Significance
Bardet-Biedl syndrome (BBS) is a rare human ciliopathy associated with blindness, obesity, and kidney anomalies. It is caused by defects in the assembly or transport of the BBSome, a conserved octamer of BBS proteins, which assists intraflagellar transport (IFT) with the removal of signaling proteins from cilia. Here, we show that the GTPase component of IFT-B1, IFT22, autonomous of IFT-B1 association, binds and stabilizes BBS3 in the cell body. When both are in their GTP-bound state, IFT22 and BBS3 recruit the BBSome to the basal body for loading onto the IFT machinery for ciliary entry. IFT22 recruits the BBSome to the basal body through BBS3 in a nucleotide-dependent manner providing a regulatory mechanism for BBSome entry into the cilium.
Keywords: IFT22, BBS3, BBSome, basal body, cilia
Abstract
Bardet-Biedl syndrome (BBS) is a ciliopathy caused by defects in the assembly or distribution of the BBSome, a conserved protein complex. The BBSome cycles via intraflagellar transport (IFT) through cilia to transport signaling proteins. How the BBSome is recruited to the basal body for binding to IFT trains for ciliary entry remains unknown. Here, we show that the Rab-like 5 GTPase IFT22 regulates basal body targeting of the BBSome in Chlamydomonas reinhardtii. Our functional, biochemical and single particle in vivo imaging assays show that IFT22 is an active GTPase with low intrinsic GTPase activity. IFT22 is part of the IFT-B1 subcomplex but is not required for ciliary assembly. Independent of its association to IFT-B1, IFT22 binds and stabilizes the Arf-like 6 GTPase BBS3, a BBS protein that is not part of the BBSome. IFT22/BBS3 associates with the BBSome through an interaction between BBS3 and the BBSome. When both IFT22 and BBS3 are in their guanosine triphosphate (GTP)-bound states they recruit the BBSome to the basal body for coupling with the IFT-B1 subcomplex. The GTP-bound BBS3 likely remains to be associated with the BBSome upon ciliary entry. In contrast, IFT22 is not required for the transport of BBSomes in cilia, indicating that the BBSome is transferred from IFT22 to the IFT trains at the ciliary base. In summary, our data propose that nucleotide-dependent recruitment of the BBSome to the basal body by IFT22 regulates BBSome entry into cilia.
The cilium is composed of a microtubule axoneme surrounded by a specialized plasma membrane (termed the ciliary membrane) and acts as a cellular signaling hub for coordination of cell physiology and extracellular stimuli through ciliary signaling proteins (1, 2). Ciliary signaling proteins include G protein-coupled receptors (GPCRs) (3–7), ion channels (8), as well as other classes of signaling proteins (6, 9). These proteins are translocated from the cell membrane to ciliary membrane or vice versa by molecular motor protein-driven intraflagellar transport (IFT) trains along the axoneme (10–12). During this process, the BBSome, an octameric complex of eight conserved BBS protein (BBS1/2/4/5/7/8/9/18) (13–16), acts as cargo-specific IFT adaptors to couple the ciliary signaling proteins to IFT trains. Therefore, defects in the composition and assembly of the BBSome and factors required for the IFT-dependent cycling of the BBSome through cilia by IFT trains result in the loss and/or abnormal accumulation of ciliary signaling proteins (17–19). This loss or accumulation eventually results in Bardet-Biedl syndrome (BBS), a human ciliopathy characterized principally by obesity, polydactyly, retinal dystrophy, and renal failure in some cases (1).
IFT trains are composed of repeating units of two multiprotein complexes called IFT-A (6 subunits) and IFT-B (16 subunits) (20–22), which are conserved among ciliated species (23). IFT-B is further divided into stable IFT-B1 and IFT-B2 subcomplexes (24–26). IFT-driven ciliary cycling of the BBSome can be dissected into four steps, including assembly at the basal body, ciliary entry (from the base to the tip, anterograde traffic), turnaround at the ciliary tip, and ciliary removal (from the tip to the base, retrograde traffic) (27–30). Fragmental evidence has identified small GTPases as critical mediators in different ciliary cycling steps of the BBSome. In the ciliary entry step, the Arf-like 6 (ARL6) GTPase BBS3 was reported, in its guanosine triphosphate (GTP)-bound conformation, to bind the N-terminal β-propeller domain of the BBSome subunit BBS1 (31) and subsequently target the BBSome to cilia (32). In the turnaround step, the BBSome is likely to be released from IFT trains to load cargo proteins first and is then reloaded to IFT trains for its ciliary exit (19). Our evidence and others’ identified the IFT-B1 proteins IFT25 and the Rab-like 4 (RABL4) GTPase IFT27 are involved in mediating ciliary exit of the BBSome (18, 19, 33, 34), probably by activating BBS3 at the ciliary tip (19). Other than those, IFT and BBS proteins are present in a pool near the basal bodies (16, 35). Since anterograde IFT trains are assembled at the basal bodies, recruitment of the BBSome to the basal bodies is a likely prerequisite for BBSome loading onto IFT trains and entry into cilia (36). However, how the BBSome is recruited to the basal body has not been established.
IFT22 is a Rab-like 5 (RABL5) GTPase and is conserved across ciliated organisms (25, 26, 37–40). In this study, we explored the interplay among IFT22, BBS3, and the BBSome at a molecular level by using a combination of functional, biochemical, and single particle in vivo imaging assays on Chlamydomonas reinhardtii. We show that IFT22 recruits the BBSome through BBS3 to the basal body for entry into cilia via IFT. IFT22 is also a subunit of the IFT-B1 subcomplex but its loss does not impair ciliary assembly.
Results
IFT22 Is an IFT-B1 Component with Low Intrinsic GTPase Activity.
Unlike most Rab proteins, the Rab-like 5 GTPase IFT22 lacks G4, one of five consensus domains (G1–G5) thought to be required for GTP/GDP binding and GTPase activity (SI Appendix, Fig. S1A). However, similar to other Rab-like GTPases, IFT22 possesses the conserved P-loop threonine residue (T19) in its G1 domain, which aids in GTP binding; and the conserved glutamine (Q69) at the GTPase active site (G3 domain) essential for GTP hydrolysis (SI Appendix, Fig. S1A) (41). To test whether IFT22 is an active GTPase, we preloaded bacterial-expressed IFT22 with a C-terminal HA/6×His fusion tag (IFT22::HA::6×His) with GTP and used a colorimetric assay to measure the release of inorganic phosphate. This assay detected low intrinsic GTPase activity of recombinant IFT22, indicating that C. reinhardtii IFT22 is an active GTPase with low intrinsic activity (SI Appendix, Fig. S1B).
