Significance
Small fragments of antibodies, called nanobodies, are emerging tools in structural biology, cell imaging, and drug discovery. Here we pioneer the use of their biocompatible spin-labeled variants as reporters of conformational dynamics of membrane proteins using DEER spectroscopy. Our study shows that it is possible to monitor the fraction of outward-facing conformation of a wild-type ABC transporter not only under turnover conditions in vitro but also in isolated cellular membranes with no need of purification or labeling of the target transporter. This paves the way for in-cell EPR on membrane proteins, a challenging task in the field.
Keywords: spin label, nanobody, ABC transporters, EPR, DEER
Abstract
Nanobodies are emerging tools in a variety of fields such as structural biology, cell imaging, and drug discovery. Here we pioneer the use of their spin-labeled variants as reporters of conformational dynamics of membrane proteins using DEER spectroscopy. At the example of the bacterial ABC transporter TM287/288, we show that two gadolinium-labeled nanobodies allow us to quantify, via analysis of the modulation depth of DEER traces, the fraction of transporters adopting the outward-facing state under different experimental conditions. Additionally, we quantitatively follow the interconversion from the outward- to the inward-facing state in the conformational ensemble under ATP turnover conditions. We finally show that the specificity of the nanobodies for the target protein allows the direct attainment of structural information on the wild-type TM287/288 expressed in cellular membranes without the need to purify or label the investigated membrane protein.
Single domain antibodies (named nanobodies) are extremely versatile tools that in recent years gained great scientific interest in different fields from basic science to drug discovery (1–4).
Their classical generation relies on immunizations of animals, which is difficult for delicate membrane proteins, and generally impedes selections in the presence of ligands typically needed to populate defined conformational states. To overcome these limitations, an in vitro selection platform based on synthetic single domain antibodies, named sybodies, was recently developed (5, 6).
Despite their low molecular weight (∼15 kDa), nanobodies and sybodies exhibit high affinity and specificity for the desired target, which make them suitable both for in vitro and in-cell studies as shown by recent publications in the fields of protein crystallization (for one example see ref. 7) and protein localization in cells with microscopy techniques (two recent examples can be found in refs. 8 and 9).
We introduce here the use of spin-labeled nanobodies for electron paramagnetic resonance (EPR) spectroscopy. The classical approach in site-directed spin-labeling EPR relies on the attachment of different types of spin labels, mostly via cysteine chemistry, to the protein of interest (for a recent review see ref. 10). The most prominent information that can be obtained via EPR to gain structural insights into biological systems is the distance distribution between pairs of spin labels. When two (or more) spin probes are present in the system (11–14), interspin mean distances and their distributions can be extracted at cryogenic temperature with high precision. The four-pulse double electron–electron resonance technique (DEER, also known as PELDOR) is the most established method to measure distances from 1.5 up to 15 nm between couples of spin labels in biomolecules (15). Through distance measurements under different experimental conditions, conformational changes and equilibria between different protein states can be monitored in vitro and used to propose functional models of biologically relevant molecular machines (a few recent examples can be found in refs. 16–19).
EPR studies in a truly native environment still represent a challenge, due to the intrinsic difficulty of labeling and/or inserting proteins in the cell. Toward this goal, several approaches have been tested, such as protein expression with unnatural amino acids carrying a stable paramagnetic center (20), insertion of a recombinantly produced spin-labeled protein inside the cell (21, 22) via hypotonic swelling or electroporation, or spin labeling of bacterial proteins directly in the outer membrane of bacteria (23, 24).
Here, we propose an approach based on protein-specific nanobodies, spin labeled with gadolinium (Gd) probes, which bind with high affinity to their target in physiological environments. The advantage of using spin-labeled nanobodies is that they can be easily inserted into cells with conventional techniques (e.g., osmotic shock, hypotonic swelling, or electroporation) and can provide structural information on their target via internanobody distances detection. This method is particularly helpful in the case of membrane proteins, that cannot be delivered inside of the cell with the above-mentioned techniques.
Two scenarios are possible: if one spin-labeled nanobody is bound to each target protein, the interspin distance between nanobodies report on protein interaction networks, or alternatively, if two nanobodies bind to the same target, they can report on conformational changes of the protein. In this work, we used a dodecane tetraacetic acid (DOTA)-chelated gadolinium ion (Fig. 1), a contrast agent already employed in magnetic resonance imaging (25, 26), which is biocompatible and nontoxic due to its negligible rate of metal release. This property is also a prerequisite for a good spin label for protein science, because losing the metal ion from the chelator decreases the maximal achievable modulation depth in DEER and consequently the signal-to-noise ratio. In contrast to nitroxide-based spin labels, gadolinium labels do not suffer from chemical reduction in a cellular medium and were proven to be valuable for in-cell EPR studies (see for example refs. 21, 27, and 28). In addition to that, Gd-based labels are spectroscopically orthogonal to nitroxides (i.e., they are characterized by different spectra, relaxation properties, and transition moments). Therefore, if both labels are present in the same sample, it is possible to independently obtain the gadolinium–gadolinium, nitroxide–nitroxide, and nitroxide–gadolinium distances via three different DEER channels, increasing the information content achievable in one sample (see for example refs. 29 and 30).
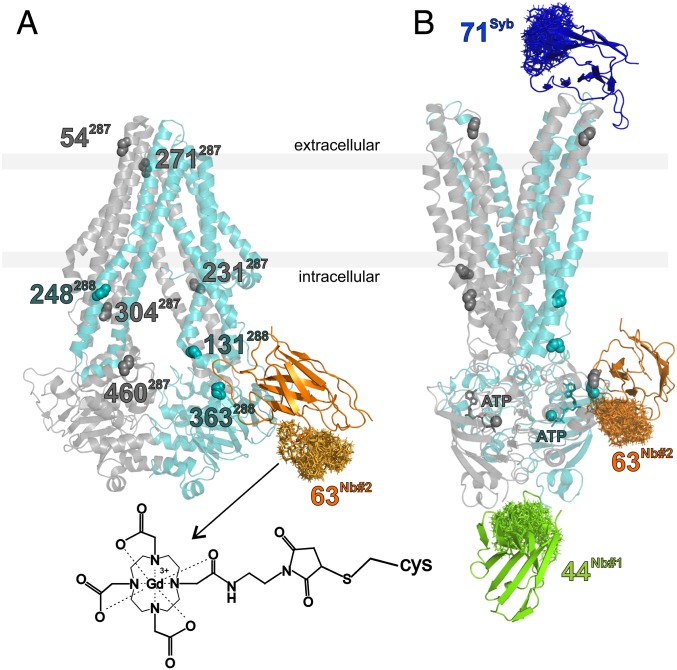
Fig. 1.
Binding epitopes and labeling sites of the three sybody/nanobodies under investigation. (A) TM287/288 in the apo-state, adopting an IF conformation (PDB file: 4Q4H) with the nonstate-specific nanobody Nb_TM#2 (in orange) docked to the structure. The backbone atoms of the eight amino acids labeled for control experiments with MTSL in TM287 (gray) and TM288 (cyan) are highlighted. (B) Crystal structure of the OF conformation of the protein obtained from the superposition of the PDB files 6QUZ, 6QV1, and 6QV2, to highlight the binding sites of the three nanobodies in the presence of ATP. The clouds in the nanobodies’ structures represent the rotamers of the Gd-maleimide-DOTA spin labels attached to the engineered cysteines obtained with the Matlab-based package MMM (48). The Inset shows the structure of the label bound to a cysteine.
