Significance
Postnatal development of the human gut microbiota is linked to healthy growth. Because the number of potential interactions between community components is vast, an unanswered question is what mechanisms determine the form of community assembly (succession). We created a simplified, manipulable in vivo model where bacterial strains cultured from an infant were divided into consortia, representing earlier and later periods in assembly, and introduced in different order into germ-free mice fed infant formula. Measuring strain abundances, bacterial gene expression and levels of gut nutrients, and applying computational tools to deduce interacting features, we identify genomic and metabolic correlates of how strains establish and maintain themselves. This approach may facilitate discoveries of how communities respond to various perturbations and microbiota-directed therapeutics.
Keywords: gut microbiome, microbial community assembly/succession, feature-reduction algorithms, metabolic pathways
Abstract
Human gut microbiota development has been associated with healthy growth but understanding the determinants of community assembly and composition is a formidable challenge. We cultured bacteria from serially collected fecal samples from a healthy infant; 34 sequenced strains containing 103,102 genes were divided into two consortia representing earlier and later stages in community assembly during the first six postnatal months. The two consortia were introduced alone (singly), or sequentially in different order, or simultaneously into young germ-free mice fed human infant formula. The pattern of fitness of bacterial strains observed across the different colonization conditions indicated that later-phase strains substantially outcompete earlier-phase strains, although four early-phase members persist. Persistence was not determined by order of introduction, suggesting that priority effects are not prominent in this model. To characterize succession in the context of the metabolic potential of consortium members, we performed in silico reconstructions of metabolic pathways involved in carbohydrate utilization and amino acid and B-vitamin biosynthesis, then quantified the fitness (abundance) of strains in serially collected fecal samples and their transcriptional responses to different histories of colonization. Applying feature-reduction methods disclosed a set of metabolic pathways whose presence and/or expression correlates with strain fitness and that enable early-stage colonizers to survive during introduction of later colonizers. The approach described can be used to test the magnitude of the contribution of identified metabolic pathways to fitness in different community contexts, study various ecological processes thought to govern community assembly, and facilitate development of microbiota-directed therapeutics.
Studies performed during the past decade provide a rapidly expanding body of evidence that the gut microbiota is an important determinant of health status. These insights have arisen from correlative studies between host physiologic, metabolic, immune, and/or other phenotypes and microbial community configuration (defined at varying levels of resolution). Tests of whether microbiota configurations are causally related to host phenotypes of interest include assessments of the ability of communities to transmit these phenotypes to recipient germ-free animals, or whether transplantation of microbiota from healthy humans can ameliorate disease in affected humans. The increasing number of associations between the microbiota and host biological features highlights the need to identify mechanisms that determine how microbial communities function, for example how they assemble following birth, how they adapt to various environmental perturbations, how they maintain their robustness/resiliency, and how they influence various aspects of host physiology and pathophysiology. Obtaining these insights is complicated by the complexity of the microbiota (1–3); in addition to harboring strain-level variants of many microbial species, the system is very dynamic with genetic and metabolic features and patterns of gene expression varying over time, space, and at different scales. Organisms, genes, and gene products interact with each other, with the number of possible pairwise and higher-order interactions becoming so large as to befuddle interpretation of community organization and dynamics. The pressing nature of the problem is underscored by the fact that the ease and cost of generating genomic, transcriptional, and metabolomic datasets are changing in ways that are yielding a tsunami of “multiomic” data that describe ever-expanding lists of community components (4–7).
The history of fields that study complex phenomena and collect vast amounts of data emphasizes how establishing relationships between the properties of a system and the properties of its constituent parts is often a formidable challenge. Key advances occur when groups of interacting components are identified and the resulting dimension reduction generates testable hypotheses about mechanisms that determine how a system operates (8–15). Assembly of the gut microbiota (succession) illustrates the challenges encountered as well as the insights that can be gleaned from this type of approach. There are several reasons for using succession as a case study. First, studies of birth cohorts representing different geographic locales and anthropologic features have revealed shared features of community assembly across biologically unrelated healthy individuals (15). This finding suggests that there are organizing principles that underlie the process of succession. Second, studies of children with moderate and severe acute malnutrition have shown that this process is interrupted, yielding communities that appear younger (more immature) than those of chronologically age-matched children with healthy growth. Initial studies indicate that repair of this immaturity with microbiota-directed complementary foods is associated with marked changes in numerous biomarkers and mediators of healthy growth, including those related to bone biology, immune function, metabolic regulation, and neurodevelopment (15, 16). The latter observation suggests that healthy microbiota development is linked to healthy growth. Third, it is possible to model succession in gnotobiotic mice so that the number of system components and variables can be constrained.
Here, we characterize microbial succession in gnotobiotic mice colonized with 34 gut bacterial strains from a single infant. These strains, which contain a total of 103,102 known or putative protein-coding genes, were divided into two consortia representing earlier and later periods in microbiota assembly during the first six postnatal months. Each consortium was introduced alone, in a different order, or together, into groups of young, germ-free animals fed a compositionally defined human infant formula (IF) diet. Employing techniques originally used for feature reduction in signal processing (17), we identify a small set of genomic features whose presence and/or pattern of expression is associated with the fitness of consortium members, including determinants of the establishment and persistence of a subset of early colonizers. In principle, the experimental and computational approaches described in this paper should be generally applicable for studies of competition, niche partitioning, and other processes that govern succession. They may also help guide development of prebiotic, probiotic, and synbiotic approaches for treating or preventing defects in community development.
Results
A Manipulable In Vivo Model of Human Gut Microbial Succession.
Generating consortia of cultured bacterial strains representing stages of microbiota development during predominant milk feeding.
We began by generating a clonally arrayed collection of 68 bacterial strains isolated from six fecal samples obtained on postnatal days 38, 67, 133, 247, 339, and 683 from a healthy, vaginally delivered member of a US birth cohort comprised of 40 twin pairs (18). This infant consumed both breast milk and formula, with exclusive formula feeding beginning during the second postnatal month and complementary feeding commencing at 19 wk of age (Dataset S1). The genomes of bacterial isolates were sequenced. To identify associations between the bacterial strains that were recovered (Dataset S2) and temporal stages of microbiota development, we performed an indicator species analysis (19) on a bacterial V4-16S ribosomal RNA dataset generated from the 40 twin pairs (n = 21 ± 6 [mean ± SD] samples per individual; total of 1,670 samples; see Materials and Methods). Microbiota developmental trajectory was divided into three stages: stage 1 (S1), encompassing the first two postnatal months, stage 2 (S2), months 2 through 6, and stage 3 (S3), months 6 through 24. S1 was intended to capture organisms that colonize early, either due to their specific fitness advantages in exclusively milk-fed infants or their enhanced ability to colonize naïve habitats. S2 was intended to capture organisms that may still have preferences for nutrients in milk, but also reflects the period during which the infant gut community is rapidly acquiring new taxa and is first exposed to complementary foods. S3 was intended to capture microbes with preferences for nutrients represented in an expanding “menu” of complementary foods and in fully weaned diets. Colonization by S2 and S3 organisms could reflect environmental conditions engineered by earlier colonizers and their ability to either coexist with or displace earlier colonizers. Guided by the indicator species analysis, the 68 cultured strains were assigned to developmental stages based on their taxonomic relationships to S1-, S2-, or S3-associated operational taxonomic units (OTUs) from the twin pair dataset (Dataset S2).
We focused on the period of colonization encompassed by predominant milk feeding (S1 and S2), allowing us to model, in gnotobiotic mice, interactions between members of a given stage consortium, as well as between different stage consortium members, in a commonly used human IF diet context. Young (3- to 4-wk-old) germ-free C57BL/6J mice were weaned directly onto the IF diet and then maintained on this diet ad libitum. Different groups of animals were subjected to different histories of exposure to the S1 consortium (composed of 21 members) and the S2 consortium (13 members); one group was only colonized with the S1 consortium, another with only the S2 consortium, a third with the S1 consortium followed by the S2 consortium 13 d later (S1→S2), a fourth group with the S2 consortium followed by S1 consortium 13 d later (S2→S1), and a fifth group where both sets of strains were administered simultaneously (S1+S2) (n = 5 mice per treatment group).
S2 organisms substantially and reproducibly outcompete S1 organisms.