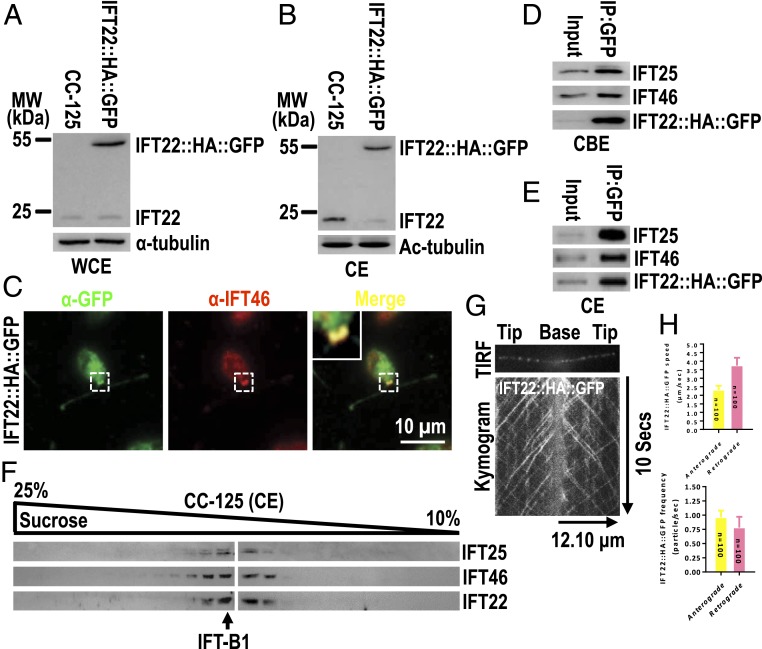
To determine the subcellular localization and dynamics of IFT22, we expressed IFT22 fused at its C terminus to a hemagglutinin (HA) tag and green fluorescent protein (GFP) (IFT22::HA::GFP) in wild-type (WT) C. reinhardtii cells (CC-125) (Fig. 1A). Expression of IFT22::HA::GFP strongly reduced the abundance of endogenous IFT22 in cilia, suggesting that tagged IFT22 outcompetes the endogenous IFT22 in ciliary entry (Fig. 1B). IFT22::HA::GFP colocalized with the IFT-B1 subunit IFT46 at the basal bodies and in a spotted pattern along the length of cilia (Fig. 1C); the IFT-B1 subunits IFT25 and IFT46 immunoprecipitated with the tagged IFT22 from cell body and ciliary extracts of IFT22::HA::GFP cells (Fig. 1 D and E); the endogenous IFT22 cosedimented with IFT25 and IFT46 in sucrose density gradients of ciliary extracts of CC-125 cells (Fig. 1F). Finally, total internal reflection fluorescence (TIRF) imaging identified IFT22::HA::GFP moving bidirectionally in cilia at velocities and frequencies characteristic for Chlamydomonas IFT (Fig. 1 G and H and Movie S1) (33, 42, 43). Thus, C. reinhardtii IFT22 is an IFT-B1 component and is stably bound to IFT while cycling through cilia.
Fig. 1.
IFT22 is an IFT-B1 component. (A and B) Immunoblots of whole cell extracts (WCE) (A) and ciliary extracts (CE) (B) of CC-125 and the IFT22::HA::GFP cells probed with α-IFT22. α-Tubulin and acetylated-tubulin (Ac-tubulin) were used to adjust the loading for WCE and CE, respectively. MW, molecular weight. (C) IFT22::HA::GFP cells stained with α-GFP (green) and α-IFT46 (red). Inset shows the basal bodies. (Scale bar: 10 µm.) (D and E) Immunoblots of α-GFP captured proteins from cell body extracts (CBE) (D) and CE (E) of IFT22::HA::GFP cells probed with α-IFT25, α-IFT46, and α-HA. (F) Immunoblots of CE of CC-125 cells separated by sucrose density gradient centrifugation probed for IFT25, IFT46, and IFT22. (G) TIRF image and corresponding kymogram of IFT22::HA::GFP cells. In the kymogram, the time and transport length are indicated on the Right and the Bottom, respectively. (H) The graph lists speeds of IFT22::HA::GFP in cilia of the IFT22::HA::GFP cells to be 2.31 ± 0.25 and 3.74 ± 0.46 μm/s for anterograde and retrograde, respectively, and their frequencies to be 0.96 ± 0.12 and 0.78 ± 0.19 particle/s for anterograde and retrograde, respectively.
Nucleotide State Controls IFT22 Basal Body Recruitment and Binding to IFT-B1.
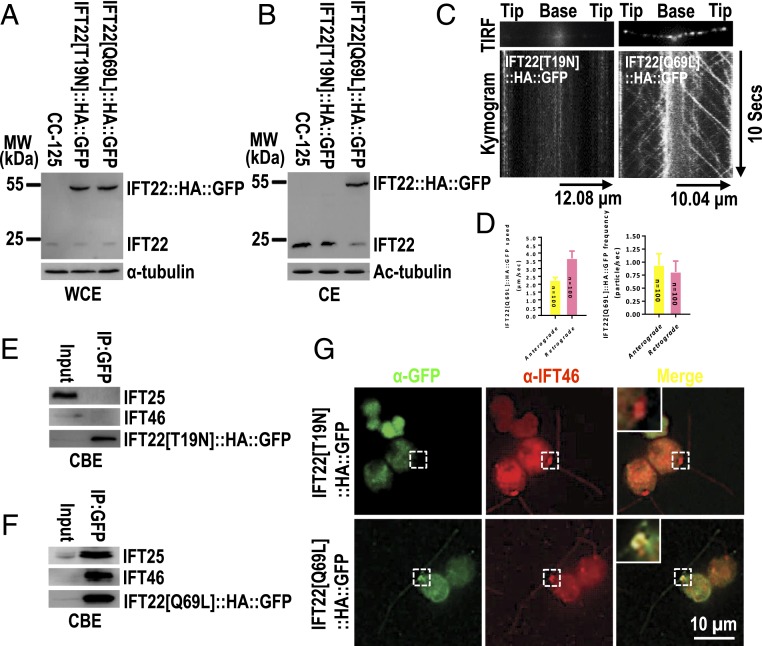
To “lock” IFT22 in either the GDP-bound (inactive) or GTP-bound (active) state, we introduced point mutations into IFT22 that are predicted to either disrupt GTP binding while allowing limited GDP binding (IFT22[T19N]) or prevent GTP hydrolysis (IFT22[Q69L]) (SI Appendix, Fig. S1A) (39, 41). GTP hydrolysis of the recombinant IFT22 was accelerated in the presence of cell body extracts while ciliary extracts had no effect (SI Appendix, Fig. S1B). As expected, IFT22[Q69L] had minimal GTPase activity, even in the presence of cell body extracts, suggesting that IFT22[Q69L] is a constitutive-active variant (SI Appendix, Fig. S1C). IFT22[T19N] did not show any GTPase activity, indicating that IFT22[T19N] is an inactive GDP-bound variant or simply a dominant-negative variant (SI Appendix, Fig. S1C). With these tools in hand, we tested how the GTP state of IFT22 as simulated by the altered constructs affects the distribution of the protein. When expressed in similar amounts in control cells, IFT22[Q69L]::HA::GFP entered cilia while IFT22[T19N]::HA::GFP failed to enter cilia (Fig. 2 A and B and SI Appendix, Fig. S2A). TIRF imaging showed bidirectional IFT of IFT22[Q69L]::HA::GFP in cilia with velocities and frequencies similar to those of IFT22::HA::GFP, whereas no signal was detected for IFT22[T19N]::HA::GFP (Figs. 1 G and H and 2 C and D and Movie S2). Further, IFT22[Q69L]::HA::GFP immunoprecipitated and cosedimented in sucrose density gradients of ciliary extracts with IFT25 and IFT46 (SI Appendix, Fig. S2 B and C). We conclude that IFT22[Q69L] is stably bound to IFT-B1 while cycling through cilia. To test if this association is already established in the cell body, we used immunoprecipitation from cell body extracts. IFT25 and IFT46 coimmunoprecipitated with IFT22[Q69L]::HA::GFP but not IFT22[T19N]::HA::GFP, revealing that IFT22[Q69L] but not IFT22[T19N] binds to IFT-B1 already in the cell body (Fig. 2 E and F). Staining with anti-GFP revealed that IFT22[Q69L]::HA::GFP colocalized with IFT46 at the basal bodies while IFT22[T19N]::HA::GFP was absent from the IFT pool at the basal bodies (Fig. 2G). Thus, we conclude that the nucleotide state of IFT22 controls its recruitment to the basal body and its association with IFT-B1.
Fig. 2.
