Significance
Plants have evolved unique membrane receptor kinases with extracellular leucine-rich repeat domains that regulate diverse developmental processes and that form the first layer of the plant immune system. Here it is shown that 2 sequence-related receptor kinases and their shape-complementary coreceptors selectively sense members of a small family of secreted peptide hormones to control formation of an important diffusion barrier in the plant root.
Keywords: Arabidopsis, receptor kinase, peptide hormone, root development, coreceptor
Abstract
Plants use leucine-rich repeat receptor kinases (LRR-RKs) to sense sequence diverse peptide hormones at the cell surface. A 3.0-Å crystal structure of the LRR-RK GSO1/SGN3 regulating Casparian strip formation in the endodermis reveals a large spiral-shaped ectodomain. The domain provides a binding platform for 21 amino acid CIF peptide ligands, which are tyrosine sulfated by the tyrosylprotein sulfotransferase TPST/SGN2. GSO1/SGN3 harbors a binding pocket for sulfotyrosine and makes extended backbone interactions with CIF2. Quantitative biochemical comparisons reveal that GSO1/SGN3–CIF2 represents one of the strongest receptor–ligand pairs known in plants. Multiple missense mutations are required to block CIF2 binding in vitro and GSO1/SGN3 function in vivo. Using structure-guided sequence analysis we uncover previously uncharacterized CIF peptides conserved among higher plants. Quantitative binding assays with known and novel CIFs suggest that the homologous LRR-RKs GSO1/SGN3 and GSO2 have evolved unique peptide binding properties to control different developmental processes. A quantitative biochemical interaction screen, a CIF peptide antagonist and genetic analyses together implicate SERK proteins as essential coreceptor kinases required for GSO1/SGN3 and GSO2 receptor activation. Our work provides a mechanistic framework for the recognition of sequence-divergent peptide hormones in plants.
Plant membrane receptor kinases with leucine-rich repeat ectodomains (LRR-RKs) form the first layer of the plant immune system and are key regulators of plant growth and development (1). LRR-RKs have evolved to sense small molecule, peptide and protein ligands, with linear peptides representing a large class of sequence-diverse signaling molecules in plants (1, 2). These peptides are processed from larger preproteins and subsequently posttranslationally modified (3). The size of the final, bioactive peptide ranges from 5 (phytosulfokine [PSK]) (2) to ∼21 to 23 amino acids (PEP1; CASPARIAN STRIP INTEGRITY FACTORS, CIF1/2) (4–6). Posttranslational peptide modifications include proline hydroxylation (Hyp), hydroxyproline arabinosylation, and tyrosine sulfation (sTyr) (2), and these modifications may allow for specific ligand recognition by the cognate LRR-RK (7–9). The disulfated PSK peptide binds to a pocket that is formed by the LRR domain of the receptor PSKR and a small “island domain” (9). PSK binding stabilizes the island domain and enables PSKR to interact with a SERK coreceptor, which is shared between many LRR-RK signaling pathways (1, 9). Unsulfated PSK variants bound the receptor with ∼25-fold reduced affinity when compared to wild type (9). Subsequently, other tyrosine sulfated peptides were discovered, including the ROOT MERISTEM GROWTH FACTORs (RGFs), 13-amino acid peptides containing an N-terminal Asp-Tyr (DY) motif (10), which is recognized by the sole tyrosylprotein sulfotransferase TPST in Arabidopsis (11). RGFs are sensed by a class of SERK-dependent LRR-RKs termed RGFRs (12, 13). RGFs bind the LRR ectodomain of RGFRs with dissociation constants in the high nanomolar range (13). Nonsulfated variants of the linear peptides showed an ∼200-fold reduction in binding affinity (13). The N-terminal sTyr in RGFs maps to a hydrophobic pocket located at the inner face of the LRR solenoid in RGF–RGFR complex structures, with the peptide adopting an extended conformation (13). A His/Asn motif forms the C terminus of RGFs and many other plant peptide hormones, such as IDA/IDLs involved in organ abscission and CLE peptides controlling plant stem cell maintenance (1, 7). The C-terminal His/Asn motif is recognized by 2 conserved arginines (the RxR motif) located at the inner surface of the LRR cores of different peptide sensing LRR-RKs (7, 13–16).
The LRR-RKs GASSHO1/SCHENGEN 3 (GSO1/SGN3) and GASSHO2 (GSO2) carry a RxR motif and were initially shown to be redundantly required for embryonic development (17, 18). Subsequently, a nonredundant role for GSO1/SGN3 was identified through a genetic screen for Casparian strip formation, an endodermal barrier allowing for selective nutrient uptake in the root (19, 20). The presence of the RxR motif suggested that GSO1/SGN3 and GSO2 may bind peptide ligands in planta, but the identify of these peptides remained unknown. The discovery that tpst/sgn2 loss-of-function mutants display Casparian strip phenotypes similar to sgn3 resulted in the identification of two 21-amino acid long, tyrosine sulfated peptides CIF1/2 as ligands for GSO1/SGN3 (6). A complementary biochemical interaction screen identified GSO1/SGN3 and GSO2 as bona fide receptors for these peptide hormones (5). Here we report the crystal structure of the GSO1/SGN3–CIF complex and dissect its ligand binding mode. We report previously uncharacterized CIF peptides differentially sensed by GSO1/SGN3 and GSO2 and reveal that GSO1 and GSO2 require SERK coreceptor kinases for receptor activation.
