Significance
Phytochromes and related photoreceptors distinguish themselves for their long-wavelength absorption and large spectral shift between parental state and photoproduct. Both features are not well understood, partly due to lack of high-resolution structural data and insufficient support from quantum-chemical calculations. The red–green switching cyanobacteriochrome Slr1393g3 shows an absorption shift larger than 110 nm. Both parental state and photoproduct could be crystallized with high resolution, together with a “hybrid” form carrying the chromophore in parental state geometry, whereas the protein remained in the photoproduct conformation. The crystal structures reveal how chromophore and protein mutually regulate their conformational changes, yielding the observed spectral shift. Quantum-chemical calculations, based on these structures, provide a deeper understanding of the spectral tuning mechanisms.
Keywords: crystal structure, photochromicity, phytochrome, theoretical chemistry
Abstract
The three-dimensional (3D) crystal structures of the GAF3 domain of cyanobacteriochrome Slr1393 (Synechocystis PCC6803) carrying a phycocyanobilin chromophore could be solved in both 15-Z dark-adapted state, Pr, λmax = 649 nm, and 15-E photoproduct, Pg, λmax = 536 nm (resolution, 1.6 and 1.86 Å, respectively). The structural data allowed identifying the large spectral shift of the Pr-to-Pg conversion as resulting from an out-of-plane rotation of the chromophore’s peripheral rings and an outward movement of a short helix formed from a formerly unstructured loop. In addition, a third structure (2.1-Å resolution) starting from the photoproduct crystals allowed identification of elements that regulate the absorption maxima. In this peculiar form, generated during X-ray exposition, protein and chromophore conformation still resemble the photoproduct state, except for the D-ring already in 15-Z configuration and tilted out of plane akin the dark state. Due to its formation from the photoproduct, it might be considered an early conformational change initiating the parental state-recovering photocycle. The high quality and the distinct features of the three forms allowed for applying quantum-chemical calculations in the framework of multiscale modeling to rationalize the absorption maxima changes. A systematic analysis of the PCB chromophore in the presence and absence of the protein environment showed that the direct electrostatic effect is negligible on the spectral tuning. However, the protein forces the outer pyrrole rings of the chromophore to deviate from coplanarity, which is identified as the dominating factor for the color regulation.
Cyanobacteriochromes (CBCRs) constitute a subgroup in the bilin-binding protein superfamily of phytochromes (1–3). These photoreceptors exhibit a remarkable photochromicity, in case of the plant photoreceptors the absorption shifts by 65 nm upon irradiation. The dark state is described as a red-absorbing parental form (Pr) with λmax = 665 nm that is converted into a far-red–absorbing photoproduct (Pfr) with λmax = 730 nm (4). The light-driven conversion between the two states is common to all phytochromes and is initiated by a site-specific photoisomerization of the double bond between the rings C and D of the chromophore, converting the 15-Z isomer to the 15-E conformation (Fig. 1A, shown for CBCRs). The photoisomerization is completed within a few picoseconds (5–8) and is followed by protein motions expanding into the long milliseconds time range (9, 10). In the “classical” phytochromes, a three-domain (PAS–GAF–PHY) arrangement is essential to maintain the spectral and kinetic properties. In this three-domain arrangement, the GAF domain holds the chromophore through a covalent bond to a cysteine residue (Fig. 1 A and B) and other noncovalent interactions in the binding pocket. CBCRs unite these properties—autocatalytic chromophore binding and photochromicity—within a single GAF domain even when expressed autonomously as a small protein (19 kDa) (11, 12). Whereas canonical phytochromes switch between a red- and a far-red–absorbing state, the absorbance maxima of CBCRs (parental and photoproduct states) cover the entire visible spectrum, even expanding into the near-ultraviolet range (1, 13, 14). Therefore, CBCRs have been classified into several subfamilies according to their absorbance range and amino acid sequence (3, 15). The most prevalent and best-studied subfamily is the red/green switching one with a red-absorbing parental state (Pr, λmax around 650 nm) and a green-absorbing photoproduct (Pg, λmax around 540 nm) showing a remarkably large hypsochromic shift of ∼110 nm. This subfamily moved into the focus because their compact size and variable position of absorption bands approve them as useful tools for optogenetics applications (12, 16). In addition, CBCRs show a noticeable fluorescence for both photoswitchable states, making them also applicable in superresolution microscopy (17, 18). Various mechanisms of spectral tuning have been proposed, among others a steric hindrance that strongly distorts the chromophore or a change in polarity in the chromophore-binding cavity upon protein conformational changes (18). However, a solid support on the molecular level by, e.g., quantum-chemical calculations underpinning either of these mechanisms, is still missing.
