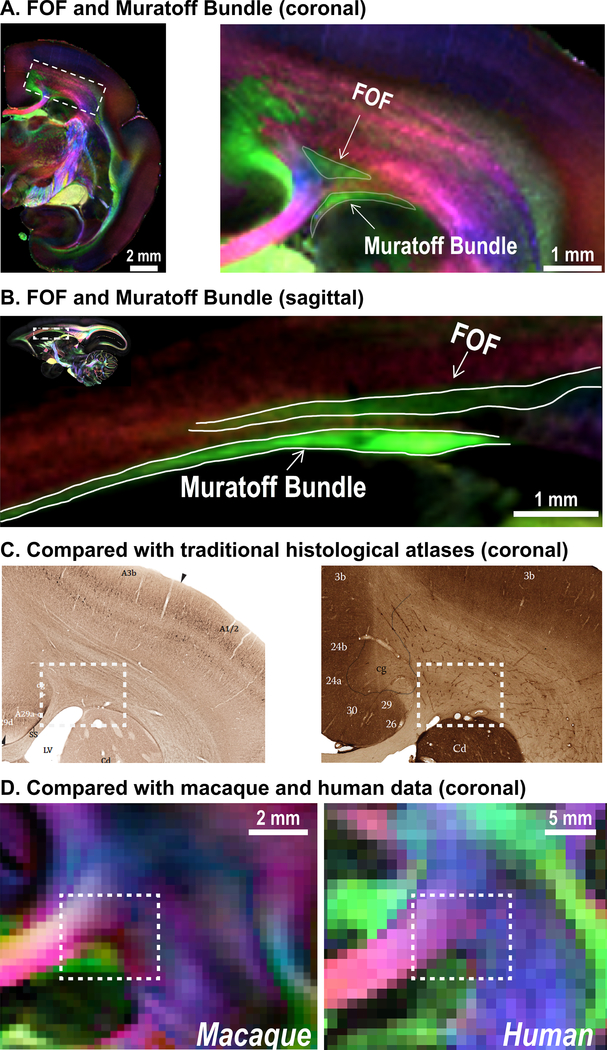
Fig. 3 highlights two major fiber pathways of primates that are indicated by red arrows-4 in Fig. 2, one belonging to the fronto-occipital fasciculus (FOF) and the other to the Muratoff bundle 3. Although these two fibers run in a similar direction (DEC-green; an anterior-posterior direction) under the corona radiata (Fig. 3A–B), they are entirely distinct white matter pathways. The FOF is a long association fiber pathway that connects parietooccipital regions with the dorsolateral prefrontal and premotor cortices, while the Muratoff bundle is a major striatal fasciculus that supports corticostriatal connections 3.
Figure 3. Fronto-occipital fasciculus (FOF) and the Muratoff bundle.
(A) Coronal slice (64 μm) through the corona radiata of the marmoset, where the FOF and the Muratoff bundle are highlighted. (B) Sagittal slice (80 μm) showing the FOF and Muratoff bundle. Two white matter fiber pathways run in a similar direction (green: anterior-posterior) and are separated by a small fiber running left-to-right (red). (C) Non-phosphorylated neurofilament staining of coronal slices from the Paxinos atlas41 and the Hardman atlas42 (modified with permission from the original authors) show that previous histological marmoset atlases cannot distinguish the two fiber bundles due to the lack of fiber-orientation contrasts. (D) As well, the macaque (250 μm) and human dMRI data (1050 μm) cannot distinguish the two fiber bundles due to their insufficient spatial resolution.
These two pathways have been discovered and described by anterograde neuronal tracing techniques 3. Despite their existence, previously published primate atlases (including human atlases) either have not been able to identify the two fibers or have labeled only the FOF 6–8. Traditional histology-based atlases, which use two-dimensional slices, can hardly distinguish these two fiber bundles, due to their inability to reveal fiber orientation (Fig. 3C). Meanwhile, modern dMRI-based atlases were acquired at too low of a resolution to identify the thin commissural fibers that separate the FOF and the Muratoff bundle (Fig. 3D). Owing to the high resolution at 80 μm and 64 μm, we could identify and distinguish the Muratoff bundle from the FOF.
2). Fiber composition of the occipital white matter
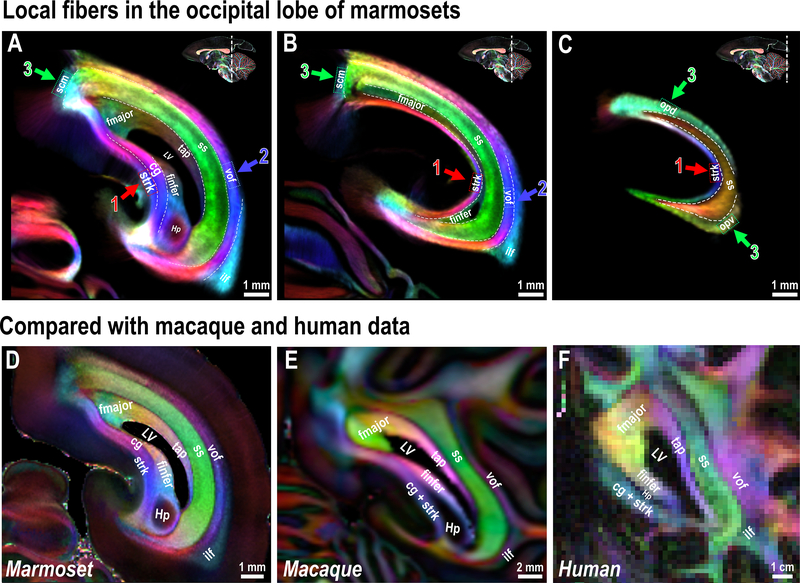
The primate visual cortex is one of the most studied brain regions in neuroscience research, but existing atlases fail to identify the local fibers in the occipital lobe that support complex visual processing. Gross dissection studies of the human brain, which were performed more than one hundred years ago by Sachs (1892)23 and Dejerine (1895)24, provided early descriptions of these local occipital fibers (Supplementary Fig. 2A). However, very few studies investigated these local occipital fibers until recently25, 26. As a result, most modern brain atlases of primate species, including the marmoset, provide only a coarse description of the white matter fibers in the occipital lobe (Supplementary Fig. 2B).
Our high-resolution dMRI demonstrates the layer-like structures of the occipital white matter of the marmoset brain (Fig. 4A–C). The caudal end of the (Cg) disappears at a coronal level near the rostral limit of the calcarine sulcus (Fig. 4A), where area prostriata 27 occupies the dorsal bank. At this same level, the stratum calcarinum (Strk) emerges, which circumvents the calcarine sulcus (Fig. 4, red arrows-1). Although the Cg and the Strk share similar inferior-superior fiber directions across several coronal slices (Fig. 4A), we could observe a clear boundary between the two fibers at 80 μm (Supplementary Fig. 3B) and 64 μm data (Fig. 4A and 4D). This boundary is invisible in the marmoset 150 μm dataset (Supplementary Fig. 3A), the 250 μm macaque dataset (Fig. 4E), or the 1050 μm human connectome dataset (Fig. 4F).