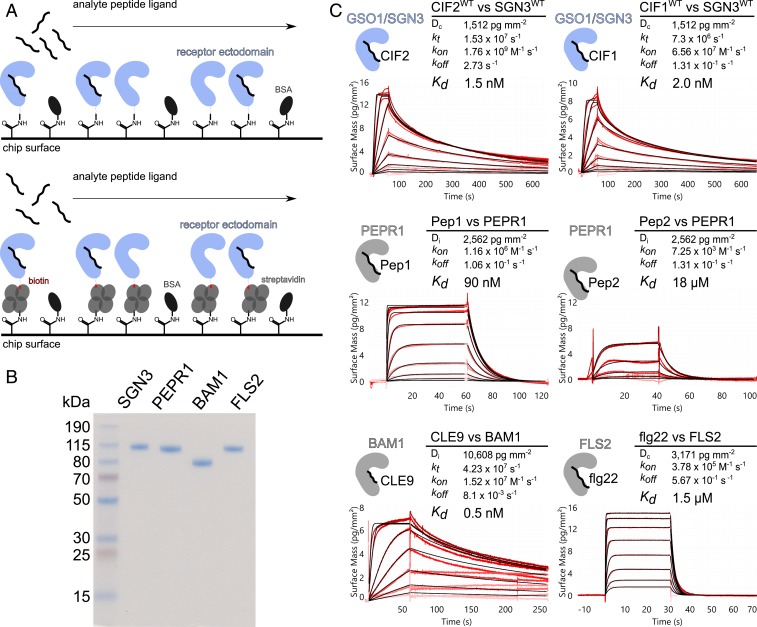
Results
The interaction between the GSO1/SGN3 ectodomain and synthetic sTyr-containing CIF1/2 peptides has been previously characterized in quantitative isothermal titration calorimetry (ITC) steady-state binding assays, yielding dissociation constants (Kds) ranging from ∼2 to 50 nM (6). We performed grating-coupled interferometry (GCI) kinetic binding assays (21) and found that GSO1/SGN3 binds the CIF1 and CIF2 peptides with Kds of ∼5 and ∼1 nM, respectively (Fig. 1), in agreement with the earlier report (6). Next, we compared the binding kinetics of GSO1/SGN3–CIF1/CIF2 to other, known receptor–peptide ligand pairs from Arabidopsis: The 23-amino acid PEP1 and PEP2 danger signal peptides bind the LRR-RK PEPR1 (4, 14) with drastically different binding affinities of ∼100 nM and ∼20 μM, respectively (Fig. 1). The CLE9 peptide (12 amino acids) binds the ectodomain of the LRR-RK BAM1 (22) with a Kd of ∼1 nM, similar to GSO1/SGN3–CIF2 (Fig. 1), and in agreement with a previously reported ITC experiment (23). The well-characterized immune elicitor peptide flg22 (24) binds the isolated FLS2 ectodomain (25) with a dissociation constant of 1.5 μM (Fig. 1). Together, our comparison reveals that plant LRR-RKs can sense peptide ligands with drastically different binding affinities and kinetics, with the GSO1/SGN3–CIF1/2 interaction ranking among the strongest receptor–ligand pairs.
Fig. 1.
GSO1/SGN3–CIF2 represents one of the strongest LRR-RK peptide–ligand pairs in Arabidopsis. (A) Schematic overview of the GCI binding assay. (A, Top) For direct amine coupling, receptor ectodomains (in blue) were immobilized onto the GCI chip, followed by passivation and quenching of the surface with BSA (in dark gray). Peptide ligands (in black) were applied as analyte in different concentrations to derive binding kinetics. (A, Bottom) For Avi-tag based coupling, streptavidin (in gray) was immobilized using the same amine-coupling method as shown above. Next, the biotinylated ectodomain of the respective receptor was captured by streptavidin. (B) Purity of the recombinantly expressed and purified LRR-RK ectodomains used in the GCI experiments. Shown is a Coomassie-stained SDS/PAGE, 1 µg of the respective LRR ectodomain was loaded per lane. Purified proteins were isolated from monomeric peak fractions in size-exclusion chromatography experiments. (C) Quantitative comparison of GSO1/SGN3–CIF2 with other known LRR-RK peptide–ligand pairs by GCI. The flow rate was 100 µL min−1 on each channel, except for FLS2 where the flow rate was adjusted to 55 µL min−1. Shown are sensorgrams with raw data in red and their respective fits in black. Binding kinetics were analyzed by a 1-to-1 binding model with mass transport in the case of GSO1/SGN3–CIF1/2 and CLE9–BAM1; a 1-to-1 binding model was used for the remaining interactions. Table summaries of kinetic parameters are shown alongside (Dc/Di, density of captured/immobilized protein; kt, mass transport coefficient; kon, association rate constant; koff, dissociation rate constant; Kd, dissociation constant).
To gain mechanistic insight into the GSO1/SGN3–CIF1/2 interaction, we determined the crystal structure of a GSO1/SGN3–CIF2 complex. The native protein did not yield diffraction quality crystals and hence we partially deglycosylated GSO1/SGN3 using a mix of endoglycosidases H, F1, and F3 (Materials and Methods). Crystals obtained in the presence of a synthetic sTyr-containing CIF2 peptide diffracted to ∼3.0-Å resolution and the structure was solved using the molecular replacement method. The final model contains 2 GSO1/SGN3–CIF2 complexes in the asymmetric unit, with a solvent content of ∼70%. The GSO1/SGN3 ectodomain contains 32 LRRs folding into a superhelical assembly previously seen in other plant LRR-RKs (Fig. 2A and SI Appendix, Fig. S1) (1). The structure completes ∼1.5 helical turns, forming the largest LRR ectodomain currently known in plants (Fig. 2A). The GSO1/SGN3 LRR core is sandwiched between canonical, disulfide bond-stabilized capping domains (Fig. 2A and SI Appendix, Fig. S1). Sixteen N-glyosylation sites are evident in the electron density maps of the partially deglycosylated protein, evenly distributed along the ectodomain (Fig. 2A and SI Appendix, Fig. S1). One CIF2 peptide binds in a fully extended conformation to the GSO1/SGN3 LRR core (LRRs 3 to 23) (Fig. 2 and SI Appendix, Fig. S1). In agreement with the crystal structure, GSO1/SGN3 behaves as a monomer in right-angle light scattering experiments in the presence and absence of the CIF2 ligand (Fig. 2B).
Fig. 2.
GSO1/SGN3 harbors a large spiral-shaped LRR domain providing the CIF peptide binding surface. (A) Structural comparison of the SGN3–CIF2 complex (Right) and the RGFR1–RGF1 complex (Left; PDB ID 5hyx, ref. 13). LRR domains (ribbon diagram) are shown in blue, peptide ligands in yellow (in bonds representation), N- and C-terminal capping domains in magenta, disulfide bonds in green, and N-glycans in gray. (B) Isolated and CIF2-bound GSO1/SGN3 ectdomains behave as monomers in solution. (B, Top) Analytical size-exclusion chromatography traces of the SGN3 ectodomain in the absence (blue line) or presence (red dotted line) of CIF2 peptides. (B, Bottom) Right-angle light scattering (RALS) traces in the absence (blue, Left) or presence (red, Right) of CIF2 peptides and including the derived molecular masses (black) of GSO1/SGN3 apo or SGN3–CIF2. Table summaries report the observed molecular weight (MW) and the dispersity (Mw/Mn). The theoretical molecular weight is 94.1 kDa for GSO1/SGN3 (residues 19 to 870) and ∼2.5 kDa for the synthetic CIF2 peptide.
We compared our GSO1/SGN3–CIF2 complex to the previously reported structure of the sTyr-peptide binding receptor RGFR (13) and found that the RGF peptide and the RGFR ectodomain are much smaller compared to CIF2 and GSO1/SGN3 (Fig. 2A). However, both RGFR and GSO1/SGN3 provide a binding pocket for the N-terminal sTyr residue and a RxR motif in close proximity to the C terminus of the respective peptide ligand (SI Appendix, Fig. S2). In our structure we find sTyr64 located in a hydrophobic pocket formed by GSO1/SGN3 residues originating from LRRs 3 to 5 (Fig. 3A and SI Appendix, Fig. S2). It has been previously established that the tyrosylprotein sulfotransferase TPST/SGN2 is genetically required for Casparian strip formation (6). In line with this, recombinant TPST/SGN2 obtained by secreted expression from insect cells has specific tyrosylprotein sulfotransferase activity toward CIF2, using 3′-phosphoadenosine-5′-phosphosulfate as substrate (SI Appendix, Fig. S3).
