Significance
The paper proposes and demonstrates an approach for collecting solution NMR spectra from RNAs with high sensitivity, based on the chemical exchanges that labile hydrogens in the imino groups of nucleic acids can undergo with hyperpolarized water. This can lead to hundred-fold sensitivity enhancements for these diagnostic resonances, providing a tool to monitor complex RNA-based interconversions via 1-dimensional or 2-dimensional spectroscopy. The potential of this technology is demonstrated here by following the folding of the guanine-sensing riboswitch aptamer domain upon injection of hypoxanthine. This is a refolding process occurring within ∼1-s timescales, whose intermediates can be followed with atomic-level resolution under physiological temperatures and conditions, thanks to the enhancements brought by hyperpolarized NMR.
Keywords: RNA structure, riboswitch refolding, nuclear magnetic resonance, hyperpolarized NMR, sensitivity enhancement
Abstract
NMR sensitivity-enhancement methods involving hyperpolarized water could be of importance for solution-state biophysical investigations. Hyperpolarized water (HyperW) can enhance the 1H NMR signals of exchangeable sites by orders of magnitude over their thermal counterparts, while providing insight into chemical exchange and solvent accessibility at a site-resolved level. As HyperW’s enhancements are achieved by exploiting fast solvent exchanges associated with minimal interscan delays, possibilities for the rapid monitoring of chemical reactions and biomolecular (re)folding are opened. HyperW NMR can also accommodate heteronuclear transfers, facilitating the rapid acquisition of 2-dimensional (2D) 15N-1H NMR correlations, and thereby combining an enhanced spectral resolution with speed and sensitivity. This work demonstrates how these qualities can come together for the study of nucleic acids. HyperW injections were used to target the guanine-sensing riboswitch aptamer domain (GSRapt) of the xpt-pbuX operon in Bacillus subtilis. Unlike what had been observed in proteins, where residues benefited of HyperW NMR only if/when sufficiently exposed to water, these enhancements applied to every imino resonance throughout the RNA. The >300-fold enhancements observed in the resulting 1H NMR spectra allowed us to monitor in real time the changes that GSRapt undergoes upon binding hypoxanthine, a high-affinity interaction leading to conformational refolding on a ∼1-s timescale at 36 °C. Structural responses could be identified for several nucleotides by 1-dimensional (1D) imino 1H NMR as well as by 2D HyperW NMR spectra acquired upon simultaneous injection of hyperpolarized water and hypoxanthine. The folding landscape revealed by this HyperW strategy for GSRapt, is briefly discussed.
Dissolution dynamic nuclear polarization (dDNP) can provide significant sensitivity enhancements to solution-state NMR (1–5). dDNP takes advantage of the nearly complete electron spin polarization available under cryogenic high magnetic field conditions, transferring this to the surrounding nuclei before rapidly melting and shuttling the resulting hyperpolarized mixture to an NMR spectrometer for observation with dramatically improved sensitivity. While this strategy is successfully used to hyperpolarize a variety of small molecules for in vivo and in vitro studies (6–11), the direct hyperpolarization of biomacromolecules is limited due to the relaxation losses occurring during the transfer between the polarizer and the NMR spectrometer (4, 5, 12, 13). To cope with this limitation, the use of hyperpolarized water (“HyperW”) has been proposed as a means to enhance biomolecular solution NMR (14–23). The ensuing HyperW NMR experiment relies on exploiting the high solution-state polarizations that dDNP can transfer to the protons of water, and the relatively long (10 to 30 s) T1 relaxation times that these 1Hs can achieve under suitable dissolution conditions. This combination allows one to enhance, via spontaneous chemical exchanges, the NMR of biomacromolecules that are waiting in a magnet and that receive a sudden injection of HyperW. HyperW has been successfully used to polarize a variety of amine and amide sites in peptides, intrinsically disordered and folded proteins, and to obtain high-resolution 1-dimensional (1D) and 2-dimensional (2D) 15N-1H correlations of exposed residues exhibiting up to 400× enhancements over their thermal counterparts at a detection magnetic field strength of 14.1T.