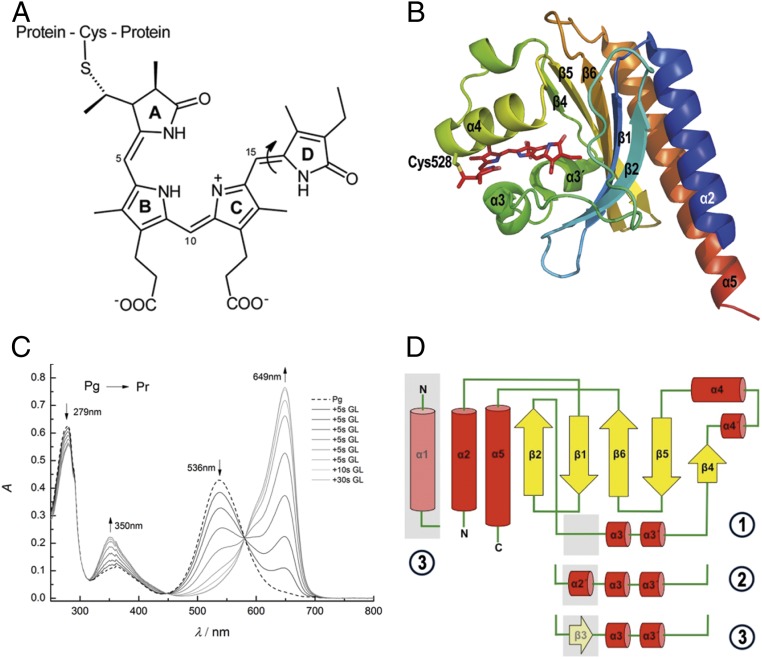
Fig. 1.
Chromophore structure and absorption spectra. (A) Chemical structure of phycocyanobilin chromophore of Slr1393g3. The chromophore is bound covalently to the protein via a thioether bond between Cys528 and the 31 position of PCB. The molecule is shown in the parental-state configuration (Z,Z,Z,s,s,a). The double-bond photoisomerization (double bond between rings C and D) is indicated by an arrow. (B) Cartoon representation of Slr1393g3 structure in its in vitro-assembled parental Pr state (PDB ID code 5DFY). Secondary-structure elements are labeled according to the AnPixJg2 structure (PDB ID code 3W2Z). The PCB chromophore is shown in stick representation; the covalent bond to Cys528 is highlighted. (C) Illumination-induced conversion between the red- (Pr) and green-absorbing (Pg) forms of Slr1393g3. Formation of the red-absorbing form follows stepwise irradiation of Pg (total irradiation time, 80 s; irradiation source, 670-nm LED). (D) Topology of Slr1393g3 in comparison to AnPixJg2: ① Slr1393g3-Pr state, ② Slr1393g3-Pg and the hybrid form, and ③ AnPixJg2 topology. The topology depicts Slr1393g3 in the parental, red-absorbing state. The gray box between β2 and α3 (part of an unstructured loop) converts into a short helical element in the photoproduct and in the photoisomerization hybrid (coined α2′ in the main text). In the parental state of AnPixJg2, this adapts a β-sheet conformation (β3).
Quantum-chemical calculations for phytochromes are challenging and sparse for two reasons: 1) the large size of the bilin chromophore and 2) the lack of high-resolution structural information of the proteins. Regarding the latter aspect, in the red/green switching subfamily, Narikawa et al. (19) have solved the structure of the red form of AnPixJg2, and Lim et al. (20) have recently obtained NMR-based data on both, the parental and photoproduct forms for NpR6012g4. Overall, there are still very few cases, in which both states of a phytochrome-related protein have been structurally determined with resolution good enough to serve as basis for precise quantum-chemical calculations. The photosensory domain from the bacteriophytochrome of Deinococcus radiodurans has been crystallized in both states (21, 22) with a resolution of nearly 1 Å for the parental state, although with much lower resolution (>3 Å) for the photoproduct. Structural information for both states of a phytochrome-related photoreceptor was reported for the CBCR TePixJg from Thermosynechococcus elongatus (2.4 and 2.0 Å) (19, 23). However, in contrast to canonical phytochromes and red/green switching CBCRs, TePixJg shows a very blue-shifted parental state (λmax = 433 nm) and a green-absorbing photoproduct (λmax = 531 nm). These short-wavelength absorptions of both states of TePixJg are caused by atypical chromophores, a PCB-to-PVB (phycoviolobilin) rearrangement (green-absorbing state), and the short absorption maximum of the blue-absorbing parental state results from covalent attachment of cysteine 494 to the central C10 position, thus restricting the conjugated π-system to only two rings (C and D) (13).