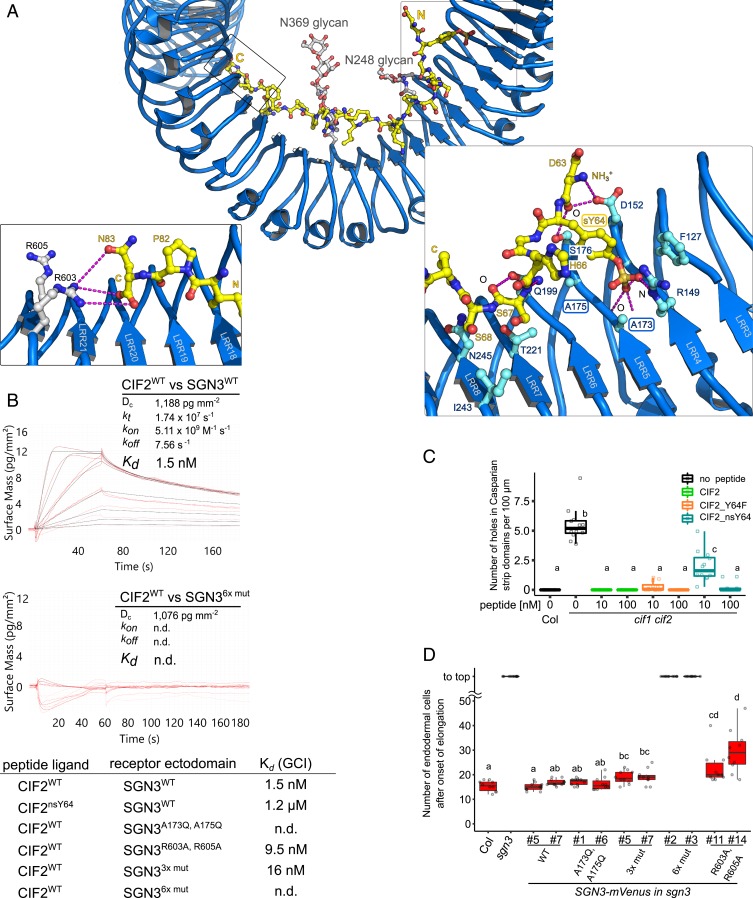
Fig. 3.
Many peptide–receptor interactions enable high-affinity CIF2 binding by GSO1/SGN3. (A, Top) Overview of the CIF2 binding site in GSO1/SGN3; colors are as in Fig. 2. (A, Left) Close-up view of the C terminus of the CIF peptide (in yellow) and the GSO1/SGN3 RxR motif (in gray). Potential hydrogen bonds are indicated as dotted lines (in magenta). (A, Right) Close-up view of the sTyr binding pocket in GSO1/SGN3. (B) GCI binding assays of CIF2 variants versus wild-type and mutant GSO1/SGN3 ectodomains. Peptides were supplied at 100 µL min−1 flow rate. Raw sensorgrams are shown in red, fitted data in black. A 1-to-1 binding model with mass transport was used in case of GSO1/SGN3–CIF2WT, and a 1-to-1 binding model for GSO1/SGN36x mut–CIF2WT. Table summaries of kinetic parameters are shown alongside (n.d., no detectable binding). (C) Quantitative analyses for the number of holes in Casparian strip domains per 100 µm in cif1 cif2 double mutants with CIF2 peptide-variant treatments. Shown are box plots spanning the first to third quartiles, with the bold line representing the median, and circles indicating the raw data. Whiskers indicate maximum and minimum values, except outliers (b and c, statistically significant difference from a, with P < 0.05, one-way ANOVA and Tukey test). (D) Quantification of propidium iodide (PI) staining on sgn3 mutants complemented with wild-type or mutant SGN3–mVenus under the control of the SGN3 promoter (bc, cd, and d, statistically significant from a, with P < 0.05, with one-way ANOVA and Tukey test).
The GSO1/SGN3 ectodomain bound synthetic sTyr-containing CIF2 (CIF2WT) with Kds of ∼2 nM and ∼40 nM in GCI and ITC assays, respectively (Fig. 3B and SI Appendix, Fig. S4). The binding stoichiometry is ∼1 in our ITC assays, in agreement with the GSO1/SGN3–CIF2 complex structure (Fig. 2 and SI Appendix, Fig. S4). Nonsulfated CIF2nsY64 interacted with the GSO1/SGN3 ectodomain with ∼100- to 1,000-fold reduced binding affinity, depending on the assay used (Fig. 3B and SI Appendix, Fig. S4). This suggests that the sTyr moiety formed by TPST/SGN2 in planta contributes to the specific recognition of CIF2 by GSO1/SGN3.
To validate our GSO1/SGN3–CIF2 complex structure, we replaced the conserved Ala173 and Ala175 from the sTyr binding pocket with glutamine (Fig. 3A and SI Appendix, Fig. S1). We found that the GSO1/SGN3A173Q/A175Q mutant protein bound CIF2WT and CIF2nsY64 with low micromolar affinity in ITC experiments (SI Appendix, Fig. S4). In kinetic GCI assays, no specific binding was detected for CIF2WT or CIF2nsY64 to GSO1/SGN3A173Q/A175Q (Fig. 3B and SI Appendix, Fig. S4). However, while replacement of sTyr with tyrosine in the synthetic peptide, or mutation of the sTyr binding pocket in the receptor strongly decreased CIF2 binding (∼100- to 1,000-fold) in vitro, the nonsulfated CIF2 peptide and the GSO1/SGN3A173Q/A175Q mutant protein could complement cif1 cif2 and sgn3 loss-of-function phenotypes in Casparian strip formation, respectively (Fig. 3 C and D and SI Appendix, Fig. S5).
We thus analyzed how other amino acids in the large CIF2 binding site in GSO1/SGN3 (∼1,500-Å2 buried surface area) (26) would contribute to the specific recognition of the peptide hormone (Fig. 3A). We first mutated the conserved RxR motif in GSO1/SGN3 LRR23, which is involved in the coordination of the C-terminal Asn83 in CIF1/CIF2 (Fig. 3A) and in many other plant peptide hormones (1, 7, 13, 16). Replacing Arg603 and/or Arg605 with alanine had only a moderate effect on CIF2 binding by GSO1/SGN3 (2- to 10-fold reduction) (Fig. 3B and SI Appendix, Fig. S4). In line with this, we find Arg603 and Arg605 not in direct hydrogen bonding distance with either the side chain of Asn83 or the C-terminal carboxyl group of the CIF2 peptide (Fig. 3A). Despite their moderate contribution to CIF2 binding, a GSO1/SGN3R603A/R605A mutant only partially complemented the sgn3 Casparian strip phenotype (Fig. 3D).