Nucleotide state controls IFT22 basal body recruitment and binding to IFT-B1. (A and B) Immunoblots of WCE (A) and CE (B) of CC-125, IFT22[T19N]::HA::GFP and IFT22[Q69L]::HA::GFP cells probed with α-IFT22. α-Tubulin and Ac-tubulin were used to adjust the loading for WCE and CE, respectively. MW, molecular weight. (C) TIRF images and corresponding kymograms of IFT22[T19N]::HA::GFP and IFT22[Q69L]::HA::GFP cells. In the kymograms, the time and transport length are indicated on the Right and on the Bottom, respectively. (D) The graph lists speeds of IFT22[Q69L]::HA::GFP in cilia to be 2.25 ± 0.18 and 3.65 ± 0.47 μm/s for anterograde and retrograde, respectively, and their frequencies to be 0.94 ± 0.22 and 0.81 ± 0.21 particle/s for anterograde and retrograde, respectively. (E and F) Immunoblots of α-GFP captured proteins from CBE of IFT22[T19N]::HA::GFP (E) and IFT22[Q69L]::HA::GFP cells (F) probed with α-IFT25, α-IFT46, and α-HA. (G) IFT22[T19N]::HA::GFP and IFT22[Q69L]::HA::GFP cells stained with α-GFP (green) and α-IFT46 (red). Inset shows the basal bodies. (Scale bar: 10 µm.)
GTP-Bound IFT22 Is Required for Recruiting the BBSome to the Basal Body.
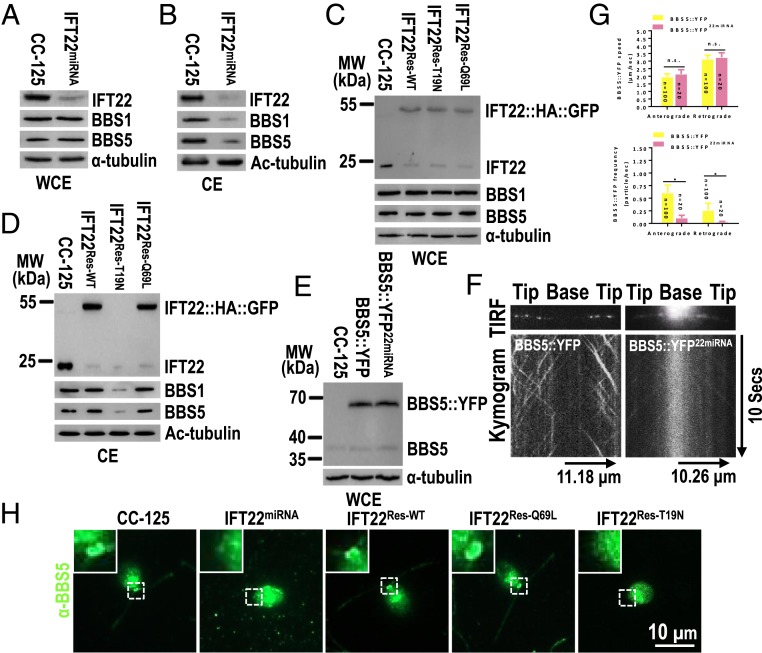
To elucidate the cellular function of IFT22, we used vector-based miRNA to deplete endogenous IFT22 to ∼8% of wild-type level; we refer to this strain as IFT22miRNA (Fig. 3A). Reflecting its cellular reduction, IFT22 was strongly reduced to ∼11% of wild-type level in IFT22miRNA cilia (Fig. 3B). However, IFT22miRNA cells possessed cilia of normal length, suggesting that IFT22 is not essential for ciliary assembly (SI Appendix, Fig. S3A). To analyze the role of IFT22 within the IFT pathway, we surveyed the presence of IFT and BBS proteins in cilia and observed normal levels of the IFT proteins IFT25, IFT46, IFT57 (IFT-B), and IFT43 and IFT139 (IFT-A) (SI Appendix, Fig. S3B) but a marked reduction of the BBSome subunits BBS1 and BBS5 (∼23% of control levels; Fig. 3B). Whole cell extracts of IFT22miRNA showed normal levels of both of those IFT and BBS proteins, revealing that the ciliary presence of the BBSome but not IFT train requires IFT22 (Fig. 3A and SI Appendix, Fig. S3C). To test whether the GTP state of IFT22 is critical for the ciliary presence of the BBSome, we expressed IFT22::HA::GFP, IFT22[Q69L]::HA::GFP, and IFT22[T19N]::HA::GFP in similar amounts in IFT22miRNA cells (resulting strains IFT22Res-WT, IFT22Res-Q69L, and IFT22Res-T19N) (Fig. 3C). All three strains had near normal levels of BBS1 and BBS5 in whole cell samples (Fig. 3C). IFT22::HA::GFP and IFT22[Q69L] but not IFT22[T19N] were present in cilia and restored the abundance of BBS1 and BBS5 in cilia to wild-type levels (Fig. 3D). Thus, the GTP-bound IFT22 is required to maintain normal ciliary level of the BBSome.
Fig. 3.
GTP-bound IFT22 is required for recruiting the BBSome to the basal body. (A and B) Immunoblots of WCE (A) and CE (B) of CC-125 and IFT22miRNA cells probed for IFT22, BBS1, and BBS5. (C and D) Immunoblots of WCE (C) and CE (D) of CC-125, IFT22Res-WT, IFT22Res-T19N, and IFT22Res-Q69L cells probed with α-IFT22, α-BBS1, and α-BBS5. (E) Immunoblots of WCE of CC-125, BBS5::YFP, and BBS5::YFP22miRNA cells probed with α-BBS5. MW, molecular weight. (F) TIRF images and corresponding kymograms of BBS5::YFP and BBS5::YFP22miRNA cells. In the kymograms, the time and transport lengths are indicated on the Right and the Bottom, respectively. (G) In the graph, the speeds of the BBS5::YFP in cilia of the BBS5::YFP cells were 1.96 ± 0.20 and 3.14 ± 0.26 μm/s for anterograde and retrograde, respectively, and their frequencies to be 0.61 ± 0.15 and 0.26 ± 0.14 particle/s for anterograde and retrograde, respectively. BBS5::YFP traffics at 2.15 ± 0.28 and 3.25 ± 0.30 μm/s for anterograde and retrograde, respectively, in BBS5::YFP22miRNA cilia, while its frequencies were dramatically decreased in both anterograde (0.11± 0.05 particles/s) and retrograde (0.03 ± 0.01 particle/s) directions as compared to that in BBS5::YFP cilia. (H) CC-125, IFT22miRNA, IFT22Res-WT, IFT22Res-Q69L, and IFT22Res-T19N cells stained with α-BBS5 (green). Inset shows the basal bodies. (Scale bar: 10 µm.) For A–E, α-tubulin and Ac-tubulin were used to adjust the loading for WCE and CE, respectively.
To determine whether the reduction of the BBSome in cilia is caused by its decreased ciliary entry, increased ciliary exit, or both, we expressed BBS5 fused at its C terminus to yellow fluorescent protein (YFP) (BBS5::YFP) in both control and IFT22miRNA cells (resulting strains BBS5::YFP and BBS5::YFP22miRNA) (Fig. 3E). Anterograde and retrograde IFT of BBS5::YFP were frequently observed in the control cells but were dramatically reduced in the IFT22miRNA cells; the velocity of BBS5::YFP transport was similar in both strains (Fig. 3 F and G and Movies S3 and S4) (9, 16, 33). Since similar amounts of tagged BBS5 were present in both strains (Fig. 3E), we conclude that IFT22 knockdown affects the efficiency of BBSome entry into cilia.
In C. reinhardtii, BBS proteins accumulate near the basal bodies (16) and we wondered if IFT22 knockdown affects this pool. In control cells, immunostaining with anti-BBS5 showed an accumulation of the endogenous BBS5 at the basal bodies while the signal was diminished in the IFT22miRNA cells (Fig. 3H). The BBS5 signal at the basal bodies was restored to wild-type levels in the IFT22Res-WT and IFT22Res-Q69L rescue cells but not in the IFT22Res-T19N strain (Fig. 3H). Therefore, we conclude that IFT22 in its GTP-bound form recruits the BBSome to the basal body; in IFT22miRNA cells, recruiting fails, largely impairing BBSome entry into cilia.