Figure 4. White matter fiber pathways in the marmoset occipital lobe.
(A-C) White matter structures in the marmoset occipital lobe. Three different coronal slices are presented with the color-coded tract density maps generated from the 64 μm dataset. Colored boxes and arrows highlight local fibers. (D-F) Cross-species comparison of the occipital white matter, which shows highly homologous structures across marmosets (64 μm) (D), macaques (250 μm) (E) and humans (1050 μm) (F). Small fiber branches and local U fibers formed by sulci are not shown. Hp - hippocampus.
From medial to lateral, we identified commissural fibers of the inferior forceps (finfer), the forceps majors (fmajor), and the caudal part of the tapetum of the corpus callosum (tap). Lateral to these commissural fibers is the most prominent white matter region in the occipital lobe, the sagittal stratum (ss), which includes, but is not limited to, fibers from the optic radiation 3. The ss shows a clear boundary and a different fiber orientation in comparison with the occipital vertical fasciculus (vof; Fig. 4, blue arrows-2). The ss runs in an anterior-posterior direction (DEC-green) while the vof runs mostly in an inferior-superior direction (DEC-blue and purple), connecting the dorsal and ventral visual cortices 26, 28. In the most lateral part of the occipital white matter is the inferior longitudinal fasciculus (ilf) that connects the occipital lobe with the temporal lobe 3. In addition to these major white matter fibers, three small white matter regions showed clear boundaries with the tracts mentioned above (Fig. 4, green arrows-3). One is a local short association fiber of the callosomarginal sulcus (scm), which is located at the medial parietal and occipital transitional areas, while the other two (opd and opv) circumvent around the lateral V1 and disappear where V2 and the vof emerge. All the above white matter regions can be identified and separated in our dMRI data (Fig. 4A–D).
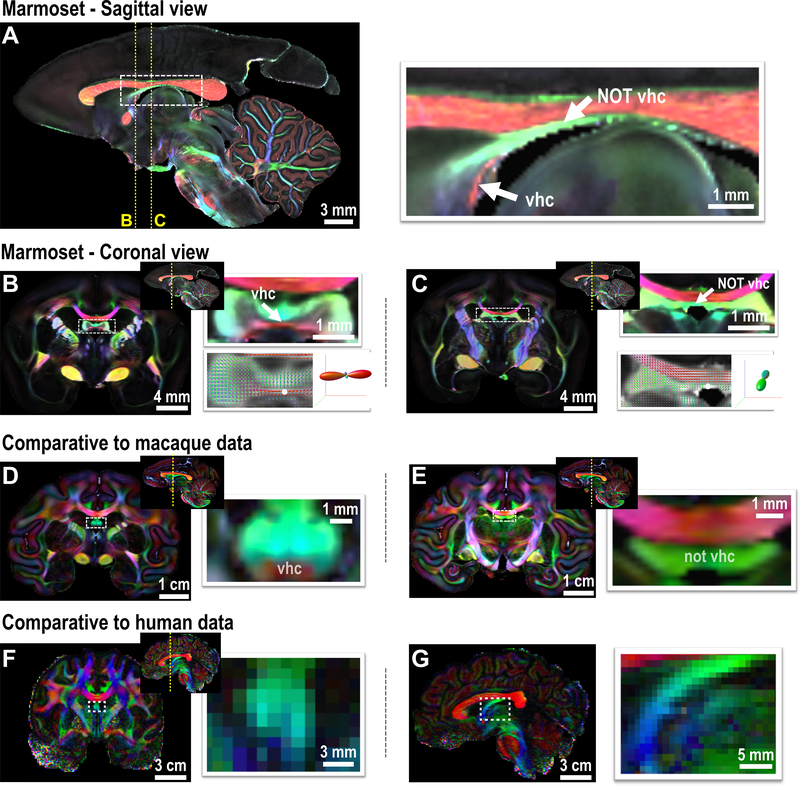
3). Ventral hippocampal commissure
The ventral hippocampal commissure (vhc) is an essential and primitive commissural fiber tract connecting the hippocampi of the two hemispheres. In rodents, the vhc is described as a prominent “mustache-shaped” transverse pathway in the coronal plane (Supplementary Fig. 4A). In primate atlases, including those of the marmoset, the vhc often points to a white matter tract running just beneath the corpus callosum (Supplementary Fig. 4B). As neuroanatomists charted these atlases without the benefit of either diffusion orientation information or conventional tract-tracing, they inferred the location of the primate vhc based on a similar gross appearance to the rodent vhc in coronal sections. We show here that this subcallosal structure is not, in fact, the vhc of the primate brain (Fig. 5) since the principal fiber orientation is anterior-posterior, rather than left-right. The structure usually labeled the vhc is the fornix, whereas the real vhc is much smaller and lies more ventrally near the ventral-most portion of the commissural plate.
Figure 5. The ventral hippocampal commissure (vhc) of primates.
(A) Middle sagittal slice of the FA-weighted DEC image of the marmoset brain (80 μm). White arrows highlight the vhc and the area mistakenly labeled as the vhc in most atlases. (B) A coronal slice of the marmoset that shows dominant left-right fiber orientation in the vhc. The bottom right is the fiber orientation distribution image for each voxel and overlaid on the FA image, and the fiber orientation of the voxel highlighted by the white dot is presented. (C) A coronal slice of the marmoset brain showing the non-vhc region (fornix area), in which fiber orientation is dominated by the anterior-posterior (DEC-green) direction. (D-E) The vhc and non-vhc white matter in the macaque brain, which shows a relatively smaller vhc. (F-G) The vhc of the human brain is too small to be identified by the in-vivo connectome data (1050 μm). A coronal image is shown in F, and a middle sagittal image is shown in G.
The disproportionately larger vhc in rodents compared to primates is a source of confusion. For example, in the mouse brain, the vhc is thicker than the corpus callosum, and the area of the vhc at the midsagittal slice is about 0.30 mm2. In primates, including humans, the vhc is much smaller than in rodents. For example, while the marmoset brain size is about 20x bigger in volume than the mouse brain size, the area of its vhc at the midsagittal slice is about 0.19mm2. Similarly, in the macaque, the size of the vhc is even smaller, considering the enlarged brain size, and in the 250 μm dataset, it appears to be a small spot underneath the fornix (Fig. 5D). In humans, the relative size of the vhc may be even smaller, and thus it cannot be detected in the in-vivo human connectome data (1050 μm, Fig. 5F,G), nor any other published dataset of humans. The difference in the relative size of the vhc highlights a significant evolutionary distinction in the interhemispheric connections of the hippocampus between rodents and primates. However, mainly due to the limitations of histological data, many existing primate atlases have failed to describe this difference and mislabeled the location of the ventral hippocampal commissure.
