Significance
We describe a mechanism by which the obligate intracellular pathogen Chlamydia pneumoniae induces curvature of the host-cell plasma membrane and recruits a central component of the endocytotic machinery. We demonstrate that a type III-secreted C. pneumoniae effector protein named CPn0678 binds via its N-terminal amphipathic helix to negatively charged phospholipids in the inner leaflet of the host plasma membrane at the site of entry, and induces membrane curvature. Its proline-rich region then recruits SNX9 (sorting nexin 9), a key regulator of endocytosis, and the complex facilitates uptake of C. pneumoniae into host cells.
Keywords: effector protein, membrane modulation, lipid binding, endocytosis
Abstract
During invasion of host cells, Chlamydia pneumoniae secretes the effector protein CPn0678, which facilitates internalization of the pathogen by remodeling the target cell’s plasma membrane and recruiting sorting nexin 9 (SNX9), a central multifunctional endocytic scaffold protein. We show here that the strongly amphipathic N-terminal helix of CPn0678 mediates binding to phospholipids in both the plasma membrane and synthetic membranes, and is sufficient to induce extensive membrane tubulations. CPn0678 interacts via its conserved C-terminal polyproline sequence with the Src homology 3 domain of SNX9. Thus, SNX9 is found at bacterial entry sites, where C. pneumoniae is internalized via EGFR-mediated endocytosis. Moreover, depletion of human SNX9 significantly reduces internalization, whereas ectopic overexpression of CPn0678–GFP results in a dominant-negative effect on endocytotic processes in general, leading to the uptake of fewer chlamydial elementary bodies and diminished turnover of EGFR. Thus, CPn0678 is an early effector involved in regulating the endocytosis of C. pneumoniae in an EGFR- and SNX9-dependent manner.
All members of the Chlamydiaceae are obligate intracellular pathogens of humans and animals, and cause a variety of diseases depending on the tissues they target (1). The two species that affect humans are Chlamydia trachomatis and Chlamydia pneumoniae, which are the causative agents of severe urogenital conditions, including pelvic inflammation and ectopic pregnancies, and respiratory disorders such as pneumonia and bronchitis, respectively (2, 3). C. pneumoniae infections are also associated with several chronic diseases, including asthma, Alzheimer’s disease, multiple sclerosis, and even lung cancer (4–7).
The most critical step in the life cycle of an obligate intracellular bacterium is internalization into the host cell. The most common entry strategies are 1) the “zipper” and 2) the “trigger” mechanisms. In the former, a bacterial adhesin/invasin interacts with a surface receptor, thereby activating its downstream signaling machinery and effectively hijacking receptor endocytosis for bacterial internalization. In the latter, an initial and rather weak interaction between pathogen and host is rapidly followed by translocation of bacterial proteins, called effectors, into the host cytoplasm. These effectors modulate the host cytoskeleton and induce extensive ruffling of the plasma membrane (PM) to facilitate pathogen entry (8). Both mechanisms involve the use of bacterial proteins to manipulate essential components of the endocytic machinery, such as the phosphoinositide-converting enzymes that regulate the lipid composition (and hence the curvature) of the PM (9, 10), adaptors and regulators like sorting nexin 9 (SNX9) that control endocytosis and vesicle trafficking (11, 12), and finally actin polymerization, which facilitates bacterial uptake (13, 14). Interestingly, SNX9 harbors a membrane-curvature–sensing bin-amphiphysin-rvs (BAR) domain and binds preferentially to membranes of high curvature (15). Using an in vitro system it has been proposed that binding to PI(4,5)P2, the early endosome marker PI(3)P, and domains of high membrane curvature recruits SNX9 in order to trigger the actin machinery and complete endocytosis (16).
Internalization is preceded by stable adhesion to the host cell, which induces intracellular signaling and recruitment of endocytosis-related proteins. Recently, host receptors like the ephrin receptor (EPHA2) or EGFR have been shown to promote adhesion of C. trachomatis, but the chlamydial interaction partner remains undefined, as does the mechanism of entry (17, 18). For C. pneumoniae we have shown that the pathogen uses one of its highly diverse polymorphic membrane proteins, Pmp21, to bind and activate the EGFR (19). EGFR activation triggers the PI3 kinase, which in turn recruits specific endocytic adaptor proteins to facilitate the EGFR-mediated endocytosis of C. pneumoniae (20). In addition to this zipper mechanism, C. pneumoniae employs the trigger approach to enter host cells. Simultaneously with the Pmp21–EGFR interaction, C. pneumoniae secretes its TarP ortholog CPn0572 via a type III secretion (T3S) system. CPn0572 then binds and polymerizes actin to enforce bacterial uptake into actin-rich structures (21).
To determine whether C. pneumoniae employs other mechanisms to achieve efficient internalization, we searched for new early effector proteins involved in these processes. Here we show that the effector CPn0678 is also a T3S substrate, and localizes to the PM at bacterial entry sites. CPn0678 can bind phospholipids in both natural and synthetic membranes, and upon expression in human cells it generates membrane tubulations, and interacts with the host protein SNX9, a multifunctional protein involved in clathrin-mediated endocytosis, membrane remodeling, and actin dynamics (12). Our data suggest that, during host-cell entry, secreted CPn0678 binds to and curves the PM, which recruits SNX9 to the late stages of endocytosis. These findings show that CPn0678-induced membrane curvature plays a central role in the uptake of C. pneumoniae.
Result
CPn0678: Prototype of a T3S Chlamydial Effector Protein.
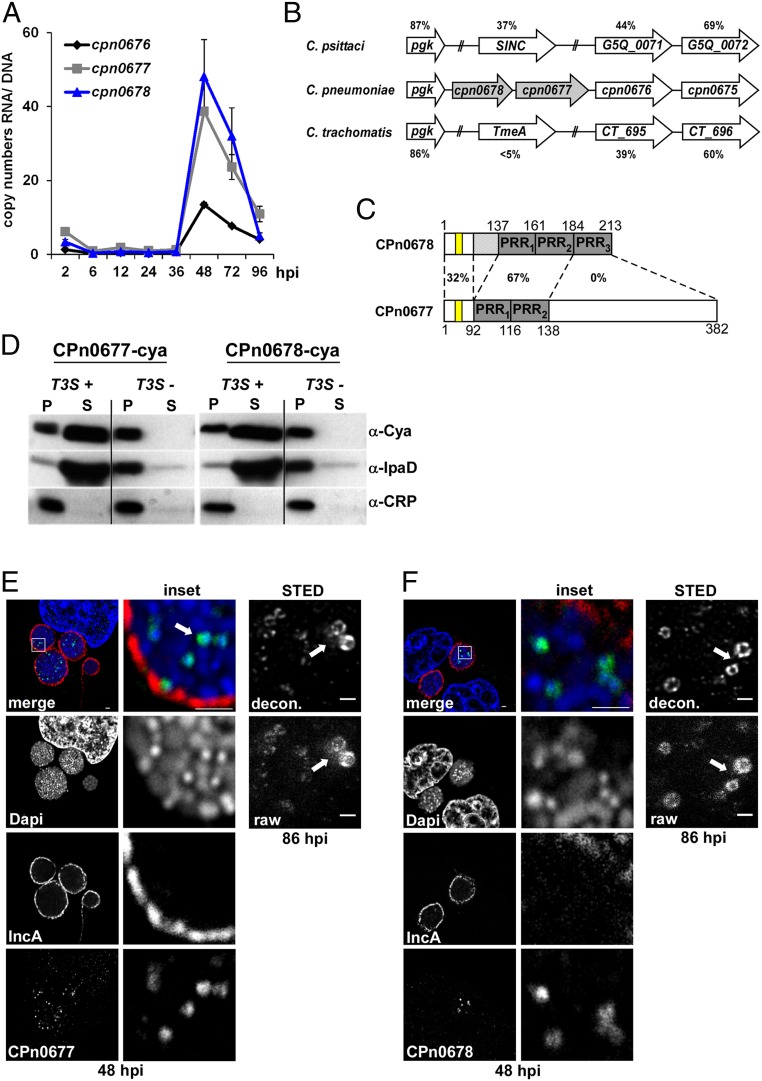
A common feature of both chlamydial adhesins and early effectors is that they are expressed late in the preceding infection, and displayed on the elementary body (EB) or stored within it for early secretion. Therefore, we performed a genome-wide transcriptional analysis to screen for such genes and found 88 genes to be significantly up-regulated (SI Appendix, Table S1). This set included genes for previously described adhesins, components, and potential substrates of the T3S system, and hypothetical genes. Among the latter class, we identified a highly up-regulated cluster of three genes comprising cpn0676, cpn0677, and cpn0678. RT-PCR confirmed that their transcript levels increased significantly from 36 h onward, with cpn0677 and cpn0678 RNAs becoming more abundant than cpn0676 (Fig. 1A).