To provide a proof of principle, we focus here on different state- and nonstate-specific nanobodies binding to a wild-type ABC transporter and show that they can report its conformational dynamics in vitro and in cellular membranes.
ABC transporters are complex molecular machines that couple the energy derived from binding and hydrolysis of ATP with large conformational changes that alternate the transporter’s conformation between an inward- and an outward-facing state (IF and OF, respectively), in order to translocate substrates across the membrane. In this study, we investigate TM287/288, a bacterial heterodimeric exporter extensively studied in our groups (17, 31–34) for which we recently solved three outward-facing crystal structures with the help of sy- and nanobodies (17). The sybody (called Sb_TM#35) was found to be state specific toward the outward-facing conformation of TM287/288 (KD <100 nM). One of the two nanobodies binding to the nucleotide-binding domains (Nb_TM#1) was found to have a single-digit nanomolar KD for the outward-facing state of TM287/288, and no complex could be formed with the apo-state of the transporter up to submicromolar concentrations, as shown by surface plasmon resonance (SPR); in contrast, the second nanobody (Nb_TM#2) showed no binding preference for the IF or OF state of the transporter, therefore it is nonstate specific. The availability of the nanobody-transporter structures was highly useful to determine the positions of the engineered cysteines required to spin label the sy- and nanobodies for DEER studies.
The careful design of the spin-labeled nanobodies allowed us to monitor the switch of the wild-type transporters from the OF to the IF state under turnover conditions and to exploit the specificity of the selected nanobodies toward TM287/288 to perform DEER measurements in intact inner membranes of Escherichia coli.
Results
The three single domain antibodies (17) and the selected spin-labeling sites chosen for this study are shown in Fig. 1. Their binding epitopes in the IF and OF states are presented. Sb_TM#35 inhibits ATPase activity of the transporter and binds only in the presence of ATP to the extracellular region, thereby shifting the IF/OF equilibrium of the transporter toward the OF state. In contrast, Nb_TM#2 binds sideways to NBD288 regardless of the presence or the absence of ATP. Although Nb_TM#2 does not appear to be conformation specific, it slightly inhibits the ATPase activity as well (17).
Nb_TM#1 decreases the ATPase activity and binds specifically to the transporter in the OF state to an epitope that is shared between the two nucleotide-binding domains (NBDs) (17). For this study, Sb_TM#35 was labeled with gadolinium-maleimide-DOTA at position 71, Nb_TM#1 at position 44, and Nb_TM#2 at position 63 (Fig. 1). Labeling efficiencies based on mass spectrometry analysis were found to be 95%, 100%, and 65%, respectively (SI Appendix, Fig. S1).
ATPase activity and SPR experiments revealed that labeling of the nanobodies only marginally affected their properties (SI Appendix, Table S1). However, we noted that the maximal response units in the SPR measurements were consistently reduced for the gadolinium-maleimide-DOTA–labeled binders.
Using the available structures, the labeling sites in Nb_TM#1 and #2 were carefully chosen on the basis of rotamer calculations using the Matlab-based package MMM (35), in order to obtain a distance distribution centered around 5 nm between the two spin labels when both nanobodies are bound to the transporter (Fig. 1). Due to the state specificity of Nb_TM#1, the detection of this distance between the Nb_TM#1 and Nb_TM#2 will provide the fingerprint of the OF state.
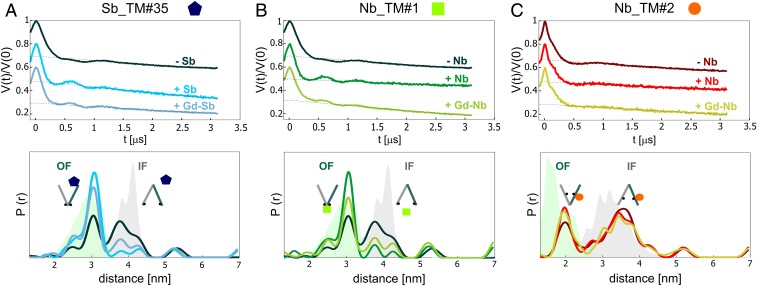
Previously we showed that the unlabeled sybody shifts the conformational equilibrium of TM287/288 toward the OF state (17); therefore, we investigated here the effect on the equilibrium upon binding of the unlabeled and Gd-labeled sy/nanobodies. To this end, we monitored nitroxide–nitroxide distances between pairs of nitroxide spin labels [(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSL)] engineered at the NBDs and in the intracellular region of the transporter (Fig. 1). In particular, we focused on the pair 460TM287/363TM288, which has been investigated in a previous study (31), to monitor the effect of Sb_TM#35 and Nb_TM#1 (Fig. 2). In the case of Nb_TM#2, the presence of a spin label at position 363TM288 prevented nanobody binding due to a steric clash. Therefore, we used the pair 131TM288/248TM288 instead, which is located at the intracellular portion of the transmembrane domains (Fig. 2 in the presence of Nb_TM#2 and SI Appendix, Fig. S2 with Sb_TM#35).
Fig. 2.
Effects of sybody/nanobody binding to the IF/OF equilibrium. (Upper) Primary DEER data for the nitroxide–nitroxide (NO–NO) distances of the nucleotide-binding domain pair 460TM287-363TM288 in the ATP-bound state in the absence (−) and presence (+) of Sb_TM#35 (A) and Nb_TM#1 (B). (C) Primary DEER data for the nitroxide–nitroxide distances of the intracellular pair 131TM288-248TM288 in the ATP-bound state in the absence and presence of Nb_TM#2. (Lower) Corresponding distance distributions obtained with Tikhonov regularization together with MMM simulations for the OF and the IF conformations (green and gray shaded areas, respectively). For each pair, the NO–NO distances were measured without the sybody or nanobody (−Sb/−Nb), with the unlabeled sybody or nanobody (+Sb/+Nb), and with the Gd-labeled sybody or nanobody (+Gd-Sb/+Gd-Nb). The corresponding form factors and the error estimation of the distance distributions are shown in SI Appendix, Figs. S3 and S4. The IF/OF equilibrium shift of the unlabeled sybody was shown previously (17).
None of the nanobodies affected the protein conformation in the apo-state (SI Appendix, Fig. S3). The effects of nanobody binding on the IF/OF conformational equilibrium in the presence of ATP-EDTA (ethylenediaminetetraacetic acid) are shown in Fig. 2 and SI Appendix, Fig. S2. The chelator EDTA was used to sequester the magnesium ions present in the sample, which are necessary to catalyze ATP hydrolysis.
We observed an almost complete shift of the equilibrium toward the OF state in the presence of Sb_TM#35 and Nb_TM#1. Notably, labeling with Gd-maleimide-DOTA was found to slightly reduce the equilibrium shift. For the nonstate-specific Nb_TM#2, only a negligible equilibrium shift was observed.