Fine-detailed white matter atlas and its applications
The examples above demonstrate the levels of detail of white matter structures of the marmoset brain that our dMRI dataset can identify. Based on the data, we digitally dissected and manually drew 82 white matter tracts on multi-modal MRI templates, which covered most of the white matter area of the marmoset brain, including both cortical and non-cortical white matter (Supplementary Table S1 for a complete list). The high-resolution DEC maps directly identified most major white matter tracts, but many required additional information, including estimation from probabilistic tractography and previous knowledge from neuronal tracing data, histological data, and comparative evidence. In the Methods section, we provide examples of how we combined different evidence to dissect the white matter.
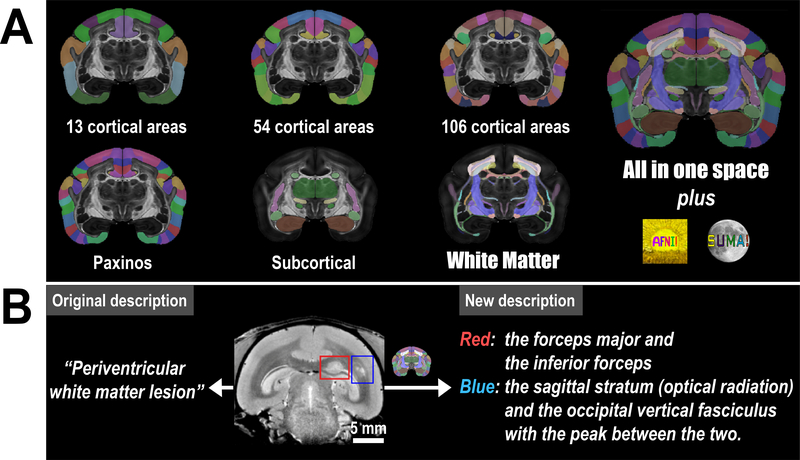
The white matter atlas is a product from the second stage (V2) of the MRI-based Marmoset Brain Mapping Project. In the previous stage (V1)16, we released a gray matter atlas that provided detailed cortical parcellations and a beta version of the subcortical atlas (Supplementary Table S2 for a complete list). We used the same brain sample to generate both our previous (V1) atlas16 and the current (V2) white matter atlas. To improve the usability of our new resource, we fused the V1 atlas into the new high-resolution MRI templates, and thus we have complete labeling of both gray and white matter in the V2 atlas (Fig. 6A).
Figure 6. Atlas version 2 and its applications in preclinical research.
(A) Version 2 of the Marmoset Brain Mapping Atlas includes not only the new white matter atlas but also atlases of different cortical parcellation and subcortical gray matter structures from version 116. All these atlases are provided in the same coordinate space at 80 μm and 50 μm with multi-modal MRI templates. These atlases and high-resolution templates are integrated into the AFNI software to provide a fully featured atlas utility. (B) An example from a marmoset disease model that demonstrates how our atlas can improve the localization of the white matter lesion. The original description is cited from29.
Similar to our previous V1 atlas16, the new V2 atlas also features multi-modal MRI templates, including multiple DTI contrasts, T2, T2*, and MTR templates to facilitate registration of our atlas to different imaging modalities such as MRI, PET, CT, histology, etc. For most MRI studies, our atlas allows users to select a template contrast that is matched with their MRI images to ensure accurate registration. As for histological images, for example, Nissl-stained images, the MTR template allows for a better registration than the other template modalities. The V2 atlas and associated MRI templates can be downloaded in NIFTI format from our website (marmosetbrainmapping.org/atlas.html) and implemented in the widely distributed neuroimaging data analysis software AFNI (AFNI command: @Install_NIH_marmoset). The following section presents demonstrative applications.
1). Localization of white matter regions
In a recently published preclinical study of a marmoset model of multiple sclerosis29, multiple white matter lesions were detected in the marmoset brain (Fig. 6B). Due to the lack of a white matter atlas of the marmoset brain, the study could only provide a general descriptions of the location of the lesions, such as “periventricular white matter lesions” by using the ventricles as landmarks. Now, with the availability of our atlas, we can accurately describe the two featured lesions: one involves the forceps major (or splenium) of the corpus callosum and the inferior forceps (red box in Fig. 6B), while the other is located between the sagittal stratum (or optical radiation) and the occipital vertical fasciculus (blue box in Fig. 6B). By combining the white matter atlas with our neuronal tracing database, we may infer which cortical regions are associated with the lesioned tracts, and predict the functional deficit caused by the lesions. This ability makes for a much more useful information than the original description of the white matter lesions.
2). White matter atlas guided tractography
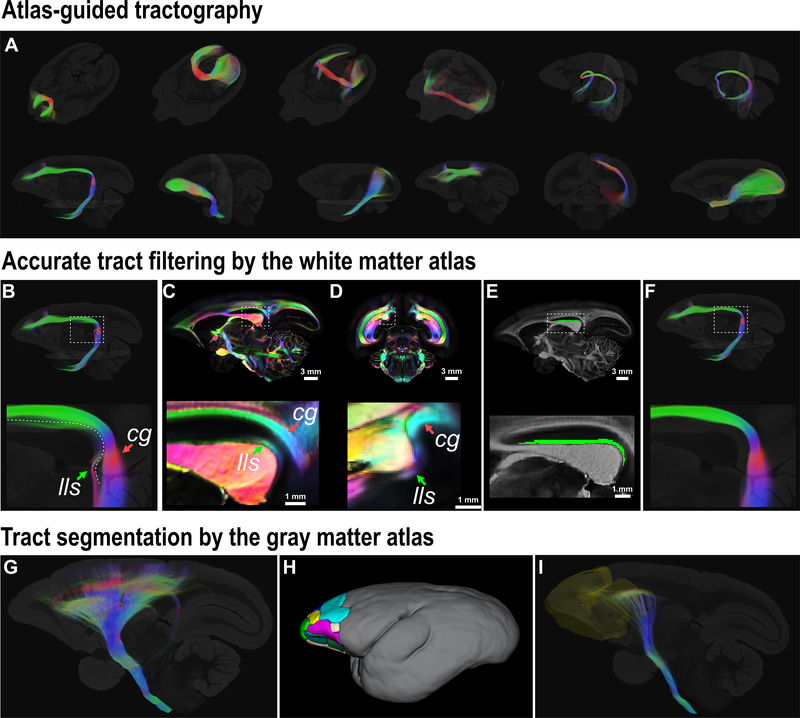
In diffusion tractography, prior knowledge of neuroanatomy and connections is routinely used to constrain tractography algorithms, e.g. by using white matter waypoint ROIs. The fully labeled 3D whole-brain white matter regions (ROIs) provide a versatile solution to constrain dMRI tractography (Fig. 7) and relieves operators from the tedious task of drawing ROIs.
Figure 7. Atlas-guided dMRI tractography.