The scope of the present study is to close the gap between the biological and photochemical studies by providing the high-resolution crystal structures of both parental and photoproduct states. These structures were analyzed by applying quantum-chemical calculations in order to explain the spectral tuning in the red/green CBCR subfamily. We have selected Slr1393 from Synechocystis PCC6803 (λmax of Pr = 649 nm, λmax of Pg = 536 nm) (9, 17). The full-length protein is composed of three successive GAF domains and a C-terminally located histidine kinase, making it a member of the two component signaling system (17). Out of the three GAF domains, only GAF3 (Slr1393g3) binds a bilin chromophore. This property and all photochemical features are also preserved, if Slr1393g3 is expressed independently as an autonomous protein (Fig. 1B) (9). The absorption maxima of both states are sufficiently separated to allow nearly complete photoconversion of one form into the other, and the photoproduct shows a remarkable thermal stability (reconversion in the dark into the parental state by less than 10% over 24 h, 20 °C) (9).
These properties allowed crystallization and structure determination in the parental and the photoproduct states of Slr1393g3 with high resolution and therefore the identification of conformational changes in the protein and in the chromophore. The quality of the obtained structures encouraged us to apply recently developed high-level ab initio quantum-chemical calculations that enabled us to obtain accurate excitation energies and to provide insight into the origin of the large blue shift in Slr1393g3.
Results
Crystallization of GAF3 Domain of Slr1393.
Crystals of in vivo assembled Slr1393g3, i.e., PCB incorporation during biosynthesis of the apoprotein (following a two-plasmid protocol; Methods) in its parental (Pr) and photoproduct (Pg) states diffracted up to 1.8 and 1.86 Å, respectively. In addition, the apo-form of the protein, assembled with PCB in vitro (i.e., after the immobilized metal ion affinity chromatography [IMAC] purification step) with PCB, was crystallized as Pr with 1.6-Å resolution (data statistics and refinement in SI Appendix, Table S1). An additional structure was obtained with properties in-between the parental and the photoproduct states (diffracted down to 2.1 Å) carrying the chromophore already in the 15-Z conformation as in the Pr state with an absorption around 640 nm, while the protein still remains in the Pg conformation. We refer to this structure as a hybrid form; besides both thermally stable forms, calculations starting from this hybrid form were instrumental to understand the factors controlling the spectral tuning.
Structure of the Red-Absorbing State (Pr State).
Structures of the Pr state of the in vivo- and the in vitro-assembled GAF3 domain from Synechocystis (Protein Data Bank [PDB] ID codes 5DFX and 5DFY) are nearly identical (0.13 Å rmsd over 158 Cα atoms, 1 Å over all atoms). We refer to the in vitro structure in the following, as its resolution is slightly higher. Crystals of the in vitro-assembled Slr1393g3 diffracted to 1.6 Å and contained one monomer per asymmetric unit (ASU). The entire sequence of Slr1393g3 could be modeled into the electron density (residues 441 to 596, full-length numbering of Slr1393) with two N-terminal residues originating from the expression tag (Gly439 to Ser440). The R values after refinement were Rwork 15.5% and Rfree 18.1%, respectively.
Slr1393g3 shows a typical GAF domain folding with a central twisted antiparallel β-sheet sandwiched by α-helices (Fig. 1B). The overall fold is very similar to that from AnPixJg2 (PDB ID code 3W2Z) (19) with 1.87 Å rmsd over 154 Cα atoms and also to the GAF domain structure of the canonical phytochrome Cph1 (PDB ID code 2VEA) with 1.98 Å rmsd over 142 Cα atoms (SI Appendix, Fig. S1 A and B). In the following discussion, the names of canonical and bacterial phytochromes, e.g., Cph1, refer to only their chromophore-binding GAF domain.