The central part of the CIF peptide binding groove in GSO1/SGN3 is mainly formed by hydrophobic residues and by selected hydrogen bond interactions between residues originating from LRRs 6 to 17 and backbone atoms from CIF2 (Fig. 3A and SI Appendix, Fig. S4). CIF peptides have been previously demonstrated to be hydroxyprolinated (5) and the corresponding Pro69 and Pro71 residues in CIF2 form part of the central binding site (SI Appendix, Fig. S4). While the hydroxyl group of Hyp71 may establish a hydrogen bond with GSO1/SGN3 residue Asp293, we found that CIF2Hyp69,71 and CIF2WT bound GSO1/SGN3 with very similar dissociation constants and both could complement the cif1 cif2 Casparian strip phenotype in a same concentration range (SI Appendix, Fig. S6).
We next replaced 3 conserved aromatic residues Tyr416, Phe438, and Tyr440 in the central binding groove by alanine (hereafter called SGN33xmut), and again observed a moderate reduction in CIF2 binding (∼10-fold) (Fig. 3B and SI Appendix, Fig. S4). Transgenic plants recapitulating these mutations partially rescued the sgn3 phenotype in planta (Fig. 3D). However, when we combined this triple mutant with the mutations targeting the sTyr binding pocket in GSO1/SGN3 (SGN36xmut) (Fig. 3A), CIF2 binding was disrupted (Fig. 3B and SI Appendix, Fig. S4) and the GSO1/SGN36xmut mutant failed to complement the sgn3 phenotype (Fig. 3D and SI Appendix, Fig. S5). Together, our structural and mutational analysis suggests that GSO1/SGN3 uses a large number of interactions to specifically recognize CIF peptides, requiring numerous receptor–peptide contacts to be altered in order to disrupt CIF peptide binding in vitro and GSO1/SGN3 function in vivo.
We noted in our structure that outside the sTyr binding pocket, CIF2 mainly uses main-chain atoms to contact the GSO1/SGN3 LRR domain. Thus, sequence-divergent tyrosine sulfated peptides may represent bona fide ligands for GSO1/SGN3. We identified additional, putative CIF peptides in Arabidopsis and in other plant species, harboring an N-terminal Asp-Tyr motif required for TPST/SGN2 substrate recognition (10), 2 central proline residues, and a C-terminal His/Asn residue (SI Appendix, Fig. S7). From these candidates we selected the closely related, previously uncharacterized At5G04030 (CIF3 hereafter) and At1G28375 (CIF4) for further analysis (Fig. 4A). GCI experiments revealed that tryosine sulfated but not the nonsulfated CIF3 synthetic peptide bound to the GSO1/SGN3 ectodomain with nanomolar affinity (Fig. 4B). Due to its hydrophobicity, we could not dissolve the CIF4 peptide in our GCI buffer, and thus performed ITC experiments instead, titrating CIF4 into a GSO1/SGN3 solution containing 5% (vol/vol) dimethyl sulfoxide (DMSO). In these buffer conditions, CIF4 binds GSO1/SGN3 with 300 nM affinity and with 1:1 binding stoichiometry (Fig. 4C). DMSO appears to negatively affect binding, as the CIF2 control bound with ∼6-fold reduced binding affinity when compared to aqueous buffer conditions (Fig. 4C and SI Appendix, Fig. S4). Together, the newly identified CIF3 and CIF4 peptides bind to GSO1/SGN3 with high affinity in vitro.
Fig. 4.
Structure-guided identification of CIF peptides. (A) Multiple sequence alignment of the CIF1–4 peptides. The conserved sTyr is highlighted in red, hydroxyprolines are in yellow, and the C-terminal asparagine/histidine is shown in blue. (B) GCI assays of CIF3WT or CIF3nsY46 versus the wild-type GSO1/SGN3 ectodomain. Flow rate was 100 µL min−1. Sensorgrams are presented with raw data in red and their respective fits in black. Binding kinetic analyses were performed with a 1-to-1 binding model. Table summaries of kinetic parameters are shown alongside. (C) ITC assays of CIF2 or CIF4 wild-type peptides versus the SGN3 wild-type ectodomain. Table summaries for dissociation constants (Kd) and binding stoichiometries (N) are shown (±fitting error).
We next tested if CIFs can also bind to the LRR-RK GSO2, which together with GSO1/SGN3 controls plant embryo development (17). We found that CIF3 but neither CIF1 or CIF2 bound to the recombinant GSO2 ectodomain (Fig. 4B). CIF3 binds both GSO1/SGN3 and GSO2 with a Kd of ∼4 nM (Fig. 4B). Due to its hydrophobicity, we could not assess binding of CIF4 to GSO2. Together, GSO1/SGN3 and GSO2 display different CIF peptide binding preferences in vitro.
In line with our biochemical findings, application of synthetic CIF3 and CIF4 peptides could rescue the cif1 cif2 Casparian strip phenotypes (Fig. 5A). However, CIF3 and CIF4 marker lines showed no expression in roots and a cif3 cif4 double mutant had no apparent Casparian strip phenotype (Fig. 5 B and C). The cif3 cif4 double mutant in addition did not display the known seed shape and cuticle permeability phenotypes previously characterized for gso1 gso2 mutant plants (Fig. 5 D and E and SI Appendix, Fig. S8) (17). Given the fact that we could identify CIF3 and CIF4 orthologs in other plant species (SI Appendix, Fig. S7), we speculate these CIF peptides to be involved in yet unidentified GSO1/GSO2 regulated signaling events.
Fig. 5.
CIF3 and CIF4 are not involved in Casparian strip formation. (A) Quantitative analyses of number of holes in Casparian strip domains per 100 µm in Col (WT) or cif1 cif2 mutant plants treated with synthetic CIF2, CIF3, or CIF4 peptide, respectively (n = 12 [experiment with CIF3] and for n ≥ 12 [experiment with CIF4] for each condition). Shown are box plots spanning the first to third quartiles, with the bold line representing the median, and circles indicating the raw data. Whiskers indicate maximum and minimum values, except outliers. Different letters indicate statistically significant differences (b and c, statistically significant difference from a, with P < 0.05, one-way ANOVA and Tukey test). Note that due to the solubility of CIF4, the experiment with CIF4 was done with 0.05% (vol/vol) DMSO in all conditions including the control. (B) Promoter activities around onset of Casparian strip formation. Each promoter drives a nuclear localization signal (NLS)–3xVenus reporter gene. Cell walls were stained with PI. Cell layers are labeled as Epi (epidermis), Cor (cortex), En (endodermis), and Ste (stele). (Scale bar, 40 μm.) (C) cif3 cif4 double mutants do not show Casparian strip barrier defects. Lignin images were taken around 10 cells after onset of Casparian strip formation. (Scale bar, 20 μm.) (D) CIF peptides do not display gso1 gso2 seed shape phenotypes. Shown are mature seeds from Col, cif1cif2, cif3 cif4-1, cif3 cif4-2, and sgn3/gso1 gso2. The seeds from sgn3/gso1 gso2 had aberrant shapes (indicated by a *) but seeds from other genotypes showed the normal shapes as did the Col wild-type control. (Scale bars, 0.5 mm.) (E) Cuticle permeability to toluidine blue in etiolated seedlings. Shown are representatives of each genotype. (Scale bar, 1 mm.)