This work extends these principles to the observation of hyperpolarized resonances in nucleic acids. Imino RNA protons are especially attractive targets from a HyperW point of view, since they are known to exchange rapidly with water, while being well separated spectrally from potential interferences arising from the intense and broad hyperpolarized water peak. Furthermore, imino resonances are known to be sensitive to changes in conformation of a nucleic acid chain, making them particularly valuable probes to follow the strand’s structure, dynamics, binding, and potential subsequent refolding (24). The latter is particularly important in RNA studies, since although the energetic preferences for specific conformations tend to be small, their equilibrium distributions will often be strongly affected by the binding of small metabolites (25–27), ions (28), and proteins (29). The present study targeted one such system by 1D 1H and 2D 1H-15N HyperW NMR: the guanine-sensing riboswitch (GSR) aptamer domain (GSRapt) of the Bacillus subtilis xpt-pbuX operon (30). This riboswitch aptamer domain is a regulatory element located in the 5′-untranslated region of mRNA that binds the small metabolites guanine and hypoxanthine (24, 30–35). This GSR belongs to the group of transcriptional “off-switches,” where the binding of a specific metabolite to the aptamer domain induces a structural reorientation that stops the transcription of mRNA. In B. subtilis, this mRNA transcriptional control is affected by the selective, high-affinity binding of guanine or hypoxanthine, which, in turn, will lead to an allosteric conformational rearrangement. We have previously monitored this aptamer domain’s refolding processes by using photocaged hypoxanthine and triggering the dynamics by the action of light (24). A number of ligand binding-induced RNA rearrangements were thus noted, including the refolding of a binding pocket and the establishment of tertiary “kissing loop” interactions (28), within kinetic timescales of ∼20 to 30 s at 10 °C. Delineating additional ligand-induced structural rearrangements occurring on subsecond to second timescales—faster than what has been possible to date, but too slow for relaxation-dispersion (36, 37) or chemical exchange saturation transfer-based (38, 39) experiments—requires devising real-time NMR methods.
The present study explores the possibility of accessing such rearrangements and timescales, utilizing a combination of water-based dDNP and sudden injections of refolding agents. To this end a fast, robust setup for the injection of HyperW was applied on GSRapt, leading to significant enhancements (∼300-fold at 14.1T) of the majority of imino peaks in the aptamer. The observed enhancements were heterogeneous within the construct, but they reproduced well the water exchange rates exhibited by the different nucleobases. Thereafter, HyperW was injected in combination with hypoxanthine, to trigger a folding of the RNA that could be followed by time resolved 1D and 2D HyperW NMR acquisitions within the folding timescale (<10 s), to extract insights into the structural transitions undergone by the aptamer domain upon binding hypoxanthine.
Results and Discussion
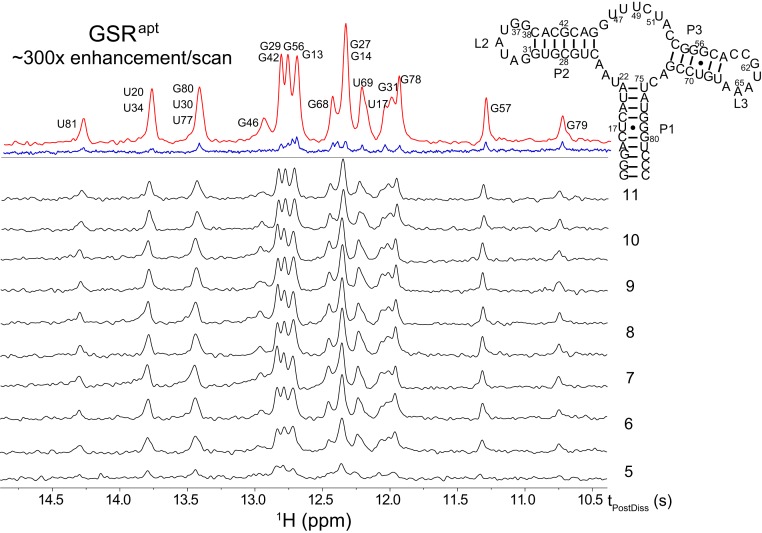
Fig. 1 compares a series of HyperW selective spin-echo–based 1D 1H NMR spectra collected in a single shot on a natural-abundance GSRapt sample following dissolution, against a thermal spectrum acquired for the same sample using a slightly more optimized (excitation sculpted) (40, 41) sequence in 2,048 scans. These results indicate ∼300-fold enhancements for the imino resonances vs. their thermal 14.1T counterparts; this is similar to the highest enhancements we have observed with this hyperpolarization setup on fully unfolded peptides and proteins (17, 20). Aiding in achieving these enhancements are the fast solvent-exchange rates characterizing RNA’s imino protons: while in proteins, only water-exposed residues will exchange sufficiently fast to exhibit substantial HyperW enhancements, steric or electrostatic hindrances rarely affect the exchanges of RNA’s imino protons. In fact, resonances arising from nucleotides in exposed loops may be altogether invisible, as their imino peaks may be broadened beyond detectability by exchange with the solvent. For the sample studied here, exchange rates appeared to be in a range (kex ≥ 20 s–1) that is conducive to large HyperW enhancements for most of the nucleobases. Such rates are expected to be common for RNAs under physiological temperatures and pHs, making this HyperW experiment of general usefulness for the study of nucleic acids. (SI Appendix, Fig. S2 and its associated discussion examine in closer detail how these solvent exchange rates are expected to influence HyperW enhancements for 1D and 2D NMR acquisitions.) These experiments also benefit from the substantial separation between the imino resonances and that of water, enabling the former’s effective excitation while minimizing perturbations of the hyperpolarization “reservoir.”
Fig. 1.