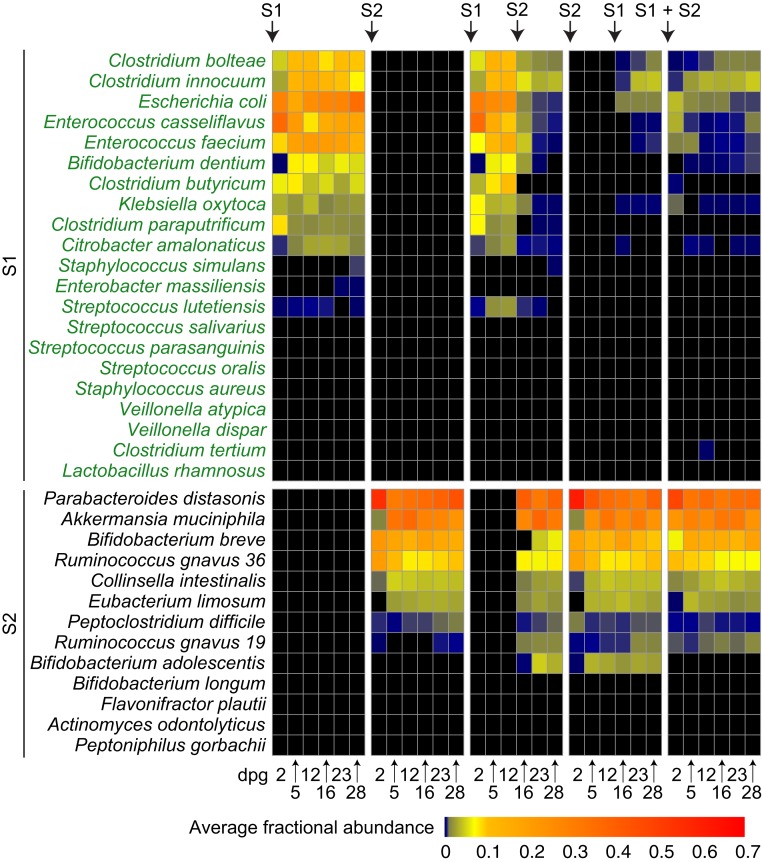
We performed COmmunity PROfiling by short-read shotgun sequencing (COPRO-Seq; Materials and Methods) of fecal DNA samples collected over time from animals with the five different colonization histories (Dataset S3A). Fig. 1 shows the fitness (fractional abundance in the fecal microbiota) of members of the S1 and S2 consortia as a function of time and order of introduction.
Fig. 1.
Modeling microbial succession in gnotobiotic mice. Heat map of the fractional abundances of bacterial strains in the feces of gnotobiotic mice fed human IF and colonized with S1, S2, S1→S2, S2→S1, or S1+S2 consortia. Abundances were defined by shotgun sequencing of fecal DNAs prepared from samples collected on the indicated days post gavage (dpg). Average fractional abundances for S1 and S2 strains represented in the microbiota of mice belonging to each treatment group at each time point are shown (n = 5 mice per group; see Dataset S3A for values from individual animals).
The pattern of fitness of bacterial strains observed across the different colonization conditions indicated that S2 organisms substantially outcompete S1 organisms, although four S1 members persist: Clostridium bolteae, Clostridium innocuum, Escherichia coli, and Enterococcus casseliflavus (≥0.01 fractional representation in at least one of the three colonization conditions with S2 consortium members) (Fig. 1 and Dataset S3A). Thus, colonization success does not seem to be driven by “arrival” order but rather by fitness differences (i.e., in this model, priority effects are not prominent). The results obtained in this initial study of five types of colonization treatments and in a follow-up experiment involving mice that received the S1 consortium alone, the S2 consortium alone, or S1 followed by S2 (n = 5 mice per treatment group) established that the patterns of colonization/fitness were highly reproducible (SI Appendix, Fig. S1 and Dataset S3B).
To determine whether there were also reproducible effects on the gut nutrient/metabolic environment, we used targeted mass spectrometry to measure the concentrations of 27 carbohydrates, 19 amino acids, and 10 B vitamins in cecal contents harvested from animals subjected to colonization with the S1 consortium alone, the S2 consortium alone, or the S1→S2 sequence. The extent to which levels of a given analyte were increased for a particular colonization condition relative to germ-free controls was calculated (SI Appendix, Fig. S2A and Dataset S4). The results provided additional evidence for the reproducibility of the two independent experiments involving the three different types of colonization treatments (SI Appendix, Fig. S3). Increases in the relative levels of nine analytes (isomaltose, gluconate, glucuronate, maltitol, raffinose, riboflavin, pyridoxal, pyridoxamine, and nicotinic acid) reflected the effect of colonization per se rather than consortium-specific effects (SI Appendix, Fig. S2A). Principal components analysis (PCA) was subsequently performed on the 47 analytes whose levels were consortium-specific. Plotting the changes in levels of these 47 analytes in a given mouse subjected to a given colonization history disclosed that the cecal metabolic landscape of animals colonized with S2 members was distinct from that produced by the S1 consortium alone (SI Appendix, Fig. S2B and Dataset S4D).
Identifying Determinants of Fitness.
Analytic approach.
These observations raise the question of what are the determinants of fitness of organisms subjected to these deliberately orchestrated colonization sequences, including the four persistent S1 members. Addressing this question is challenging for a number of reasons. Given the very large number of potential pairwise and higher-order interactions that may occur between organisms, performing a systematic series of “leave one or more bacterial strains out prior to gavage” experiments involving S1 and S2 consortia and noting the effects on the fitness of the remaining consortium members in vivo is not feasible. An analogous in vitro approach is not tenable for similar reasons and is compounded by 1) current limitations in the ability to culture defined consortia of microbes in bioreactors in a manner that yields reproducible community structures, 2) the lack of media that reproduce the nutrient/metabolic landscapes of different regions of a naïve (germ-free) gastrointestinal tract, and 3) the lack of representation of other habitat features (e.g., adhesive surfaces including partially digested food particles that could serve as sites of attachment of community members in ways that foster metabolic exchange/syntropic relationships, the mixing/agitative/propulsive forces experienced by gut microbes as they transit the gut, components of the adaptive and innate immune system, etc.). Moreover, tools for genome-wide forward genetic screens of fitness determinants are not available for a majority of the strains included in the S1 and S2 consortia. Even if these tools were available, a multiplicity of genes/pathways could contribute to the fitness of a given organism, thus limiting the effect size produced by single mutations in a given genome. Moreover, these genetic screens would, ideally, be conducted simultaneously in more than one community member to identify fitness determinants that reflect key interactions between organisms (20, 21).