While Nb_TM#1 and Nb_TM#2 did not form homo- or heterodimers in aqueous solutions, the sybody exhibited a long Gd–Gd distance centered at 6 nm, indicative of dimer formation (SI Appendix, Fig. S5), which seems to be an intrinsic characteristic of Sb_TM#35, already shown with the MTSL-labeled variant (17). In order to exclude the dimerization as a possible cause for the reduced shift in equilibrium, we added a three-fold excess of the Gd-labeled sybody and found no difference in the DEER traces (SI Appendix, Fig. S5).
As mentioned in the introduction, one of the advantages of using gadolinium as a spin probe in the nanobody relies on its spectroscopic orthogonality with MTSL. Therefore, by placing a pair of nitroxides at different positions within the transporter, one can simultaneously investigate the effects of nanobody binding on the transporter’s conformation via nitroxide–nitroxide distances, the nanobody binding to the transporter via Gd–nitroxide distances, and the nanobody–nanobody interactions via Gd–Gd distances.
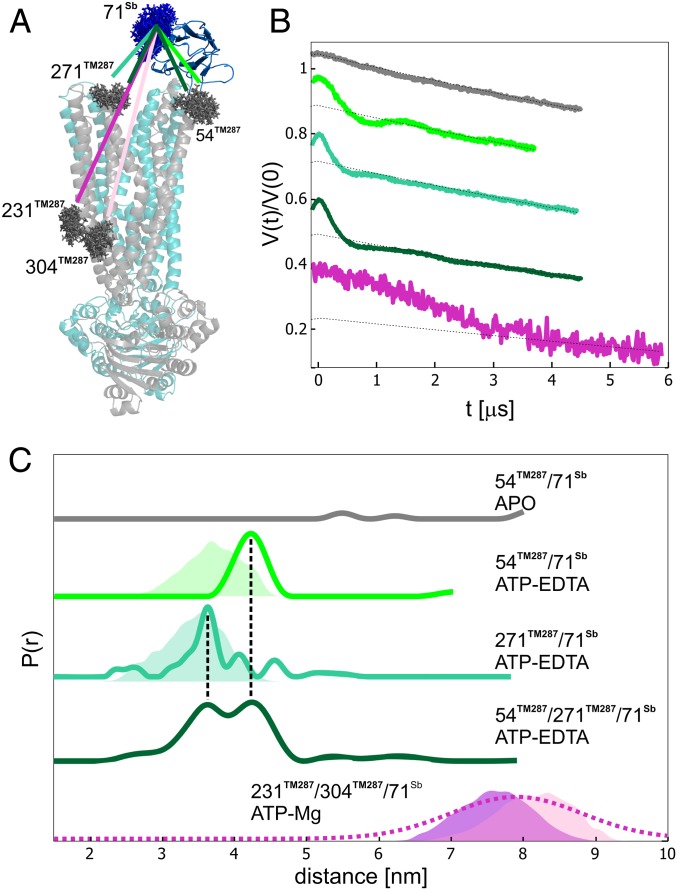
We show the advantages of the nitroxide–Gd DEER channel by spin labeling TM287/288 at position 54TM287 with MTSL and the sybody at position 71 with Gd-maleimide-DOTA. We performed DEER experiments on the sample in the absence and in the presence of ATP (Fig. 3). In the apo-state, we did not observe dipolar coupling between the gadolinium and nitroxide spin labels, confirming that the state specificity observed by SPR at submicromolar concentrations at room temperature is maintained for Sb_TM#35 in frozen conditions up to the concentration used in the DEER experiment (25 µM). Upon addition of ATP-EDTA, the appearance of a dipolar modulation with a distance distribution centered at 4.3 nm provided a clear indication of the binding of the sybody at the extracellular wing (Fig. 3 A and B). The experimental distance was found to be slightly longer than the predicted distance based on the OF structure (Fig. 3C).
Fig. 3.
Sybody binding to the transporter via Gd–nitroxide distances. (A) Sybody-bound crystal structure of TM287/288 in the OF state (PDB file: 6QV0). The spin-labeled residues (MTSL on the transporter, in gray, and Gd-maleimide-DOTA, in blue, on the sybody) are depicted with the rotamers predicted by MMM2018. (B) Primary Gd–nitroxide DEER data. (C) Distance distributions obtained with Tikhonov regularization for the different transporter/sybody systems and simulated distance distributions (shaded areas). For the 231TM287-304TM287-71Sb complex, the predicted distance distribution for 231TM287-71Sb is displayed in purple, while the 304TM287-71Sb distance is colored in pink. The extracted distance distribution for this pair was obtained by Gaussian model fit (dashed line) to stabilize the output distances with respect to a Tikhonov model-free fit. The corresponding form factors and error estimations are shown in SI Appendix, Fig. S6.
When another position on the transporter, 271TM287, was labeled with MTSL, we detected a 3.5-nm distance between sybody and transporter, in good agreement with the MMM predictions.
Two distinct peaks at 3.5 and 4.3 nm could also be distinguished when the Gd-labeled sybody was bound to the doubly labeled transporter (54TM287-271TM287) in the ATP-bound state, which shows the sensitivity of DEER to obtain precise structural information from multimodal distance distributions. Moreover, we attempted to monitor the long 7- to 9-nm distance predicted between the Gd spin label and two nitroxide labels (231TM287-304TM287) across the transmembrane region, opposite from the binding site of the sybody. The limited length of the DEER time trace is related to large uncertainties in the obtained distance distribution (Fig. 3C and SI Appendix, Fig. S6); therefore, a maximal mean distance of 7 nm is the upper experimental limit for the spin type and concentration used.
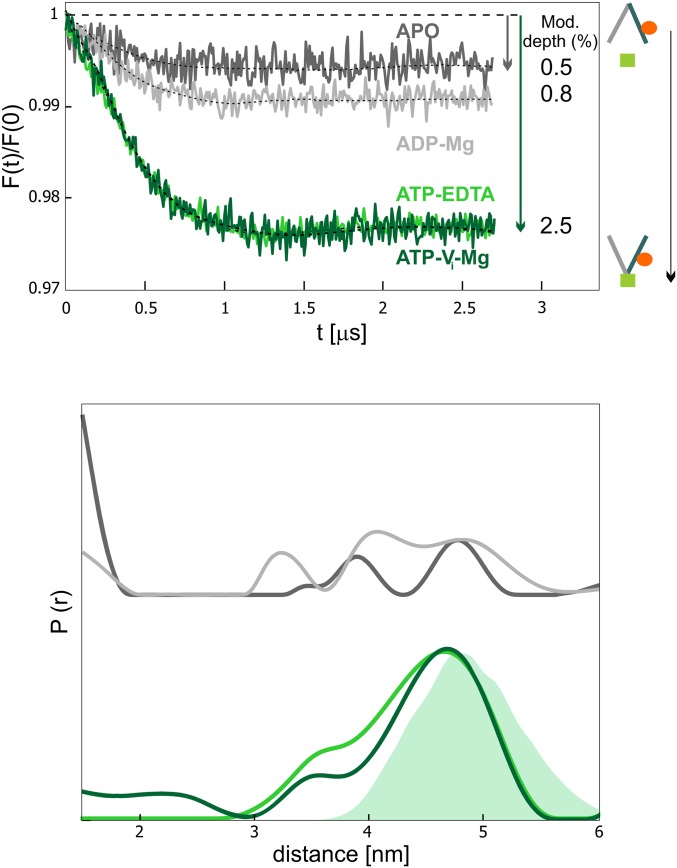
We then further proceeded with the study of the unlabeled wild-type transporter using the two Gd-labeled nanobodies Nb_TM#1 and Nb_TM#2 as reporters of the protein’s conformational states. Based on the previous knowledge on the conformational specificity of Nb_TM#1, a dipolar frequency should only be observed when both nanobodies are bound to the outward-facing transporter. The modulation depth of the dipolar frequency is directly proportional to the percentage of the OF state present in the conformational ensemble. In the absence of nucleotides, the OF state is not populated, whereas the addition of ATP is sufficient to partially populate it. The maximal shift toward the OF state is observed with ATP-vanadate (ATP-Vi-Mg), which mimics the posthydrolytic transition state intermediate, while in the ADP-bound state the equilibrium is shifted back toward the IF state.