In black, 1D selective spin-echo single-shot spectra acquired using the sequence in SI Appendix, Fig. S1A, upon injection of HyperW at 37 °C into a nonlabeled GSRapt sample. The final concentration of RNA was ∼300 µM, and data were recorded with a time resolution of 0.685 s at the postdissolution times indicated on the right. These times represent the seconds elapsed after the water dissolution started and include the transfer to the magnet and a mixing period (notice how turbulence, still present in the first spectrum, affects the peaks’ intensities). The top trace in the figure compares the sum of the 10 HyperW spectra collected from 4.80 to 10.96 s (red), against a thermal spectrum acquired for the same sample using 2,048 fully relaxed transients (in blue). Shown above the traces is the aptamer’s secondary structure and its numbering, together with selective peak assignments.
The significant enhancement of the water, its fast exchange with the iminos, and its relatively long T1 (∼15 s) under the employed dissolution conditions, also enable the acquisition of multiscan dDNP-enhanced 2D 15N-1H correlations with superior sensitivities. Fig. 2 compares such a HyperW heteronuclear multiple quantum coherence (HMQC) spectrum acquired under typical postdissolution conditions for the GSR aptamer, against a thermal spectrum acquired for an approximately twofold more concentrated sample prepared in 90% protonated buffer—i.e., effectively, in a 100-fold more concentrated sample. Apparent spectral similarities notwithstanding, a few differences between the two sets are also clear. One concerns the peaks’ linewidths along the indirect dimension: in the HyperW case, these are affected by the water’s depolarization over the course of the experiment, leading to a decaying signal over the course of successive t1 increments and thus to an apparent broadening along the 15N axis. Also evidenced by the spectra is that hyperpolarization applies an exchange filter to the data: since different imino protons have different solvent-exchange rates, their relative peak ratios between the 2D HyperW and thermal HMQC spectra are also different. This difference provides additional insight about the exchange rates for the different bases, favoring and in some cases making visible labile protons that are normally broadened beyond detection in the thermal spectrum due to fast solvent exchange. This is the case, for instance, for the nucleobase resonances G46, U75, and U77; these are nucleotides residing at the termini of the helices facing the ligand-binding pocket, and are thus most likely not single-stranded but rather adopting labile structures and sampling open conformations more frequently than nucleotides within the helices (24). Similarly, G31, positioned in the L2 loop, experiences substantial enhancements upon injecting the HyperW—presumably as a result of faster exchanges with the solvent.
Fig. 2.
(A) Thermal 15N-1H imino HMQC correlations acquired for a reference sample containing 350 μM GSR aptamer in 90% H2O buffer; these data were acquired using four scans per t1 increment, 128 complex t1 points, and a recycling delay of 1 s to allow relaxation of imino protons, leading to a 19-min 36-s acquisition. (B) HyperW HMQC spectrum acquired upon sudden injection of hyperpolarized water, using two scans and 37-ms relaxation delay per t1, yielding a total acquisition time of 43 s for 64 complex t1 points. The experiment utilized band-selective 90° and 180° 1H pulses as shown in SI Appendix, Fig. S1B. The final postdissolution RNA concentration was ∼170 μM, and the spectrum was acquired in a ∼2% hyperpolarized H2O/98% D2O buffer solution at pH 6.2 and 37 °C. Labeled in red are residues that were undetectable in the thermal experiment—presumably as a result of broadening due to fast exchanges with the solvent.
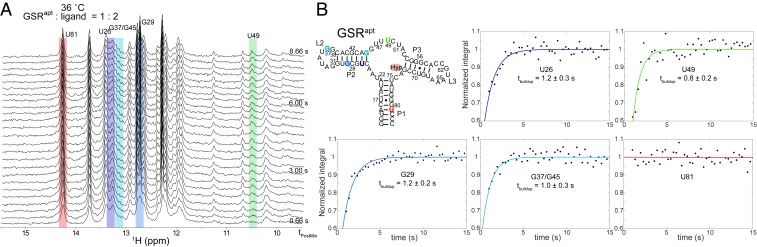
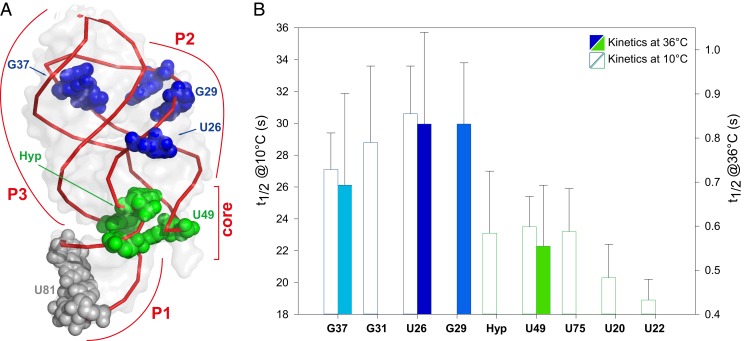