IFT22 Stabilizes BBS3 and Is Required for Recruiting BBS3 to the Basal Body.
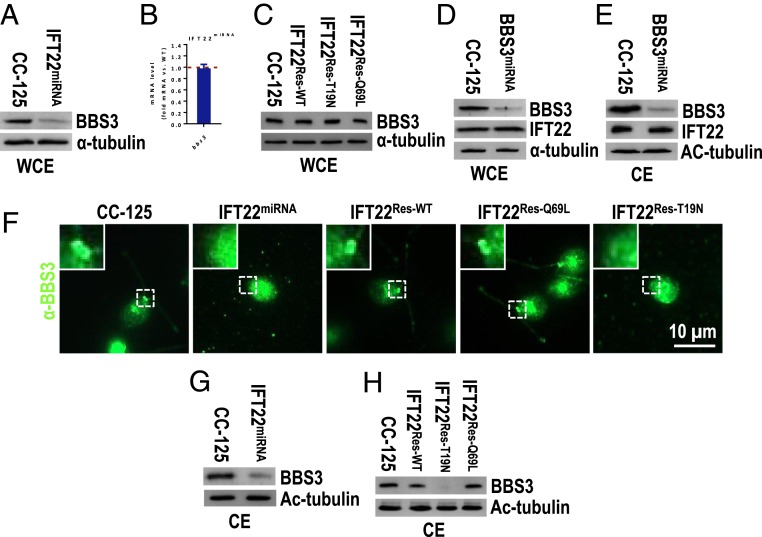
In vertebrates, the BBSome targeting to cilia depends on BBS3 (32, 44). To gain insights into the interplay between IFT22 and BBS3, we determined the abundance of BBS3 in control and IFT22miRNA cells. Knockdown of IFT22 reduced cellular BBS3 to ∼15% of wild-type level (Fig. 4A). Since the transcripts representing BBS3 remained unaltered in the IFT22miRNA strain (Fig. 4B), we conclude that IFT22 is required to maintain cellular BBS3 level likely by stabilizing the protein. This notion is further supported by our observation that expression of IFT22::HA::GFP in the IFT22miRNA strain restored the wild-type level of BBS3 (Fig. 4C). The loss of BBS3 in the IFT22miRNA strain was also rescued by expression of the IFT22[Q69L] and IFT22[T19N] variants, revealing that BBS3 is stabilized by IFT22 independent of its nucleotide state (Fig. 4C). To determine whether BBS3 is conversely required for IFT22 stability, we knocked down BBS3 using vector-based BBS3 miRNA (resulting strain BBS3miRNA). BBS3 was reduced to ∼13% of the wild-type level in both the cell body and the cilia (Fig. 4 D and E). The knockdown of BBS3 did not affect the abundance of IFT22 in the cell body or in cilia (Fig. 4 D and E). In conclusion, IFT22 affects the stability of BBS3 but not vice versa.
Fig. 4.
IFT22 stabilizes BBS3 and is required for recruiting BBS3 to the basal body. (A) Immunoblots of WCE of CC-125 and IFT22miRNA cells probed for BBS3. (B) Quantification of BBS3 mRNA level in IFT22miRNA cells. Messenger RNA level was normalized to the GBLP housekeeping gene and presented as fold change relative to CC-125 mRNA. (C) Immunoblots of WCE of CC-125, IFT22Res-WT, IFT22Res-T19N, and IFT22Res-Q69L cells probed for BBS3. (D and E) Immunoblots of WCE (D) and CE (E) of CC-125 and BBS3miRNA cells probed for BBS3 and IFT22. (F) CC-125, IFT22miRNA, IFT22Res-WT, IFT22Res-Q69L, and IFT22Res-T19N cells stained with α-BBS3 (green). Inset shows the basal bodies. (Scale bar: 10 µm.) (G) Immunoblots of CE of CC-125 and IFT22miRNA cells for BBS3. (H) Immunoblots of CE of CC-125, IFT22Res-WT, IFT22Res-T19N, and IFT22Res-Q69L cells probed for BBS3. For A, C–E, G, and H, α-tubulin and Ac-tubulin were used to adjust the loading for WCE and CE, respectively.
Similar to other BBS proteins, BBS3 was enriched at the basal bodies in control cells but was hardly detected at the basal bodies in IFT22miRNA cells (Fig. 4F). Since the basal body localization of BBS3 in the IFT22miRNA strain was restored by the expression of IFT22::HA::GFP and IFT22[Q69L] but not IFT22[T19N] (Fig. 4F), we conclude that the GTP-bound IFT22 is needed to recruit BBS3 to the basal body. As a result, knockdown of IFT22 reduced ciliary BBS3 to ∼22% of wild-type level (Fig. 4G). Since ciliary BBS3 level was restored by IFT22::HA::GFP and IFT22[Q69L] but not IFT22[T19N] (Fig. 4H), we then conclude that recruitment of BBS3 to the basal body by GTP-bound IFT22 is a prerequisite for efficient entry of BBS3 into cilia.
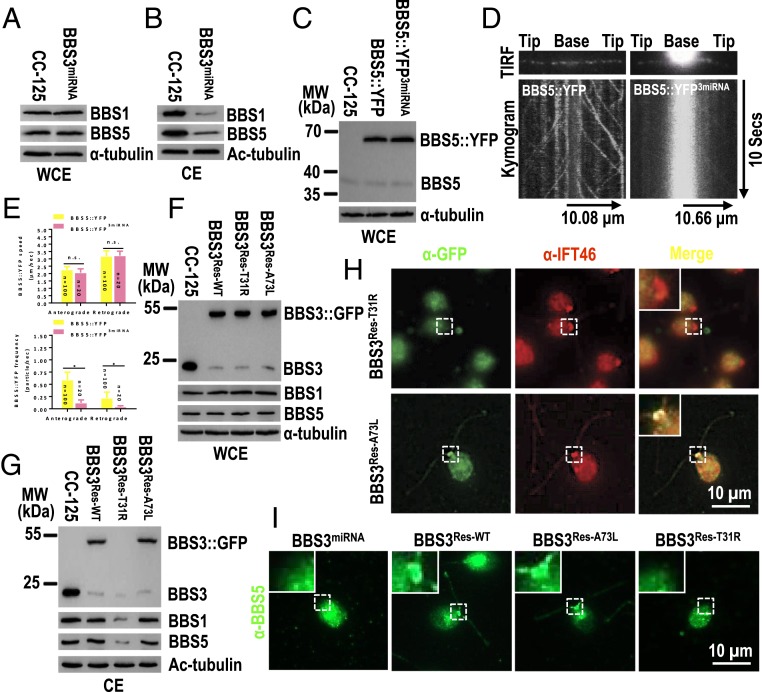
Nucleotide State of BBS3 Controls Basal Body Recruitment of BBS3 and the BBSome.