(A) Several major pathways reconstructed by atlas-guided tractography are presented. From left-to-right and top-to-bottom are: the forceps minor; the forceps major; the inferior forceps; the anterior commissure; the circuit tract connecting hippocampus of two hemispheres via the vhc; the hippocampus-fornix-mammillary bodies circuit; the cingulum; the uncinate fasciculus; the inferior longitudinal fasciculus; the occipitofrontal fasciculus; the occipital vertical fasciculus; the optic tract - LGN - optic radiation circuit tract. (B-F) An example of accurate filtering by the atlas. (B) shows the reconstructed cingulum (cg) that is contaminated by the lateral longitudinal striae (lls); (C-D) shows the cg and lls on a sagittal slice and a coronal slice (80 μm), respectively. (E) shows the ROI of the lls from the atlas on a sagittal slice. (F) shows the filtered cg in which the lls is removed by using the atlas. (G-I) shows an example of a tract segmentation by the cortical atlas. (G) shows the reconstructed projection fibers via the internal capsule. (H) shows the frontal cortical regions of the cortical atlas (ROIs 1–16 of the MRI-based 54-area cortical parcellations in our atlas16, Supplementary Table 2). (I) shows the frontopontine fibers from the projection fibers in (G) by using the frontal cortical ROIs in (H) as the filtering mask.
Here, we use the reconstruction of the Cg as an example (Fig. 7B–F). The Cg is a major association fiber pathway of the limbic system. The main bundle of the Cg stretches from the medial frontal cortex, extends laterally along the cingulate gyrus (superior cingulum (Scg)), curves ventrally around the splenium and runs into the parahippocampal areas (parahippocampal cingulum). The cingulum is a well-known and exemplary tract in dMRI studies of human and animal models3, 8. However, most studies of the Cg failed to warn that a small white matter tract, called the lateral longitudinal striae (lls), can easily contaminate dMRI tractography of the Cg. The lls is one of the fiber tracts embedded in the supracallosal gyrus (or the indusium griseum), which is located underneath the cingulate gyrus, and thus, is spatially close to the Cg bundles. The lls, with a similar fiber orientation as the Cg, curves into the hippocampus around a location that is very close to where the Cg curves into medial temporal areas. As our atlas has the small white matter tracts labeled in a 3D manner, we can directly use the ROI of the lls as a filtering mask to separate the lls from the Cg.
In addition to ROIs from the white matter atlas itself, we can also apply gray matter ROIs to filter any segment tract of interest, such as the frontopontine fibers (Fig. 7G–I). For this example, we first reconstructed gross projection tracts by using the ROIs of the internal capsule, the cerebral peduncle, and the longitudinal pontine fasciculus as seeds and masks. We then picked all prefrontal and premotor ROIs from the cortical atlas to segment the frontopontine tract from the whole projection tracts. With a different combination of ROIs from our complete atlas, the atlas-guided dMRI tractography greatly facilitates dMRI and white matter studies of the marmoset brain.
3). Mapping anatomical connections with neuronal tracing data
Marmoset Brain Connectivity Atlas17 is the most comprehensive retrograde tracing database for marmoset cortical connections (Fig. 8A, left panel). Although the retrograde tracer provides accurate mapping of cortical connections by staining cell bodies, it cannot reveal white matter fiber-pathways connecting the regions. However, in many applications, we want to know and analyze the white matter underlying these connections, which is valuable information missing from the tracing database.
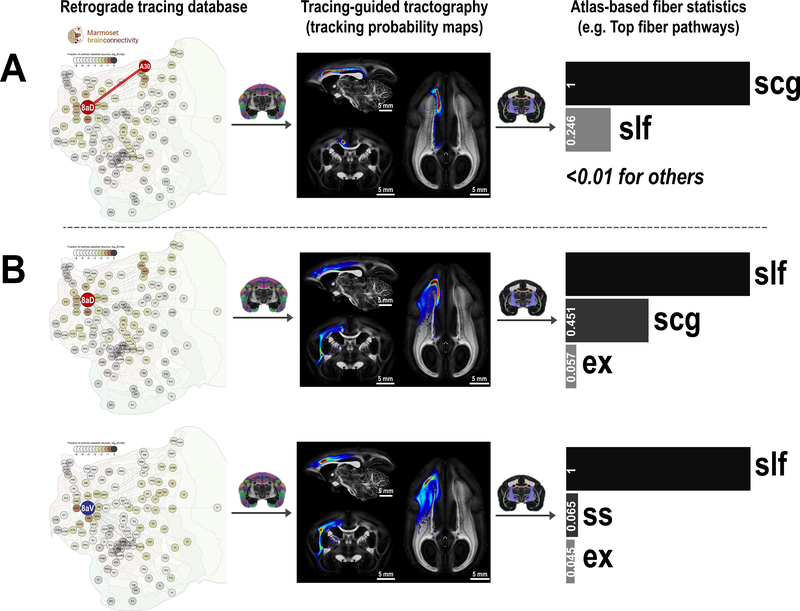
Figure 8. Fiber-pathway profiles of cortical connections and regions.
(A) Fiber pathway of a connection (e.g., the connection between A8aD and A30). The tracing database (Marmoset Brain Connectivity Atlas) provides information about which regions are connected (first column). Probabilistic tractography between two connected regions generates a tracking probability map (second column). By ranking the probability map with our white matter atlas, we estimate the fiber pathway supporting the connection (third column). (B) Fiber pathway profiles of a cortical region (A8aD and A8aV). All tracking maps of one region are averaged and then ranked by the white matter atlas to estimate the fiber pathway profiles of the region. All fiber profiles are scaled so that the top-ranking fiber pathway has a value of 1. slf: superior longitudinal fasciculus; scg: superior cingulum; ex: external capsule; ss: sagittal stratum.
Here, we combined high-resolution dMRI tractography with neuronal tracing data to fill the missing information. By performing probabilistic tractography between any two connected regions, we generated a tracking probability map as an estimation of the white matter pathways of the connection (Fig. 8A, middle panel). After obtaining the tracking probability maps, we calculated the sum of probability in each white matter ROIs from our atlas. We ranked the white matter ROIs based on their sum of probabilities, and the top ROIs represented the most probable fiber-pathways that support the selected connection. For example, the analysis revealed the superior cingulum is a major tract connecting area 8aD and A30 (Fig. 8A, right panel).
Additionally, we averaged all tracking probability maps associated with one specific cortical region to examine fiber pathways supporting one region. For example, this approach revealed that different fiber pathways support areas 8aD and 8aV (Fig. 8B). Area 8aD connects to the parietal and posterior areas of the brain using both a medial cortical pathway via the superior cingulum (scg) and a lateral pathway via the superior longitudinal fasciculus (slf), whereas area 8aV connects mainly via the slf. Altogether, these tracking probability maps form a comprehensive fiber-pathway mapping of marmoset cortical connections, which are downloadable and can be directly viewed online via our website (2dviewer.marmosetbrainmapping.org).