These 1D and 2D NMR results confirm that HyperW experiments relying on fast exchanges between the imino and the water protons can yield substantial enhancements and sensitivity improvements, even if using short or no interscan delays. This, in turn, makes them attractive options for the exploration of kinetic molecular details such as those associated with a ligand-induced refolding of the aptamer domain, via real-time NMR. Previously, we have shown that ligand-induced conformational changes in GSRapt could be followed at atomic resolution in this manner, utilizing real-time NMR methods in combination with the laser-triggered release of the ligand (24). These ligand-induced refolding studies, however, had to coadd several kinetic NMR experiments (arising from physically different samples) to reach a sufficient signal-to-noise ratio (SNR), and had to be performed at relatively low temperatures (10 °C) to accommodate the signal-averaging timescale. While bypassing the challenges arising under more physiologically affine conditions, these experimental and temperature conditions are naturally suboptimal. In particular, higher temperatures would “bleach” some of the resonances as a result of faster solvent exchange (SI Appendix, Fig. S3), while the increases in Kd values at higher temperatures would demand higher [ligand]:[RNA] stoichiometries to perform the experiments. The enhancement and timescales provided by HyperW NMR, by contrast, make these higher-temperature experiments feasible. Fig. 3A demonstrates this with an array of 1D spectra, acquired upon injecting hyperpolarized water together with the ligand hypoxanthine (comixed in the dissolution buffer at approximately 37 °C), to a GSRapt solution at 36 °C. Differences emerge between this spectral array and that presented in Fig. 1, reflecting the effects of the ligand binding. Although multiple peaks undergo changes in this kinetic time series, a quantitative analysis of the HyperW spectra needs to consider that resonances in the ligand-bound holo form will initially grow due to the chemical kinetics—but eventually decay together with all remaining peaks due to the T1 relaxation of the hyperpolarized water pool. In order to enable an analysis of the binding kinetics while accounting for these effects, all spectra were individually phased, apodized, and weighted according to the water T1 decay before an individual peak’s time evolution was computed. Data processed in this fashion could be reliably fitted to monoexponential functions, which then revealed the kinetic rates of interest. Fig. 3B summarizes such ligand-induced refolding results for several imino protons. These are consistent with what had been observed at lower temperatures using laser-triggered binding, according to which hypoxanthine binds to this aptamer in a two-step process, with rates falling into two slightly different time regimes that reflect different conformational refoldings. Ligand binding precedes the formation of tertiary loop–loop interactions as previously observed. Thus, for example, U49 (highlighted in green), a base involved in the formation of the ligand-binding pocket, shows a faster signal rise than bases U26, G37, and G29 (in blue), as the latter are affected by the slower helix–helix packing after closure of the ligand binding pocket (GSRapt secondary structure; Fig. 3B). Also consistent with previous observations, peaks such as those arising from U81 (in red), a nucleotide belonging to the P1 helix, remain constant throughout the experiment. Overall, the kinetic rates observed for these processes by the HyperW NMR series are also consistent with the 10 °C literature reports, after factoring in the effects of thermal activation. This is summarized in Fig. 4, which illustrates the kinetic changes experienced by these and other nucleobases, as measured at 10 °C by Buck et al. (24) and by the 36 °C HyperW experiments reported in this study, and shows the location of these bases within GSR’s tertiary structure.
Fig. 3.
(A) Array of 1D selective spin-echo spectra obtained upon injecting hyperpolarized water and hypoxanthine into a solution of the GSR aptamer. The final concentration of RNA was ∼170 μM and of ligand ∼340 μM. Spectra were recorded as in Fig. 1 with a time resolution of 0.33 s; the first data point was discarded due to turbulence present at the start of acquisition. Multiple peaks display site-specific kinetics associated with structural changes induced by the binding of the hypoxanthine; some of these are highlighted with different colors. (B) Buildup plots for several bases indicating folding kinetics on the time scale of ∼1 s. As a control, evolution of U81 is plotted to show that some bases do not experience changes upon binding. Buildup curves with different colors correspond to peaks highlighted accordingly in the spectral time series.
Fig. 4.
(A) Tertiary structure of the GSR aptamer, highlighting the bases experiencing faster ligand-binding kinetics (green), slower tertiary loop–loop rearrangement (blue), and no change (gray). (B) Histogram plots summarizing the half-life times extracted from the 36 °C HyperW measurements and from the kinetics measured at 10 °C by Buck et al. (24), highlighting the distinct kinetics observed.