As expected, we retrieved this information quantitatively from the analysis of the modulation depth of the Gd–Gd traces (nanobody–nanobody distances) for the wild-type transporter trapped in different states (Fig. 4). By ranking the OF percentage according to increasing modulation depths, we observed that apo < ADP < ATP-EDTA = ATP-Vi-Mg. This rank order is in line with the data from nitroxide–nitroxide distances in the spin-labeled transporter (31), with the exception that we observed the same percentage of OF states in the ATP-EDTA and the ATP-vanadate state (Fig. 4). This is due to the shift toward the OF state induced by the state-specific nanobody Nb_TM#1 in the presence of ATP-EDTA (Fig. 2).
Fig. 4.
Fingerprint nanobody–nanobody distances obtained in the presence and absence of nucleotides. Background-corrected DEER data (Upper) and corresponding distance distributions normalized by modulation depth (Lower) characteristic of the Gd–Gd distances between Nb_TM#1 and Nb_TM#2 in the different conformational states of the transporter. The values of the modulation depth are given on the right of the corresponding trace. The error in the modulation depth, calculated as SD of six repetition experiments is 0.1% (SI Appendix, Fig. S7). An increase in the modulation depth relates to an increase of the population of the OF state. (Lower) The shaded area represents the MMM simulation. The corresponding primary data are shown in SI Appendix, Fig. S9.
The 5 nm mean Gd–Gd distance obtained from DEER data analysis (Fig. 4) is in good agreement with the predictions, indicating that both nanobodies bind simultaneously to their different binding epitopes. With our Q-band setup (see Materials and Methods), the maximal modulation depth detected between two Gd-DOTA–labeled sites with 100% labeling efficiency is ∼3.5 to 4.0%, in line with previously published data obtained with a slightly larger excitation bandwidth and a similar setup (36). We observed a smaller maximal modulation depth of ∼2.5% (Fig. 4), which is due to the 65% labeling efficiency of Nb_TM#2 (SI Appendix, Fig. S1) and the 20% molar excess of each nanobody with respect to the transporter concentration (Materials and Methods). To ensure the reliability of the information extracted from the contrast in the modulation depths in the apo- and ATP samples, we performed a statistical analysis with several repetitions on the ATP-EDTA sample in detergent. Using the same fitting parameters for the background, we obtained a mean modulation depth of 2.5% with a SD of 0.1% (SI Appendix, Fig. S7).
The internanobody experimental distance distribution is broad (2-nm full width at half maximum) and is in line with the 1.5-nm width predicted by the MMM simulation. Therefore, we reasonably ascribe this characteristic to the intrinsic flexibility of the Gd-DOTA linker and not to the disorder in the nanobody–transporter interface. Notably, different linkers which are under development in the field might alleviate this problem (37). To verify the reliability of the distance distribution for the fraction of distances >5 nm, we measured a longer trace (4.5 µs) on the ATP-EDTA sample and performed error analysis (SI Appendix, Fig. S8). The distance distribution was found to be very similar to that obtained with the 3-µs trace. The main focus of this part of the study is the reliable extraction of the modulation depth parameter; therefore, to optimize the signal-to-noise and to minimize the accumulation time, 3-µs traces were recorded.
All DEER measurements presented in Fig. 4 were carried out at a transporter concentration of 5 µM. This comparatively low spin concentration was chosen to minimize errors in the determination of the modulation depth due to background fitting uncertainties. In contrast to our previous findings on the state specificity toward the OF state of Nb_TM#1, we noticed a detectable and reproducible residual dipolar coupling in the apo-state (modulation [mod.] depth 0.5 ± 0.1%), indicating that Nb_TM#1 weakly binds to the apo-transporter.
To further investigate the origin of this feature, nitroxide–nitroxide DEER data on the intracellular pair 131TM288/248TM288 labeled with MTSL in the presence of Nb_TM#1 and Nb_TM#2 labeled with Gd-DOTA were carried out, proving that, in the absence of nucleotides, the OF state is not populated (SI Appendix, Fig. S10). As the state-specific nanobody occupies an epitope that is shared between the two NBDs, which do not largely dissociate in the absence of nucleotides (33, 34), we reasoned that a small fraction of Nb_TM#1 might bind with weak affinity to the transporter in the apo-state as well. Unspecific binding of Nb_TM#1 to the apo-transporter was not previously assessed with SPR (17), where the maximal concentration was one order of magnitude lower than the one used in DEER. To resolve this discrepancy, SPR measurements were performed at micromolar concentrations of Nb_TM#1 using immobilized wild-type apo-TM287/288. In line with the DEER data, Nb_TM#1 was found to weakly bind to the apo-state (KD > 10 μM, SI Appendix, Fig. S11). SPR measurement artifacts were excluded by the absence of binding signal when probing against the control protein MsbA. We further corroborated these findings via detection of residual Gd–NO dipolar coupling between the Gd–nanobody and the nitroxide–transporter in the apo-state (SI Appendix, Fig. S12). Hence, despite the two orders of magnitude difference in the affinities toward the OF and IF conformation of the wild-type transporter, Nb_TM#1 can be considered fully state specific for the OF state only if added in a 1:1 stoichiometric ratio to transporters below single-digit micromolar concentrations.
In summary, with spin-labeled nanobodies we can quantitatively measure the different fractions of the OF state in an ensemble of wild-type transporters in response to ATP binding, and the contrast between the modulation depths in the apo- and ATP-EDTA state (0.5 ± 0.1 vs. 2.5 ± 0.1%, respectively) is satisfactory at single-digit micromolar concentrations of the transporter–nanobody complex.