Discussion
There are three commonly used approaches to identify and map white matter fiber pathways: 1) histological methods to stain axons or myelin; 2) case-based neuronal tract-tracing methods; 3) dMRI tractography. Although the traditional histological staining methods can provide higher resolution than the dMRI, these methods utilize 2D sections of the brain, making it challenging to reconstruct complex white matter pathways in 3D. By revealing the real axons of fiber pathways, the neuronal tracing methods provide vital information about specific anatomical connections, but their case-based procedure is not efficient in mapping white matter pathways over the entire brain. With the value of preserving fiber orientation information in 3D and the intact mapping of the brain, the dMRI-based method employed here provides a unique opportunity to describe anatomical features of white matter pathways that were previously out of reach. Also, MRI contrasts can be directly used as templates for registration and normalization, which dramatically improves the usability of the brain atlases. Most importantly, the same MRI techniques can be used in both humans and animal models, and thus, MRI-based atlases have advantages in comparative and translational research 30.
One limitation of dMRI is the difficulty in achieving fine enough spatial resolution that allows mapping of small but important fiber pathways, and in dissecting complex white matter regions, such as the corona radiata. Hardware constraints set the limits for collecting ultra-high-resolution dMRI data in humans and NHPs with large brains that hardly fit into a high-field magnet with a strong gradient set. Here, we chose to image the small marmoset brain, which fit in ultra-high field strength vertical magnets equipped with stronger gradient sets and more sensitive RF coils. These technical advantages outweighed the disadvantages of the small size in terms of MRI data quality, which made the acquisition of our ultra-high-resolution dataset and the construction of the fine-detailed 3D white matter atlas possible.
Another limitation of the dMRI is the questionable accuracy of dMRI tractography in examining the anatomical connectivity of the brain19, 20. One approach to improve accuracy is resorting to prior knowledge from neuronal tracing data. In this aspect, the marmoset has the most comprehensive neuronal tracing databases among all primate species, including the Marmoset Brain Connectivity Atlas17 and the Marmoset Brain Architecture Project31. Taking advantage of one of these neuronal tracing databases, we used the tracing data to constrain dMRI tractography. Not only this approach improved the accuracy of our tractography, but it also provided a comprehensive map of the white matter fiber pathways connecting different cortical regions. However, we still need to be cautious when interpreting the results of dMRI tractography. DMRI tractography is a macroscale method, whereas neuronal tracing is a mesoscale method. The comparison of techniques at different scales hardly yields absolute agreement. Although our high-resolution resource may improve the accuracy of mapping fiber-pathways, it could not resolve the fundamental inherent ambiguities of dMRI tractography19, 20, 32, 33. In addition, our tracing-guided tractography used entire cortical areas as seeds, but white matter connections from different subregions within large cortical areas often take different routes to reach their target4, 34. One future direction will be to develop atlas-free maps of white matter fiber pathways by using each individual injection site as a seed for dMRI tractography, which will further improve their accuracy.
The small and smooth brain of the marmoset provides many technical advantages in mapping fiber pathways and studying brain connectivity. As demonstrated in this study, white matter bundles share common features across different primate species. These similarities were also the reason why we were able to identify many marmoset white matter fibers based on comparative evidence from macaques3. In addition to anatomical structures, recent studies have also revealed functional brain networks are largely preserved between marmosets and other primates, including humans11–14. The marmoset brain provides an easier way to describe and understand the fundamental brain architectures of the primate brain, as the core laminar organization of the white matter pathways is more apparent in the marmoset due to fewer influences from gyrification.
However, caution should be exercised when making direct inference about other primates, especially regarding those brain regions that highly evolved in humans12, 35, 36. Here, we demonstrated the resolution limitations of existing MRI data of macaques and humans, which requires higher resolution to fully describe their neuroanatomy features. Our scanning protocols of marmoset brain samples were a useful experience to design and optimize the scanning protocols for other primates. Together with recent development in MRI hardware and rapid imaging, such as the multi-band sequence and compressing-sensing techniques37, 38, acquiring ultra-high-resolution data of other primates will be feasible, and by then the marmoset data and atlases released here will be valuable comparative references to reveal common connectional and evolutional principles of the primate brains.
Methods
Data acquisition and preprocessing
1). Brain samples
All experiments were approved by the Animal Care and Use Committee of the National Institute of Neurological Disorders and Stroke. Formalin-fixed brain samples from two male common marmosets (Callithrix jacchus) were scanned for ultra-high-resolution MRI. One brain sample was from a 4.5-year-old marmoset, and the other was a 10.9-year-old. These samples were used in all procedures described here. Before MRI scanning, brain samples were soaked with 0.2% gadolinium (1 mmol/mL, Gadavist, Bayer) in 1x PBS for 2 weeks and then with 0.2% gadolinium in pure water for 1 day. The gadolinium effectively reduced the T1 relaxation time of tissues, and thus, reduced total acquisition time, and the final day pure-water protocol helped restore the T2 relaxation time by reducing formalin in the tissues. Customized 3D-printed brain holders were made to maintain the samples in a convenient orientation and reduce vibration during scanning. More information can be found in the Life Sciences Reporting Summary.
2). MRI data collection
One brain sample (4.5-year-old) was scanned on a 7T/30cm horizontal bore MRI spectrometer (Bruker Biospin) with a 30-mm ID quadrature millipede coil (ExtendMR, LLC, Milpitas, CA). This millipede coil was tailored for scanning adult marmoset brain samples. Its sensitivity was about 2.5 times better than a 35-mm linear birdcage coil from Bruker used in our previous publication16. The white matter atlas was built on data collected from the brain sample. Another brain sample (10.9-year-old) was scanned on a 14T/8.9cm vertical bore MRI spectrometer (Bruker Biospin) with a 20-mm linear millipede coil (m2m Imaging). The 14T microimager had a stronger magnetic field and gradient strength, but the bore size can only fit half marmoset brain. Thus, we acquired multi-shell dMRI data at 64 μm for one half of the brain (the right hemisphere).
On the 7T scanner, whole-brain multi-modal MRI data were collected, including 80 μm multi-shell dMRI, 80 μm MTR, 50 μm T2*-weighted and T2-weighted images:
The dMRI data were collected with an 3D diffusion-weighted multi-shot spin-echo EPI sequence: TR = 200 ms, TE = 29 ms, number of averages = 1, number of segments = 88, FOV = 38×29.76×29.76 mm, matrix size = 474×372×372, resolution = 80 μm isotropic, a total of 204 DWI images with three b values (8 b = 0, 6 b = 30, 64 b =2400, and 126 b = 4800), and the total acquisition time was about 15 days. Multi-shell gradient sampling schemes of the dMRI were generated by the IMOC method of DMRITool43.
The MTR image was collected with a 3D FLASH sequence: TR = 29.5 ms, TE = 4.4 ms, flip angle = 20o, FOV = 39×28×25.6 mm, matrix size = 487×350×320, resolution = 80 μm isotropic, number of averages = 2, with (Ms) and without (M0) an offset magnetization transfer (±2000 Hz off-resonance, Gaussian-shaped), acquisition time for each image is 2.5 hours. One MTR image was calculated from four images (two M0 and two Ms). We acquired five MTR images and averaged them into one final image (total acquisition time was 50 hours).