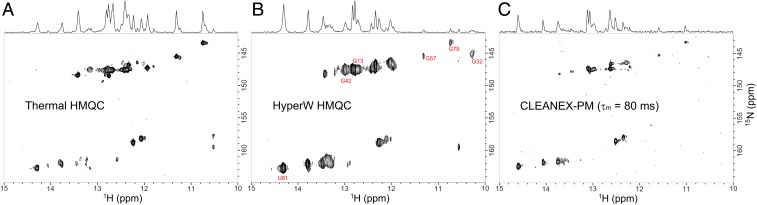
Fig. 5 presents the extension of these hypoxanthine-binding analyses to heteronuclear 2D correlations. Fig. 5A shows a thermal 2D HMQC spectrum acquired for the same reference sample as in Fig. 2A, but after the system has equilibrated with hypoxanthine. Distinct spectral changes can be seen when comparing these two spectra, reflecting the different structure of the ligand-bound state. Fig. 5B shows the HyperW 2D HMQC spectrum arising right after injection of HyperW and hypoxanthine, into an awaiting GSRapt sample; when compared with the HyperW 2D spectrum recorded in the absence of hypoxanthine (Fig. 2B), the fast refolding induced by the purine’s coinjection is confirmed. It is interesting to note that while in the apo form (Fig. 2B), most of the peaks are uniformly well enhanced; in the holo state (Fig. 5B), a heterogeneity among the enhancements is noticeable. For example, whereas in the hypoxanthine-bound aptamer, G57 and G79 are weakly enhanced (∼65×), G13, G42, and U81 experience a high relative enhancement (>320×) typical of a fast chemically exchanging site. To illustrate the close correlation between HyperW enhancements and solvent exchange rates, Fig. 5C presents a clean chemical exchange (CLEANEX)-filtered 15N-1H HMQC imino spectrum acquired for a similar sample at the same temperature. CLEANEX is an experiment specifically designed to highlight the labile protons that exchange fast enough with the solvent, over the course of a preassigned mixing time (42, 43). The close correspondence between the relative intensities of selected peaks in the HyperW and CLEANEX spectra evidence the strong dependence of the hyperpolarized enhancements on the solvent chemical exchange rates. Furthermore, based on the differences in HyperW enhancements between ligand-free and ligand-bound states, one can conclude that solvent exchange rates for certain imino sites change after refolding of the GSRapt holo form. This is the case for instance with G57, G79, and especially with G32, a nucleotide in loop L2 that is detected when the hyperpolarized experiment is realized with hypoxanthine but that apparently broadens beyond detection in HyperW experiments performed without ligand. This would suggest that its exchanges with water are fast in the ligand-free apo form, but appear to slow down in the ligand-bound state.
Fig. 5.
(A) Thermal 15N-1H imino HMQC spectrum acquired for a reference sample of 350 μM GSR aptamer in 90% H2O buffer in the presence of two equivalents of the ligand hypoxanthine. The spectrum was acquired with 64 complex t1 points in 5 min 25 s, using four scans per t1 and a relaxation delay of 0.5 s. (B) HyperW 15N-1H imino correlation acquired within 45 s after injection of HyperW and ligand into an RNA sample waiting in the magnet. The final RNA and ligand concentrations were ∼170 μM and ∼340 μM, respectively, and the postinjection solution contained ∼2% hyperpolarized H2O diluted in 98% D2O buffer. The spectrum was acquired using two scans per t1, 50-ms delay, and 64 complex t1 points. (C) Thermal 15N-1H imino CLEANEX-filtered HSQC spectrum acquired for the same reference sample as in A, containing 350 μM aptamer in 90% H2O buffer in the presence of two equivalents of the ligand hypoxanthine using 192 scans per t1, a CLEANEX mixing time of 80 ms, and a relaxation delay of 1.5 s (total acquisition time: 11 h, 44 min).
Conclusions
This study explored some of the potential applications enabled by hyperpolarized water injections, when studying the imino 1H NMR resonances in nucleic acids. Sizable enhancements of fast exchanging protons could be observed in these biopolymers, enabling rapid acquisitions with short recycling delays. This opens opportunities to perform real-time kinetic 1D and 2D NMR measurements of RNA folding events, with an enhanced sensitivity that allows one to access subsecond timescales; it also provides a means to assess changes in solvent accessibility, associated with ligand binding or other RNA structural rearrangements. This potential of the HyperW NMR approach was demonstrated by studying the binding of the small cognate metabolite hypoxanthine to its target riboswitch GSRapt, comprising 78 nucleotides. Ligand-induced RNA refolding could thus be assessed at a physiologically relevant temperature (36 °C), after deconvolving the hyperpolarized water relaxation/decay effects, from the observed kinetics. Hitherto unavailable refolding kinetics/values parameters could thus be obtained from the 1D data: simple SNR calculations indicate that approximately 16 scans per spectrum would be needed to obtain from thermal per-protio NMR spectra the kind of SNR afforded by the HyperW single-scan experiment for the 170 μM RNA solution—too long to catch the fast conformational changes occurring during the binding at the studied temperatures. With time scales of a few seconds for RNA binding and refolding kinetics, the time resolution that HyperW offers is well-suited for monitoring these processes at 36 °C. HyperW 2D HMQC NMR spectra also were collected, providing additional resolution enhancement and revealing valuable spectral changes that occurred upon binding. Even more detailed kinetic measurements can arise from combining HyperW 2D NMR with nonuniform sampling (44–48) and/or with ultrafast 2D methods (49–51), which could deliver multiple snapshots of the reaction with enhanced site resolution and enable the probing of even