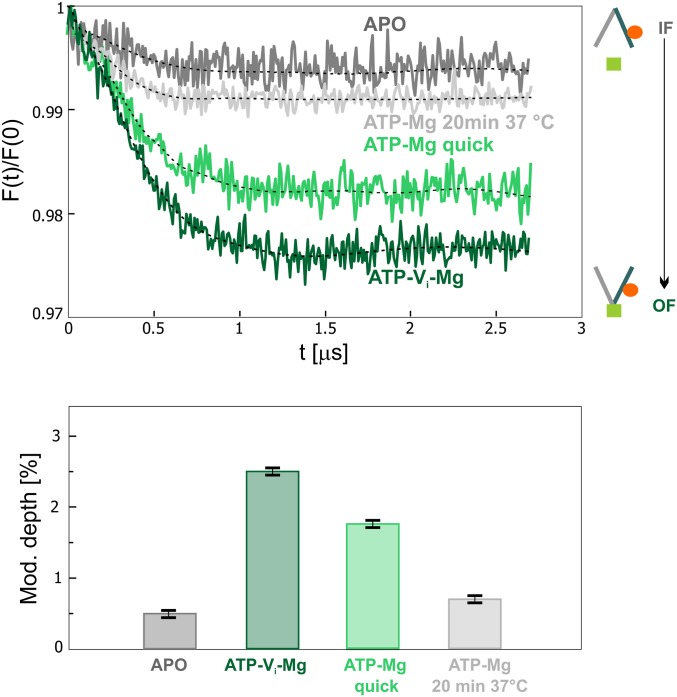
Next, we monitored the transporter during its working cycle in the presence of ATP-Mg. When the sample was quickly shock frozen in liquid nitrogen within 40 s after ATP-Mg addition, the Gd–Gd modulation depth was smaller (1.8 ± 0.1%) compared to the ATP-vanadate state (2.5 ± 0.1%), indicating that there is a smaller fraction of OF states in the ensemble (Fig. 5). The sample was then thawed and incubated for 20 min at 37 °C before being shock frozen and measured again. Based on the residual ATPase activity of the wild-type transporter in detergent (SI Appendix, Fig. S1D), this incubation is sufficient to hydrolyze nearly all ATP molecules in the sample. Indeed, we observed a significant decrease in the modulation depth, which reached a value similar to the one obtained in the apo-state (Fig. 5). Therefore, as expected, the incubation resulted in an almost complete switch of the transporter to the inward-facing conformation, which cannot be probed by the Gd-labeled nanobodies. In the bottom panel of Fig. 5, a bar chart representing the modulation depth (related to the fraction of OF state) in the different conditions is reported.
Fig. 5.
IF/OF interconversion under turnover conditions of wild-type transporter. (Upper) Background-corrected DEER data during the Mg-dependent hydrolysis of ATP. The traces obtained in the apo- and ATP-vanadate states from Fig. 4 are shown as lower and upper limits of the modulation depths. (Lower) Bar chart showing the values of the modulation depths in the four different samples, given as a mean value. The error calculated as SD of six repetition experiments is 0.1% (SI Appendix, Fig. S7). The primary data are shown in SI Appendix, Fig. S9.
To corroborate our interpretation, we compared the trend in the OF fractions obtained by Gd–Gd modulation depths with the IF/OF ratio extracted from the nitroxide–nitroxide distance distributions using an orthogonally spin-labeled nanobodies/transporter complex. To this end, the transporter was labeled with MTSL at the intracellular pair 131TM288/248TM288 and incubated with Nb_TM#1 and Nb_TM#2 labeled with Gd-maleimide-DOTA. We compared the modulation depth of the gadolinium–gadolinium DEER traces (related to the amount of OF state) and the distance distributions obtained from the nitroxide–nitroxide DEER traces (indicative of the relative amount of IF and OF in the sample) before and after incubation with ATP-Mg (SI Appendix, Fig. S13). Indeed, we found a high degree of correlation between the decrease of the modulation depth in the Gd–Gd DEER traces during hydrolysis and the decrease in the OF/IF intensity ratio in the distance distributions obtained from the nitroxide–nitroxide DEER channel.
Finally, to explore the specificity of the nanobodies toward their target in a cellular context, we overexpressed TM287/288 in E. coli cells, and prepared inside-out vesicles (ISOVs) as described in Materials and Methods. This type of cellular preparation allows for monitoring how different substrates, inhibitors, and other ligands affect membrane proteins, which expose their cytoplasmic domains toward the exterior of the vesicles. Importantly, the two nanobodies Nb_TM#1 and #2 are expected to bind to the accessible NBDs of the transporter in ISOVs. Hence, the population of outward-facing TM287/288 can in principle be monitored in the cellular membrane environment without the need to purify and reconstitute the transporter.
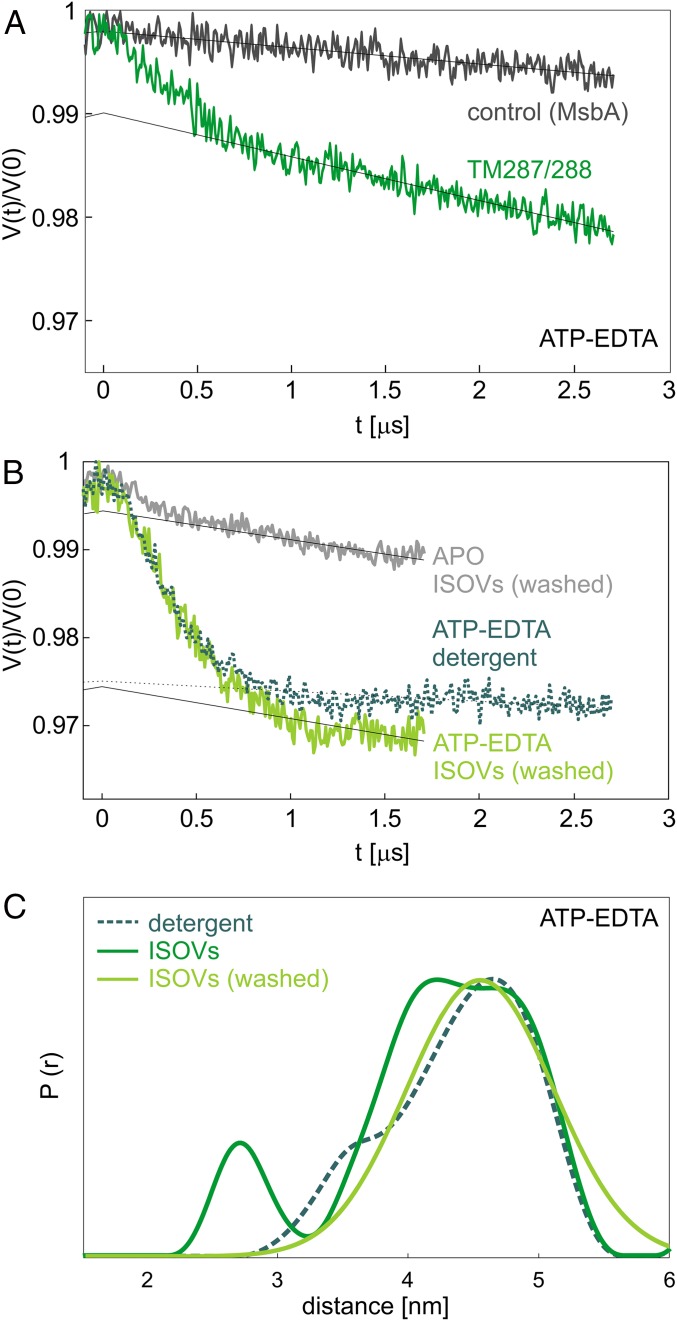
Based on the yield of TM287/288 from cells harvested after 4 h of overexpression, the mean transporter concentration in the ISOVs preparation was estimated to be around 5 µM, similar to the bulk concentration used in the detergent samples. In the presence of ATP-EDTA and upon addition of the two nanobodies at a concentration of 6 µM each, we obtained a clear dipolar modulation in the primary DEER traces (Fig. 6A, dark green trace) characterized by a modulation depth of about 1%, indicating that the transporter concentration is in the estimated range.