GTP-bound BBS3 is a known regulator of BBSome ciliary targeting (32, 44). In BBS3miRNA cells, the total cellular abundance of BBS1 and BBS5 was unaffected while BBS1 and BBS5 were strongly reduced (to ∼11% of wild-type levels) in cilia, confirming a role for BBS3 in BBSome ciliary targeting (Fig. 5 A and B) (32, 44). Indeed, when BBS5::YFP was expressed in similar amounts in control and BBS3miRNA cells (resulting in strain BBS5::YFP3miRNA) (Fig. 5C), the anterograde and retrograde frequencies of BBS5::YFP transport were strongly reduced while the transport velocities were not affected when compared to those of a BBS5::YFP control strain (Fig. 5 D and E and Movies S5 and S6) (33). Thus, BBS3 and IFT22 knockdowns appear to affect BBSome traffic in a similar manner, raising the question of whether the GTP state of BBS3 is critical for BBSome entry into cilia. To answer this question, we expressed BBS3::GFP and the variants BBS3[A73L]::GFP and BBS3[T31R]::GFP, which have been reported to lock BBS3 in wild-type, GTP-bound, and GDP-bound states, respectively (31, 32), in BBS3miRNA cells (resulting strains BBS3Res-WT, BBS3Res-A73L, and BBS3Res-T31R). All three strains had near normal levels of BBS3 (i.e., the endogenous BBS3 plus BBS3::GFP or its variants) and of BBS1 and BBS5 in whole cell samples (Fig. 5F). Remarkably, BBS3::GFP and BBS3[A73L] but not BBS3[T31R] were present in cilia and restored the ciliary abundance of BBS1 and BBS5 to wild-type levels (Fig. 5G). This suggests that the GTP-bound BBS3 is required for maintaining the ciliary abundance of the BBSome.
Fig. 5.
Nucleotide state of BBS3 controls basal body recruitment of BBS3 and the BBSome. (A and B) Immunoblots of WCE (A) and CE (B) of CC-125 and BBS3miRNA cells probed for BBS1 and BBS5. (C) Immunoblots of WCE of CC-125, BBS5::YFP, and BBS5::YFP3miRNA cells probed with α-BBS5. (D) TIRF images and corresponding kymograms of BBS5::YFP and BBS5::YFP3miRNA cells. In the kymograms, the time and transport lengths are indicated on the Right and on the Bottom, respectively. (E) In the graph, BBS5::YFP3miRNA cells showed dramatically decreased frequencies of BBS5::YFP in both anterograde (0.12 ± 0.06 particles/s) and retrograde (0.04 ± 0.02 particle/s) directions as compared to BBS5::YFP cells (0.59 ± 0.16 μm/s for anterograde and 0.22 ± 0.12 μm/s for retrograde, respectively). The speeds of BBS5::YFP in cilia of BBS5::YFP3miRNA cells were 2.06 ± 0.26 and 3.22 ± 0.30 μm/s for anterograde and retrograde, respectively, similar to that (2.26 ± 0.22 μm/s and 3.18 ± 0.36 μm/s for anterograde and retrograde, respectively) in cilia of BBS5::YFP cells. (F and G) Immunoblots of WCE (F) and CE (G) of CC-125, BBS3Res-WT, BBS3Res-T31R, and BBS3Res-A73L cells probed with α-BBS3, α-BBS1, and α-BBS5. MW, molecular weight. (H) BBS3Res-T31R and BBS3Res-A73L cells stained with α-GFP (green) and α-IFT46 (red). (I) BBS3miRNA, BBS3Res-WT, BBS3Res-A73L, and BBS3Res-T31R cells stained with α-BBS5 (green). For A–C, F, and G, α-tubulin and Ac-tubulin were used to adjust the loading for WCE and CE, respectively. For H and I, Inset shows the basal bodies. (Scale bar: 10 µm.)
IFT22 mediates both BBS3 and BBSome recruitment to the ciliary base. Since the BBSome fails to enter cilia of BBS3 knockdown cells despite normal BBS protein levels in the cell body, we wondered if BBS3 knockdown affects its recruitment and that of the BBSome to the basal bodies. The BBS3::GFP variants differed strongly in their ability to accumulate at the basal bodies: BBS3[A73L]::GFP colocalized with IFT46 at the basal bodies while BBS3[T31R]::GFP was not observed near the basal bodies in the BBS3Res-A73L and BBS3Res-T31R strains, respectively (Fig. 5H). Further, the BBSome subunit BBS5 was barely detected at the basal bodies in BBS3miRNA cells (Fig. 5I) but its basal body localization was restored by the expression of BBS3::GFP and BBS3[A73L]::GFP but not BBS3[T31R]::GFP (Fig. 5I). In conclusion, GTP-bound BBS3 is required to recruit the BBSome to the ciliary base and only GTP-bound BBS3 localizes to the basal bodies.
IFT22 Binds BBS3 and Recruits the BBSome to the Basal Body for IFT-B1 Coupling.
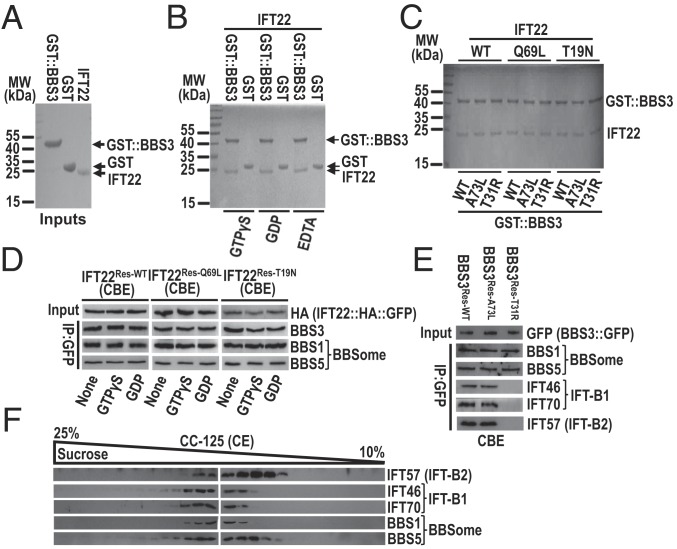
To explore if IFT22 binds BBS3 directly, we performed an in vitro protein–protein interaction assay using recombinant N-terminal GST-tagged BBS3 (GST::BBS3) and IFT22 (Fig. 6A); GTPγS, GDP, or EDTA were added to lock the two GTPases in a GTP-bound, a GDP-bound, or a nucleotide-free state, respectively (19). While IFT22 did not interact with GST along, IFT22 and GST::BBS3 bind to each other in the presence of GTPγS, GDP, or EDTA, indicating that the two proteins bind to each other independently of their nucleotide states (Fig. 6B). The same purification procedure was performed on all combinations of wild-type, GTP-bound, or GDP-bound variants of IFT22 and BBS3, and under all conditions they bound each other, confirming that the interaction between the two GTPases is independent of their nucleotide state (Fig. 6C). We next performed immunoprecipitation assays on cell body extracts of IFT22Res-WT, IFT22Res-Q69L, and IFT22Res-T19N cells in the presence of GTPγS, GDP, or without added nucleotides, which are proposed to lock the endogenous BBS3 in a GTP-bound, GDP-bound, or wild-type state, respectively (31). In agreement with the in vitro assay, immunoprecipitation with GFP-tagged IFT22 and its variants recovered BBS3 under all three conditions (Fig. 6D), further confirming that IFT22 and BBS3 bind to each other independent of their nucleotide states in the cell body.
Fig. 6.
IFT22 binds BBS3 and recruits the BBSome to the basal body for IFT-B1 coupling. (A) IFT22, GST, and GST::BBS3 were purified before SDS/PAGE and Coomassie staining. (B) IFT22 mixed with GST::BBS3 or GST in the presence of GTPγS, GDP, or EDTA and complexes recovered on glutathione beads before SDS/PAGE and Coomassie staining. (C) IFT22, GST::BBS3, GTP-locked, or GDP-locked variants of BBS3 and IFT22 mixed to form nine combinations and complexes recovered on glutathione beads before SDS/PAGE and Coomassie staining. MW, molecular weight. (D) Immunoblots of α-GFP captured proteins from CBE of IFT22Res-WT, IFT22Res-Q69L, and IFT22Res-T19N cells in the presence of GTPγS, GDP, or neither probed for BBS3, BBS1, and BBS5. Input was quantified with α-HA by immunoblotting. (E) Immunoblots of α-GFP captured proteins from CBE of BBS3Res-WT, BBS3Res-A73L, and BBS3Res-T31R cells probed for BBS1, BBS5, IFT46, IFT70, and IFT57. Input was quantified with α-GFP by immunoblotting. (F) Immunoblots of CE of CC-125 cells separated by sucrose density gradient centrifugation probed for IFT46, IFT70, IFT57, BBS1, and BBS5.