faster, more complex RNA rearrangements and folding pathways than the ones explored in a system that, like the GSRapt, had already been explored at low temperatures. We also envision HyperW NMR to open vistas in the detection of minority RNA states (52, 53) and/or of RNAs at the concentrations in which they are found under typical cellular conditions. Despite these advantages, HyperW NMR is still limited by a number of factors. One is that most dissolutions are performed with deuterated buffers; while this helps prolong the hyperpolarization lifetime, the concomitant dilution of available protons results in a substantial reduction in the effective sample contribution to the 1H NMR signal. Efforts are thus being made to improve the dissolution process and reduce the dilution of the experiment. The exponential decay of the hyperpolarized signals as a result of water’s T1 relaxation is also a problem, resulting in biased kinetic results and in additional broadening superimposed along the indirect dimension of 2D acquisitions. The former problem was solved here by referencing the behavior observed for resonances that undergo exchange with the solvent but are not affected by the kinetic refolding; nonuniform sampling, t1 shuffling (54), and variable-angle excitation (55) techniques could help to alleviate this problem, and are also currently under investigation. With regard to fast kinetic measurements, HyperW’s built-in rapid mixing offers a natural starting point for triggering a reaction’s initiation. Still, this process has a nonzero dead-time associated with injection and turbulence settling times (∼1 s in our pressurized, Arduino-controlled setup; ref. 56) that will restrict applications of this method and needs further refinements to study faster, subsecond processes.
Materials and Methods
Sample Preparation.
Isotopically labeled GSR aptamer RNA was synthesized via in vitro transcription using nucleotides from Silantes GmbH and purified as described by Fürtig et al. (57). The RNA was obtained in milligram yield and dissolved in standard NMR buffer (25 mM potassium phosphate, 30 mM potassium chloride, pH 6.2) containing H2O:heavy water (D2O) (90%:10%) for reference samples, or 99.9% D2O for samples waiting in the magnet prior to dissolution experiments.
Dynamic Nuclear Polarization.
An Oxford Instrument Hypersense polarizer equipped with a 3.35T magnet was employed to hyperpolarize water. An EH-500 Edwards booster was added to the existing Oxford-supplied E2M80 vacuum pump, in order to further reduce operating pressures to below 1 torr. Thus, DNP was usually done at ∼1.15 to 1.30 K, by irradiating a 10 mM 4-amino-tetramethylpiperidinyloxy nitroxide radical dissolved in ∼150 μL of a solution consisting of 15% glycerol and 85% H2O (vol/vol) at ∼94.1 GHz. Microwave power levels and pumping times were set to 100 mW and 150 min, respectively. The ensuing hyperpolarized water pellets were dissolved with 2.6 mL of 99.9% D2O buffer with and without hypoxanthine; ∼300 μL of the melted, hyperpolarized samples were then transferred using a preheated (50 °C) tubing line and injected into a medium wall 5-mm tube containing the RNA dissolved in buffered D2O waiting in the NMR magnet. After the mixing, final H2O:D2O ratio was ∼2%:98%.
Injection Setup.
An automated injection system was utilized to achieve robust, reproducible transfers, assuring minimum bubble and foam formation (56, 58). The design of the injection system was previously described (20). In short, it relies on a two-state valve operation using both forward and backward gas pressures to regulate the volume of hyperpolarized sample reaching the NMR tube, and an Arduino-based software to control the injection setup. Previous optimization of the injection setup yielded driving and equilibration pressures of 17 and 3.5 bar.
NMR Spectroscopy.
Postdissolution NMR experiments were conducted using a 5-mm liquid nitrogen-cooled Prodigy probe in a 14.1T Bruker magnet interfaced to an Avance III console. These experiments included 1D and 2D NMR acquisitions, which were triggered upon injecting the HyperW sample into NMR tubes waiting with RNA samples inside the magnet bore. Unless otherwise indicated (e.g., SI Appendix, Fig. S3A), experiments were carried out at 36 °C; 1D NMR spectra were acquired using the selective spin-echo pulse sequence given in SI Appendix, Fig. S1A, while 2D HyperW NMR spectra were acquired using the HMQC-based sequence shown in SI Appendix, Fig. S1B. These sequences excite and echo the imino region selectively and employ minimal recycle delays to minimize water relaxation losses. Thermal NMR measurements were carried out on the same sample with the same hardware and using the same pulse sequence but with longer recycling delays. In order to obtain more reliable measures of the HyperW site-specific enhancements, water suppression was used in the acquisition of the 1D thermal spectra. For further accuracy, reference thermal spectra (Figs. 2 and 5 and SI Appendix, Fig. S3) were acquired using both the postdissolution samples and samples containing 90% water and higher concentrations of GSRapt. All NMR data were processed using the Bruker Topspin and MestreNova softwares and subsequently plotted and analyzed using Matlab. This manuscript’s experimental data are available free of charge at the Weizmann Institute of Science Library public repository (https://weizmann.alma.exlibrisgroup.com/view/delivery/972WIS_INST/1258056990003596).