Based on these considerations, we focused on identifying metabolic pathways whose presence/absence and/or levels of expression correlated with the different degrees of fitness of S1 and S2 organisms. Our goal was to address the question of why S2 organisms generally outcompete S1 organisms and what allows the subset of S1 organisms to persist in the climax communities resulting from the three different tandem colonization sequences. We reasoned that referencing metabolic pathway presence/expression to the climax community resulting from colonization with the S1 consortium alone or the S2 consortium alone would be an inadequate way of relating the state of the transcriptome to the fitness of organisms that have experienced a history of S1→S2, S2→S1, or S1+S2 colonization. Rather, an alternative strategy would be to embrace a “within-treatment group” analysis and use a data-driven approach to identify collective sets of metabolic pathways whose presence and/or levels of expression vary between organisms of varying fitness over each of the S1→S2, S2→S1, and S1+S2 treatment conditions. To do so, we generated microbial RNA-sequencing (RNA-Seq) datasets using cecal contents harvested at day postgavage 28 (dpg 28) from all animals in all treatment groups. Dataset S5A presents correlations between the number of reads that mapped to each organism’s genome and the abundance of that organism across all conditions tested in the two independent sets of colonization experiments. The results show that the strength of the Pearson correlation varies considerably between the organisms (range of r2 values 0.13 to 0.98). However, because a substantial number of organisms did have statistically significant correlations (P < 0.05), transcripts counts were normalized as reads per kilobase of a given gene’s length per million transcript reads (TPM) for each organism in each colonization condition in order to contextualize its transcriptional profile relative to the total abundance (number) of its expressed transcripts (Dataset S5B). We then aggregated the normalized RNA-Seq counts according to functional annotations of metabolic pathways represented in the genomes of S1 and S2 consortium members, focusing on pathways involved in carbohydrate utilization and fermentation, biosynthesis of amino acids, and vitamins/cofactors. Our rationale for this focus was that 1) carbohydrates comprise a key source of carbon and energy for a variety of heterotrophic microbes via central carbon metabolism, 2) protein production is critical for fitness, and 3) B vitamins play a critical role as precursors of essential cofactors for myriad metabolic reactions (B1 [thiamine], B2 [riboflavin], B3 [nicotinic acid], B5 [pantothenate], and B6 [pyridoxine] are precursors of cofactors that drive hundreds of indispensable biochemical transformations, while B7 [biotin], B9 [folate], and B12 [cobalamin] function as cofactors of enzymes involved in fatty acid biosynthesis, single-carbon metabolism, methionine biosynthesis, and several other pathways). Pathway annotations and in silico metabolic reconstructions were based on the RAST/SEED platform. This platform combines homology- and genome context-based evidence with known sets of enzymatic reactions and nutrient transporters to group genes into “microbial community (mc) subsystems” (mcSEED subsystems) that capture and project variations in particular metabolic pathways/modules across thousands of microbial genomes (22, 23). Genome annotations categorized by mcSEED subsystems are presented in Dataset S5B and summarized in SI Appendix, Fig. S4 in the form of a binary phenotype matrix (BPM) that plots predicted metabolic phenotypes, such as biosynthetic capabilities for amino acids, vitamin/cofactors, and other essential metabolites (e.g., queuosine and menaquinone), as well as the ability or inability to utilize specific carbohydrates and/or generate short-chain fatty acid (SCFA) products of fermentation (for details see Bacterial Genome Sequencing, Assembly, in Silico Metabolic Reconstructions, and Phenotype Predictions). For analysis of expression data, we utilized a broader and more granular set of curated mcSEED metabolic pathways/modules (24). After aggregating TPM-normalized RNA-Seq counts according to these mcSEED functional annotations, we generated tables of mcSEED metabolic pathway/module-normalized transcript counts for all S1 and S2 organisms. The summed expression of all genes comprising a given metabolic pathway/module provided a quantitative measure of its activity. Expression of a specific mcSEED pathway in an organism was then averaged over mice that had been subjected to a given colonization sequence.
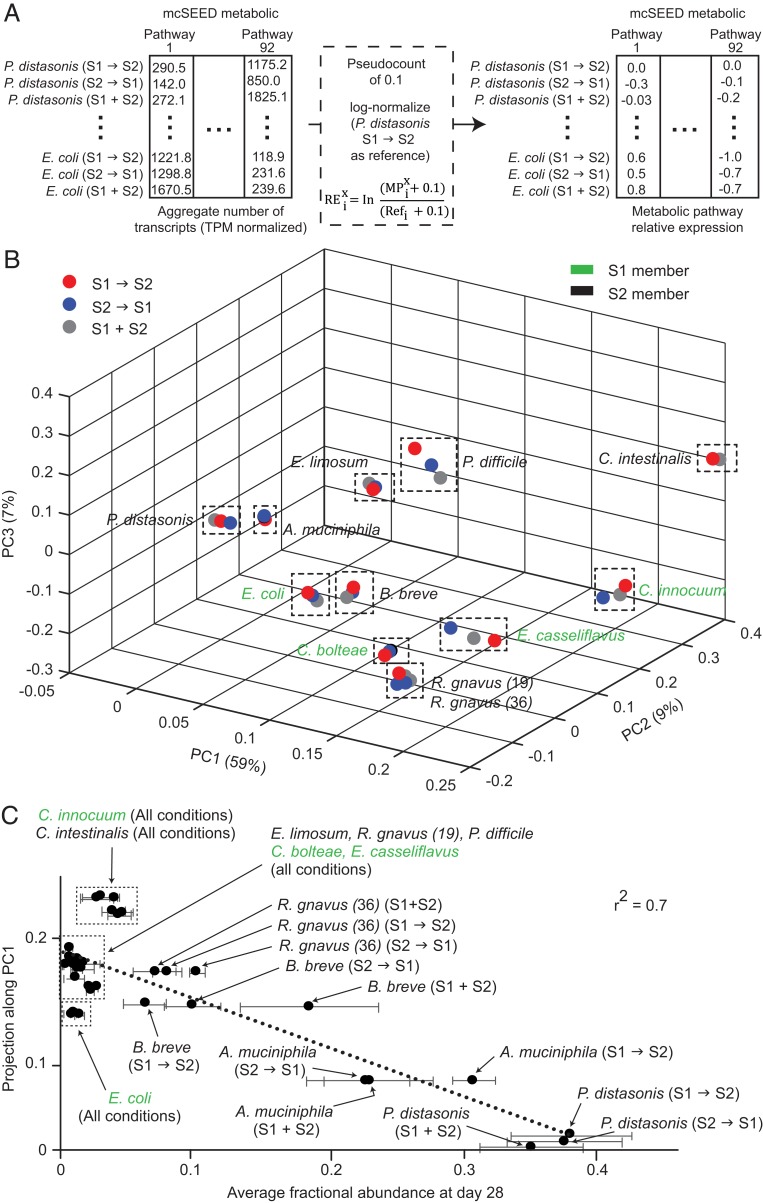
As the S1 and S2 consortia comprise organisms that are genetically diverse and therefore contain different sets of encoded metabolic pathways/modules, we evaluated the extent to which the transcriptomes of community members resembled that of the highest-fitness organism at the end of each of the three types of tandem colonization experiments—Parabacteroides distasonis. To do so, we log-normalized expression of a given pathway to the same pathway in P. distasonis. A “relative expression score” (REix in Eq. 1) was calculated for each metabolic pathway i in a given organism subject to a specific colonization condition (MPix) relative to expression of the metabolic pathway i in P. distasonis under the S1→S2 colonization condition (Refi):
| [1] |
For this calculation, TPM-normalized aggregated transcript counts for mcSEED metabolic pathways that were either 1) absent or 2) not expressed were ascribed a 0. To enable a quantitative comparison to the reference, a pseudocount of 0.1 was added to the metabolic pathway MPix and Refi.
The set of relative expression scores calculated for a given organism (Fig. 2A) was termed that organism’s “mcSEED pathway relative expression profile.” We subsequently created a matrix of mcSEED pathway relative expression profiles for all four S1 organisms that survived introduction of the S2 consortium plus each of the eight S2 organisms that had successfully colonized under all treatment conditions and whose summed contribution to the cecal metatranscriptome represented ≥96% of total reads averaged over all mice in each treatment group (Dataset S6 A and B).
Fig. 2.
Expressed metabolic pathways related to the fitness of S1 and S2 members in tandem colonization experiments. (A) Analysis of TPM-normalized microbial RNA-Seq datasets. A matrix of mcSEED metabolic pathways (columns) for all S1 and S2 organisms subject to the S1→S2, S2→S1, and S1+S2 colonization sequences (rows) is created where each element is the aggregated number of transcripts for a given pathway (92 pathways in total). A pseudocount of 0.1 is added to each element of the matrix. Each row is log-normalized against the reference row (P. distasonis in the S1→S2 colonization sequence) to create an mcSEED pathway/module relative expression profile for each organism in the climax community resulting from each of the three colonization treatments. See main text for further information about the terms used in the equation shown for relative expression score (REix). (B) mcSEED pathway/module relative expression profiles plotted in PCA space for the indicated colonization conditions using P. distasonis under the S1→S2 colonization condition as a reference. Red, blue, and black indicate the different colonization treatments. Strain names are colored green and black based on whether they are members of the S1 or S2 consortium, respectively. (C) Projection along PC1 in B is plotted against average fractional abundance for the indicated organisms on the last day of fecal sampling (dpg 28) for each of the three tandem colonization conditions. Names of organisms are color-coded as in B. Horizontal lines denote the SD of mean fractional abundance for the indicated organisms in a particular colonization condition. See Dataset S7 for details.
We performed PCA on the matrix of mcSEED pathway relative expression profiles of individual strains under each tandem colonization sequence tested to identify groups of covarying metabolic pathways/modules that cluster organisms along principal components (eigenvectors) based on the similarity of their transcriptional profiles (Dataset S6C and see Materials and Methods). The first three principal components explained 75% of variation in the mcSEED relative expression profiles of S1 and S2 member species (Fig. 2B and Dataset S6D). Moreover, the mcSEED relative expression profiles of each organism clustered together in the PCA space irrespective of whether the climax community reflected a history of S1→S2, S2→S1, or S1+S2 colonization (including the two Ruminococcus gnavus strains) (Fig. 2B). Notably, there was a linear relationship between the projection of mcSEED relative expression profiles for a given strain along PC1 and its fitness (fractional abundance) (r2 [Pearson] = 0.7; Fig. 2C). Dataset S7 shows that the order of strain projection along PC1 was the same in each of the three different tandem colonization conditions evaluated. Importantly, because transcript counts were TPM-normalized prior to analysis of relative pathway expression, the linear relationship observed could not be ascribed simply to abundance differences between organisms, but rather pointed to differences in pathway presence and/or expression as fitness determinants.