Fig. 6.
Recognition of protein target and conformational states via nanobody-mediated DEER measurements in cellular membranes. (A) Primary DEER data between Nb_TM#1 and Nb_TM#2 in inside-out vesicles obtained from cells in which TM287/288 is overexpressed for 4 h (dark green), compared to a control sample containing overexpressed MsbA (black). Both samples were incubated with ATP-EDTA. (B) Contrast between the apo- and ATP-EDTA state in ISOVs containing TM287/288 (1-h overexpression), incubated with an excess of nanobodies (15 µM), and washed with the corresponding buffers. For comparison, the ATP-EDTA trace in detergent is shown as a dotted line. (C) Internanobody distance distributions obtained in the presence of ATP-EDTA in detergent-purified protein (dashed line, teal) and inside-out vesicles before and after washing (solid line, dark and light green).
The smaller modulation depth found in ISOVs compared to the one in detergent (2.5 ± 0.01%, see Fig. 4) accounts for the fact that the transporter concentration cannot be easily estimated and that the preparation of the ISOVs has variable efficiencies in the range of 70 to 90%, meaning that not all transporters have their NBDs facing the outside of the vesicles. This introduces an additional complication in the estimation of the transporters interacting with the nanobodies. The internanobody distance is very similar to that obtained using detergent-solubilized wild-type transporters (Fig. 6C, green dotted). Hence, the nanobodies recognize the same epitopes in the cellular membrane, and outward-facing TM287/288 exhibits the same relative distance between the nucleotide-binding domains in membranes and in detergent. As a control, we prepared ISOVs containing overexpressed MsbA (an ABC exporter with considerable sequence homology to TM287/288 in the nucleotide-binding domains, where the nanobodies’ epitopes are located, and similar expression levels in E. coli) and no dipolar modulation could be retrieved under the same experimental conditions (Fig. 6A, black trace). In summary, these experiments demonstrate the selectivity of the nanobodies toward their target in cellular membranes.
Unexpectedly, we detected a very poor contrast in the modulation depths when the ISOVs were measured in the absence of ATP, and the DEER signal detected in the apo-state persisted also after prolonged incubation with Mg2+, proving that there is no residual ATP in the sample (SI Appendix, Fig. S14A). This indicates that the weak affinity of Nb_TM#1 to the apo-state (KD > 10 µM), together with high local concentrations of the overexpressed transporters in the membranes, is detrimental for the selective recognition of the OF state from the analysis of the DEER traces. However, we found that incubating the ISOVs with an excess of nanobodies (10 µM) and washing them by ultracentrifugation to remove the nanobodies added in excess as well as those weakly binding to the transporter, we could retrieve a modulation depth of about 2% in the ATP state (comparable to the detergent sample) and a satisfactory contrast between the apo- and the ATP states (SI Appendix, Fig. S14B).
To verify if the high local concentrations of transporters is the reason for the poor contrast in modulation depths observed in the ISOVs, we prepared a second batch of cells with a lower overexpression level (induction for 1 instead of 4 h), which showed an improved apo–ATP contrast (SI Appendix, Fig. S14A). The DEER traces detected on these ISOVs incubated with excess of nanobodies (15 µM) and subjected to a washing step, show an apo–ATP contrast and a modulation depth in the ATP state, which are consistent with those obtained in detergent (Fig. 6B and SI Appendix, Fig. S14B). In agreement with the better performance in terms of modulation depth contrast, we found that reducing the overexpression level also reduces the steepness of the background of the DEER traces (SI Appendix, Fig. S14C) due to lower local concentration of the transporters in the membrane.
In conclusion, the experiments with ISOVs provide clear evidence that the two nanobodies show a distinct contrast in modulation depths in the absence and presence of ATP-EDTA, thereby selectively recognizing the OF state of TM287/288 in cellular membranes.
Discussion
Here we show a proof-of-principle study on the use of spin-labeled nanobodies as valuable tools for EPR structural investigations of proteins in vitro and in cellular membranes. In particular, we discuss their applicability on a specific class of membrane proteins, namely ABC transporters, which are investigated in great detail by EPR techniques (16, 17, 31, 38–42). Notably, the main novelty of this approach is that with the spin-labeled nanobodies, we can explore unlabeled wild-type membrane proteins in cellular context, as the spin label is placed on the nanobody. The technique requires one or more spin-labeled nanobodies with nanomolar or higher affinity toward the protein of interest, which can be obtained by immunization techniques or selected in vitro.
Spin-labeled nanobodies as conformational reporters for EPR share the closest analogies to fluorescent nanobodies targeting endogenous proteins in cells (9), which were recently introduced for advanced light microscopy and can also possibly be used for Förster resonance energy transfer (FRET) studies. For technical reasons, the minimal spin concentration detectable by EPR is in the low micromolar range (1 to 5 μM), while for fluorescence studies, nanomolar to single molecule detection is possible. However, in contrast to FRET, with DEER we can obtain with high precision the distance distributions between the same labels for mean distances in a 1.5- to 16-nm range (the high distance limit was obtained with protein and solvent deuteration, ref. 43). Realistically, for gadolinium-labeled nanobodies, the maximum detectable distance in cellular membranes may be restricted to 6 to 8 nm at single-digit micromolar spin concentration in the absence of extensive protein and solvent deuteration (Fig. 6). Furthermore, spin-labeled nanobodies offer additional advantages. In EPR we are not restricted to the use of two different spin labels to retrieve interspin distances, thereby the labeling schemes are simpler. In analogy to different colors of fluorescent proteins, different types of spin labels, such as gadolinium, trityl, copper, and nitroxide, could be covalently attached to nanobodies as well. Notably, for in-cell studies the spin labels need to be fully biocompatible (44, 45).
An important advantage of nanobodies directly targeting the investigated protein is that there are no flexible linkers between the protein and the label, which is usually the case for genetic fusion of fluorescent proteins, that may result in an uncoupled motion of the “label” with respect to the protein. In our study, we decorated TM287/288 with nanobodies that are rigidly bound to the protein, as demonstrated by the similar width of the experimental distance distribution and the one simulated on a rigid protein structure with both nanobodies bound to their epitopes (Figs. 1 and 4). Therefore, the conformational freedom of the nanobody is minimal and does not add up to the internal disorder of the gadolinium spin label for EPR studies. Due to their characteristics, these molecules are perfect candidates to investigate conformational dynamics and protein–protein interactions on wild-type proteins down to spin concentrations of a few micromolar.