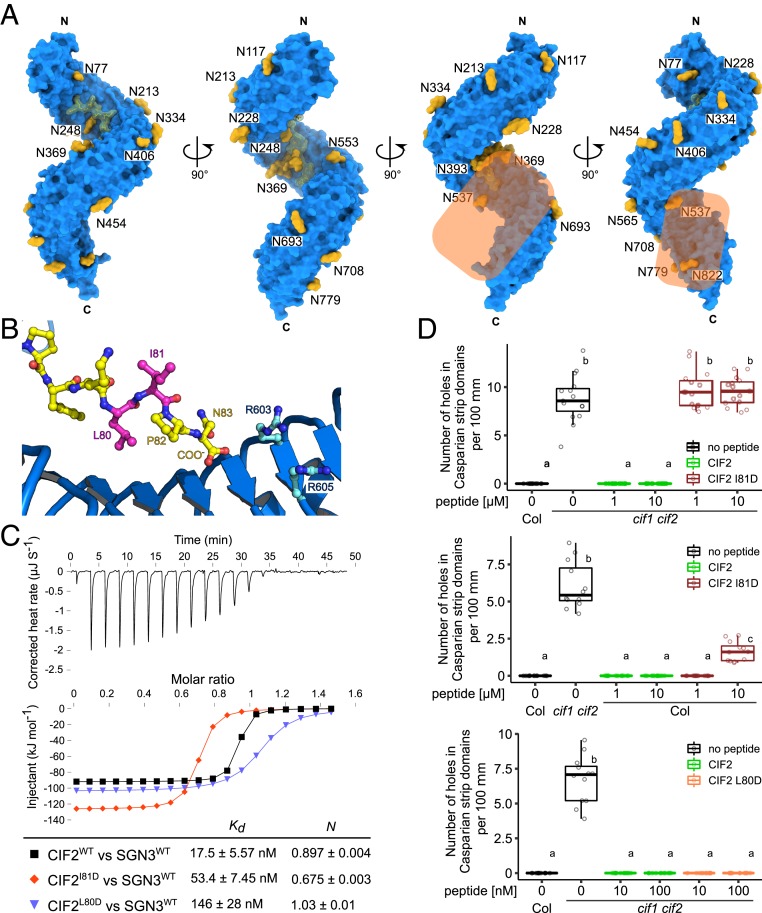
Many of the currently known LRR-RKs require the interaction with a shape-complementary coreceptor kinase for high-affinity ligand binding and for receptor activation (1, 21). In contrast, for example, to the peptide hormone IDA, CIF1–4 bind to GSO1/SGN3 with nanomolar affinity already in the absence of a coreceptor kinase (Figs. 1 and 3) (6, 7). This could in principle suggest that GSO1/SGN3 does not require a coreceptor (6). However, we found that both apo and CIF2-bound GSO1/SGN3 ectodomains behaved as monomers in analytical size-exclusion chromatography (SEC) and right-angle light scattering experiments, respectively (Fig. 2B). This makes it unlikely that CIF2 binding alters the oligomeric state of GSO1/SGN3, an activation mechanism used by the LRR domain-containing animal Toll-like receptors (27). However, structural features in the GSO1/SGN3–CIF2 complex suggest that a shape-complementary coreceptor kinase may be required for receptor activation: First, CIF2 contains a C-terminal asparagine residue in close proximity to the GSO1/SGN3 RxR motif (Fig. 3A). Both motifs are involved in the recruitment of a SERK coreceptor kinase in the structurally related IDA–HAESA and RGF–RGFR complexes (7, 13). Second, mutation of the RxR motif to alanine has no apparent effect on CIF2 binding in vitro, but the mutant receptor can only partially complement the sgn3 Casparian strip phenotype (Fig. 3 B and D). Thus, the GSO1/SGN3 RxR motif may not be essential for CIF peptide binding, but may instead be part of a putative receptor–coreceptor complex interface. Third, a surface area covering the C terminus of the CIF2 peptide and the C-terminal LRRs in GSO1/SGN3 is not masked by carbohydrate, thus representing a potential protein–protein interaction surface (Fig. 6A). The corresponding region in SERK-dependent LRR-RKs has been previously shown to represent the receptor–coreceptor complex interface (1).
Fig. 6.
Structural and biochemical evidence for a coreceptor kinase required for GSO1/SGN3 activation. (A) The GSO1/SGN3–CIF complex structure reveals a potential coreceptor binding site. Shown is the GSO1/SGN3 ectodomain (surface representation, in blue) in complex with the CIF2 peptide (surface view and bonds representation, in yellow), N-glycans (surface representation in yellow). The potential coreceptor binding surface not masked by carbohydrate is highlighted in orange. (B) Close-up view of CIF2 C terminus bound the GSO1/SGN3, indicating the positions of the side chains of Leu80 (pointing toward the receptor) and Ile81 (pointing to the solvent) (in magenta). (C) ITC assays of CIF2 mutant peptides versus the SGN3 wild-type ectodomain. (D) Quantitative analyses of number of holes in Casparian strip domains per 100 µm in cif1 cif2 double mutants upon treatment with CIF2 peptide variants (n = 15 for the Top, n = 12 for the Middle, and n ≥ 11 for the Bottom). Shown are box plots spanning the first to third quartiles, with the bold line representing the median, and circles indicating the raw data. Whiskers indicate maximum and minimum values, except outliers (b and c, statistically significant difference from a, with P < 0.05, one-way ANOVA and Tukey test).
We thus sought to obtain evidence for the involvement of a coreceptor kinase in SGN3 signal transduction. We hypothesized that a coreceptor may bind to the CIF2 C terminus, coordinated by the GSO1/SGN3 RxR motif (Fig. 6B). We replaced CIF2 Ile81, which faces the solvent in our structure, with aspartate (CIF2I81D) (Fig. 6B) and found that while the mutant peptide still binds GSO1/SGN3 with nanomolar affinity in vitro (Fig. 6C), it cannot rescue Casparian strip membrane domain formation in cif1 cif2 mutant plants (Fig. 6D). Importantly, wild-type (WT) plants treated with micromolar concentrations of CIF2I81D displayed dominant negative Casparian strip integrity phenotypes, while treatment with CIF2WT had no apparent effect (Fig. 6D). Mutation of the neighboring Leu80 to aspartate more strongly reduced binding to GSO1/SNG3 when compared to CIF2I81D, in agreement with our complex structure, which reveals Leu80 to be part of the CIF2–GSO1/SGN3 complex interface (Fig. 6 B and C). CIF2L80D application did not reveal a dominant negative effect but rather rescued the cif1 cif2 double mutant phenotype (Fig. 6D). Based on these findings, we speculate that CIF2I81D and CIF2L80D both can bind GSO1/SGN3 in vivo, but CIF2I81D specifically blocks interaction with an essential adapter protein required for GSO1/SGN3 activation.