The T2*-w image was collected with a 3D FLASH sequence: TR = 50 ms, TE = 22 ms, flip angle = 80o, FOV = 39×28×25.6 mm, matrix size = 780×560×512, resolution = 50 μm isotropic, number of averages = 1, and acquisition time for one image was about 4h. We acquired ten images and averaged them into one (total acquisition time was 40 hours).
The T2-w image was collected with a 3D spin-echo sequence: TR = 250 ms, TE = 17.5 ms, FOV = 39×28×25.6 mm, matrix size = 780×560×512, resolution = 50 μm isotropic, number of averages = 1, and acquisition time for one image was about 20h. We acquired three images and averaged them into one (total acquisition time was 60 hours).
On the 14T scanner, half-brain multi-shell dMRI data were collected using a 3D diffusion-weighted multi-shot spin-echo EPI sequence: TR = 160 ms, TE = 23 ms, number of averages = 3, number of segments = 51, FOV = 25.6×17.4 ×13.3 mm, matrix size = 400×272×208, resolution = 64 μm isotropic, a total of 191 DWI images with three b values (1 b = 0, 64 b =2400, and 126 b = 4800), and the total acquisition time was about 11.3 days.
3). MRI data preprocessing
All data processing was performed on computing clusters of the High Performing Computation (HPC) systems at the National Institutes of Health (hpc.nih.gov). The dMRI data were denoised by the dwidenoise function implemented in MRtrix344 and eddy-current corrected by the eddy_correct function of FSL45. Besides, as a separate processing pipeline, the 80 μm dMRI data were also preprocessed using the DIFFPREP of TORTOISE, which allowed using T2-w image for EPI-distortion correction and DWI reorientation46. The preprocessed data were fitted with the diffusion tensor model (DTI)10, 47, 48 by the DTIFIT of FSL45 and the multi-shell multi-tissue constrained spherical deconvolution by MRtrix349, 50. These fittings generated fractional anisotropy (FA)10, and fiber orientation distribution (FOD)50 weighted MRI contrast with principle fiber direction information (DEC) that guided manual dissection of the white matter22, 51. Probabilistic tractography was conducted using the iFOD2 method of MRtrix349, which produced tracking probability maps that assisted delineating boundaries of several white matter tracts and estimated fiber-pathway profiles of cortical connections. These raw DWI data, preprocessed DWI data, and detailed preprocessing and tractography pipelines were provided in our website (“detailed info” pages of each dataset) (marmosetbrainmapping.org/data.html).
Statistics
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in our previous publication16. No statistical tests were used in the preparation of the atlas and analysis of the data obtained. Data distribution was assumed to be normal, but this was not formally tested. Data collection and analysis were not performed blind to the conditions of the experiments. No animals or data points were excluded for any reason.
White Matter Dissection
We manually labeled 82 ROIs on multi-model MRI templates at 80 μm isotropic, which covered most white matter area of the marmoset brain. Most major white matter could be identified in the DEC maps, but some required additional information. The following examples demonstrate the use of additional information to identify the white matter of interests.
1). Dissection based on DEC maps
The high spatial resolution (80 μm and 64 μm) and color-coded information of the DEC maps from our dMRI data provided essential fiber orientation contrasts to dissect complex white matter areas. The three examples presented in the Results section demonstrate the capability of the DEC to dissect the most complicated white matter areas of the primate brain.
For example, in the corona radiata (Fig. 2 and 3), DEC maps show “green-color” spots among branches of the dominating “purple-color” fibers. These spots are the superior longitudinal fasciculus that runs in an anterior-posterior direction (perpendicular to the coronal section), which was hard to identify in previous dMRI data and histological atlases. At the bottom of the corona radiata, a small branch (“red-color”) from the corpus callosum separates the FOF and the Muratoff bundle, both of which have a similar anterior-posterior direction (“green-color”). This branch was not visible in previous dMRI or histological data.
The primate occipital lobe is another brain region with a complex white matter structure (Fig. 4). Unlike the crossing-fiber structure of the corona radiata, the white matter of the occipital lobe clearly has a layered structure. Each layer represents different fiber-pathways with different fiber orientations, which are identifiable in the DEC maps. The high-resolution and high-contrast DEC maps also made it easier to identify small white matter tracts such as the ventral hippocampal commissure (vhc), which has been incorrectly labeled in many previous atlases (Fig. 5).
2). Dissection based on probabilistic tractography
DEC allowed us to identify and delineate tract boundaries directly on the images themselves. However, many adjacent structures may be branches from one vast white matter region and may not present a sharp boundary in DEC images. For example, the corpus callosum curves backward into the occipital lobe to form the forceps major (fmajor) and curve laterally into the temporal areas to form the tapetum (tap). Due to different fiber orientations, the two tracts show distinct colors in the DEC images (Fig. 4). However, since they are two different branches of the corpus callosum, they show a gradually-changing boundary that require an arbitrarily defined threshold to make white matter ROIs. To improve this arbitrary solution, we performed probabilistic tractography here to reconstruct the fmajor by using the visual cortices of the two hemispheres as the seeds and targets (inclusive mask). The tracking probability map of the fmajor provided a more precise contrast than the DEC to define the boundary between the fmajor and the tap arbitrarily.
We also used probabilistic tractography in other situations. The DEC maps provide local contrast, but many white matter tracts are defined based on the cortical regions they connect, not solely based on their local contrast. For example, the superior longitudinal fasciculus (slf), which connects the frontal area and the parietal area, merges into the dorsal part of the sagittal stratum (ss) in the occipital lobe. There is no clear boundary between the slf and the ss since both tracts run in the anterior-posterior direction (green in DEC map). In this case, we performed probabilistic tractography to reconstruct the slf by using the frontal cortical region as the seed, the parietal region as the target, and the occipital lobe as the exclusive mask. The probabilistic tractography maps thus provide a complementary contrast to guide and manually delineate the slf.
3). Dissection based on neuro-tracing data and comparative evidence
Both DEC and probabilistic tractography maps provide rich information and complementary contrasts to delineate white matter ROIs manually, but how to label these white matter ROIs anatomically poses another challenge. Despite most of the major white matter tracts being well known, some marmoset white matter tracts have few references available. Due to the homology of the white matter structures across primates, previous macaque studies provided useful comparative evidence that helped us label many white matter structures. For example, the FOF and the Muratoff bundle had been described in previous macaque tracing studies3. In marmosets, these two tracts matched their descriptions in macaques well. In addition to the comparative evidence, we also resorted to the current ongoing anterograde tracing database of the marmoset brain, the Riken Marmoset Brain Architecture Project31, to search for evidence for our labeling. For example, the FOF and Muratoff bundle were identifiable in tracing maps from anterograde AAV injections in the premotor regions and posterior parietal cortex (Supplementary Fig. S5).