However, as attaching a spin label at a specific position in a protein can interfere with its conformational equilibrium and/or its function (for example the ATPase activity in a spin-labeled ABC transporter depends on the site chosen for the labeling), nanobodies can also cause similar effects. Here we show that the nanobodies Nb_TM#1 and Nb_TM#2 indeed affect the IF/OF equilibrium and decrease the ATPase activity, but still allow the transporter to hydrolyze ATP, as shown by the turnover experiments. Interestingly, we also found that, while Sb_TM#35 retains its state specificity also at double digit micromolar concentrations, Nb_TM#1 partially loses this characteristic already at single-digit micromolar concentrations due to its weak affinity toward the apo-state. Clearly, the state specificity is reflected in the contrast (in this case in terms of modulation depth) between the fraction of bound nanobodies to the transporter in different states, and it is a concentration-dependent parameter. Based on the value of KD obtained by SPR (>10 µM), we estimated that, with 5 μM transporter and stoichiometric ratios of nanobodies (conditions used for DEER in detergent samples), around 20% of the nanobody is bound in the apo-state (in line with the mod. depth of 0.5 vs. 2.5% in the apo- and ATP-EDTA samples). Notably, in native membranes, we found that high local concentrations of the overexpressed target membrane proteins further reduce the contrast. Therefore, lower overexpression levels are preferable to perform DEER in cellular membranes.
For all these reasons, each nanobody must be carefully characterized before being chosen for the desired use. While the use of state-specific nanobodies can be advantageous as they can provide fingerprint distances related to a particular conformational state of a protein, our results suggest that nonstate-specific nanobodies may be more suitable as pure conformational reporters, as shown for Nb_TM#2, because they minimally affect the protein’s activity, and do not suffer from concentration-dependent state-specificity issues. Having two such nanobodies binding to different epitopes would allow for monitoring of different conformations directly on the wild-type target protein via distinct distances (i.e., distinct dipolar frequencies), which will produce a more reliable and richer analysis.
In our study, we made extensive use of orthogonal labeling strategies: 1) we performed different experiments on the same sample (e.g., effect of nanobody binding on the protein’s equilibrium, nanobody binding to the protein, nanobody self-dimerization); and 2) we validated the findings on the wild-type protein by comparison with the nitroxide-labeled variant. Therefore, we show that it is possible to establish in vitro the optimal nanobodies for in-cell EPR studies.
Additionally, we demonstrate that spin-labeled nanobodies can selectively bind to their target protein in the inner membranes of E. coli, and thereby permitting the investigation of conformational changes under near-native conditions in membrane vesicles.
Our work sets the stage for using biocompatible Gd-labeled nanobodies as a tool in EPR spectroscopy for the structural and biochemical analysis of wild-type proteins embedded in cellular membranes. As discussed in the introduction, electroporation methods to introduce small proteins into cells have already proven to be successful.
Our results show that standard high-power Q-band arbitrary waveform generator (AWG) spectrometers already have the potential to investigate Gd-labeled proteins at concentrations down to the single-digit micromolar range in cellular membranes. Exploiting in the future the advantages of ultrawideband excitation schemes, more sophisticated DEER sequences and higher frequencies will further increase the signal-to-noise ratio of the Gd traces, thereby fostering applications of this method. In-cell DEER experiments will face new challenges and will require the optimization of tools to verify the correct subcellular localization of the nanobodies (via superresolution microscopy) as well as to achieve stoichiometric nanobody/target ratios to maximize the measured signals.
Materials and Methods
The two nanobodies and the sybody were identified as part of a previous study (17). Single cysteines were introduced at positions S71 (Sb_TM#35), Q44 (Nb_TM#1), and S63 (Nb_TM#2) using site-directed mutagenesis.
The proteins were produced fused to maltose-binding protein (MBP) in the periplasm of E. coli, purified under reducing conditions in the presence of DTT (dithiothreitol), cleaved off from MBP by 3C protease, again purified by reverse Immobilized-Metal Affinity Chromatography and separated using size-exclusion chromatography as described in detail in a previous study (17). Purified Sb_TM#35_S71C and Nb_TM#1_Q44C were stored in 20 mM Tris⋅HCl pH 7.5, 150 mM NaCl with 2 mM DTT at −80 °C until labeling with Gd-maleimide-DOTA was performed.
The DEER measurements were performed at 50 K (nitroxide–nitroxide) or 10 K (nitroxide–Gd, Gd–Gd) on a Bruker ELEXSYS E580Q AWG dedicated-pulse Q-band spectrometer equipped with a 150 W traveling-wave tube amplifier. A 4-pulse DEER sequence with Gaussian (46), nonselective observer and pump pulses of 32-ns length (corresponding to 13.6 ns full width at half maximum) with 100-MHz frequency separation was used for NO–NO and Gd–Gd measurements; in the case of Gd–NO measurements, pump pulses of 24-ns length (10.2 ns FWHM) were used in combination with 32-ns observer pulses, with 280-MHz separation. For the Gd–Gd measurements, we found that a small zero-time artifact due to pulse overlap could be removed by using a 10-dB attenuation in the general attenuator. DEER experiments were performed using the dead-time free four-pulse DEER sequence with 16-step phase cycling. Experimental details regarding the pump and observer positions in the different experiments are given in SI Appendix, Fig. S15, while SI Appendix, Fig. S16 shows, as an example, two echo decays for the wild-type transporter in the presence of Nb_TM#1 and Nb_TM#2, both labeled with gadolinium-maleimide-DOTA, in the presence of ATP in detergent and in ISOVs. The evaluation of the DEER data was performed using DeerAnalysis2019 (47). Interspin distance simulations were performed with MMM2018 (48).
The final concentration of the detergent-solubilized spin-labeled transporter was 25 µM, and was 5 µM for the wild-type unlabeled transporter. A total of 40 µL of sample was loaded in quartz tubes with 3-mm outer diameter. The ATP-EDTA sample contained 2.5 mM ATP and 2.5 mM EDTA to completely inhibit ATP hydrolysis; samples were incubated at 25 °C and snap frozen in liquid nitrogen. For vanadate trapping, samples were incubated with 5 mM sodium orthovanadate, 2.5 mM ATP, and 2.5 mM MgCl2 for 3 min at 50 °C and snap frozen in liquid nitrogen. For DEER measurements under turnover conditions, 2.5 mM ATP and 2.5 mM MgCl2 were added to the sample and snap frozen in liquid nitrogen. Each nanobody was added to the TM287/288 in 1.2:1 stoichiometric ratio, if not elsewhere specified. The ISOVs were incubated with both nanobodies in the absence or presence of 2.5 mM ATP-EDTA and washed via ultracentrifugation at 120,000 × g.
Additional information can be found in SI Appendix.
Data Availability.
All data are available upon request from the corresponding authors.
Supplementary Material
Acknowledgments
This work was funded by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy, EXC-2033, Projektnummer 390677874, BO3000/1-2 (to E.B.), DFG grant INST 130/972-1 FUGG (to E.B.), and by a Swiss National Science Foundation (SNF) Professorship (PP00P3_144823, to M.A.S.). E.B. thanks Gunnar Jeschke (ETH Zürich), for the home-made Q-band resonator and Maxim Yulikov for the stock of the Gd labels. G.M. and M.A.S. thank the Functional Genomics Center Zürich for their assistance with the SPR measurements on the Biacore TM-100.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data related to this paper are available at https://gitlab.com/haditim/2019_spin-labeled_nanobodies__materials (Project ID: 16061223).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913737117/-/DCSupplemental.