Pathways associated with the two S2 organisms having highest fitness in all tandem colonization experiments.
Singular value decomposition (SVD) is a mathematical technique used to relate the eigenspectrum of the rows of a matrix to the eigenspectrum of the columns of the matrix (Fig. 3A) (17). We employed SVD to identify those mcSEED metabolic pathways/modules that most contribute to variance along PC1 in the relative pathway expression profiles of the four S1 and eight S2 organisms that survived in the three different types of tandem colonization experiments (Dataset S8A and see Materials and Methods). Specifically, we focused on the positive and negative 10th percentiles of mcSEED pathway/module projections along the first right singular vector shown in Fig. 3B (Dataset S8A). Hierarchical clustering of strains by relative expression scores for these 18 pathways shows that the two strains with the highest and second highest fractional abundance values in all tandem colonization experiments (P. distasonis and Akkermansia muciniphila, respectively) have similar relative pathway expression profiles that are distinct from the other six S2 and four S1 organisms that we were evaluating (Dataset S8B). The 18 mcSEED metabolic pathways/modules that distinguish the S2 organisms P. distasonis and A. muciniphila from the other bacterial strains include those involved in carbohydrate utilization (N-acetylgalactosamine/galactosamine [GAlNAc/GalN], fructoselysine/glucoselysine, and tagatose), metabolism of amino acids (branched-chain amino acid [BCAA] uptake, tryptophan uptake, and biosynthesis), vitamin/cofactors (riboflavin uptake), respiratory electron carriers (menaquinone biosynthesis), and fermentation products (l-lactate) (Fig. 3C).
Fig. 3.
Using SVD to identify pathways distinguishing bacterial strains with different fitness characteristics. (A) The mathematical relationship between the correlation structure of strains and mcSEED pathways/modules is depicted. The relationship between S1 and S2 strains (n = 36) is given by the 36 × 36 correlation matrix Xij and between mcSEED pathway/modules (n = 18) by the 18 × 18 correlation matrix Fij. The equation for eigendecomposition of each correlation matrix is shown within the matrix. SVD relates the two correlation matrices by transforming the relative expression matrix (ME) into a product of three different matrices, U, V, and Σ1/2. U and V are matrices of the left and right singular vectors from the strain and mcSEED pathway/modules correlation matrices, respectively; they are related by the singular values contained within Σ1/2. (B) Histogram of the projection of mcSEED metabolic pathway/modules along the first right singular vector computed by SVD. (C) Heat map of mcSEED metabolic pathway/module relative expression relative to the reference condition highlighted in boldface (P. distasonis in the S1→S2 colonization condition) (Dataset S8B). Strains are hierarchically clustered according to the relative expression profile of the mcSEED metabolic pathways/modules that project within the 10th percentile of the histogram shown in B. Strain names are colored based on their membership in the S1 or S2 consortium as in Fig. 2B. (D) The source of the relative expression score for each organism/metabolic pathway pair from C is indicated by the coded key. “Reference pathway” refers to the mcSEED metabolic pathway/module in the reference organism, P. distasonis in S1→S2 colonization condition. “Test pathway” refers to the pathway/module of a test organism in the indicated colonization condition. See Dataset S9 for values associated with each symbol in the matrix. Symbol key:  , indicated metabolic pathway/module present and expressed in both test and reference organisms with expression in the test organism being statistically significantly different from the reference organism;
, indicated metabolic pathway/module present and expressed in both test and reference organisms with expression in the test organism being statistically significantly different from the reference organism;  , pathway/module present and expressed in test and reference strains but not at statistically significantly different levels;
, pathway/module present and expressed in test and reference strains but not at statistically significantly different levels;  , present but not expressed in test, but present and expressed in reference strain;
, present but not expressed in test, but present and expressed in reference strain;  , absent in test strain, but present and expressed in reference strain;
, absent in test strain, but present and expressed in reference strain;  , present and expressed in test but absent in reference strain;
, present and expressed in test but absent in reference strain;  , present but not expressed in test strain and absent in reference strain;
, present but not expressed in test strain and absent in reference strain;  , absent in both test and reference strains.
, absent in both test and reference strains.
Fig. 3D deconvolutes the relative expression scores for the 18 metabolic pathways/modules in the four S1 and eight S2 organisms in each of the three colonization conditions into whether the pathway was present or absent, if present whether it was expressed, and if expressed whether expression was significantly different from in the reference P. distasonis strain present in the community that had experienced the indicated colonization history (i.e., S1→S2, S2→S1, or S1+S2). [Nonparametric statistical comparisons were performed between mcSEED-aggregated expression data by Dunn’s multiple comparisons test versus the corresponding reference P. distasonis, following a significant Kruskal–Wallis test (P < 0.05) (Dataset S9)]. Fig. 3 C and D present 648-cell matrices of 18 metabolic pathways/modules by 12 organisms by three tandem colonization conditions. Fig. 3D shows the nine mcSEED pathways/modules comprising the negative 10th percentile of pathway/module projections along the first right singular vector (ethanol fermentation, BCAA uptake, GalNAc/GalN utilization, l-lactate fermentation, riboflavin uptake, pantothenate [vitamin B5] uptake, tryptophan uptake, and tagatose utilization) and the nine pathways/modules comprising the positive 10th percentile of projections (propionate fermentation, lipoate biosynthesis, cobalt uptake for cobalamin [B12] biosynthesis, tryptophan biosynthesis, gluconate utilization, inositol utilization, menaquinone biosynthesis, proline degradation, and histidine degradation). Comparing Fig. 3D with the scores provided in Fig. 3C reveals that the source of the positive relative expression score of the pathways comprising the negative 10th percentile in the test organisms is largely due to their absence in P. distasonis (227 of 324 cells [70%] with the remainder of cells reflecting absence in both the reference and test organisms). In contrast, for the nine pathways comprising the positive 10th percentile of projections, 79 of 324 cells (24%) reflect statistically significant differences in expression while 144 of 324 (44%) reflect pathway absence. Importantly, with the exception of the lipoate and tryptophan biosynthesis pathway in Peptoclostrium difficile (the taxonomic assignment now applied to what was formerly Clostridium difficile; ref. 25) and the tryptophan biosynthesis and gluconate utilization pathways in E. casseliflavus, the pattern of “expressed/not expressed” across the nine pathways/modules in each of the 11 test organisms was robust to order of colonization. Based on this criterion, priority effects appear to be minimal in this model of succession.
A. muciniphila is phylogenetically unrelated to P. distasonis but exhibits the second-highest fitness in each of the tandem colonizations tested (Fig. 2C). Therefore, using A. muciniphila as an alternative reference strain for interrogating the source of relative expression scores shown in Fig. 3C provides an opportunity to test whether the results displayed in Fig. 3D are robust to the choice of reference strain. We find that using A. muciniphila recapitulates a pattern of relative expression scores very similar to when P. distasonis is employed as the reference with the exception of four pathways: ethanol fermentation, pantothenate uptake, gluconate utilization, and histidine degradation (compare Fig. 3D, SI Appendix, Fig. S5, and Dataset S10). Genes comprising these four pathways have opposite patterns of presence/absence in the two organisms: A. muciniphila has the apparatus for ethanol fermentation and pantothenate uptake while P. distasonis does not; P. distasonis contains the pathway for degradation of histidine and a potential gluconate transporter while A. muciniphila does not (Fig. 3D).
Fitness determinants in the four S1 organisms that persist in the presence of S2 consortium members.