Fig. 1.
CPn0678 is a C. pneumoniae-specific T3S effector. (A) HEp-2 cells were infected with C. pneumoniae (multiplicity of infection [MOI] 100 for the period 2 to 12 hpi, MOI 5 for 24 to 96 hpi) and harvested at the indicated time points for nucleic acid isolation. Levels of cpn0677 and cpn0678 mRNA were determined by qRT-PCR using a segment of the ompA gene as the genomic reference. Data are represented as means ± SEM. (B) Schematic representation of the C. pneumoniae gene cluster cpn0676-cpn0678 and the syntenic loci of C. trachomatis and C. psittaci. Percentage amino acid sequence identity between homologs (C. psittaci vs. C. pneumoniae, C. trachomatis vs. C. pneumoniae) is indicated. (C) Domain structure and sequence comparison of CPn0677 and CPn0678. The position of the APH is highlighted in yellow. (D) Chimeric proteins consisting of the first 25 amino acids of CPn0677 or CPn0678 fused to the reporter Cya were expressed in ipaB (T3S+) and mxiD (T3S−) mutant strains of S. flexneri. Supernatant (S) and pellet (P) preparations were fractionated by SDS/PAGE and probed with antibodies against Cya, IpaD, or CRP. (E and F, Left) Confocal images of HEp-2 cells infected for 48 h with C. pneumoniae were fixed with PFA, permeabilized with methanol, and stained with specific antibodies against CPn0677 (E) or CPn0678 (F) and anti-rabbit Alexa488. The inclusion membrane was stained with anti-IncA and anti-mouse Alexa594. DNA of host and bacteria was labeled with DAPI. The area in the white box is shown at higher magnification in the Inset (Top two rows, Center). White arrows indicate stained EBs. (Scale bars, 5 µm.) (Right) EBs released from HEp-2 cells infected for 84 h were fixed with PFA, permeabilized with methanol and stained with anti-CPn0677 (E) or CPn0678 (F) and anti-rabbit Alexa488. STED images of EBs were generated with a Leica TCS SP8 STED 3× microscopy system. Deconvolution was performed using Huygens Professional with standard parameters. (Scale bars, 0.5 µm.)
Comparison of the C. pneumoniae cpn0676-cpn0678 locus with other chlamydial genomes showed that the genes are part of a syntenic locus. CPn0676 is conserved among all Chlamydia species with approximately 39 to 44% identity, while comparison of CPn0677 and CPn0678 revealed a more complex pattern. All other species have only one gene at this locus, while C. pneumoniae carries two genes, cpn0677 and cpn0678 (Fig. 1B). CPn0678 itself harbors three conserved proline-rich repeats (PRR), two of which are conserved within the N-terminal segment of CPn0677 and are 67% identical to each other (Fig. 1C). In-depth comparisons with other chlamydial species revealed that these two proteins are C. pneumoniae-specific, displaying no or only low homology to other chlamydial proteomes. Thus, we generated a phylogenetic tree showing the relationships across the chlamydial kingdom by comparing full-length CPn0678 with the syntenic proteins of various species (SI Appendix, Fig. S1A). The most distantly related proteins, with less than 5% identity to CPn0678, are found in C. trachomatis, Chlamydia muridarum, and Chlamydia suis (Fig. 1B and SI Appendix, Fig. S1A). One of these is C. trachomatis TmeA (CT_694), a known T3 effector protein, which interacts with the host protein AHNAK and is involved in early steps in the C. trachomatis infection (22, 23). Although the three species are closely related, C. trachomatis TmeA shares only 50 to 55% identity with its homologs in C. suis and C. muridarum, possibly suggesting differences in their functions. The syntenic proteins from Chlamydia abortus, Chlamydia felis, Chlamydia psittaci, and Chlamydia caviae share higher levels of identity with CPn0678 (SI Appendix, Fig. S1A), ranging between 34% and 40%. This is because the PRR1 and PRR2 of CPn0678 are conserved among the homologous proteins in these four species (SI Appendix, Fig. S1A). Among them is SinC (G5Q_0070), an effector expressed by C. psittaci, which associates with the nuclear membrane during late stages of infection (24). These findings clearly demonstrate that this genomic locus encodes effector proteins with diverse functions in different chlamydial species.
To confirm that CPn0677 and CPn0678 are T3S effector proteins, we used a heterologous Shigella flexneri T3S assay (25). The first 25 N-terminal amino acids of each protein were fused to the calmodulin-dependent adenylate cyclase (Cya) reporter protein. These constructs were then separately expressed in a S. flexneri ipaB (constitutive T3S) or mxiD (deficient in T3S)-null strain (26, 27). Fractionation experiments confirmed that both proteins contained a T3S-competent signal sequence, as the reporter constructs were detectable in the supernatants, as was the positive control IpaD, whereas the intracellular protein CRP was retained in the pellet (Fig. 1D). In addition, confocal microscopy of inclusions labeled with specific antibodies at 48 h postinfection (hpi) revealed that both CPn0677 (Fig. 1E and SI Appendix, Fig. S1C) and CPn0678 (Fig. 1F and SI Appendix, Fig. S1C) were associated with the condensed DAPI staining characteristic of EBs and not with the nuclear envelope, as shown for SinC (24). In agreement with our transcriptional studies, the two proteins were detected from 48 to 84 hpi in lysates of infected cells (SI Appendix, Fig. S1B). Deconvolution of stimulated emission depletion microscopy (STED) images of EBs released from bursting inclusions at 86 hpi showed a ring-like structure for both proteins (Fig. 1 E and F). The protein signals in these rings do not look continuous, but seem to be enriched in certain areas, which perhaps indicates the clustered presence of the T3S system preloaded with the effector proteins. Taken together, these findings indicate that both proteins are T3S effectors, which are secreted in the early phase of infection.
CPn0678 Is a Lipid-Binding, Membrane-Tubulating Effector Protein.