We initially used a reverse genetic approach to identify coreceptors for GSO1/SGN3, based on previous studies on SERKs and SERK-related LRR-RKs (1, 23, 28, 29). However, analysis of known serk and cik/nik/clerk loss-of-function mutant combinations revealed no apparent Casparian strip phenotype (SI Appendix, Fig. S9). We thus performed a biochemical interaction screen, using the known SERK1 and SERK3 coreceptors as well as other GSO1/SGN3 interacting LRR-RKs, recently identified in a high-throughput biochemical screen (30). From the LRR-RK candidates identified in this screen, we selected putative coreceptors with small LRR ectodomains, including SERK5 (1), CIK/NIK/CLERK proteins recently reported as coreceptors for CLE peptide sensing LRR-RKs (23, 28, 29), the SRF receptor kinases (31), and the immune receptor kinase SOBIR1 (32). We expressed and purified the LRR ectodomains of SERK1, SERK3, SERK5, NIK3, NIK4, SRF3, SRF9, and SOBIR1 and tested for CIF-dependent interaction with the GSO1/SGN3 ectodomain in quantitative GCI assays (Fig. 7 A and B and SI Appendix, Fig. S10). We observed specific binding of SERK1 to GSO1/SGN3 in the presence of either CIF1, 2, or 3, with dissociation constants ranging from ∼20 to 300 nM (Fig. 7C and SI Appendix, Fig. S10). No SERK1 binding to SGN3 was observed in the absence of CIF peptide (SI Appendix, Fig. S10), and the coreceptor did not bind the GSO1/SGN36xmut mutant (Fig. 7C, see above). CIF2-dependent formation of a GSO1/SGN3–SERK1 complex could also be observed in analytical SEC experiments (Fig. 7D). In line with our structural and physiological assays, the CIF2I81D peptide specifically blocked GSO1/SGN3–SERK1 interaction in GCI and SEC experiments, rationalizing its dominant negative effect on Casparian strip formation (Fig. 7 C and D, compare Fig. 6 C and D). We found that GSO1/SGN3 also interacts with SERK3 in GCI assays, but not with SERK5 or any of the other coreceptor candidates derived from the high-throughput screen (SI Appendix, Fig. S10) (30). We observed specific SERK1 and SERK3 binding to GSO2 in the presence of CIF3 (Kd ∼20 to 80 nM) (SI Appendix, Fig. S10).
Fig. 7.
A quantitative interaction screen identifies SERK proteins as putative coreceptors for GSO1/SGN3. (A) Schematic overview of the biochemical screen for a GSO1/SGN3 coreceptor. Streptavidin (in gray) was immobilized by using amine coupling as described in Fig. 1A. The GSO1/SGN3 ectodomain was captured by streptavidin on the GCI chip surface (in blue), the CIF peptide is provided in access in the running buffer (in black), and different recombinantly purified coreceptor candidates are assayed for binding (in orange). (B) Coomassie-stained SDS/PAGE of the receptor and coreceptor candidates used in the screen. Each lane depicts 1 µg of the LRR ectodomain of each indicated candidate. Shown are isolated monomeric peak fractions from size-exclusion chromatography experiments. (C) GCI assays of SERK1 LRR-RK ectodomain versus the SGN3 wild-type and mutant ectodomains in the presence of CIF2 variant peptides. The remaining candidates are shown in SI Appendix, Fig. S10. Coreceptor candidates were supplied at a flow rate of 25 µL min−1. Sensorgrams are shown with raw data in red and their respective fits in black. A 1-to-1 binding model was used for analysis. Table summaries of kinetic parameters are shown. (D) Complex formation of SERK1 and SGN3 ectodomains. (D, Left) Analytical size-exclusion chromatography traces of the SGN3 ectodomain in the absence (blue line) or presence of CIF2WT peptides (red dotted line) or CIF2I81D antagonistic peptides (black dotted line). An SDS/PAGE analysis of the corresponding fractions is shown alongside. The theoretical molecular weight is 94.1 kDa for SGN3 (residues 19 to 870) and 21.5 kDa for SERK1 (residues 24 to 213), respectively. (E) Induced barrier defect in inducible SERK3 dominant negative lines. Quantification of barrier permeability was done using the PI assay (n ≥ 8 for each condition). Shown are box plots spanning the first to third quartiles, with the bold line representing the median and circles indicating the raw data. Whiskers indicate maximum and minimum values, except outliers (b statistically significant difference from a, with P < 0.05, one-way ANOVA and Tukey test).
To our surprise, the interaction of SERKs with ligand-associated GSO1 and GSO2 was much tighter than previously reported for the LRR-RKs BRI1 and HAESA (21). GCI analysis of PEPR1–Pep1–SERK1/3 complex formation, however, revealed an even tighter interaction (Kds 1 to 4 nM), while the related LRR-RK immune receptors FLS2 and EFR bound SERK3 with low micromolar affinity (SI Appendix, Fig. S11). Together, our quantitative receptor–coreceptor interaction screen revealed SERK1 and 3 as bona fide coreceptors for GSO1/SGN3 and GSO2. We hypothesized that different SERKs may act redundantly as coreceptor kinases for GSO1/SGN3 in the endodermis, complicating the analysis of serk loss-of-function alleles (SI Appendix, Fig. S9). We thus generated an estradiol-inducible, dominant negative SERK3 line (33) and found that it significantly delays Casparian strip formation (Fig. 7E). While the effect is not as strong as observed for sgn3 loss-of-function alleles, this provides initial in vivo evidence for a role of SERK3 and other SERKs in GSO1/SGN3-mediated Casparian strip formation. Taken together, our biochemical and genetic experiments implicate SERK proteins as coreceptors for the receptor kinases GSO1/SGN3 and GSO2.
Discussion
Plants harbor many different classes of signaling peptide hormones, the bioactive forms of which are generated by proteolytic processing from larger preproteins and by posttranslational modifications, including hydroxyprolination and tyrosine sulfation (2). The 21-amino acid CIF1 and CIF2 peptides carry a sulfated tyrosine residue in position 64 in vivo (5) and have been shown to represent ligands for the LRR-RK GSO1/SGN3 (5, 6). GSO1/SGN3 tightly interacts with CIF1 and CIF2 with dissociation constants in the low nanomolar range (Fig. 1) (6). The sTyr-containing peptide hormone PSK binds its cognate receptor PSKR with a Kd of ∼1 μM (9). RGF peptides that share the N-terminal Asp-Tyr motif with CIF1/2, interact with different RGFRs with dissociation constants in the high nanomolar-to-midmicromolar range (13). Recently, the tyrosine sulfate RaXX peptide from Xanthomonas oryzae has been shown to bind the rice LRR-RK XA21 with a Kd of ∼15 nM (34). Thus, GSO1/SGN3–CIF1/2 represents the strongest receptor–ligand pair for sTyr-modified signaling peptides currently known in plants. Comparing GSO1/SGN3–CIF1/2 to known LRR-RK peptide–ligand pairs reveals that plant membrane receptor kinases can sense their cognate peptide ligands with drastically different binding affinities (spanning the micro- to nanomolar range) (Fig. 1) (1, 7). The binding affinity does not, however, simply correlate with the size of the respective peptide ligand, as both the 21-amino acid CIF2 and 12-amino acid CLE9 peptide can bind their cognate LRR-RKs with very high affinity (Fig. 1). It is of note, however, that high-affinity ligand sensing may require both the receptor and a SERK coreceptor, with the coreceptor completing the ligand binding pocket and slowing down ligand dissociation (7, 21).