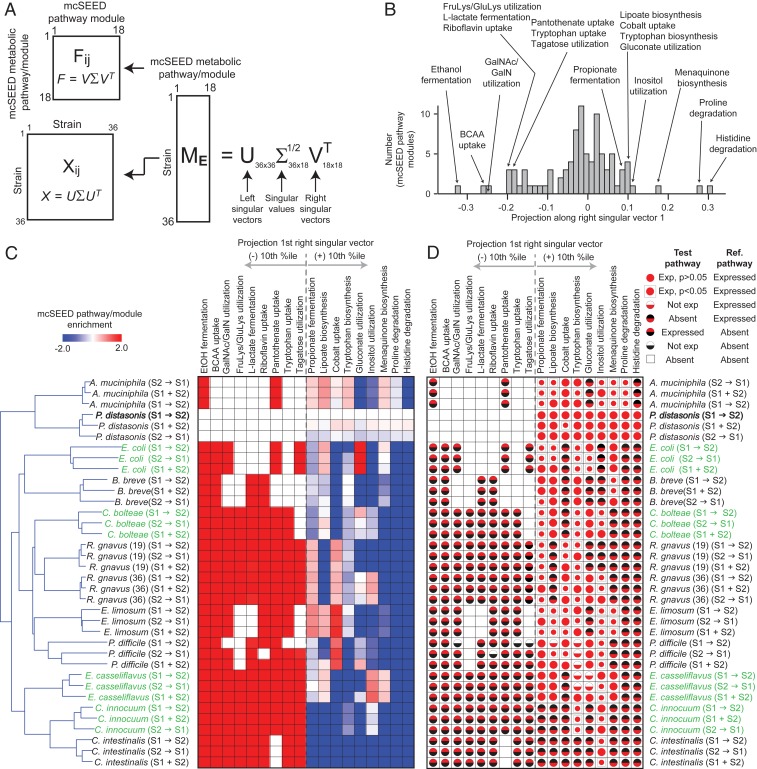
Having identified 18 fitness-associated metabolic pathways among the 92 evaluated, we focused on the four S1 organisms (C. innocuum, E. casseliflavus, C. bolteae, and E. coli) that survive introduction of the S2 consortium as well as the S1 consortium member, Enterococcus faecium, that suffers the greatest mean reduction in its fractional abundance after introduction of S2 organisms (fractional abundance 0.14 in the S1-only colonization condition compared to 0.002, 0.006, and 0.007 in the S1→S2, S2→S1, and S1+S2 conditions, respectively [SI Appendix, Fig. S5 and Dataset S3A]). To address the question of why C. innocuum, E. casseliflavus, C. bolteae, and E. coli are able to survive while E. faecium exhibited a pronounced reduction in fitness, mcSEED relative expression profiles were created for each of these organisms for all four colonization conditions (S1 alone, S1→S2, S2→S1, and S1+S2). E. faecium in the “S1 consortium only” treatment group served as the reference to create a “mcSEED relative expression matrix” spanning the 18 mcSEED metabolic pathways described above (Dataset S11 A and B). Using E. faecium as the reference allowed us to determine 1) the extent to which its transcriptome changed in the presence of the S2 consortium members compared to the S1 alone colonization condition where its fitness in the climax community was greater than that of C. innocuum, E. coli, and C. bolteae and 2) the extent to which its transcriptome in the tandem colonization conditions resembled that of the four survivors. Coverage of genes represented in the E. faecium transcriptome in the S2→S1 and S1+S2 colonization conditions was comparable to the S1 alone condition, while coverage in S1→S2 was decreased from ∼83% for S2→S1 compared to S1 alone to 38% for S1→S2 compared to S1 alone with the coverage threshold for inclusion of a gene in the analysis set at 50 reads for that gene (note that this reduced coverage trend is preserved if the threshold is raised to 100, 500, or 1,000 reads; see Dataset S12). Therefore, the E. faecium transcriptome from the S1→ S2 colonization condition was omitted from the analysis.
Considering the S1 alone, S1+S2, S1→S2, and S2→S1 colonization conditions, PCA revealed that the first three principal components explain 88% of variability in the mcSEED relative metabolic pathway/module expression profiles of C. innocuum, E. casseliflavus, C. bolteae, and E. coli and E. faecium (Fig. 4A, Dataset S11C, and see Materials and Methods). The relative expression profiles of C. innocuum, E. casseliflavus, C. bolteae, and E. coli were not affected by exposure to the S2 consortium as defined by their positions along any of the three principal components, while that of E. faecium changed primarily along PC3. This was true regardless of the order presentation of the S2 consortium.
Fig. 4.
S1 members that survive after introduction of the S2 consortium retain their mcSEED metabolic pathway expression pattern. (A) mcSEED metabolic pathway/module relative expression profiles defined by the 18 mcSEED pathways/modules identified by SVD in Fig. 3B for each organism under all colonization conditions are plotted on a PCA space. The relative expression profile of E. faecium (S1 alone) is used as the reference. The space on the right of the panel has been rotated by 60° for ease of visualization. (B–D) SVD performed on the PCA space in A yields projections of mcSEED metabolic pathways/modules onto right singular vectors 1 to 3. Histograms of these projections onto right singular vector 1 (B), vector 2 (C), and vector 3 (D) are shown. Projections within the positive and negative 10th percentiles for the first and second singular vectors are labeled and included in E. As the third right singular vector is negatively skewed, the projections within the negative 20th percentile are labeled and also included in E. (E) The mcSEED metabolic pathways/modules that define separation of species’ relative expression profiles along PC1, PC2, and PC3 are shown in heat-map form for all organisms in all colonization conditions. mcSEED metabolic pathway/module relative expression scores are shown as are a breakdown of the elements of that pathway/module’s score (i.e., pathway present/absent; whether or not expressed; whether expressed at levels significantly different from the pathway in the reference organism). See the legend to Fig. 3D for the key to the symbols used in each cell in the lower portion of the panel and Dataset S13 for the values associated with the symbols. (F) Box plots of levels of N-acetylgalactosamine and tagatose in cecal contents harvested from mice belonging to the indicated treatment group.
By performing SVD on the mcSEED relative expression matrix, we found that the position of each organism’s mcSEED pathway relative expression profile along the three principal components is determined by 11 of the 18 metabolic pathways (Fig. 4 B–D, Dataset S11D, and see Materials and Methods). The mcSEED pathway relative expression profile for E. coli under all colonization conditions is positioned on the negative extreme of PC1 (Fig. 4A). Fig. 4E shows that this position in PCA space corresponds to positive relative expression scores for metabolic pathways/modules involved in BCAA (isoleucine/leucine/valine) uptake, tryptophan biosynthesis, and menaquinone biosynthesis relative to E. faecium in the S1-alone colonization condition and negative scores for the l-lactate fermentation pathway (Dataset S11A). The positive relative expression score for the BCAA biosynthesis pathway reflects its increased expression compared to the reference organism (statistically significant in the S1+S2 and S2→S1 treatment groups and but not reaching statistical significance in the S1-alone and S1→S2 conditions [Dataset S13]). For the tryptophan and menaquinone biosynthesis pathways, the positive score originates from its expression in E. coli and pathway absence in the reference E. faecium strain. The negative score for the l-lactate fermentation pathway reflects its absence from E. coli and presence in the reference E. faecium strain (Dataset S11A). The mcSEED pathway relative expression profile for C. innocuum under all colonization conditions is positioned on the positive extreme of PC2 (Fig. 4A). This corresponds to positive relative expression scores for the BCAA uptake, tryptophan biosynthesis, and fructoselysine/glucoselyine utilization pathways and negative scores for the gluconate utilization, lipoate biosynthesis, and propionate production (fermentation) pathways (Fig. 4E). The negative score of the latter pathways reflects their absence in C. innocuum (Dataset S11A). The positive scores for the BCAA uptake and fructoselysine/glucoselysine utilization pathways reflects their statistically significant increases in expression in C. innocuum relative to E. faecium (Dataset S13), while the positive score for the tryptophan biosynthetic pathway is due to its absence in the genome of E. faecium (Dataset S11A). The mcSEED metabolic pathway/module relative expression profile for C. bolteae under all colonization conditions is positioned on the negative extreme of PC3 (Fig. 4A). This corresponds to negative scores for its tagatose and GalNAc/GalN utilization pathways, both of which share a common intermediate, tagatose-6-phosphate (Fig. 4E and Dataset S11A). The negative score for the tagatose pathway is due to its absence in C. bolteae (Dataset S13).