Chlamydomonas BBS3 binds the BBSome through a direct interaction between BBS3 and BBS1 (31). Consistent with this, BBS1 and BBS5 were immunoprecipitated by BBS3::GFP in cell body extracts (Fig. 6E). Whereas only the GTP-bound BBS3 is able to bind the BBSome in rodent cells (32), immunoprecipitation with the variants BBS3[A73L] and BBS3[T31R] both recovered BBS1 and BBS5 in cell body extracts, suggesting that the nucleotide state does not affect BBS3 for binding the BBSome in the cell body of C. reinhardtii (Fig. 6E). This was confirmed as BBS1 and BBS5 were also immunoprecipitated from the cell body extracts by IFT22 in the presence of various nucleotides (Fig. 6D). As shown above, when both are in their GTP-bound states, IFT22 and BBS3 recruit the BBSome to the basal body (Figs. 3H and 5I). BBS3[A73L]::GFP but not BBS3[T31R]::GFP immunoprecipitated the IFT-B1 subunits IFT46 and IFT70 but not the IFT-B2 subunit IFT57 in cell body extracts (Fig. 6E), suggesting that the BBSome couples with IFT-B1 at the basal body. BBS1 and BBS5 remained to be associated with IFT-B1 in cilia as they cosedimented in sucrose density gradients of ciliary extracts with IFT46 and IFT70 but not IFT57 (Fig. 6F).
Ciliary Entry and Cycling of the BBSome Does Not Require IFT22.
During the course of this study, insertional mutant in the IFT22 gene became available from the CLiP mutant collection (45); the strain LMJ.RY0402.055518 has the ∼3.3-kb paromomycin-resistant gene cassette inserted at the 3′-proximal region of the third exon of IFT22 and we refer to this strain as ift22 (SI Appendix, Fig. S4 A and B). This insertion caused generation of a mutated ift22 cDNA (SI Appendix, Fig. S4 C and D), which is predicted to encode a mutant IFT22 protein (IFT22-mut) in which the 9-amino acid sequence 127EGGGLGGWQ135 is replaced with the 17-amino acid sequence GWTQEQLVCQKARRQSA (SI Appendix, Fig. S4 E and F). Interestingly, IFT22-mut was detected in ift22 cells by Western blotting but not in cilia (Fig. 7 A and B). When expressed in IFT22miRNA cells (resulting strain IFT22Res-22-mut) (Fig. 7C), IFT22-mut::HA::GFP failed to enter cilia of the IFT22miRNA cells (Fig. 7D). In addition, IFT22-mut::HA::GFP failed to immunoprecipitate IFT25 and IFT46 from cell body extracts of the IFT22Res-22-mut strain (Fig. 7E), indicating that the altered IFT22 has lost its ability to bind to the IFT-B1 subcomplex likely explaining its absence from cilia (Fig. 7 B and D).
Fig. 7.
Ciliary entry and cycling of the BBSome does not require IFT22. (A and B) Immunoblots of WCE (A) and CE (B) of CC-5325 and ift22 cells probed for IFT22, BBS3, BBS1, and BBS5. (C and D) Immunoblots of WCE (C) and CE (D) of CC-125, IFT22miRNA, and IFT22Res-22-mut cells probed with α-IFT22, α-BBS3, α-BBS1, and α-BBS5. MW, molecular weight. (E) Immunoblots of α-GFP captured proteins from CBE of IFT22Res-22-mut cells probed for IFT25, IFT46, BBS3, BBS1, and BBS5. Input was quantified with α-GFP by immunoblotting. (F) CC-5325 and ift22 cells stained with α-IFT22 (red). (G) IFT22Res-WT and IFT22Res-22-mut cells stained with α-GFP (green). For A–D, α-tubulin and Ac-tubulin were used to adjust the loading for WCE and CE, respectively. For F and G, the corresponding phase contrast images were presented. Inset shows the basal bodies. (Scale bar: 10 µm.)
Immunofluorescence staining revealed that the endogenous IFT22-mut and the transgenic IFT22-mut::HA::GFP both are enriched at the basal bodies (Fig. 7 F and G). Next, we analyzed how IFT22-mut affects BBS3, since IFT22 is required for BBS3 stability. Expression of IFT22-mut::HA::GFP in the IFT22miRNA cells restored wild-type levels of BBS3 (Fig. 7C), IFT22-mut::HA::GFP immunoprecipitated BBS3 in cell body extracts of IFT22Res-22-mut cells (Fig. 7E), and recombinant IFT22-mut and BBS3 interact in vitro (SI Appendix, Fig. S5 A and B). Thus, IFT22-mut is defective in IFT-B1 binding and ciliary entry but still binds and stabilizes BBS3, allowing us to separate IFT22’s roles in recruiting the BBSome from its role in the IFT trains. Strikingly, BBS3, BBS1, and BBS5 entered cilia of the ift22 cells (Fig. 7B), and IFT22-mut::HA::GFP restored BBS3, BBS1, and BBS5 in cilia to wild-type levels (Fig. 7D). Also, in vivo imaging revealed that the BBSome as visualized by expression of BBS5::YFP in CC-5325 and ift22 cells (resulting strains BBS3::YFP5325 and BBS5::YFP22), cycles bidirectionally with normal velocities and frequencies (SI Appendix, Fig. S5 C–E and Movies S7 and S8). In summary, our data indicate that IFT22, independent of its IFT-B1 association, binds and stabilizes BBS3 in the cell body and, through its interaction with BBS3, recruits the BBSome to the basal bodies; BBSome entry and cycling in cilia, however, do not require IFT22.
Discussion
Defects in ciliary assembly and cycling of the BBSome results in ciliary dysfunction and diseases (17–19, 46). The BBSome loads onto IFT trains before entering cilia; loading likely occurs near the basal body region as described for other IFT cargoes (16, 35, 36, 47). However, the molecular mechanism which targets the BBSome to the basal body remains unknown. Here, we identify that RABL5/IFT22 via its interaction with BBS3 recruits the BBSome to the basal body in a GTP-dependent manner closing a gap in our understanding of the BBSome cycle through cilia.
IFT22 via Its Interaction with BBS3 Recruits the BBSome to the Basal Body.