Many plant peptides, including the CLE and IDA/IDL families, are posttranslationally modified, and in both cases these modifications have been shown to be important for high-affinity ligand recognition and for the bioactivity of the respective peptide hormone (7, 8). For CIF1, two posttranslational modifications have been identified, sulfation of tyrosine 64 and hydroxyprolination of prolines 69 and 71. Using 2 complementary quantitative binding assays, we find that the sulfation of Tyr64 in different CIF peptides is required for high-affinity ligand binding to GSO1/SGN3 in vitro, but surprisingly, removal of the sulfate group from the peptide, or mutation of the sTyr binding pocket in GSO1/SGN3, had little effect on Casparian strip formation (Fig. 3). In sharp contrast, for example, to the HAESA–IDA complex (7), both hydroxyproline residues in CIF2 do not seem to play a major role in ligand sensing, or bioactivity, at least under the conditions tested (SI Appendix, Fig. S6). Similarly, the mutation of the GSO1/SGN3 RxR motif conserved among many peptide ligand sensing LRR-RKs (13), had little effect on CIF2 binding and resulted in intermediate Casparian strip formation phenotypes (Fig. 3). We had to go all of the way to a GSO1/SGN3 sixtuple mutant to disrupt CIF2 binding in vitro, and receptor function in planta (Fig. 3). Based on these findings, we speculate that the concentration of mature CIF1 and CIF2 peptides in the Casparian strip may exceed the nanomolar range, and thus partially functional receptors can still rescue the sgn3 phenotype. In line with, application of 10 to 100 nM of nonsulfatable CIF2Y64F can still complement the cif1 cif2 phenotype, despite having a 100- to 1,000-fold reduced binding affinity to GSO1/SGN3 (Fig. 3).
Our GSO1/SGN3–CIF2 structure prompted us to search for additional CIF peptides and we indeed identified several candidates and characterized CIF3 and CIF4 (Fig. 4 and SI Appendix, Fig. S7). We found that while GSO1/SGN3 binds CIF1–4 with high affinity, the homologous LRR-RK GSO2 specifically senses CIF3 (Fig. 4). CIF3 and CIF4 are not expressed in the endodermis (Fig. 5 and SI Appendix, Fig. S12) and potentially control other, GSO1/SGN3- and GSO2-mediated developmental processes (17, 35). The partially distinct binding specificities of SGN3 and GSO2 suggest that the 2 receptors have evolved unique functions, possibly to mediate to specific signal inputs in as yet unknown tissue and organ contexts during development. However, a single mutant phenotype for GSO2 has not been described, the only currently known function being redundant with GSO1/SGN3 in embryonic cuticle formation (17). Since neither cif1 cif2 nor cif3 cif4 double mutants show an embryonic cuticle phenotype, it will be important to identify whether a combination of cif1–4, possibly a quadruple mutant is required for this developmental process, or whether it is mediated by an additional, thus far unidentified, peptide ligand.
While the high-affinity recognition of CIF peptides by GSO1/SGN3 and GSO2 does not require a coreceptor kinase, the receptor activation mechanism for these LRR-RKs remained to be identified. Despite our initial genetic analyses arguing against a role for the common SERK coreceptor kinases in GSO1/SGN3 function, a quantitative biochemical interaction screen identified SERK1 and SERK3 as bona fide coreceptors. SERKs bind GSO1/SGN3 and GSO2 only in the presence of CIF peptide ligands, suggesting that the previously established ligand-induced receptor–coreceptor heteromerization mechanism (1, 21) is conserved in GSO1/SGN3 and GSO2 (Fig. 7). CIF3 promotes a much stronger interaction of GSO1/SGN3 or GSO2 with SERK1 when compared to CIF1/2, suggesting that CIF peptides may not only have unique receptor binding specificities, but also different affinities for SERK coreceptors (SI Appendix, Fig. S10). It is of note that CIF-dependent interaction of GSO1/SGN3 or GSO2 with SERKs is ∼50 times stronger than previously described for the LRR-RKs BRI1 and HAESA (21). We speculate that minute amounts of SERK coreceptor may suffice to allow for GSO1/SGN3 receptor activation, possibly rationalizing why serk double and triple mutants show no apparent Casparian strip defects (SI Appendix, Fig. S9). The dominant negative effect of our SGN3::XVE:SERK3Δkinase-GFP line nonetheless provides genetic support for the involvement of SERK proteins in Casparian strip formation (Fig. 7). Obtaining clear-cut loss-of-function evidence might prove challenging, since multiple SERK mutants lead to highly pleiotropic phenotypes, including seedling lethality and sterility, in line with their involvement in a large number of LRR kinase-mediated signaling processes (36–39). The biochemical identification of different CIF peptides and of GSO1/2 coreceptor kinases however now offers avenues to dissect peptide hormone signaling specificity in a developmental context.
Materials and Methods
Crystallization and Data Collection.
Crystals of the deglycosylated SGN3 in complex with the CIF2 peptide developed at room temperature in hanging drops composed of 1 µL protein solution (1 mg mL−1) containing 0.5 mM CIF2 and 1 µL of crystallization buffer (17% [wt/vol] PEG 6000, 0.1 M Tris pH 7.5, 0.2 M LiCl), suspended above 1.0 mL of the latter as reservoir solution and using microseeding protocols. Crystals of SGN3 in complex with the CIF2Hyp69,71 peptide developed in crystallization buffer (16% [wt/vol] PEG 4000, 0.1 M Tris pH 8.5, 0.2 M MgCl2). Crystals were cryoprotected by serial transfer into crystallization buffer supplemented with 20% (vol/vol) glycerol (SGN3–CIF2) or 20% (vol/vol) ethylene glycol (SGN3–CIF2Hyp69,71) and cryocooled in liquid nitrogen. Sulfur single-wavelength anomalous diffraction (SAD) data to 4.0-Å resolution was collected at beam-line PXIII at the Swiss Light Source (Villigen, Switzerland), CH with λ = 2.066 Å. A native dataset to 2.95-Å resolution was collected on a crystal from the same drop cryoprotected in the same way with λ = 1.0 Å. Data processing and scaling was done in XDS (40).
Structure Solution and Refinement.