Mass spectrometry-based measurements of tagatose, N-acetylgalactosamine, proline, and histidine in cecal contents confirmed statistically significant differences in their levels in mice colonized with the S1 consortium alone compared to mice inoculated with just the S2 consortium or subjected to the various tandem colonization conditions (Dataset S4E and Fig. 4F). One conjecture is that low levels of GalNAc reflect the fact that among the most abundant strains in mice harboring the S1 consortium alone are three GalNAc utilizers (E. coli, Clostridium butyricum, and Citrobacter amalonaticus), while the S2 community only includes one GalNAc-utilzing strain (Collinsella intestinalis). Low levels of tagatose could be related to the high abundance of a tagatose utilizer, E. faecium in the S1 community, while the S2 community has only C. intestinalis as a potential tagatose utilizer. However, testing hypotheses about how these observed differences in carbohydrate and amino acid concentrations are related to differences in bacterial gene expression as well as their extracellular versus intracellular distributions, rates of uptake, and metabolic origins or fates in S1 and S2 community members will require time-series expression measurements and complex in vivo flux analyses using isotopically labeled compounds.
In summary, as shown in the PCA space in Fig. 4A, the mcSEED metabolic pathway/module relative expression profiles of the four S1 “survivors” remain unchanged in the various colonization contexts that also contain members of the S2 consortium, thus providing a measure of how these organisms are suited to the environment created by introduction of S2 organisms. Our findings indicate that, in the context of this experimental model, their fitness reflects the presence and/or expression of key genomic assets (mcSEED pathways/modules) that are not present in the nonpersisting E. faecium.
Discussion
Characterizing determinants of microbial succession is critical for defining how gut bacterial community compositions are achieved; this includes understanding 1) to what degree the sequential order of microbial introduction determines composition, 2) what determines the fitness of specific member species over others, and 3) how member species can survive perturbations to the gut ecosystem. Here, we describe a simplified model of microbial succession that involves two defined consortia of cultured, sequenced bacterial strains representing early and later colonizers of an infant human gut, introduced singly and in different order into gnotobiotic mice fed an IF diet. Our experimental and computational approaches allowed us to reduce a dataset of 103,102 genes to a lower-dimensional dataset of 18 metabolic pathways that correlate with bacterial fitness in competition experiments between S1 and S2 consortium members. Members of the S2 consortium generally outcompeted members of the S1 consortium in a reproducible fashion, independent of their order of arrival, indicating that priority effects were not deterministic in this model. The model system allowed us to dissect the underpinning of the dominance of individual S2 consortium members over S1 consortium members by determining the extent to which 18 mcSEED metabolic pathways/modules were represented in their genomes and expressed. Moreover, the representation of these pathways and their expression in the four S1 organisms that do survive in the three different types of tandem colonization treatments examined revealed how they were better suited (adapted) to the environment created by the S2 consortium than the other S1 strains.
The considerable genetic, transcriptional, and metabolic variation observed within and across the gut microbial communities of humans provides a “substrate” for gut microbiota adaptation to varying selective pressures over varying time scales (26). Relating the varied genomic features of gut community members, and expression of these features, to the overall temporal pattern of community development represents a key aspect of the formidable challenge faced when seeking to understand the determinants and form of succession. The ability to deliberately control membership of bacterial consortia and the order of their introduction into recipient gnotobiotic animals, combined with feature-reduction approaches, provides an avenue for identifying the significance of genome variation/gene expression, at strain-level resolution, in determining the “trajectory” of succession. The output of this type of experimental and computational approach is a low-dimensional description of fitness determinants.
The experimental and analytic approaches employed may be useful for a variety of future studies. In principle, they are suitable for characterizing ecological processes that govern community assembly in the gut such as competition, niche partitioning, exclusion, limiting similarity, and priority effects and testing hypotheses based on these concepts. They can be used to model a variety of types of community disturbances in order to 1) decipher the underpinnings of robustness/resiliency when such disturbances are applied and 2) identify new metrics for characterizing the severity of perturbations and the efficacy of their repair with existing and new therapeutic agents. The present study used mcSEED pathways as an exemplary annotation because these pathways are extensively curated. However, a variety of other annotation schemes can be applied to microbial genomes in order to systematically assess their value in revealing which key features in community members relate to their fitness. Finally, we do not provide evidence of the dependence of our results (identified pathways) on the choice of strains used to construct the S1 and S2 consortia. The approaches to modeling succession described in the present paper underscore the need to aggressively expand the capacity to genetically manipulate members of the gut microbiota so that the magnitude of the contribution of identified features to organismal properties in different community contexts can be tested directly.
Materials and Methods
Bacterial Culture Collections.
Deidentified fecal samples that had been collected from a vaginally delivered infant on postnatal days 38, 67, 133, 247, 339, and 683 were used to create a bacterial culture collection; these samples were obtained during the course of a previously reported and completed study that had been approved by the Human Research Protection Office of Washington University School of Medicine and described in ref. 18. No new fecal samples were collected for the current study. All fecal samples had been frozen at −80 °C shortly after their production and maintained at that temperature until further use.
To generate the culture collection, previously weighed frozen samples were brought into an anaerobic Coy chamber (atmosphere: 75% N2, 20% CO2, and 5% H2) and placed in sterile 50-mL conical tubes containing 5 mL of 2-mm-diameter sterile soda lime glass beads (26396-58; VWR). A sufficient amount of reduced phosphate-buffered saline (PBS) (PBS with 0.05% l-cysteine-HCl) was added to each sample so that the final concentration of fecal material was 100 mg/mL buffer. The tube was then subjected to four cycles of vortexing (30 s at 3,000 rpm with a 30-s pause per cycle) to disrupt clumps in the fecal sample. The resulting suspension was passed through a sterile 100-μm-pore-diameter cell strainer (352360; Corning Life Sciences). An equal volume of sterile reduced PBS/30% glycerol was added to the resulting filtrate and the tube was inverted several times to ensure mixing. Aliquots of the clarified fecal sample were stored in 2-mL Crimp-Top EZ Vials (Wheaton) that were then sealed and frozen at −80 °C. Other aliquots were serially diluted in reduced PBS and plated on brain heart infusion agar (BHI; Becton-Dickinson) supplemented with 10% horse blood and Mega33.1 agar (Dataset S2). Plates were incubated at 37 °C for 2 to 3 d in the anaerobic Coy chamber. A total of 3,000 colonies were picked and each inoculated into Mega33.1 medium in 96 deep-well plates (260251; Thermo Fisher). Plates were covered with aluminum foil seals and incubated at 37 °C for 2 to 3 d in the anaerobic Coy chamber. A BioTek Precision XS robot, located within the Coy chamber, was used to transfer a 50-μL aliquot of each of culture into wells of a 96 shallow-well plate (92696; Midwest Scientific) containing 50 μL of reduced PBS/30% glycerol. Replicate plates were generated, sealed, and stored at −80 °C until use.
Genomic DNA was isolated from the remaining material from the deep-well plate and subjected to V4-16S ribosomal DNA (rDNA) amplicon sequencing (primers 515F and 806R). Isolates were grouped into 97% ID OTUs. Three to four isolates representing each OTU were then subjected to full-length 16S rDNA gene sequencing (primers 8F and 1391R). Isolates were grouped into a total of 68 unique isolates; those sharing ≥99% nucleotide sequence identity in their 16S rDNA genes were considered to represent a unique isolate. The 68 isolates were then cultured in Mega33.1 broth, LYHBHI broth, or LYHBHI broth supplemented with 0.1% (wt/vol) soluble starch (21780; Difco) and 0.5% (wt/vol) partially purified porcine stomach mucin (M1778; Sigma) (Dataset S2). Stocks were made in culture media containing 15% glycerol and stored at −80 °C.
Bacterial Genome Sequencing, Assembly, in Silico Metabolic Reconstructions, and Phenotype Predictions.
DNA was purified from each isolate. Barcoded libraries were prepared (Illumina TruSeq Nano DNA Library Prep Kit or Illumina Nextera DNA Library Prep Kit.) and sequenced (Illumina MiSeq instrument; paired-end 150-nt or 250-nt reads). Reads were demultiplexed and assembled (Spades version 3.5.0). Genes were initially annotated using Prokka (v1.11). Additional annotations were based on SEED, a genomic integration platform that includes a growing collection of complete and nearly complete microbial genomes with draft annotations performed by the RAST server (22). SEED contains a set of tools for comparative genomic analysis, annotation, curation, and in silico reconstruction of microbial metabolism. Microbial Community SEED (mcSEED) is an application of the SEED platform used for manual curation of a large and growing set of bacterial genomes representing members of the human gut microbiota (currently ∼2,600). mcSEED subsystems are user-curated lists/tables of specific functions (enzymes, transporters, and transcriptional regulators) that capture current (and ever-expanding) knowledge of specific metabolic pathways, or groups of pathways, projected onto this set of ∼2,600 genomes (22, 23). mcSEED metabolic pathways are lists of genes comprising a particular metabolic pathway or module; they may be more granular than a subsystem splitting it into certain aspects (e.g., uptake of a nutrient separately from its metabolism) (24). mcSEED pathways are presented as lists of assigned genes and their annotations in Dataset S5B. Predicted phenotypes are generated from the collection of mcSEED subsystems represented in a microbial genome and the results described in the form of a BPM (prototrophy or auxotrophy for an amino acid or a vitamin/cofactor; the ability to utilize specific carbohydrates and/or generate SCFA products of fermentation).