Several white matter ROIs can be segmented into smaller sub-regions in different ways. Here, we segmented white matter ROIs only if (1) the sub-regions had a clear boundary in the contrasts by DEC maps or tracking probability maps; or (2) there was consensus about the segmentation. For example, one may choose to divide the corpus callosum into three (the genu, the body, and the splenium) or four segments (the rostrum, the genu, the body, and the splenium). Not only their boundaries are hard to identify in our data, but also there is no consensus about how to divide the corpus callosum. Thus, we kept the central part of the corpus callosum as a single ROI (unsegmented), to allow users to choose how to divide the corpus callosum.
Atlas Construction
All white matter ROIs were manually drawn directly on the 80 μm data, which provided multi-modal MRI templates, including multiple dMRI contrasts,T2, T2* and MTR images. The T2 and T2*-weighted images were down-sampled from 50 μm to 80 μm, and all MRI contrasts were co-registered into the same template space (80 μm). Additionally, we transferred all atlas labeling to the native 50 μm T2-w and T2*-w MRI templates as well. All atlas sets were constructed on the right hemisphere and mirrored to the left hemisphere.
In our previous study16, we released a gray matter atlas that provided detailed cortical parcellations and a beta version of the subcortical atlas. We manually delineated 54 cortical areas based on multi-modal MRI contrasts. The 54 areas were merged into 13 large cortical regions based on their spatial locations, and parcellated into 106 smaller cortical areas based on dMRI tractography based connectivity. In addition, we provided a parcellation in the previous atlas based on the Paxinos histological atlas41. We revealed about 25% disagreement among different published marmoset cortical atlases16. As there are no gold standards for cortical parcellations, we provided the different parcellations in our atlas and allowed the users to select the one they prefer to use16.
Our previous gray matter atlas16 and the current white matter atlas were generated from the same marmoset brain sample. To improve the usability of our new resource, we fused the previous gray matter atlas into the new high-resolution MRI templates using non-linear transformation by ANTs52 to account for sample distortions. The non-linear transformation was estimated based on the DTI-FA templates of two atlases. Thus, our new atlas (V2) has a complete set of labels for both gray and white matter. The V2 atlas described here can be automatically installed in the latest version of the software AFNI53 via the AFNI command @Install_NIH_Marmoset. Alternatively, it can be directly downloaded from our website (marmosetbrainmapping.org/atlas.html).
Atlas and Data Application
1). Application in diffusion tractography
All diffusion tracking was performed using the iFOD2 method of the software Mrtrix349. Reconstruction of different fiber-pathways was achieved via a combination of atlas ROIs for tracking seeds, inclusion, and/or exclusion masks (Fig. 7). For example, we used the following ROI combination to generate the tracking map of the cingulum (Fig. 7B). The ROIs of the superior cingulum and the parahippocampal cingulum were used as the seeds. Two coronal slices of the cingulum were used as the inclusive masks, one located at the anterior end of the superior cingulum and the other at the end of the parahippocampal cingulum. The superior longitudinal fasciculus and the lateral longitudinal striae were used as the exclusive masks. In another example (Fig. 7C), we combined ROIs of the internal capsule, the cerebral peduncle, and the longitudinal pontine fasciculus as the seeds. One coronal slice of the internal capsule and one slice at the end of the frontopontine tract were selected as the inclusive masks. The ROI combination guided the reconstruction of the whole projection fiber system via the internal capsule. The resulting projection tract can be segmented using the gray matter ROIs. For example, we used the frontal cortical ROIs as the filtering mask to segment the frontopontine tract from the whole projection tract. The detailed tracking command pipeline is described on our website (marmosetbrainmapping.org/80um.html#tracking).
2). Application in the analysis of neuronal tracing databases
The Marmoset Brain Connectivity Atlas has 143 cortical injections using retrograde tracers that provide a dense matrix of directional connections17. Since the tracers only labeled cell bodies, we did not know which white matter pathways support each pair of cortico-cortical connections. Here, we used our high-resolution diffusion tractography and the white matter atlas to estimate the missing information. For each pair of connected cortical regions in the tracing database, we conducted diffusion tractography by using one region as the seed and the other as the target, and vice-versa, to generate two sets of tracking probability maps. The two probability maps were averaged into a single map to represent the final map of the connection. We then calculated the sum of probabilities in each white matter ROI and ranked them according to their sum of probabilities. The top ROIs represented the most probable fiber-pathways that support the selected brain connection. Similarly, for each cortical region, all tracking maps of the region were averaged and then ranked by the white matter atlas to estimate the fiber pathway profiles of the region.
Data Availability
The resource presented here includes: (1) diffusion MRI (dMRI) data with the highest resolution available to date revealing white matter features never previously described; (2) a comprehensive 3D white matter atlas depicting fiber pathways that were either omitted or misidentified in previous atlases; and (3) comprehensive fiber-pathway maps of cortical connections combining dMRI tractography and neuronal tracing data. The raw data and all other features of the resource can be downloaded in NIFTI format from our website (marmosetbrainmapping.org).
Code Availability
The resource presented here includes code and analysis routines for each dataset that are available for downloading under the corresponding “Detailed Info” section in our website: https://marmosetbrainmapping.org/data.html. More information is available in the Supplementary Software.