The structure was solved using the molecular replacement method as implemented in the program PHASER (41), and using the isolated ectodomain of the LRR-RK PEPR1 as search model (PDB ID 5gr8). The solution comprised a dimer in the asymmetric unit and the structure was completed in alternative cycles of manual model building in COOT (42) and restrained TLS refinement in phenix.refine (43). A phased anomalous difference electron density map calculated with the program ANODE (44) was used to assign the position of disulfide bonds and free cysteines/methionines in the structure. Analysis with phenix.molprobity (45) reveal good stereochemistry of the final model. Structural diagrams were prepared using Pymol (https://sourceforge.net/projects/pymol/) and povray (http://www.povray.org/).
Grating-Coupled Interferometry.
GCI experiments were performed with the Creoptix WAVE system (Creoptix AG) using either 4PCP or 4PCH WAVE chips (thin quasiplanar polycarboxylate surface or thick polycarboxylate hydrogel surface with high capacity, respectively; Creoptix). For direct amine coupling, chips were conditioned with borate buffer (100 mM sodium borate pH 9.0, 1 M NaCl; Xantec) and the respective ligands were immobilized on the chip surface using standard amine coupling; 7 min activation (1:1 mix of 400 mM N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride and 100 mM N-hydroxysuccinimide; Xantec), followed by injection of the ligands (50 to 100 µg mL−1) in 10 mM sodium acetate pH 5.0 (Sigma) until the desired density was reached, passivation of the surface (0.5% BSA [Roche] in 10 mM sodium acetate pH 5.0), and final quenching with 1 M ethanolamine pH 8.0 for 7 min (Xantec). For biotinylated ligand capturing, streptavidin (50 µg mL−1; Sigma) was immobilized on the chip surfaces by the same method with direct amine coupling, followed by capturing respective biotinylated ligands (50 to 100 µg mL−1) until the desired density was reached. Kinetic analyses for peptide ligands were performed at 25 °C with a 1:2 dilution series from 100 nM for CIF variants in the presence of sulfation or 10 µM in the absence of sulfation, for a coreceptor screen using the biotinylated ligand-captured chips with a 1:3 dilution series from 6.7 µM for SERK1 and SERK3 or 20 µM for the others in 20 mM citrate pH 5.0, 250 mM NaCl, 0.01% Tween 20. Blank injections were used for double referencing and a DMSO calibration curve for bulk correction. Analysis and correction of the obtained data were performed using the Creoptix WAVE control software (correction applied: X and Y offset; DMSO calibration; double referencing). Mass transport binding models with bulk correction were used for the experiments of SGN3–CIF peptide binding and 1-to-1 binding models for the other experiments.
Isothermal Titration Calorimetry.
All ITC experiments were perfomed on a MicroCal PEAQ-ITC (Malvern Panalytical) with a 200-µL sample cell and a 40-µL injection syringe at 25 °C. Proteins were dialyzed into ITC buffer (20 mM sodium citrate pH 5.0, 250 mM NaCl, exceptionally containing 5% [vol/vol] DMSO for CIF4 experiments) prior to all titrations. A typical experiment consisted of injecting 200 µM CIF peptide in 2-µL intervals into the cell containing 20 µM GSO1/SGN3 receptor. The MicroCal PEAQ-ITC analysis software (version 1.21) was used for data analysis.
Data Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under ID code 6S6Q.
Supplementary Material
Acknowledgments
We thank the staff of Beam Line PXIII of the Swiss Light Source (Villigen, Switzerland) for their technical assistance during data collection. This work was supported by grants 31003A_176237 and 31CP30_180213 (M.H.), 31003A_156261 and 310030E_176090 (N.G.) from the Swiss National Science Foundation, a European Research Council Consolidator Grant (616228-ENDOFUN) (N.G.), Human Frontier Science Program Organization postdoctoral fellowship LT000567/2016-L (S.O.), a Japan Society for the Promotion of Science (JSPS) fellowship (S.F.), and by an International Research Scholar Award from the Howard Hughes Medical Institute (M.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6S6Q).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911553117/-/DCSupplemental.
References
- 1.Hohmann U., Lau K., Hothorn M., The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Matsubayashi Y., Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Schardon K., et al. , Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Huffaker A., Pearce G., Ryan C. A., An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakayama T., et al. , A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355, 284–286 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Doblas V. G., et al. , Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Santiago J., et al. , Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5, e15075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y., A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Wang J., et al. , Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265–268 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y., Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y., Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 15067–15072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinohara H., Mori A., Yasue N., Sumida K., Matsubayashi Y., Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 3897–3902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song W., et al. , Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26, 674–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang J., et al. , Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Lin X., Han Z., Qu L.-J., Chai J., Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 26, 543–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazak O., et al. , Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep. 18, 1367–1381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuwamoto R., Fukuoka H., Takahata Y., GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 54, 30–42 (2008). [DOI] [PubMed] [Google Scholar]
- 18.San-Bento R., Farcot E., Galletti R., Creff A., Ingram G., Epidermal identity is maintained by cell-cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J. 77, 46–58 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Pfister A., et al. , A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3, e03115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doblas V. G., Geldner N., Barberon M., The endodermis, a tightly controlled barrier for nutrients. Curr. Opin. Plant Biol. 39, 136–143 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Hohmann U., et al. , Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. U.S.A. 115, 3488–3493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinohara H., Matsubayashi Y., Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: Dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82, 328–336 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Anne P., et al. , CLERK is a novel receptor kinase required for sensing of root-active CLE peptides in Arabidopsis. Development 145, dev162354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felix G., Duran J. D., Volko S., Boller T., Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Sun Y., et al. , Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Leonard J. N., et al. , The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 258–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu C., et al. , A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 4, 205–211 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Cui Y., et al. , CIK receptor kinases determine cell fate specification during early anther development in Arabidopsis. Plant Cell 30, 2383–2401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smakowska-Luzan E., et al. , An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyüboglu B., et al. , Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol. 7, 16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hohmann U., Hothorn M., Crystal structure of the leucine-rich repeat ectodomain of the plant immune receptor kinase SOBIR1. Acta Crystallogr. D Struct. Biol. 75, 488–497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domínguez-Ferreras A., Kiss-Papp M., Jehle A. K., Felix G., Chinchilla D., An overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1-1-dependent manner. Plant Physiol. 168, 1106–1121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luu D. D., et al. , Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. U.S.A. 116, 8525–8534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Racolta A., Bryan A. C., Tax F. E., The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev. Dyn. 243, 257–278 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Colcombet J., Boisson-Dernier A., Ros-Palau R., Vera C. E., Schroeder J. I., Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17, 3350–3361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He K., et al. , BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Meng X., et al. , Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., et al. , SERK family receptor-like kinases function as Co-receptors with PXY for plant vascular development. Mol. Plant 9, 1406–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W., Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993). [Google Scholar]
- 41.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorn A., Sheldrick G. M., ANODE: Anomalous and heavy-atom density calculation. J. Appl. Cryst. 44, 1285–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis I. W., et al. , MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under ID code 6S6Q.