References
- 1.Ries J., Kaplan C., Platonova E., Eghlidi H., Ewers H., A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Rothbauer U., et al. , Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Manglik A., Kobilka B. K., Steyaert J., Nanobodies to study G protein-coupled receptor structure and function. Annu. Rev. Pharmacol. Toxicol. 57, 19–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmansa S., Alborelli I., Bieli D., Caussinus E., Affolter M., A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife 6, e22549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmermann I., et al. , Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7, e34317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon C., et al. , Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bräuer P., et al. , Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science 363, 1103–1107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virant D., et al. , A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 9, 930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein A., et al. , Live-cell labeling of endogenous proteins with nanometer precision by transduced nanobodies. Chem. Sci. (Camb.) 9, 7835–7842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bordignon E., “EPR spectroscopy of nitroxide spin probes” in eMagRes, Stoll S., Goldfarb D., Eds. (Wiley, 2017), Vol. 6. [Google Scholar]
- 11.Yagi H., et al. , Gadolinium tagging for high-precision measurements of 6 nm distances in protein assemblies by EPR. J. Am. Chem. Soc. 133, 10418–10421 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Reginsson G. W., Kunjir N. C., Sigurdsson S. T., Schiemann O., Trityl radicals: Spin labels for nanometer-distance measurements. Chemistry 18, 13580–13584 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Garbuio L., et al. , Orthogonal spin labeling and Gd(III)-nitroxide distance measurements on bacteriophage T4-lysozyme. J. Phys. Chem. B 117, 3145–3153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham T. F., Putterman M. R., Desai A., Horne W. S., Saxena S., The double-histidine Cu2+-binding motif: A highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angew. Chem. Int. Ed. Engl. 54, 6330–6334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeschke G., “EPR spectroscopy- fundamentals and methods” in eMagRes, Stoll S., Goldfarb D., Eds. (Wiley, 2018), pp. 401–423. [Google Scholar]
- 16.Dastvan R., Mishra S., Peskova Y. B., Nakamoto R. K., Mchaourab H. S., Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors. Science 364, 689–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutter C. A. J., et al. , The extracellular gate shapes the energy profile of an ABC exporter. Nat. Commun. 10, 2260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Y. N., et al. , Structural basis for functional interactions in dimers of SLC26 transporters. Nat. Commun. 10, 2032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingler L. M., et al. , Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell 176, 468–478.11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt M. J., Fedoseev A., Summerer D., Drescher M., Genetically encoded spin labels for in vitro and in-cell EPR studies of native proteins. Methods Enzymol. 563, 483–502 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Theillet F. X., et al. , Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 530, 45–50 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Martorana A., et al. , Probing protein conformation in cells by EPR distance measurements using Gd3+ spin labeling. J. Am. Chem. Soc. 136, 13458–13465 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Joseph B., Sikora A., Cafiso D. S., Ligand induced conformational changes of a membrane transporter in E. coli cells observed with DEER/PELDOR. J. Am. Chem. Soc. 138, 1844–1847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph B., et al. , Distance measurement on an endogenous membrane transporter in E. coli cells and native membranes using EPR spectroscopy. Angew. Chem. Int. Ed. Engl. 54, 6196–6199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law G. L., Wong W. T., “An introduction to molecular imaging” in The Chemistry of Molecular Imaging, Long N., Wong W. T., Eds. (John Wiley & Sons, 2015), pp. 1–24. [Google Scholar]
- 26.Sherry A. D., Caravan P., Lenkinski R. E., Primer on gadolinium chemistry. J. Magn. Reson. Imaging 30, 1240–1248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalaloyan A., et al. , Tracking conformational changes in calmodulin in vitro, in cell extract, and in cells by electron paramagnetic resonance distance measurements. ChemPhysChem 20, 1860–1868 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Yang Y., et al. , High sensitivity in-cell EPR distance measurements on proteins using an optimized Gd(III) spin label. J. Phys. Chem. Lett. 9, 6119–6123 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Yulikov M., “Spectroscopically orthogonal spin labels and distance measurements in biomolecules” in Electron Paramagnetic Resonance, Chechik V., Murphy D. M., Gilbert B., Eds. (2015), Vol. 24, pp. 1–31. [Google Scholar]
- 30.Teucher M., et al. , A new perspective on membrane-embedded Bax oligomers using DEER and bioresistant orthogonal spin labels. Sci. Rep. 9, 13013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timachi M. H., et al. , Exploring conformational equilibria of a heterodimeric ABC transporter. eLife 6, e20236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göddeke H., et al. , Atomistic mechanism of large-scale conformational transition in a heterodimeric ABC exporter. J. Am. Chem. Soc. 140, 4543–4551 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Hohl M., Briand C., Grütter M. G., Seeger M. A., Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Hohl M., et al. , Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U.S.A. 111, 11025–11030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeschke G., MMM: A toolbox for integrative structure modeling. Protein Sci. 27, 76–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyhach Y., Bordignon E., Jeschke G., Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 13, 2356–2366 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Gmeiner C., Dorn G., Allain F. H. T., Jeschke G., Yulikov M., Spin labelling for integrative structure modelling: A case study of the polypyrimidine-tract binding protein 1 domains in complexes with short RNAs. Phys. Chem. Chem. Phys. 19, 28360–28380 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Yang Y., et al. , A reactive, rigid GdIII labeling tag for in-cell EPR distance measurements in proteins. Angew. Chem. Int. Ed. Engl. 56, 2914–2918 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Verhalen B., et al. , Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borbat P. P., et al. , Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5, e271 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittal A., Böhm S., Grütter M. G., Bordignon E., Seeger M. A., Asymmetry in the homodimeric ABC transporter MsbA recognized by a DARPin. J. Biol. Chem. 287, 20395–20406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barth K., et al. , Conformational coupling and trans-inhibition in the human antigen transporter ortholog TmrAB resolved with dipolar EPR spectroscopy. J. Am. Chem. Soc. 140, 4527–4533 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Hellmich U. A., et al. , Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 134, 5857–5862 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Schmidt T., Wälti M. A., Baber J. L., Hustedt E. J., Clore G. M., Long distance measurements up to 160 Å in the GroEL tetradecamer using Q-band DEER EPR spectroscopy. Angew. Chem. Int. Ed. Engl. 55, 15905–15909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jassoy J. J., et al. , Versatile trityl spin labels for nanometer distance measurements on biomolecules in vitro and within cells. Angew. Chem. Int. Ed. Engl. 56, 177–181 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Bleicken S., et al. , gem-Diethyl pyrroline nitroxide spin labels: Synthesis, EPR characterization, rotamer libraries and biocompatibility. ChemistryOpen 8, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teucher M., Bordignon E., Improved signal fidelity in 4-pulse DEER with Gaussian pulses. J. Magn. Reson. 296, 103–111 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Jeschke G., et al. , DeerAnalysis2006–A comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the corresponding authors.