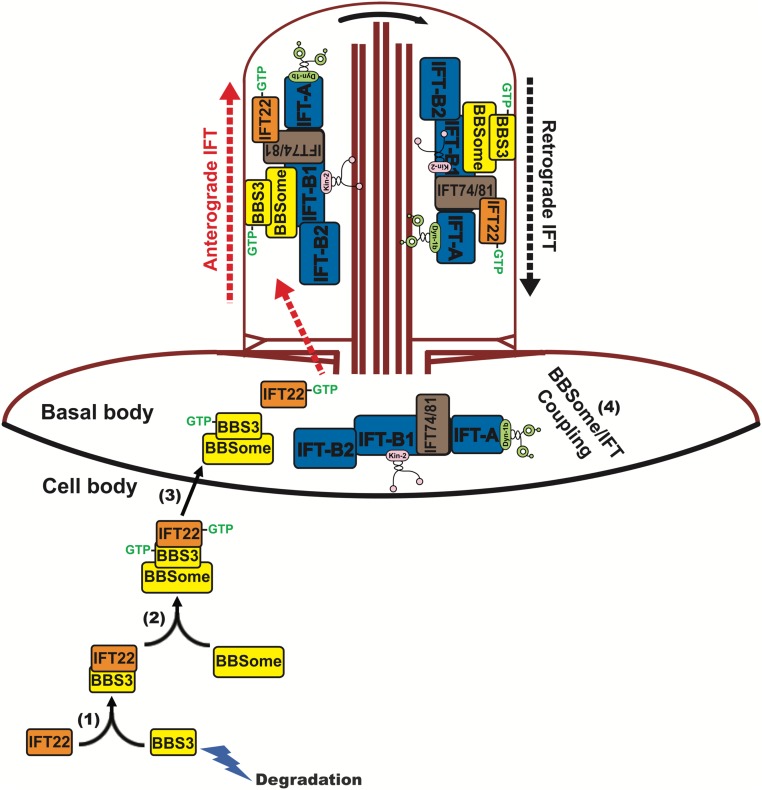
In nematodes, C. reinhardtii, and mammals, the BBSome undergoes bidirectional intraflagellar movements at the same rate as IFT trains, suggesting its association with IFT trains (16, 33, 48, 49). Here, we show that recruitment of the BBSome to the basal body region by IFT22 is a prerequisite for the BBSome to load onto IFT trains and enter cilia. Based on observations, we postulate the following model of IFT22-based recruitment of the BBSome to the basal body (Fig. 8). IFT22 binds and stabilizes BBS3 in the cell body independent of their nucleotide states. The analysis of mutant proteins suggests that the recruitment of the IFT22/BBS3 complex to the basal body requires both proteins to be in their GTP-bound states. In its GTP-bound state, the basal body-associated IFT22/BBS3 complex recruits BBSomes to the ciliary base. BBSome basal body recruitment and ciliary entry function normally in an ift22 mutant that expresses an altered IFT22 protein, which does not enter cilia but still accumulates at the ciliary base. While we cannot exclude that wild-type IFT22 remains associated with BBS3/BBSome during ciliary entry in control cells, this evidence suggests that IFT22 does not need to associate with the BBSome, that the BBSome will enter cilia without IFT22 as long as recruitment to the basal body is functional, and likely that IFT22 fulfills its role in BBSome basal body recruitment independently from its potential role as an IFT subunit.
Fig. 8.
Hypothetical model of how IFT22 mediates basal body recruitment of the BBSome through BBS3. (1) IFT22 binds BBS3 to stabilize the latter in the cell body; this binding is independent of their nucleotide states. (2) BBS3 binds the BBSome independent of its nucleotide state. (3) When both are in the GTP-bound state, IFT22 and BBS3 recruits the BBSome to the basal body. (4) At the basal body, the BBSome associates with IFT-B1 and then enters cilia. As an IFT-B1 subunit, GTP-bound IFT22 is likely released from BBS3 and binds IFT-B1 through direct interaction between IFT22 and the IFT74/81 subcomplex for ciliary entry (23, 25, 62). The GTP-bound BBS3 remains associated with the BBSome and enters cilia via IFT as reported previously (32). After entering cilia, the GTP-bound BBS3 become attached to the ciliary membrane (31, 32). The kinesin-II anterograde motor and the cytoplasmic dynein-1b retrograde motor were shown (20, 63).
The model raises the question of how IFT22-dependent recruitment of BBSome to the ciliary base promotes BBSome entry into cilia. In a parsimonious model, basal body-associated IFT22 provides a docking site for the BBSome, locally increasing the concentration of BBSomes, and thereby facilitating its interaction with IFT trains and ciliary entry. Indeed, basal body-associated pools have been also described for other IFT cargoes (47). However, our data indicate that IFT22 does not enter cilia together with the BBSome, suggesting that the BBSome is transferred from IFT22 to load onto IFT trains with IFT22 associated with IFT-B1. IFT trains assemble in a step-by-step process becoming more complex and ordered the closer they get to entering cilia. IFT22 could facilitate BBSome transfer to IFT trains by positioning it near the maturing IFT trains.
How Could IFT22 and BBS3 Regulate Basal Body Targeting of the BBSome?
In concordance with a recent trypanosome IFT22 study (39), Chlamydomonas IFT22 is an active GTPase with low intrinsic GTPase activity (SI Appendix, Fig. S1B). Since GTP hydrolysis rate of IFT22 was increased in the presence of cell body extracts but not ciliary extracts (SI Appendix, Fig. S1B), we cannot exclude that GDP–GTP exchange and BBSome basal body targeting are promoted by other factors in the cell body. Thus, basal body recruitment of the BBSome could be regulated by the nucleotide state of IFT22. Hypothetically, such a scenario would allow cells to regulate the number of BBSomes available for pickup by IFT trains at the basal body and in turn could regulate the presence and amount of signaling proteins in cilia, which rely on the BBSome for ciliary export and import via the IFT pathway (3–9).
GTP-bound BBS3 associates with the ciliary membrane in cilia (19, 31) and recruits the BBSome to the ciliary membrane to interact with its cargoes in cilia (31, 32, 50). Thus, BBS3 was proposed to undergo a GTPase cycle at the ciliary tip to regulate the ciliary presence and amount of signaling proteins by mediating their export from cilia (19). Here, we show that the BBS3 together with IFT22 recruits the BBSome to the basal body in a GTP-dependent manner, suggesting that BBS3 also could regulate the presence and amount of the signaling proteins in cilia by controlling the number of BBSomes available at the basal body for entering cilia. In addition, assuming that BBS3-bound configuration is indeed related to the BBSome’s ability to interact with cargoes (50), IFT22 via binding BBS3 in the cell body could prevent BBS3 from associating with the membrane and prevent the BBSome from binding its cargoes and carrying them along into cilia. Before ciliary entry, IFT22 is released from BBS3 at the basal body, which will convert the BBS3-bound BBSome to be in a state in cilia to bind to its cargoes and removes them from cilia. Thus, the IFT22/BBS3-dependent recruitment to the basal body could also function to “activate” the BBSome, ensuring a spatial restriction of its binding to cargo to the ciliary compartment.
Chlamydomonas IFT22 Is Not Required for Ciliary Assembly or IFT.
Depletion of IFT-B1 subunits typically causes the disassembly of IFT-B, blocks the anterograde IFT, and eventually leads to loss of cilia (51–58). However, IFT22 knockdown strain, as well as an ift22 mutant, which lacks IFT22 from cilia, have normal-length cilia and apparently normal IFT but for the lack of BBSomes in the knockdown strain (SI Appendix, Fig. S3 A–F). Thus, Chlamydomonas IFT22 is not needed for ciliary assembly or IFT. This is consistent with studies in worm and mammalian cells (37, 59), whereas depletion of IFT22 in Trypanosoma leads to the loss of flagella (38, 39). Similarly, IFT25 and IFT27 are dispensable for ciliary assembly in mammalian and Chlamydomonas cells (18, 19, 33, 34) but essential for flagellar construction in Trypanosoma, adding the notion that numerous species-specific differences in the IFT machinery have developed (60, 61). In mammalian IFT25 and IFT27 knockouts, BBSome subunits and putative BBSome cargoes accumulate in cilia, indicating that these small IFT proteins mediate BBSome binding to retrograde train (18, 19, 33, 34). Our data reveal that IFT22 is required for BBSome entry into cilia, albeit not because IFT22 provides an essential BBSome docking site on anterograde trains, but by recruiting BBSomes to the ciliary base. Since the BBSome associates with IFT-B1 at the basal body and remains associated with IFT-B1 in cilia, we thus predict that anterograde IFT of BBSomes involves additional yet to be determined IFT-B1 proteins. Finally, whereas IFT22 does not influence the ciliary assembly and IFT, it is indeed present in cilia. It would be desirable to investigate the possible role(s) of IFT22 in cilia in the future.
Materials and Methods
Chlamydomonas Strains.
C. reinhardtii strain CC-125 and CC-5325 were used throughout this study. All strains, including the IFT22 CLiP strain and the miRNA strains, are listed in SI Appendix, Table S1. Cells were cultured at room temperature in liquid Tris acetic acid phosphate medium under continuous light with constant aeration.