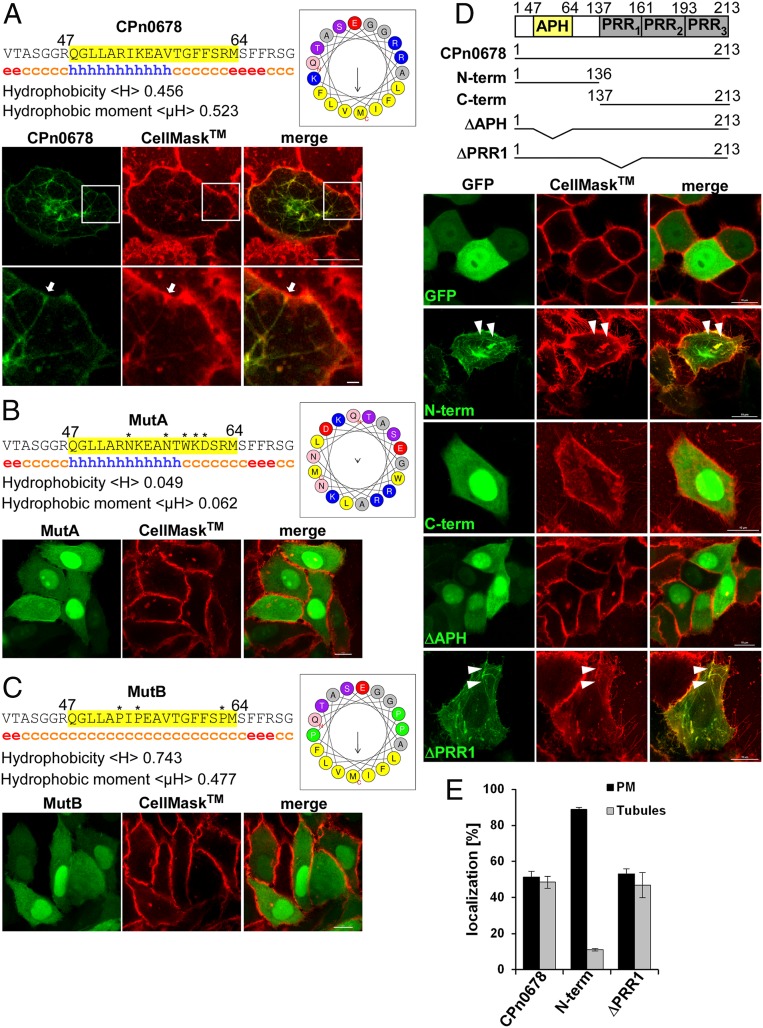
Based on our sequence comparisons and initial localization studies, we concluded that CPn0678 could be the prototype for the C. pneumoniae effector proteins CPn0678 and CPn0677 encoded by the syntenic locus, and in the following we describe our efforts to uncover the function of CPn0678. As both TmeA and SinC interact with host membranes (24, 28), we searched for potential membrane-interacting domains and found an N-terminal region (amino acids 47 to 64) with a strongly amphipathic character. Secondary-structure predictions revealed that this region also carries an α-helix, so we refer to this sequence as the amphipathic helix (APH). Projected into a helical wheel, the amphipathic amino acids of the APH all face to one side of the helix (Fig. 2A). The APH is conserved, albeit weakly, in CPn0677 (amino acids 48 to 65) (Fig. 1C). To understand the function of the APH, we first studied the localization of CPn678–GFP ectopically expressed in living HEp-2 cells treated with the PM dye CellMask (Fig. 2A). Imaging revealed that CPn678–GFP localizes to the PM and induces a strong tubulation phenotype, with CPn678-lined tubules emanating from the cell membrane into the cytosol (Fig. 2A). Next, we introduced amino acid exchanges into the APH in the full-length protein to elucidate whether the predicted α-helix or the amphipathic amino acids are functionally involved in the observed membrane tubulation (Fig. 2 B and C). In MutA the amino acid exchanges result in an APH with a 10-fold reduction in hydrophobicity, while the predicted α-helix is retained (Fig. 2B); in the converse mutant MutB the α-helix is destroyed while the hydrophobicity is increased (Fig. 2C). Ectopic expression and live imaging in the presence of CellMask revealed that both mutants have lost their tubulation phenotypes and no PM localization could be detected (Fig. 2 B and C).
Fig. 2.
The N-terminal APH of CPn0678 mediates binding to the PM and triggers tubulation of the PM. (A) The region encompassing residues 40 to 70 of CPn0678 was analyzed for hydrophobicity, hydrophobic moment and amino acid composition using HeliQuest (heliquest.ipmc.cnrs.fr) and a segment displaying a with strongly amphipathic character was identified (amino acids 47 to 64, highlighted in yellow), which is presented as a helical wheel (APH) with amphipathic amino acids in yellow and polar residues in blue. Secondary-structure prediction with GORIV (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_server.html) revealed the presence of an α-helix (amino acids 47 to 58, represented by blue “h”). (B and C) Mutational analyses were performed using both predictions to generate a mutant with a major loss of hydrophobicity (MutA, B) and one in which the α-helical structure was disrupted (MutB, C). Fluorescence images of living cells expressing CPn0678-GFP (A), MutA (B), or MutB (C) treated with the PM dye CellMask. (Scale bars, 10 µm; 1 µm in Insets.) (D) Schematic representation of the fragments of CPn0678 which were C terminally fused with GFP and used in live-cell imaging following treatment with CellMask. White arrowheads show membrane tubulations emanating from the PM. (Scale bars, 10 µm.) (E) Quantification of subcellular localization of Cpn0678, N-term and ΔPRR1 fused with GFP. Data are represented as means ± SD (n = 3).
To verify that the N-terminal APH is responsible for the tubulation phenotype, we generated different deletion variants and analyzed them under the same conditions in live imaging and for general protein expression (Fig. 2 D and E and SI Appendix, Fig. S2A). All variants harboring the APH (CPn0678, N-terminal fragment [N-term], and ΔPRR1) show both localization to the PM and a membrane-tubulating phenotype (Fig. 2D). Quantification of phenotypes revealed that cells expressing CPn0678 or ΔPRR1 show an ∼50% distribution of both phenotypes, while cells expressing the N-term variant show membrane tubules in 10% of cells; otherwise we found the protein at the PM (Fig. 2E). In contrast, localization to the PM and tubule formation were both lost when we deleted the APH (ΔAPH) or expressed a C-terminal fragment (C-term), indicating that the APH is indeed essential for membrane binding (Fig. 2D).
In proteins containing a BAR domain, membrane curvature is mediated by insertion of the APHs, which exert a bending force on the lipid bilayer (29, 30). These membrane tubules colocalize with the cortical actin cytoskeleton and can be enhanced or stabilized by its chemical depolymerization (31, 32). Similarly, we observed that the membrane tubules induced by CPn0678–GFP expression colocalized with cortical actin (SI Appendix, Fig. S2B). When these cells were treated with Cytochalasin D, an actin-depolymerizing drug, 50% more cells exhibited tubules than in the control (SI Appendix, Fig. S2C). In addition, we detected colocalization of CPn0678–GFP tubules with microtubules (SI Appendix, Fig. S2B), and inhibition of microtubule polymerization by nocodazole treatment completely suppressed tubule formation (SI Appendix, Fig. S2C). These findings indicate that cortical actin and the microtubule cytoskeleton play an important role in CPn0678-mediated membrane tubulation.
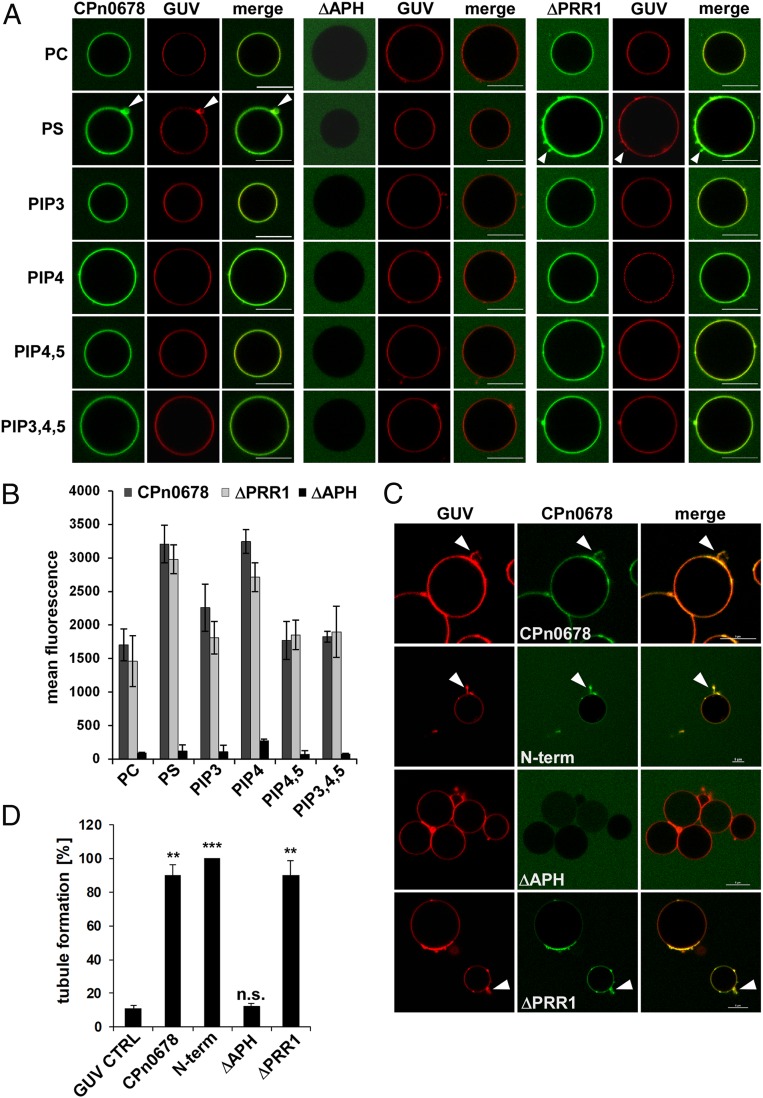
Using giant unilaminar vesicles (GUVs), a model system for biological membranes, we analyzed the membrane and lipid interactions of CPn0678 in more detail (Fig. 3). These GUVs are based on 69.75 mol% dipalmitoylphosphatidylcholine (DOPC) and 25 mol% cholesterol stained with Texas red (0.25 mol%). During vesicle assembly, additional selected phosphatidylinositols (PIPs) are incorporated, while in phosphatidylserine (PS)-containing GUVs the lipid composition is changed to 49.75 mol% DOPC/25 mol% cholesterol/0.25 mol% Texas red and 20 mol% PS. Quantification of confocal images revealed that FITC-labeled recombinant CPn0678 binds to all of the tested lipids, and shows the highest affinities for PS, the most abundant negatively charged phospholipid in the PM (33), and PIP4, a phospholipid found in Golgi membranes which is the precursor of PIP4,5 in the PM (34) (Fig. 3 A and B). Deletion of the PRR1 did not significantly affect binding, whereas deletion of the APH completely abolished lipid interaction (Fig. 3 A and B), indicating that the APH mediates the interaction of CPn0678 with lipids, especially those found in the inner leaflet of the PM (35). During these analyses, we observed that GUVs incubated with binding-competent CPn0678 variants developed membrane tubules on their surfaces, especially those containing the negatively charged PS (Fig. 3A, white arrowheads). Thus, we generated GUVs composed of PIP4,5 and PS to more closely approximate the phospholipid mixture on the PM, and imaged them in the absence or presence of different FITC-labeled CPn0678 variants. All variants harboring the APH domain (CPn0678, N-term, and ΔPRR1) elicited membrane tubulation on the GUVs, while ΔAPH did not bind to the GUVs, as shown previously (Fig. 3 C and D and Movies S1–S5). Quantification of GUVs incubated with CPn0678, N-term, and ΔPRR1 revealed that 90 to 100% of them showed membrane tubulations, while only 12% of GUVs incubated with ΔAPH showed tubulation which is close to the value for control GUVs (11%) (Fig. 3D). Furthermore, we observed that GUVs incubated with N-term showed a marked tendency to burst during incubation; bursting also occurred in GUVs treated with CPn0678, but with a lower frequency (Movies S1–S5), indicating that the tubulation elicited by the APH-carrying variants is strong enough to disturb the integrity of the GUVs.