Supplementary Material
Acknowledgments
We thank the Scientific and Statistical Computing Core of the NIMH Intramural Research Program for their support of the software AFNI, and the NIH Fellows Editorial Board and NIH library for language-editing services. We also thank Joe Guy for providing optimized T2* sequence. This work utilized the computational resources of the NIH HPC Biowulf cluster (hpc.nih.gov). This research was supported by the Intramural Research Program of the NIH, NINDS (ZIA NS003041), including the Neurophysiology Imaging Facility Core (NIMH, NINDS, NEI, ZIC MH002899). The neuronal tracing data is from the Australian Research Council (DP110101200, DP140101968, CE140100007) and International Neuroinformatics Coordinating Facility Seed Funding Grant.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Van Essen DC & Glasser MF Parcellating Cerebral Cortex: How Invasive Animal Studies Inform Noninvasive Mapmaking in Humans. Neuron 99, 640–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields RD White matter in learning, cognition and psychiatric disorders. Trends Neurosci 31, 361–370 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmahmann J & Pandya D Fiber pathways of the brain (OUP; USA, 2009). [Google Scholar]
- 4.Lehman JF, Greenberg BD, McIntyre CC, Rasmussen SA & Haber SN Rules ventral prefrontal cortical axons use to reach their targets: implications for diffusion tensor imaging tractography and deep brain stimulation for psychiatric illness. J Neurosci 31, 10392–10402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safadi Z, et al. Functional Segmentation of the Anterior Limb of the Internal Capsule: Linking White Matter Abnormalities to Specific Connections. J Neurosci 38, 2106–2117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabrese E, et al. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. NeuroImage 117, 408–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakszewski E, Adluru N, Tromp DP, Kalin N & Alexander AL A diffusion-tensor-based white matter atlas for rhesus macaques. PloS one 9, e107398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori S, Wakana S, Van Zijl PC & Nagae-Poetscher L MRI atlas of human white matter (Elsevier, 2005). [DOI] [PubMed] [Google Scholar]
- 9.Alexander AL, Lee JE, Lazar M & Field AS Diffusion tensor imaging of the brain. Neurotherapeutics 4, 316–329 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basser PJ, Pajevic S, Pierpaoli C, Duda J & Aldroubi A In vivo fiber tractography using DT‐MRI data. Magnetic resonance in medicine 44, 625–632 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Belcher AM, et al. Large-scale brain networks in the awake, truly resting marmoset monkey. J Neurosci 33, 16796–16804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, et al. Anatomical and functional investigation of the marmoset default mode network. Nat Commun 10, 1975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghahremani M, Hutchison RM, Menon RS & Everling S Frontoparietal functional connectivity in the common marmoset. Cerebral Cortex 27, 3890–3905 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Buckner RL & Margulies DS Macroscale cortical organization and a default-like apex transmodal network in the marmoset monkey. Nature Communications 10, 1976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva AC Anatomical and functional neuroimaging in awake, behaving marmosets. Dev Neurobiol 77, 373–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, et al. A digital 3D atlas of the marmoset brain based on multi-modal MRI. Neuroimage 169, 106–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majka P, et al. Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template. Journal of Comparative Neurology 524, 2161–2181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caravan P Strategies for increasing the sensitivity of gadolinium based MRI contrast agents. Chem Soc Rev 35, 512–523 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Thomas C, et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proceedings of the National Academy of Sciences 111, 16574–16579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reveley C, et al. Superficial white matter fiber systems impede detection of long-range cortical connections in diffusion MR tractography. Proc Natl Acad Sci U S A 112, E2820–2828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu AT, et al. High resolution whole brain diffusion imaging at 7T for the Human Connectome Project. Neuroimage 122, 318–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pajevic S & Pierpaoli C Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn Reson Med 42, 526–540 (1999). [PubMed] [Google Scholar]
- 23.Forkel SJ, Mahmood S, Vergani F & Catani M The white matter of the human cerebrum: part I The occipital lobe by Heinrich Sachs. Cortex 62, 182–202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dejerine J & Dejerine-Klumpke A Anatomie des centres nerveux (Rueff, 1895). [Google Scholar]
- 25.Takemura H, et al. Occipital white matter tracts in human and macaque. Cerebral Cortex 27, 3346–3359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeatman JD, et al. The vertical occipital fasciculus: a century of controversy resolved by in vivo measurements. Proc Natl Acad Sci U S A 111, E5214–5223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu HH, Chaplin TA & Rosa MG Representation of central and peripheral vision in the primate cerebral cortex: Insights from studies of the marmoset brain. Neurosci Res 93, 47–61 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Jeffs J, Ichida JM, Federer F & Angelucci A Anatomical evidence for classical and extra-classical receptive field completion across the discontinuous horizontal meridian representation of primate area V2. Cereb Cortex 19, 963–981 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee NJ, et al. Spatiotemporal distribution of fibrinogen in marmoset and human inflammatory demyelination. Brain 141, 1637–1649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Schotten MT, Croxson PL & Mars RB Large-scale comparative neuroimaging: Where are we and what do we need? Cortex (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin MK, et al. A high-throughput neurohistological pipeline for brain-wide mesoscale connectivity mapping of the common marmoset. Elife 8, e40042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maier-Hein KH, et al. The challenge of mapping the human connectome based on diffusion tractography. Nat Commun 8, 1349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jbabdi S, Sotiropoulos SN, Haber SN, Van Essen DC & Behrens TE Measuring macroscopic brain connections in vivo. Nat Neurosci 18, 1546–1555 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Petrides M & Pandya DN Efferent association pathways from the rostral prefrontal cortex in the macaque monkey. J Neurosci 27, 11573–11586 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaplin TA, Yu HH, Soares JG, Gattass R & Rosa MG A conserved pattern of differential expansion of cortical areas in simian primates. J Neurosci 33, 15120–15125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneve MH, et al. High-Expanding Regions in Primate Cortical Brain Evolution Support Supramodal Cognitive Flexibility. Cereb Cortex (2018). [DOI] [PubMed] [Google Scholar]
- 37.Setsompop K, et al. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage 63, 569–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang N, et al. Whole mouse brain structural connectomics using magnetic resonance histology. Brain Struct Funct 223, 4323–4335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seidlitz J, et al. A population MRI brain template and analysis tools for the macaque. Neuroimage 170, 121–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonov V, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54, 313–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paxinos G, Watson C, Petrides M, Rosa M & Tokuno H The marmoset brain in stereotaxic coordinates (Elsevier Academic Press, 2012). [Google Scholar]
- 42.Hardman CD & Ashwell KW Stereotaxic and chemoarchitectural atlas of the brain of the common marmoset (Callithrix jacchus) (CRC Press, 2012). [Google Scholar]
Methods-Only References
- 43.Cheng J, Shen D, Yap P-T & Basser PJ Single-and Multiple-Shell Uniform Sampling Schemes for Diffusion MRI Using Spherical Codes. IEEE transactions on medical imaging 37, 185–199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veraart J, et al. Denoising of diffusion MRI using random matrix theory. NeuroImage 142, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW & Smith SM Fsl. Neuroimage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Pierpaoli C, et al. TORTOISE: an integrated software package for processing of diffusion MRI data in 18th Scientific Meeting of the International Society for Magnetic Resonance in Medicine 1597 (2010). [Google Scholar]
- 47.Basser PJ & Pierpaoli C Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B 111, 209–219 (1996). [DOI] [PubMed] [Google Scholar]
- 48.Basser PJ, Mattiello J & LeBihan D MR diffusion tensor spectroscopy and imaging. Biophys J 66, 259–267 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tournier JD, Calamante F & Connelly A MRtrix: diffusion tractography in crossing fiber regions. International Journal of Imaging Systems 22, 53–66 (2012). [Google Scholar]
- 50.Jeurissen B, Tournier JD, Dhollander T, Connelly A & Sijbers J Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411–426 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Dhollander T, Smith RE, Tournier J-D, Jeurissen B & Connelly A Time to move on: an FOD-based DEC map to replace DTI’s trademark DEC FA. NeuroImage 59, 3976–3994 (2012).22036682 [Google Scholar]
- 52.Avants BB, Tustison N & Song G Advanced normalization tools (ANTS). Insight j 2, 1–35 (2009). [Google Scholar]
- 53.Cox RW AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29, 162–173 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The resource presented here includes: (1) diffusion MRI (dMRI) data with the highest resolution available to date revealing white matter features never previously described; (2) a comprehensive 3D white matter atlas depicting fiber pathways that were either omitted or misidentified in previous atlases; and (3) comprehensive fiber-pathway maps of cortical connections combining dMRI tractography and neuronal tracing data. The raw data and all other features of the resource can be downloaded in NIFTI format from our website (marmosetbrainmapping.org).