Indicator Species Analysis.
To identify bacterial taxa associated with different successional stages, an indicator species analysis was performed using a bacterial V4-16S rDNA dataset generated from the birth cohort of 40 healthy US twin pairs (18, 19) that included the child from whom we generated the culture collection. Prior to analysis, 97% ID OTUs that did not have a relative abundance of at least 0.1% in at least five samples were removed. The significance of each OTU’s association with S1, S2, or S3 was tested using 10,000 permutations, which were restricted within twin pairs, and P values were adjusted for false discovery rate according to the Benjamini–Hochberg method. The 68 isolated strains were then assigned to stages based on the associations of related OTUs from the twin pair dataset, according to the following criteria: 1) Each strain was assigned to the same Stage as the OTU bearing the same species assignment; 2) strains not sharing a species assignment with an OTU were assigned to the stage based on the postnatal age of the donor of the sample from which they were isolated; 3) all Enterococcaceae and Streptococcaceae were assigned to S1, because eight of nine Enterococcaceae OTUs and four of seven Streptococcaceae OTUs were indicators of S1.
Studies in Gnotobiotic Mice.
All experiments were performed according to protocols approved by the Washington University Animal Studies Committee. No inclusion or exclusion criteria were established; all animals studied were included in our analysis. Young (3- to 4-wk-old) male germ-free C57BL/6J mice were maintained in plastic flexible film gnotobiotic isolators under a strict 12-h light cycle (lights on at 0600) and weaned directly to an IF diet 1 wk before colonization with the different bacterial consortia. The IF diet consisted of a mixture of Similac Sensitive with Iron and unflavored whey protein powder (General Nutrition Corporation) mixed at a ratio of 8.5:1 (wt/wt). The powdered diet was sterilized by irradiation (20 to 50 kGy). The IF mixture (20 g) was dissolved in 50 mL of sterile water. IF was presented to each group of mice in a 50-mL-capacity liquid diet feeding tube (9019; Bioserv). Animals were able to feed ad libitum; boots were changed daily and litter (Aspen Chip; NEPCO) was changed twice a week.
Different treatment groups were maintained in separate gnotobiotic isolators (five mice per cage per treatment group per experiment; one cage per isolator). A frozen glycerol stock of a monoculture of each bacterial strain was thawed in the anaerobic Coy chamber and inoculated into medium that supported its growth (Dataset S2). Cultures were incubated under anaerobic conditions at 37 °C for 2 to 3 d. Strains were pooled at an equivalent optical density at 600 nm and then mixed with an equal volume of reduced PBS/30% glycerol. Aliquots of the pooled S1, the pooled S2, and the pooled S1+S2 consortium members were placed in 2-mL Crimp-Top EZ Vials (Wheaton), and the vials were sealed and stored at −80 °C prior to gavage. Initial gavage involved introduction of 200 μL of the pooled S1, S2, or S1+S2 consortia, via a flexible 3.8-cm-long, 20-gauge plastic tube (Fisher) into the stomach of each recipient mouse. Germ-free controls were maintained in a single gnotobiotic isolator and fed IF for 4 wk before they were killed.
COPRO-Seq.
The time points of fecal sample collection are provided in Fig. 1. Fecal DNA was purified from each sample and shotgun sequencing libraries were generated using the Illumina Nextera DNA Library Prep Kit. Short-read multiplex sequencing of barcoded libraries was performed using protocols detailed previously (16, 20, 27). COPRO-Seq data were analyzed employing software available at https://gitlab.com/hibberdm/COPRO-Seq (27, 28).
Microbial RNA-Seq.
RNA was isolated from 20- to 50-mg aliquots of cecal contents collected at the time mice were killed, immediately frozen in liquid nitrogen, and stored at −80 °C. Complementary DNA libraries were prepared, pooled, and subjected to multiplex sequencing using an Illumina NextSeq instrument (21.9 ± 13.2 million [mean ± SD] unidirectional 75-nt reads per sample) (28). Data were analyzed using methods described previously (28, 29). Briefly, reads were mapped to all bacterial genomes in the corresponding defined community using a short-read aligner appropriate for bacterial genomes (bowtie, v0.12.7; ref. 30), and allowing up to one nucleotide mismatch. Transcript abundance per gene was determined by quantifying reads that mapped within each annotated open reading frame (ORF) in each genome, followed by creation of “per-organism” tables containing sample and transcript quantitation information. Count tables and ORF length data were loaded into R (v3.6.0) and transcript counts per gene were normalized for each organism in each colonization condition using reads per kilobase per million (TPM). Normalized data were then aggregated according to mcSEED functional annotation to generate tables of mcSEED metabolic pathway/module normalized transcript count.
Following a significant Kruskal–Wallis test (P < 0.05), nonparametric statistical comparisons were performed between mcSEED-aggregated expression data by using Dunn’s multiple comparisons test (versus the corresponding reference organism). Analyses were conducted using the PMCMR (v4.3; ref. 31) in R (v3.6.0).
Targeted Mass Spectrometry.
Targeted metabolites were quantified using the external standard method based on peak areas of analytes. To measure amino acids, mono- and disaccharides, sugar acids, sugar alcohols, and amino sugars, cecal contents were homogenized in 20 volumes of high-performance liquid chromatography (HPLC)-grade water. Homogenates were centrifuged (10,000 × g for 10 min at 4 °C). A 100-μL aliquot of each supernatant was transferred to a clean 2-mL glass tube and combined with 400 μL ice-cold methanol. The mixture was vortexed and centrifuged (10,000 × g for 10 min at 4 °C) and a 450-μL aliquot of the resulting supernatant was evaporated to dryness. Dried samples were derivatized by adding 80 μL methoxylamine solution (15 mg/mL stock solution prepared in pyridine) to methoximate-reactive carbonyls (incubation for 16 h for 37 °C), followed by replacement of exchangeable protons with trimethylsilyl groups using N-methyl-N-(trimethylsilyl) trifluoroacetamide together with a 1% vol/vol catalytic admixture of trimethylchlorosilane (incubation for 1 h at 70 °C). Heptane (160 μL) was added and a 1-µL aliquot of each derivatized sample was injected into an Agilent 7890B/5977B gas chromatography–mass spectrometry system.
To measure B vitamins cecal contents were homogenized in 40 volumes per weight of 50% methanol. After centrifugation (10,000 × g, 4 °C), 200 µL of the supernatant was dried in a centrifugal evaporator. Dried samples were resuspended in 100 μL of 10% methanol and centrifuged at 10,000 × g for 2 min at 4 °C. A 80-μL aliquot of each supernatant was placed into a sample vial and 5 μL was injected into a 1290 Infinity II UHPLC system coupled to a 6470 Triple Quadrupole (QQQ) mass spectrometer equipped with a Jet Stream electrospray ionization source (Agilent Technologies). Chromatographic separation was performed on a Poroshell 120 SB-AQ, 3 × 100 mm, 2.7 µm column (Agilent Technologies), using the following gradient conditions: 5 to 95% solvent B (0 to 6 min), 95% solvent B (6 to 8 min) at a flow rate of 0.5 mL/min. Solvent A was an aqueous solution containing 0.1% formic acid and 5 mM ammonium formate. Solvent B contained methanol with 0.1% formic acid. Mass spectra were acquired in positive mode using the following conditions: capillary voltage at 2,100 V, nitrogen as the nebulizer gas (35 pounds per square inch), drying gas flow rate and temperature of 8 L/min and 300 °C, respectively, and sheath gas flow rate and temperature of 12 L/min and 300 °C. Transitions were taken from an optimized dynamic multiple reaction monitoring library that we generated.