Fig. 3.
CPn0678 binds to and tubulates artificial membranes. (A and B) Lipid binding assay using FITC-labeled recombinant CPn0678 variants and Texas red-stained GUVs containing the indicated lipids. (A) Confocal images of GUVs incubated with CPn0678 variants. White arrowheads indicate membrane deformations. (Scale bars, 10 µm.) (B) Quantification of microscopic analyses of protein binding expressed as the mean fluorescence of sets of 50 individual GUVs. Data are presented as means ± SD (n = 3). (C and D) Lipid binding assay using FITC-labeled recombinant CPn0678 variants and Texas red-stained GUVs containing PS and PIP4,5. (C) Confocal images of GUVs bound by FITC-labeled recombinant CPn0678 variants. White arrowheads indicate membrane deformations leading to disruption of GUVs, as shown in Movies S1–S5. (Scale bars 5 µm.) (D) Quantification of analyses of membrane deformation in samples containing 50 individual GUVs each. Data are presented as means ± SD (n = 2). n.s., not significantly different from CPn0678; **P value of 0.01; ***P value of 0.001.
CPn678-Induced Membrane Tubulation Enables the Protein to Interact with the Endocytic Scaffold Protein SNX9.
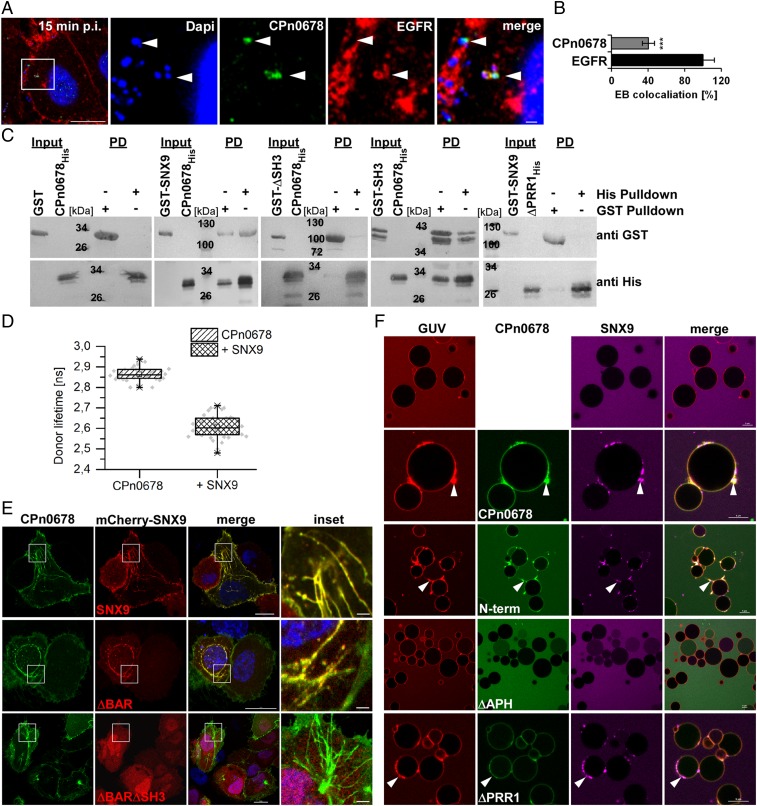
Based on our previous findings, we hypothesized that CPn0678 is an early effector secreted via T3S. Therefore, we next analyzed the localization of endogenous CPn0678 at 15 min postinfection, at which time adhesion and internalization of C. pneumoniae can be observed by colocalization of EBs with the host EGF receptor (20). As expected, we found that CPn0678 colocalized with EGFR at EB contact sites on the PM and in endocytic invaginations containing internalized EBs (Fig. 4A). We quantified and validated our imaging data and 40% of EGFR+ EBs showed colocalization with CPn0678 (Fig. 4B and SI Appendix, Fig. S3 A and B), indicating that the protein is secreted during EGFR-dependent adhesion and internalization. We then studied the function of CPn0678 during the entry process. As mentioned above, CPn0678 contains three PRR domains. Such domains are known to mediate protein–protein interactions, most commonly with proteins carrying Src homology 3 (SH3) domains (36, 37). SH3 domains are small modules of 50 to 60 amino acids with a characteristic 3D structure, which recognize the minimal motif PxxP. Extending the motif with additional prolines and charged amino acids increases the specificity of the PRR for defined target proteins (38). Interestingly, SH3 domains can be found in many proteins involved in endocytosis (39). Since C. pneumoniae utilizes EGFR-mediated endocytosis (19), we asked whether transfected CPn0678-GFP colocalized with various SH3-containing proteins. We chose proteins directly related to EGFR-mediated endocytosis, such as the endocytic adaptor proteins Grb2 (40), c-Cbl (41), and Cin85 (42). We also selected SNX9, a key regulator of dynamin assembly required for efficient clathrin-mediated endocytosis and known to be involved in actin dynamics during endocytosis (12, 43) (SI Appendix, Fig. S4 A and B). Coexpression studies revealed no colocalization of CPn0678 with Cin85, c-Cbl, or Grb2, but SNX9 colocalized with CPn0678 both at the PM and in the CPn0678-induced tubules described above (SI Appendix, Fig. S4A). In cells coexpressing both proteins, we observed that SNX9 relocalized to CPn0678+ tubules. Since the latter are phenotypically distinct from the tubules generated upon overexpression of SNX9 (SI Appendix, Fig. S4C) (44), this observation is indicative of a direct interaction. Moreover, when we immunoprecipitated CPn0678–GFP from these coexpressing cells, we only detected mCherry–SNX9 and none of the other SH3 proteins in the eluted fraction, in agreement with our imaging data (SI Appendix, Fig. S4B).
Fig. 4.