Supplementary Material
Acknowledgments
This work would not have been possible without the technical assistance of the late Koby Zibzener. This work was supported by the Kimmel Institute for Magnetic Resonance (Weizmann Institute), the European Union Horizon 2020 Program (Marie Sklodowska-Curie Grant 642773), Israel Science Foundation Grant 965/18, and the Perlman Family Foundation (L.F.). H.S. was supported by the Deutsche Forschungsgemeinschaft-funded Collaborative Research Center 902. Work at Frankfurt’s Center for Biomolecular Magnetic Resonance (BMRZ) is supported by the state of Hesse.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The experimental data reported in this paper have been deposited in the Weizmann Institute of Science Library (https://weizmann.alma.exlibrisgroup.com/view/delivery/972WIS_INST/1258056990003596).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916956117/-/DCSupplemental.
References
- 1.Ardenkjaer-Larsen J. H., et al. , Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardenkjaer-Larsen J. H., et al. , Facing and overcoming sensitivity challenges in biomolecular NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 54, 9162–9185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeschke G., Frydman L., Nuclear hyperpolarization comes of age. J. Magn. Reson. 264, 1–2 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ragavan M., Chen H.-Y., Sekar G., Hilty C., Solution NMR of polypeptides hyperpolarized by dynamic nuclear polarization. Anal. Chem. 83, 6054–6059 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Chen H. Y., Ragavan M., Hilty C., Protein folding studied by dissolution dynamic nuclear polarization. Angew. Chem. Int. Ed. Engl. 52, 9192–9195 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Golman K., Ardenkjaer-Larsen J. H., Petersson J. S., Månsson S., Leunbach I., Molecular imaging with endogenous substances. Proc. Natl. Acad. Sci. U.S.A. 100, 10435–10439 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golman K., Zandt R. I., Lerche M., Pehrson R., Ardenkjaer-Larsen J. H., Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 66, 10855–10860 (2006). [DOI] [PubMed] [Google Scholar]
- 8.von Morze C., et al. , Imaging of blood flow using hyperpolarized [(13)C]urea in preclinical cancer models. J. Magn. Reson. Imaging 33, 692–697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurhanewicz J., et al. , Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia 13, 81–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris T., Degani H., Frydman L., Hyperpolarized 13C NMR studies of glucose metabolism in living breast cancer cell cultures. NMR Biomed. 26, 1831–1843 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Brindle K. M., Imaging metabolism with hyperpolarized (13)C-labeled cell substrates. J. Am. Chem. Soc. 137, 6418–6427 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Hilty C., Determination of ligand binding epitope structures using polarization transfer from hyperpolarized ligands. J. Med. Chem. 62, 2419–2427 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Ragavan M., Iconaru L. I., Park C.-G., Kriwacki R. W., Hilty C., Real-time analysis of folding upon binding of a disordered protein by using dissolution DNP NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 56, 7070–7073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris T., Bretschneider C., Frydman L., Dissolution DNP NMR with solvent mixtures: Substrate concentration and radical extraction. J. Magn. Reson. 211, 96–100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris T., Szekely O., Frydman L., On the potential of hyperpolarized water in biomolecular NMR studies. J. Phys. Chem. B 118, 3281–3290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chappuis Q., et al. , Hyperpolarized water to study protein-ligand interactions. J. Phys. Chem. Lett. 6, 1674–1678 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Olsen G., Markhasin E., Szekely O., Bretschneider C., Frydman L., Optimizing water hyperpolarization and dissolution for sensitivity-enhanced 2D biomolecular NMR. J. Magn. Reson. 264, 49–58 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kim J., Liu M., Hilty C., Modeling of polarization transfer kinetics in protein hydration using hyperpolarized water. J. Phys. Chem. B 121, 6492–6498 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Kurzbach D., et al. , Investigation of intrinsically disordered proteins through exchange with hyperpolarized water. Angew. Chem. Int. Ed. Engl. 56, 389–392 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Szekely O., Olsen G. L., Felli I. C., Frydman L., High-resolution 2D NMR of disordered proteins enhanced by hyperpolarized water. Anal. Chem. 90, 6169–6177 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Kadeřávek P., Ferrage F., Bodenhausen G., Kurzbach D., High-resolution NMR of folded proteins in hyperpolarized physiological solvents. Chemistry 24, 13418–13423 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Sadet A., et al. , Hyperpolarized water enhances two-dimensional proton NMR correlations: A new approach for molecular interactions. J. Am. Chem. Soc. 141, 12448–12452 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Szekely O., Olsen G. L., Novakovic M., Rosenzweig R., Frydman L., “Assessing site-specific water accessibility in folded and unfolded proteins using hyperpolarization-enhanced 2D HMQC NMR” in EUROISMAR2019 Joint 15th Euromar/21st ISMAR Conference (International Society of Magnetic Resonance, 2019), p. 104. [Google Scholar]