Various experimental protocols were applied in this study, and most of the experiments were briefly introduced in the text to make the content easier to be understood. The details of each protocol are available in SI Appendix.
Statistical Analysis.
If not mentioned elsewhere, all of the data are presented as mean ± SD from three independently performed experiments, and n stands for sample numbers. Statistical analysis was performed with GraphPad Prism 7.04 (GraphPad Software). For comparisons on speeds and frequencies of the GFP (YFP)-labeled proteins, a one-sample unpaired Student t test was used. For comparisons on expression levels of protein and mRNA, a two-tailed t test was used. If not noted elsewhere, significance was set at P < 0.001 (marked as *). n.s. represents nonsignificance.
Data Availability.
Data supporting the findings of this paper are contained within the paper and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by International Center for Genetic Engineering and Biotechnology Grant CRP/CHN15-01, Tianjin Municipal Science and Technology Bureau Grant 19PTSYJC00050 to Z.-C.F., National Institutes of Health Grant GM110413 to K.F.L., and National Natural Science Foundation of China Grant 41876134 to J.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901665117/-/DCSupplemental.
References
- 1.Fliegauf M., Benzing T., Omran H., When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Drummond I. A., Cilia functions in development. Curr. Opin. Cell Biol. 24, 24–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berbari N. F., Lewis J. S., Bishop G. A., Askwith C. C., Mykytyn K., Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. U.S.A. 105, 4242–4246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta P., et al. , Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E4400–E4409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domire J. S., et al. , Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68, 2951–2960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q., Seo S., Bugge K., Stone E. M., Sheffield V. C., BBS proteins interact genetically with the IFT pathway to influence SHH-related phenotypes. Hum. Mol. Genet. 21, 1945–1953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siljee J. E., et al. , Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat. Genet. 50, 180–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valentine M. S., et al. , Paramecium BBS genes are key to presence of channels in cilia. Cilia 1, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu P., Lechtreck K. F., The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. U.S.A. 115, E934–E943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pigino G., et al. , Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187, 135–148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L., A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. U.S.A. 90, 5519–5523 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum J. L., Witman G. B., Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Nachury M. V., et al. , A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Loktev A. V., et al. , A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell 15, 854–865 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Scheidecker S., et al. , Exome sequencing of Bardet-Biedl syndrome patient identifies a null mutation in the BBSome subunit BBIP1 (BBS18). J. Med. Genet. 51, 132–136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechtreck K. F., et al. , The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117–1132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Priya S., Nampoothiri S., Sen P., Sripriya S., Bardet-Biedl syndrome: Genetics, molecular pathophysiology, and disease management. Indian J. Ophthalmol. 64, 620–627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eguether T., et al. , IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell 31, 279–290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liew G. M., et al. , The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell 31, 265–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole D. G., et al. , Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taschner M., Bhogaraju S., Vetter M., Morawetz M., Lorentzen E., Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT-B subunits. J. Biol. Chem. 286, 26344–26352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Follit J. A., Xu F., Keady B. T., Pazour G. J., Characterization of mouse IFT complex B. Cell Motil. Cytoskeleton 66, 457–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhogaraju S., Engel B. D., Lorentzen E., Intraflagellar transport complex structure and cargo interactions. Cilia 2, 10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taschner M., et al. , Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35, 773–790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taschner M., Kotsis F., Braeuer P., Kuehn E. W., Lorentzen E., Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J. Cell Biol. 207, 269–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucker B. F., et al. , Characterization of the intraflagellar transport complex B core: Direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 280, 27688–27696 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Pedersen L. B., Geimer S., Rosenbaum J. L., Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr. Biol. 16, 450–459 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Pedersen L. B., et al. , Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr. Biol. 15, 262–266 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Iomini C., Babaev-Khaimov V., Sassaroli M., Piperno G., Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13–24 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Q., et al. , The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell Biol. 14, 950–957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mourão A., Nager A. R., Nachury M. V., Lorentzen E., Structural basis for membrane targeting of the BBSome by ARL6. Nat. Struct. Mol. Biol. 21, 1035–1041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin H., et al. , The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong B., et al. , Chlamydomonas IFT25 is dispensable for flagellar assembly but required to export the BBSome from flagella. Biol. Open 6, 1680–1691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keady B. T., et al. , IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell 22, 940–951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansley S. J., et al. , Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628–633 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wingfield J. L., et al. , IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife 6, e26609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer J. C., et al. , IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J. Cell Sci. 119, 4088–4100 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Adhiambo C., Blisnick T., Toutirais G., Delannoy E., Bastin P., A novel function for the atypical small G protein Rab-like 5 in the assembly of the trypanosome flagellum. J. Cell Sci. 122, 834–841 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Wachter S., et al. , Binding of IFT22 to the intraflagellar transport complex is essential for flagellum assembly. EMBO J. 38, e101251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva D. A., Huang X., Behal R. H., Cole D. G., Qin H., The RABL5 homolog IFT22 regulates the cellular pool size and the amount of IFT particles partitioned to the flagellar compartment in Chlamydomonas reinhardtii. Cytoskeleton 69, 33–48 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Paduch M., Jeleń F., Otlewski J., Structure of small G proteins and their regulators. Acta Biochim. Pol. 48, 829–850 (2001). [PubMed] [Google Scholar]
- 42.Ishikawa H., et al. , TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. eLife 3, e01566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin H., Wang Z., Diener D., Rosenbaum J., Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17, 193–202 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q., et al. , Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc. Natl. Acad. Sci. U.S.A. 108, 20678–20683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., et al. , An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heon E., et al. , Mutations in C8ORF37 cause Bardet Biedl syndrome (BBS21). Hum. Mol. Genet. 25, 2283–2294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai J., Barbieri F., Mitchell D. R., Lechtreck K. F., In vivo analysis of outer arm dynein transport reveals cargo-specific intraflagellar transport properties. Mol. Biol. Cell 29, 2553–2565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M., Functional coordination of intraflagellar transport motors. Nature 436, 583–587 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Williams C. L., et al. , Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat. Commun. 5, 5813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klink B. U., et al. , A recombinant BBSome core complex and how it interacts with ciliary cargo. eLife 6, e27434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown J. M., Cochran D. A., Craige B., Kubo T., Witman G. B., Assembly of IFT trains at the ciliary base depends on IFT74. Curr. Biol. 25, 1583–1593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Z.-C., et al. , Chlamydomonas IFT70/CrDYF-1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol. Biol. Cell 21, 2696–2706 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I., Mitani S., IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells 12, 593–602 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Hou Y., et al. , Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653–665 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L., Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11, 1586–1590 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Brazelton W. J., Amundsen C. D., Silflow C. D., Lefebvre P. A., The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11, 1591–1594 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Pazour G. J., et al. , Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151, 709–718 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu X., Liang Y., Gao F., Pan J., IFT54 regulates IFT20 stability but is not essential for tubulin transport during ciliogenesis. Cell. Mol. Life Sci. 74, 3425–3437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takei R., Katoh Y., Nakayama K., Robust interaction of IFT70 with IFT52-IFT88 in the IFT-B complex is required for ciliogenesis. Biol. Open 7, bio033241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huet D., Blisnick T., Perrot S., Bastin P., IFT25 is required for the construction of the trypanosome flagellum. J. Cell Sci. 132, jcs228296 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Huet D., Blisnick T., Perrot S., Bastin P., The GTPase IFT27 is involved in both anterograde and retrograde intraflagellar transport. eLife 3, e02419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lucker B. F., Miller M. S., Dziedzic S. A., Blackmarr P. T., Cole D. G., Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52, and IFT46. J. Biol. Chem. 285, 21508–21518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pazour G. J., Wilkerson C. G., Witman G. B., A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979–992 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this paper are contained within the paper and SI Appendix.