CPn0678 interacts with host SNX9. (A) Colocalization of CPn0678 (stained with anti-CPn0678 and anti-rabbit Alexa488) and EGFR (stained with anti-EGFR and anti-mouse Alexa594) at bacterial entry sites at 15 min postinfection. The area in the white box is shown at higher magnification in the Inset (Right). C. pneumoniae EBs were stained with DAPI. White arrowheads indicate instances of colocalization. (Scale bar, 10 µm; 1 µm in Insets.) (B) EBs associating with EGFR were quantified for additional colocalization of CPn0678 signals shown in A. Confocal images of 300 individual EBs were analyzed. Data are presented as means ± SD (n = 3). ***P ≤ 0.001. Colocalization was validated by estimating Pearsons correlation coefficients (SI Appendix, Fig. S3 A and B). (C) Pulldown experiments using purified recombinant GST or GST–SNX9 variants (ΔSH3, SH3) and CPn067810His or CPn0678ΔPRR110His. Input and elution samples obtained from His-pulldowns and GST-pulldowns were fractionated by SDS/PAGE and probed with anti-GST and anti-His antibodies. (D) Quantification of FLIM measurements of average fluorescence lifetimes, used to monitor FRET from CPn0678–GFP to mCherry–SNX9. The lifetime of donor fluorescence measured in 30 to 40 cells with or without coexpression of the acceptor are depicted as a box plot generated by OriginPro. Data are presented as means ± SD (n = 3). (E) Confocal images of cells coexpressing CPn0678–GFP and mCherry–SNX9 variants (SNX9, ΔBAR, ΔBARΔSH3). Insets show regions in white squares. (Scale bars 10 µm; 1 µm in Insets.) (F) Confocal images of Texas red-stained GUVs containing PS and PIP4,5 preincubated for 10 min with DyLight650-labeled SNX9 and followed by addition of FITC-labeled CPn0678 variants. See Movies S6–S15 for movies of each sample. (Scale bars, 5 µm.)
We verified the SNX9–CPn0678 interaction in pulldown experiments using recombinant proteins. Here, CPn0678-His was found to interact directly with GST–SNX9 (Fig. 4C and SI Appendix, Fig. S5) in pulldowns using either His or GST resin. SNX9 contains an SH3 domain that recognizes PRRs with a positive charge at the N terminus [consensus (R/K)xφPxxP; φ = hydrophobic amino acid], as do the PRRs found in dynamin and N-WASP (43). Since this consensus sequence also occurs in the first PRR motif (RPAPPQP) of CPn0678, we deleted either the SH3 domain of SNX9 (ΔSH3) or the first PRR motif in CPn0678 (ΔPRR), and found that both variants had lost the ability to interact with their respective full-length partner (Fig. 4C and SI Appendix, Fig. S5), indicating that SNX9 and CPn0678 indeed interact via their SH3 and PRR motifs respectively. The SH3 domain alone is sufficient for interaction, as it is capable of pulling down CPn0678 on its own.
Finally, to test for a direct interaction between the two proteins, we performed fluorescence lifetime imaging microscopy (FLIM). In vivo, the average lifetime of CPn0678–GFP was measured with and without coexpression of mCherry–SNX9. Reduction of the lifetime is caused by Förster resonance energy transfer (FRET) from a donor molecule (here CPn0678–GFP) to an acceptor molecule (here mCherry–SNX9) in close proximity (<10 nm) and thus indicates interaction in vivo. The donor control showed an average lifetime of 2.86 ns (Fig. 4D). In contrast, the average lifetime of CPn0678–GFP was significantly reduced (by 256 ps) to 2.60 ns, when coexpressed with mCherry–SNX9 (Fig. 4D). Similar levels of lifetime reduction have been observed in other protein interaction studies and for fused proteins (45–47).
Taken together, these data suggest that the APH directs CPn0678 to the PM, where it can directly interact with SNX9 via PRR and SH3 domains. To elucidate the hierarchy of these interactions during recruitment of SNX9 to the secreted chlamydial effector, we performed further in vivo and in vitro assays in which we measured the interaction of different CPn0678 variants—either capable of binding and tubulating the membrane (CPn0678, N-term, and ΔPRR1) or not—with SNX9 and two deletion variants (ΔBAR, ΔBARΔSH3) (Fig. 4 E and F and Movies S6–S15). The BAR domain of SNX9 contains classic amphipathic helices to induce membrane tubulation, although SNX9 is better at sensing curvature than inducing it (44, 48). While lipid binding is maintained via the PX domain, deletion of the BAR domain disrupts membrane tubulation, and deletion of the SH3 domain inhibits all protein–protein interactions (12). Upon coexpression of CPn0678 with SNX9, colocalization of both proteins on membrane tubules was dependent on the presence of the SNX9 SH3 domain, but independent of the BAR domain (Fig. 4E and SI Appendix, Fig. S4E).
Furthermore, CPn0678 variants lacking the PRR1 motif (i.e., N-term or ΔPRR1) showed no colocalization with SNX9, whereas variants lacking the membrane localization domain, C terminus, or APH, interacted with SNX9, although interaction was restricted to the cytosol (SI Appendix, Fig. S4 D and E). This indicates that CPn0678 induces the recruitment of SNX9 to membrane structures formed by the chlamydial effector. During chlamydial endocytosis this occurs at the PM, where secreted CPn0678 locally deforms the membrane and recruits SNX9. To test our hypothesis that CPn0678-induced membrane curvature indeed recruits SNX9, we again used GUVs composed of PS and PIP4,5 (Fig. 4F and Movies S6–S15). In accordance with published data, DyLight650-labeled SNX9 did not bind to these GUVs (Fig. 4F and Movie S6); although it is capable of binding to PIP4,5 lipids (49, 50), it does so only if the membranes are already curved (16). Strikingly, when we added FITC-labeled CPn0678 to GUVs preincubated with SNX9 we observed immediate binding of CPn0678 and generation of membrane tubules, to which SNX9 was subsequently recruited (Fig. 4F and Movie S7). This recruitment is dependent on the membrane-tubulating capacity of the N-terminal APH of CPn0678, as SNX9 is recruited to the GUV surface in the presence of membrane-modulating N-term or ΔPRR1 variants but not ΔAPH (Fig. 4F and Movies S8–S10). Moreover, SNX9 recruitment to GUVs is not dependent on its own membrane-curving domain, as we observed the same pattern of recruitment of SNX9ΔBAR to CPn0678, N-term, and ΔPRR1 but not ΔAPH variants (SI Appendix, Fig. S6 and Movies S11–S15). Taken together, these results show that CPn0678 induces membrane curvature which is then sensed by SNX9.
Internalization of C. pneumoniae Depends on the Interaction between CPn0678 and SNX9.
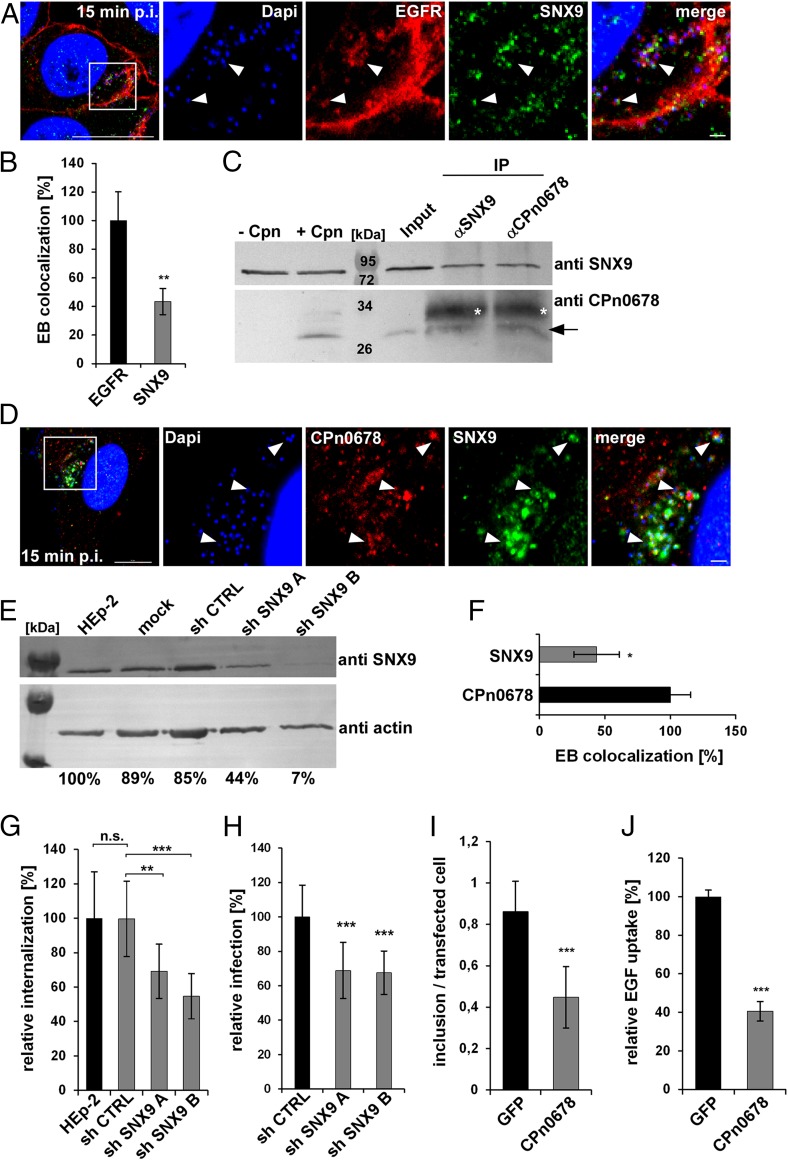
To demonstrate the interdependence of endogenous SNX9 and CPn0678 during C. pneumoniae internalization, we first analyzed whether SNX9 localizes to invading EBs. Indeed, by 15 min postinfection, we were able to detect a patch-like association of SNX9 with EBs surrounded by EGFR (Fig. 5A). Quantification and validation of the imaging data revealed that 47% of EBs that colocalize with EGFR are also positive for SNX9 (Fig. 5B and SI Appendix, Fig. S3C). Next, we performed a coimmunoprecipitation assay on cells lysed 15 min after infection, and were able to coprecipitate CPn0678 using a specific antibody against SNX9; conversely, SNX9 was recovered with an antibody directed against CPn0678 (Fig. 5C). These findings are further supported by direct colocalization of both proteins with attaching and invading C. pneumoniae EBs (Fig. 5D). We analyzed 400 EBs for CPn0678 expression, quantified, and validated how many of them colocalize with SNX9. Some 40% were found to show an overlap between SNX9 and CPn678 (Fig. 5E and SI Appendix, Fig. S3D). This indicates that, upon attachment of the EB to the host cell, CPn0678 is secreted and recruits SNX9 to the site of internalization, where both proteins together facilitate the EGFR-mediated endocytosis of C. pneumoniae.