- 24.Buck J., Fürtig B., Noeske J., Wöhnert J., Schwalbe H., Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 104, 15699–15704 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda-Ríos J., Navarro M., Soberón M., A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. U.S.A. 98, 9736–9741 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler W., Nahvi A., Breaker R. R., Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Epshtein V., Mironov A. S., Nudler E., The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. U.S.A. 100, 5052–5056 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Draper D. E., Grilley D., Soto A. M., Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34, 221–243 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Williamson J. R., Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7, 834–837 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Mandal M., et al. , Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Schwalbe H., Buck J., Fürtig B., Noeske J., Wöhnert J., Structures of RNA switches: Insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 46, 1212–1219 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Fürtig B., et al. , Time-resolved NMR studies of RNA folding. Biopolymers 86, 360–383 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Winkler W. C., Breaker R. R., Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59, 487–517 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Noeske J., et al. , Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 35, 572–583 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Game J. C., Williamson M. S., Baccari C., X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics 169, 51–63 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loria J. P., Rance M., Palmer A. G., A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999). [Google Scholar]
- 37.Orekhov V. Y., Korzhnev D. M., Kay L. E., Double- and zero-quantum NMR relaxation dispersion experiments sampling millisecond time scale dynamics in proteins. J. Am. Chem. Soc. 126, 1886–1891 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Vallurupalli P., Bouvignies G., Kay L. E., Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Bouvignies G., Kay L. E., A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: An application to protein folding. J. Biomol. NMR 53, 303–310 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Hwang T., Shaka A. J., Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 112, 275–279 (1995). [Google Scholar]
- 41.Mescher M., Tannús A., O’Neil-Johnson M., Garwood M., Solvent suppression using selective echo dephasing. J. Magn. Reson. A 229, 226–229 (1996). [Google Scholar]
- 42.Hwang T., Mori S., Shaka A. J., van Zijl P. C. M., Application of phase-modulated CLEAN Chemical EXchange Spectroscopy (CLEANEX-PM) to Detect Water−Protein Proton Exchange and Intermolecular NOEs. J. Am. Chem. Soc. 119, 6203–6204 (1997). [Google Scholar]
- 43.Hwang T. L., van Zijl P. C. M., Mori S., Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR 11, 221–226 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Mobli M., Hoch J. C., Nonuniform sampling and non-Fourier signal processing methods in multidimensional NMR. Prog. Nucl. Magn. Reson. Spectrosc. 83, 21–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuyler A. D., Maciejewski M. W., Stern A. S., Hoch J. C., Nonuniform sampling of hypercomplex multidimensional NMR experiments: Dimensionality, quadrature phase and randomization. J. Magn. Reson. 254, 121–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazimierczuk K., Orekhov V. Y., Accelerated NMR spectroscopy by using compressed sensing. Angew. Chem. Int. Ed. Engl. 50, 5556–5559 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Kazimierczuk K., Orekhov V., Non-uniform sampling: Post-fourier era of NMR data collection and processing. Magn. Reson. Chem. 53, 921–926 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Hyberts S. G., Milbradt A. G., Wagner A. B., Arthanari H., Wagner G., Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishkovsky M., Frydman L., Progress in hyperpolarized ultrafast 2D NMR spectroscopy. ChemPhysChem 9, 2340–2348 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Gal M., Mishkovsky M., Frydman L., Real-time monitoring of chemical transformations by ultrafast 2D NMR spectroscopy. J. Am. Chem. Soc. 128, 951–956 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Lee M.-K., Gal M., Frydman L., Varani G., Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 9192–9197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borkar A. N., et al. , Structure of a low-population binding intermediate in protein-RNA recognition. Proc. Natl. Acad. Sci. U.S.A. 113, 7171–7176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dethoff E. A., Petzold K., Chugh J., Casiano-Negroni A., Al-Hashimi H. M., Visualizing transient low-populated structures of RNA. Nature 491, 724–728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gołowicz D., Kazimierczuk K., Urbańczyk M., Ratajczyk T., Monitoring hydrogenation reactions using benchtop 2D NMR with extraordinary sensitivity and spectral resolution. ChemistryOpen 8, 196–200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagashima K., Optimum pulse flip angles for multi-scan acquisition of hyperpolarized NMR and MRI. J. Magn. Reson. 190, 183–188 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Katsikis S., Marin-Montesinos I., Pons M., Ludwig C., Günther U. L., Improved stability and spectral quality in ex situ dissolution DNP using an improved transfer device. Appl. Magn. Reson. 46, 723–729 (2015). [Google Scholar]
- 57.Fürtig B., Richter C., Wöhnert J., Schwalbe H., NMR spectroscopy of RNA. ChemBioChem 4, 936–962 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Bowen S., Hilty C., Rapid sample injection for hyperpolarized NMR spectroscopy. Phys. Chem. Chem. Phys. 12, 5766–5770 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.