The extent to which an analyte was increased in the cecum for a particular colonization condition relative to germ-free controls was calculated (see Dataset S4C for results from individual animals). PCA was performed on the nutrient relative abundance matrix constructed considering 1) 47 analytes (rows) whose increase was dependent on colonization sequence (orange portion of dendrogram in SI Appendix, Fig. S2A) and 2) all mice subjected to each colonization condition (25 columns). This yielded 25 principal components. PC1 and PC2, shown in SI Appendix, Fig. S2B, define two axes onto which each mouse subject to each colonization condition was projected (x and y coordinates, respectively).
Using PCA and SVD to Identify Determinants of Fitness in Figs. 2 and 3.
Microbial RNA-Seq of cecal contents was performed on mice subject to tandem colonization conditions and the data were aggregated into expression levels of mcSEED metabolic pathways from which mcSEED relative expression profiles were created for each organism in each colonization sequence. A pseudocount of 0.1 was added to all values prior to calculation of log-ratios in order to enable comparisons between profiles in which specific mcSEED pathways/modules were not encoded or not expressed. PCA was performed on the rows (32 in total) of the mcSEED relative expression matrix depicted in Fig. 2A. This calculation yielded 32 eigenvectors (principal components) onto which the mcSEED pathway/module relative expression profile of each organism subject to a specific colonization condition was projected. The first three eigenvectors defined the PCA space shown in Fig. 2B, where the projection of mcSEED relative expression profiles onto these eigenvectors comprised the x, y, and z coordinates.
SVD was performed to identify those metabolic pathways that most contribute to PC1 in Fig. 2B. The details of this calculation are as follows. The mcSEED pathway/module relative expression matrix M (dimensions of 36 rows by 92 columns; shown in Fig. 2A) was factorized using SVD into three separate matrices (Eq. 2):
| [2] |
where U is a square symmetric matrix of the “left singular vectors” signifying covariation between organisms (dimensions of 36 rows by 36 columns), E is a diagonal matrix of “singular values” relating the covariation between organisms with that of mcSEED metabolic pathways (dimensions of 36 rows by 92 columns), and V is a square symmetric matrix of the “right singular vectors” associated with the covariation between mcSEED metabolic pathways (dimensions 92 rows by 92 columns; superscript “T” indicates the matrix transpose) (Fig. 3A).
The mcSEED metabolic pathways that contribute to variance along PC1 shown on the y axis of Fig. 2C were identified by recomputing a mcSEED relative expression matrix derived from considering only the first left singular vector, singular value, and right singular vector in Eq. 3 (dimensions of each factorized matrix and recomputed matrix (MEv1) are shown in subscripts):
| [3] |
The mcSEED metabolic pathways that contribute to the top and bottom 10th percentile of the left singular vectors (matrix VT) are those shown in Fig. 3C.
Using PCA and SVD to Identify Determinants of Persistence in Fig. 4.
An mcSEED relative expression matrix was generated comprising the 18 mcSEED metabolic pathways associated with bacterial fitness identified in Fig. 3B (columns) and the four S1 “survivors” (C. bolteae, C. innocuum, E. casseliflavus, and E. coli) plus E. faecium in each of the colonization conditions involving the S1 consortium (rows). PCA was performed on the rows to create a space of mcSEED relative expression profiles associated with each organism subject to each colonization condition (Fig. 4A). To determine which mcSEED metabolic pathway/module was related to the variance observed over PC1, PC2, and PC3, the mcSEED relative expression matrix O (dimensions 19 rows by 18 columns) was subject to SVD and factorized into three separate matrices (Eq. 4):
| [4] |
Three new matrices were computed, derived from considering only the first, second, and third right singular vectors, singular values, and left singular vectors (Eqs. 5–7):
| [5] |
| [6] |
| [7] |
Histograms of the first, second, and third right singular vectors are shown in Fig. 4B. Analyses were performed using the Matlab_R2018a software package or R (v3.6.0).
Data Availability.
COPRO-Seq and microbial RNA-Seq datasets plus shotgun sequencing datasets generated from cultured bacterial strains have been deposited at the European Nucleotide Archive (ENA) under study accession no. PRJEB26512. Code is available for download from github (https://github.com/arjunsraman/Feng_et_al).
Supplementary Material
Acknowledgments
We thank Maria Karlsson, Marty Meier, Sabrina Wagoner, Su Deng, Justin Serugo, and Jessica Hoisington-López for superb technical assistance; Janaki Guruge for help with culturing human infant bacterial strains; and Barbara Warner and Philip Tarr for their earlier work in establishing a biospecimen repository of serially collected fecal samples from members of the twin birth cohort, an effort that was supported by a grant from Children’s Discovery Institute. Work described in the current study was supported by NIH grant DK30292. L.F. was the recipient of a postdoctoral fellowship from the Helen Hay Whitney Foundation and the Simons Foundation. A.S.R. was supported by the Washington University School of Medicine’s Physician Scientist Training Program. J.I.G. is the recipient of a Thought Leader award from Agilent Technologies.
Footnotes
Competing interest statement: J.I.G. is a cofounder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health.
This article is a PNAS Direct Submission. M.A.F. is a guest editor invited by the Editorial Board.
Data deposition: COPRO-sequencing and microbial RNA-sequencing datasets plus shotgun sequencing datasets generated from cultured bacterial strains have been deposited at the European Nucleotide Archive (ENA) under study accession no. PRJEB26512. Code is available for download from github (https://github.com/arjunsraman/Feng_et_al).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918951117/-/DCSupplemental.
References
- 1.Faust K., Raes J., Microbial interactions: From networks to models. Nat. Rev. Microbiol. 10, 538–550 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Layeghifard M., Hwang D. M., Guttman D. S., Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 25, 217–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proulx S. R., Promislow D. E. L., Phillips P. C., Network thinking in ecology and evolution. Trends Ecol. Evol. (Amst.) 20, 345–353 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Stewart C. J., et al. , Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vatanen T., et al. , The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd-Price J., et al. ; IBDMDB Investigators , Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou W., et al. , Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569, 663–671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höfer T., Sherratt J. A., Maini P. K., Dictyostelium discoideum: Cellular self-organization in an excitable biological medium. Proc. Biol. Sci. 259, 249–257 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Sole R., Goodwin B., Nonlinearity, Chaos, and Emergence in Signs of Life (Basic Books, New York, 2000), pp. 10–24. [Google Scholar]
- 10.Schneidman E., Berry M. J. 2nd, Segev R., Bialek W., Weak pairwise correlations imply strongly correlated network states in a neural population. Nature 440, 1007–1012 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldford J. E., et al. , Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halabi N., Rivoire O., Leibler S., Ranganathan R., Protein sectors: Evolutionary units of three-dimensional structure. Cell 138, 774–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bialek W., et al. , Statistical mechanics for natural flocks of birds. Proc. Natl. Acad. Sci. U.S.A. 109, 4786–4791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon D. M., The ecology of collective behavior. PLoS Biol. 12, e1001805 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raman A. S., et al. , A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science 365, eaau4735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehrig J. L., et al. , Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science 365, eaau4732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews H., Patterson C., Singular value decompositions and digital image processing. IEEE Trans. Acoust. 24, 26–53 (1976). [Google Scholar]
- 18.Planer J. D., et al. , Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufrêne M., Legendre P., Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366 (1997). [Google Scholar]
- 20.Wu M., et al. , Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350, aac5992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patnode M. L., et al. , Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 179, 59–73.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz R. K., et al. , The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overbeek R., et al. , The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodionov D. A., et al. , Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol. 10, 1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yutin N., Galperin M. Y., A genomic update on clostridial phylogeny: Gram-negative spore-formers and other misplaced clostria. Environ. Microbiol. 10, 2631–2641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S., et al. , Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNulty N. P., et al. , Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11, e1001637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hibberd M. C., et al. , The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci. Transl. Med. 9, eaal4069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dey N., et al. , Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell 163, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pohlert T., The pairwise multiple comparison of mean ranks package (PMCMR). R package (2014), https://cran.r-project.org/web/packages/PMCMR/index.html. Accessed 2 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
COPRO-Seq and microbial RNA-Seq datasets plus shotgun sequencing datasets generated from cultured bacterial strains have been deposited at the European Nucleotide Archive (ENA) under study accession no. PRJEB26512. Code is available for download from github (https://github.com/arjunsraman/Feng_et_al).