Fig. 5.
Interaction of SNX9 with CPn0678 is essential for infection by C. pneumonia. (A) Colocalization of SNX9 (stained with anti-SNX9 and anti-mouse Alexa488) and EGFR (stained with anti-EGFR and anti-rabbit Alexa594) at bacterial entry sites at 15 min postinfection. C. pneumoniae EBs were stained with DAPI. Insets show regions in white squares. White arrowheads indicate instances of colocalization. (Scale bars 10 µm; 1 µm in Insets.) (B) A. Confocal images of 200 individual EBs were analyzed. Data are presented as means ± SD (n = 3). **P ≤ 0.01. Colocalization was validated by estimating Pearsons correlation coefficients (SI Appendix, Fig. S3C). (C) Coimmunoprecipitation of HEp-2 cells infected or not infected with C. pneumoniae EBs (MOI 100). Cell lysates were incubated with µMACS protein G microbeads coupled to antibodies directed against SNX9 or CPn0678. Elution samples were fractionated by SDS/PAGE and detected with the appropriate antibodies. White asterisks mark the light chain of the antibody, the black arrow the specifically labeled CPn0678 band. (D) Colocalization of SNX9 (stained with anti-SNX9 and anti-mouse Alexa488) and CPn0678 (stained with anti-CPn0678 and anti-rabbit Alexa594) at bacterial entry sites at 15 min postinfection. C. pneumoniae EBs were stained with DAPI. Insets show regions outlined by white squares. White arrowheads show colocalization. (Scale bar, 10 µm; 1 µm in Insets.) (E) EBs colocalizing with CPn0678 were quantified for additional colocalization of SNX9 signals shown in D. Confocal images of 400 individual EBs were analyzed. Data are presented as means ± SD (n = 3). *P ≤ 0.1. Colocalization was validated by estimating Pearsons correlation coefficients (SI Appendix, Fig. S3D). (F–H) C. pneumoniae internalization and infection of HEp-2 cells depleted of SNX9 by stably integrated shRNA plasmids. A nontargeting shRNA served as control. (F) Immunoblot of cell lysates fractionated by SDS/PAGE and probed with antibodies against SNX9. Actin served as the loading control. Signal intensity was quantified with ImageJ and expressed as a percentage of the actin signal. (G) Quantification of internalization of EBs into cells used in F at 2 hpi based on the examination of 40 visual fields. Data are represented as means ± SD (n = 4). **P ≤0.01, ***P ≤ 0.001, n.s., not significant. (H) Quantification of infection of cells used in F at 48 hpi based on the examination of 40 visual fields. Data are represented as means ± SD (n = 4). ***P ≤ 0.001. (I and J) Infection and EGF uptake assay of HEp-2 cells transfected for 15 h with either GFP or CPn0678-GFP. (I) Transiently transfected cells were subjected to a C. pneumoniae infection (MOI 2), fixed at 24 hpi and the numbers of inclusions in 10 visual fields were counted and plotted relative to the GFP control. Data are presented as means ± SD (n = 3). ***P ≤ 0.001. (J) Transfected cells were incubated with 100 ng/mL EGF-Alexa594 for 15 min, EGF uptake was measured in 10 visual fields and plotted relative to the GFP control. Data are presented as mean ± SD (n = 3) ***P ≤ 0.001.
We subsequently used a cell line that stably expresses two different short-hairpin RNAs (shRNAs) directed against SNX9 and performed internalization and infection studies with these cells (Fig. 5 F–H). Cells expressing shRNA B retained only 9% SNX9 protein and showed a 50% reduction in internalization rate compared to control cells, while cells expressing the less effective shRNA A retained 67% of the protein, and the internalization rate felt by 30% (Fig. 5 F and G). In infection studies both cell lines showed a comparable reduction of infection (Fig. 5H), indicating that SNX9 is essential for the C. pneumoniae internalization process.
Moreover, we generated cells with a dominant-negative endocytosis phenotype by transiently overexpressing CPn0678–GFP, therefore trapping endogenous SNX9 in a CPn0678-bound state (Fig. 5 I and J) at the PM. When we analyzed their capacity for endocytosis by either exposing them to C. pneumoniae EBs or labeled EGF, we observed a 50% reduction in inclusion formation in comparison to cells expressing GFP (Fig. 5I), and a 60% reduction in uptake of EGF (Fig. 5J), showing that the chlamydial early effector protein CPn0678 recruits SNX9 to bacterial entry sites, where both proteins facilitate the uptake of C. pneumoniae into host cells in a concerted manner.
Discussion
Upon T3-mediated secretion, the effector protein CPn0678 described in this work localizes to the host PM at bacterial entry sites enriched in EGFR. CPn0678 induces membrane curvature, which is sensed by SNX9, a key regulator of endocytosis. SNX9 is then recruited by and binds to CPn0678 via the interaction of its SH3 domain with the class I polyproline sequence in the C-terminal segment of the chlamydial effector. We therefore refer to the protein hereafter as SemC (secreted effector of membrane curvature).
The BAR-PX domain of SNX9 binds preferentially to membranes of high curvature (15). Indeed, recently it was shown that SNX9 localizes to highly curved regions of late clathrin-coated invaginations, which coincide with PI(4,5)P2 and PI(3)P (50, 51). The SNX9 PX-BAR domain binds to PI(4,5)P2-containing vesicles primarily via the BAR domain, and with lower affinity to PI(3)P-binding sites via the PX domain. Based on in vitro studies, it was subsequently proposed that SNX9 is recruited via its three effectors PI(4,5)P2, PI(3)P, and membrane curvature to the clathrin-coated invagination to locally activate the actin machinery and complete endocytosis (16).
On the basis of our results, we propose that SemC acts to control the size and shape of the endocytic vesicle formed following the activation of the EGFR-mediated endocytosis machinery, which is triggered by binding of the bacterial adhesin Pmp21 to EGFR (19). During chlamydial infection, an adjustment in PM curvature is essential, because while clathrin-mediated receptor endocytosis generates vesicles with a diameter of ∼120 nm (52), chlamydial EBs have a diameter of ∼400 nm. The significantly larger size of the developing vesicle harboring the EB exhibits significantly less curvature. SNX9 binds to membranes of clathrin-coated vesicles ∼10 times better than to vesicles with lower curvature (16). Thus, we suggest that, once secreted, CPn0678 binds to the inner leaflet of the host cell PM and induces a level of curvature equivalent to that of the developing clathrin vesicle; the curved membrane recruits SNX9, and a direct interaction of SemC with SNX9 may facilitate and stabilize this process. Recruitment of SNX9 and additional endocytic components targeted by SNX9, such as dynamin for neck constriction and N-WASP, enhance actin dynamics (12, 43, 53). We have shown here that SemC uses its APH domain to induce membrane curvatures in synthetic membranes, such as GUVs, which are strong enough to disrupt these vesicles. Ectopic expression of the effector in human cells generates tubules emanating from the PM. Formation of SemC-induced tubules on GUV membranes is essential for recruitment of SNX9, which by itself does not bind to these vesicles, and tubulation is solely dependent on the presence of the APH. Formation of membrane tubules within cells is dependent on the actin and microtubule cytoskeleton. SemC-induced tubulation is intensified upon recruitment of SNX9, as revealed by our finding that, when overexpressed, SNX9 strongly enhances the SemC tubule phenotype. Furthermore, ectopic expression of SemC prior to infection negatively influences the endocytotic capacity of cells, leading to fewer internalized EBs and less endocytosed EGFR. These observations imply that, during internalization, secreted SemC redirects SNX9 function to chlamydial entry sites, which in turn promotes the uptake of EBs.
In addition, SemC may manipulate other SNX9 functions, such as vesicle maturation, activation of dynamin for vesicle scission, and the regulation of actin assembly, which is essential for membrane remodeling by interaction with N-WASP or Arp2/3 (12). Interestingly, due to this central role in endocytosis, SNX9 is targeted in different ways by other bacterial pathogens (54, 55).
Depletion of SNX9 by shRNA reduces but does not abrogate C. pneumoniae internalization. This can likely be accounted for by continuing interaction of SemC with other members of the SNX9 family, in particular SNX18 and SNX33 (56). Moreover, this might also be indicative of the robustness of the chlamydial host-cell entry process, which very likely relies on a variety of chlamydial effector protein functions yet to be identified.
In conclusion, we speculate that the chlamydial effector SemC manipulates a core process of endocytosis by binding to and inducing curvature of the PM underneath invading Chlamydiae. This membrane deformation recruits the central endocytotic scaffold protein SNX9. The SemC–SNX9 interaction is direct, and together the two proteins promote formation of a vesicle that is large enough to accommodate an EB, and stimulate its endocytosis by locally activating the actin machinery.
Materials and Methods
Antibodies and Reagents.
The primary antibody against SNX9 (OTI1E4) was purchased from Origene, anti-EGFR (PA1-1110) and anti-GFP (MA5-15256) were from Thermo Scientific, antipenta-His (#34660) from Qiagen, anti-GST (#2622) from Cell Signaling, and anti-DsRed (sc-101526) from Santa Cruz. Anti-IncA was a gift from G. Zhong, University of Texas Health Science Center at San Antonio, San Antonio, TX (57), and anti-DnaK was obtained from S. Birkelund, Aalborg University, Aalborg, Denmark (58). Antibodies against Cpn0677 and Cpn0678 were generated in our laboratory, as was anti-CPn0147. Mouse anti-Cya, rabbit anti-CRP, and rabbit anti-IpaD antibodies were generously donated by N. Guiso, A. Ullmann, and C. Parsot, Institut Pasteur, Paris, France, respectively. Secondary anti-rabbit and anti-mouse antibodies coupled to Alexa488 or Alexa594 were purchased from Thermo Scientific, and those coupled to alkaline phosphatase were sourced from Promega. CellMask (orange) and Rhodamine-Phalloidin were purchased from Thermo Scientific, and SiR-Tubulin from Spirochrome. All lipids used in this study were obtained from Avanti Lipids, and Texas red dye, NHS-FITC and DyLight650NHS from Thermo Scientific. Nocodazole and Cytochalasin D were purchased from Merck.
Growth of Chlamydia, Bacteria, and Cell Lines.
C. pneumoniae GiD was propagated in HEp-2 cells (ATCC: CCL-23). HEp-2 and HEK293-T cells stably expressing shRNA plasmids were cultured in DMEM supplemented with 10% FCS, MEM vitamins, and nonessential amino acids (Thermo Scientific). Chlamydial EBs were purified using a 30% gastrographin solution (Bayer) and stored in SPG buffer (220 mM sucrose, 3.8 mM KH2PO4, 10.8 mM Na2HPO4, 4.9 mM l-glutamine). All cloning was carried out by in vivo homologous recombination in Saccharomyces cerevisiae. The Escherichia coli strains XL-1 Blue (Stratagene) and BL21 (DE3, Invitrogen) were used for plasmid amplification and protein purification, respectively. The ipaB and mxiD strains, in which the corresponding genes (ipaB and mxiD) have been inactivated (26), are derived from the virulent wild-type M90T strain of S. flexneri, and were grown in Luria-Bertani (LB) medium supplemented with (0.1 mg/mL) ampicillin.
Shigella Heterologous Secretion Assay.
Analysis of secreted proteins was performed as described previously (25). Briefly, 1 mL of a 30 °C overnight culture of S. flexneri ipaB or mxiD that had been transformed with different Cya chimeras was added to 30 mL of LB broth containing 0.1 mg/mL ampicillin, and incubated at 37 °C for 4 h. For experiments using full-length proteins, expression was induced by adding 150 μM isfopropyl-β-d-thiogalactopyranoside (IPTG) for the 4-h growth period. Bacteria were then harvested by centrifugation and the supernatant was filtered through a Millipore filter (0.2 μm). To precipitate the proteins, a one-tenth volume of trichloroacetic acid was added to the supernatant, and both the precipitate and the bacterial pellet were resuspended in sample buffer for analysis by SDS/PAGE and immunoblot.
Preparation and Analysis of GUVs.
GUVs were prepared as described previously (59). Briefly, PIP-containing lipid mixtures contain 69.75 mol% DOPC, 25 mol% cholesterol, and 0.25 mol % Texas red to which 5 mol% PIPs was added. In PS-containing GUVs the mixture was changed to 49.75 mol% DOPC/25 mol% cholesterol/0.25 mol% Texas red/20 mol% DOPS. GUVs containing PIP4,5 and PS were prepared by mixing 44.75 mol% DOPC, 25 mol% cholesterol, 0.25 mol% Texas red, and 20 mol% DOPS with 5 mol% PIP4,5. Lipid mixtures were prepared and added to a chamber built of ITO-coated slides (Präzisions Glas & Optik) glued together with Vitrex (Vitrex Medical). The cavity between the slides was filled with 10% sucrose solution and sealed with Vitrex. The slides were connected via clamps to a frequency generator and an alternating voltage of 2.0 Vp-p was applied at a frequency of 11 Hz. The GUVs were grown for 2 to 3 h in the dark at room temperature. For microscopic analyses, µ-slides Angiogenesis (Ibidi) were coated for 5 to 10 min at room temperature with 2 mg/mL β-casein (Merck) and washed with PBS. Then 10 µL of GUV solution mixed with 30 µL of PBS was added to the slides and the GUVs were allowed to settle. Then 2 µg of NHS-FITC–labeled recombinant protein was added and incubated for 15 min at room temperature. For binding studies with two proteins, GUVs were first incubated with 2 µg of NHS-650–labeled recombinant protein and imaged for 10 min at room temperature, then 2 µg of NHS-FITC–labeled recombinant protein was added and images were acquired for additional 10 min. Images were quantified using ImageJ. For each lipid and protein combination, 50 GUVs were analyzed for their maximum fluorescence intensity on the membrane.
Microscopy and Image Processing.
General imaging was performed using an inverse Nikon TiE Live Cell Confocal C2plus equipped with a 100× TIRF objective and a C2 SH C2 Scanner. All images were generated with Nikon NIS Elements software and quantified using ImageJ.
Gated STED measurements were performed using a TCS SP8 STED 3× (Leica) equipped with an HC PL APO CS2 100× objective (NA 1.4) at a scan speed of 400 to 600 Hz. A pulsed white-light laser was used at 499 nm for AlexaFluor488 excitation and a continuous-wave fiber laser at 592 nm was used for excitation depletion. The detection range of the GaAsP hybrid detector was set from 504 nm to 580 nm to collect the emitted fluorescent signals. To further increase resolution, time gating was set from 0.5 ns to 12 ns. Finally, deconvolved STED data were calculated using standard algorithms implemented in the Huygens software (Huygens Professional, Scientific Volume Imaging) on the acquired raw data.
Statistical Analyses.
Statistical analyses of FLIM imaging data were performed using OriginPro. The data represent the mean ± SD of n experiments. For simple paired analyses between two groups, a Student’s t test was chosen. A P value of less than 0.01 was considered to be statistically significant.
Data Availability Statement.
All data are available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank M. A. McNiven for plasmids; Bernd Tebarth for implementing the Chlamydia microarray; Astrid Engel for preparation of protein samples; Elena Görres and David Shi for generating mutant constructs; and the Center of Advnced Imaging for imaging. We acknowledge grant support from the Deutsche Forschungsgemeinschaft to J.H.H. (Project-ID 267205415) as part of CRC 1208, and funding of a graduate fellowship by the Jürgen Manchot Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911528117/-/DCSupplemental.
References
- 1.Elwell C., Mirrashidi K., Engel J., Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman L., et al. , Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10, e0143304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roulis E., et al. , Comparative genomic analysis of human Chlamydia pneumoniae isolates from respiratory, brain and cardiac tissues. Genomics 106, 373–383 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Webley W. C., Hahn D. L., Infection-mediated asthma: Etiology, mechanisms and treatment options, with focus on Chlamydia pneumoniae and macrolides. Respir. Res. 18, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balin B. J., et al. , Chlamydia pneumoniae: An etiologic agent for late-onset dementia. Front. Aging Neurosci. 10, 302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossu D., Yokoyama K., Hattori N., Bacteria-host interactions in multiple sclerosis. Front. Microbiol. 9, 2966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan P., et al. , Chlamydia pneumoniae infection and lung cancer risk: A meta-analysis. Eur. J. Cancer 47, 742–747 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Cossart P., Helenius A., Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6, a016972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber M. M., Faris R., Subversion of the endocytic and secretory pathways by bacterial effector proteins. Front. Cell Dev. Biol. 6, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigele B. A., Orchard R. C., Jimenez A., Cox G. W., Alto N. M., A systematic exploration of the interactions between bacterial effector proteins and host cell membranes. Nat. Commun. 8, 532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alto N. M., et al. , The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J. Cell Biol. 178, 1265–1278 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendris N., Schmid S. L., Endocytosis, metastasis and beyond: Multiple facets of SNX9. Trends Cell Biol. 27, 189–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza Santos M., Orth K., Subversion of the cytoskeleton by intracellular bacteria: Lessons from Listeria, Salmonella and Vibrio. Cell. Microbiol. 17, 164–173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mooren O. L., Galletta B. J., Cooper J. A., Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81, 661–686 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Pylypenko O., et al. , A combinatorial approach to crystallization of PX-BAR unit of the human Sorting Nexin 9. J. Struct. Biol. 162, 356–360 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Daste F., et al. , Control of actin polymerization via the coincidence of phosphoinositides and high membrane curvature. J. Cell Biol. 216, 3745–3765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subbarayal P., et al. , EphrinA2 receptor (EphA2) is an invasion and intracellular signaling receptor for Chlamydia trachomatis. PLoS Pathog. 11, e1004846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel A. L., et al. , Activation of epidermal growth factor receptor is required for Chlamydia trachomatis development. BMC Microbiol. 14, 277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mölleken K., Becker E., Hegemann J. H., The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PLoS Pathog. 9, e1003325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mölleken K., Hegemann J. H., Acquisition of Rab11 and Rab11-Fip2-A novel strategy for Chlamydia pneumoniae early survival. PLoS Pathog. 13, e1006556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zrieq R., Braun C., Hegemann J. H., The Chlamydia pneumoniae Tarp ortholog CPn0572 stabilizes host F-actin by displacement of cofilin. Front. Cell. Infect. Microbiol. 7, 511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hower S., Wolf K., Fields K. A., Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72, 1423–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKuen M. J., Mueller K. E., Bae Y. S., Fields K. A., Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect. Immun. 85, e00640-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mojica S. A., et al. , SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol. Biol. Cell 26, 1918–1934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subtil A., Parsot C., Dautry-Varsat A., Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39, 792–800 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Allaoui A., Sansonetti P. J., Parsot C., MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol. Microbiol. 7, 59–68 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Ménard R., Sansonetti P. J., Parsot C., Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175, 5899–5906 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullock H. D., Hower S., Fields K. A., Domain analyses reveal that Chlamydia trachomatis CT694 protein belongs to the membrane-localized family of type III effector proteins. J. Biol. Chem. 287, 28078–28086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon H. T., Boucrot E., Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerberg J., Kozlov M. M., How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Itoh T., et al. , Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 9, 791–804 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Guerrier S., et al. , The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leventis P. A., Grinstein S., The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Hammond G. R., Machner M. P., Balla T., A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205, 113–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingólfsson H. I., et al. , Lipid organization of the plasma membrane. J. Am. Chem. Soc. 136, 14554–14559 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Ball L. J., Kühne R., Schneider-Mergener J., Oschkinat H., Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. Engl. 44, 2852–2869 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Kurochkina N., Guha U., SH3 domains: Modules of protein-protein interactions. Biophys. Rev. 5, 29–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saksela K., Permi P., SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 586, 2609–2614 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Marsh M., McMahon H. T., The structural era of endocytosis. Science 285, 215–220 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Jiang X., Huang F., Marusyk A., Sorkin A., Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol. Biol. Cell 14, 858–870 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visser G. D., Lill N. L., The Cbl RING finger C-terminal flank controls epidermal growth factor receptor fate downstream of receptor ubiquitination. Exp. Cell Res. 311, 281–293 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Haglund K., Shimokawa N., Szymkiewicz I., Dikic I., Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc. Natl. Acad. Sci. U.S.A. 99, 12191–12196 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundmark R., Carlsson S. R., SNX9—A prelude to vesicle release. J. Cell Sci. 122, 5–11 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Park J., Zhao H., Chang S., The unique mechanism of SNX9 BAR domain for inducing membrane tubulation. Mol. Cells 37, 753–758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llères D., et al. , Quantitative FLIM-FRET microscopy to monitor nanoscale chromatin compaction in Vivo reveals structural roles of condensin complexes. Cell Rep. 18, 1791–1803 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Long Y., et al. , Optimizing FRET-FLIM labeling conditions to detect nuclear protein interactions at native expression levels in living Arabidopsis roots. Front. Plant Sci. 9, 639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumann S., Zander S., Weidtkamp-Peters S., Feldbrügge M., Live cell imaging of septin dynamics in Ustilago maydis. Methods Cell Biol. 136, 143–159 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Neumann S., Schmid S. L., Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J. Biol. Chem. 288, 25119–25128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin N., et al. , SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J. Cell Sci. 121, 1252–1263 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Schöneberg J., et al. , Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 8, 15873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sochacki K. A., Dickey A. M., Strub M. P., Taraska J. W., Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat. Cell Biol. 19, 352–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaksonen M., Roux A., Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Soulet F., Yarar D., Leonard M., Schmid S. L., SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell 16, 2058–2067 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tapia R., Kralicek S. E., Hecht G. A., EPEC effector EspF promotes Crumbs3 endocytosis and disrupts epithelial cell polarity. Cell. Microbiol. 19, e12757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piscatelli H. L., Li M., Zhou D., Dual 4- and 5-phosphatase activities regulate SopB-dependent phosphoinositide dynamics to promote bacterial entry. Cell. Microbiol. 18, 705–719 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Park J., et al. , SNX18 shares a redundant role with SNX9 and modulates endocytic trafficking at the plasma membrane. J. Cell Sci. 123, 1742–1750 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Luo J., et al. , Characterization of hypothetical proteins Cpn0146, 0147, 0284 & 0285 that are predicted to be in the Chlamydia pneumoniae inclusion membrane. BMC Microbiol. 7, 38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birkelund S., et al. , Characterization of two conformational epitopes of the Chlamydia trachomatis serovar L2 DnaK immunogen. Infect. Immun. 64, 810–817 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathivet L., Cribier S., Devaux P. F., Shape change and physical properties of giant phospholipid vesicles prepared in the presence of an AC electric field. Biophys. J. 70, 1112–1121 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and SI Appendix.