The crystal structure of the parental state of AnPixJg2 (PDB ID code 3W2Z) and the here-reported structure are the only ones solved so far for members of the red/green-switching CBCR subfamily. A significant difference between AnPixJg2 and Slr1393g3 resides in the absence of β3 that is present as an unstructured loop in Slr1393g3. The α-helical element α4′ (Fig. 1 B and D) is also found in the AnPixJg2 structure but was not annotated in the original work (19). The central β-sheet is composed of strands β1, β2, β4, β5, and β6, flanked by helices α3, α3′, α4′, and α4 on the chromophore binding side, and α2 and α5 on the opposite side; for details about secondary-structure elements and amino acid annotation, see legend to Fig. 1. Two residues in the Slr1393g3-Pr state interact with a symmetry-related molecule in the crystal assembly (Lys487 with Asn511′ and Gln481 with Glu550′ and Gln551′, respectively) (SI Appendix, Fig. S2A). The PCB chromophore, covalently bound to Cys528 on helix α4, is embedded in a cleft between the central β-sheet array and helices α3, α3′, and α4 (Fig. 2). The protein surface charge around the chromophore binding pocket shows an overall acidic character probably facilitating attraction of the basic pyrrole rings of PCB prior to covalent bond formation. Nevertheless, the chromophore is partially exposed to the solvent (SI Appendix, Fig. S3). PCB adopts the 15–16 Z-configuration, showing an overall Z,Z,Z,s,s,a geometry for all double and single bonds in the ring-connecting bridges. The central rings B and C are in a nearly planar arrangement but with strongly out-of-plane dihedral angles for the A–B and C–D ring methine bridges: 4=5–6-NB = 21° and NC=14–15=16 = −156°. Following the nomenclature by Rockwell et al. (13), the rings A and D have α-facial disposition with respect to the plane determined by rings B and C. The D-ring is kept in place mainly by aromatic amino acids (Tyr464, 3.7 Å; Phe474, 3.6 Å; Trp496, 3.4 Å; Thr543, 2.9 Å; Tyr559, 2.6 Å) and forms an additional interaction through a hydrogen bond to a water molecule (Fig. 2A and SI Appendix, Fig. S4A).
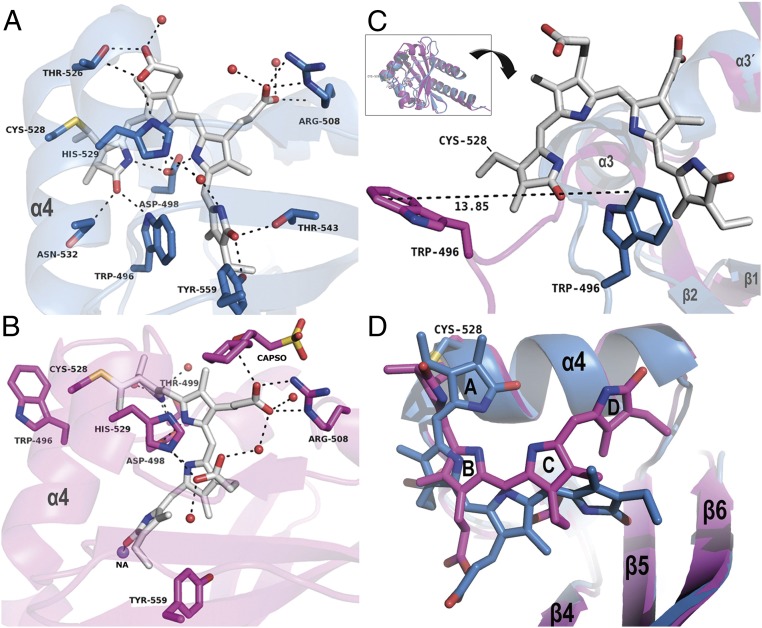
Fig. 2.
Comparison of the chromophore protein interactions in the Pr and Pg state. (A) Binding site of Slr1393g3-Pr; the chromophore (gray sticks) is in the Z,Z,Z,s,s,a configuration. (B) Binding site of Slr1393g3-Pg; the chromophore (gray sticks) is in the Z,Z,E,s,s,a configuration. In A and B, helix α4 is labeled for clarity; residues interacting with the chromophore are represented as sticks and are labeled individually, water molecules as red spheres, and sodium as purple sphere. Distances are given in SI Appendix, Table S2. (C) The Inset on Top shows an overview of the superimposed structures of Slr1393g3-Pr (blue cartoon; PDB ID code 5DFY) and Slr1393g3-Pg (magenta; PDB ID code 5M82). Below the Inset, the structure is rotated for a detailed view on the movement of Trp496; for clarity, the PCB chromophore in Pg state is omitted; the Pr-Trp is shown in blue, and Pg-Trp in magenta. (D) Detailed view on the chromophore conformation of both states with color coding adapted from C. The PCB molecule in the Pg state is, compared with the Pr state, closer to helix α4 and has moved further into the binding site.
Three residues play a key role in the incorporation of the chromophore and are found to be conserved in the red–green subfamily of CBCR: Cys528, His529, and Asp498 (Fig. 2A). His529 is located in the α-position (above the chromophore plane spanned by rings B and C; Fig. 2A) at a distance of 3.8 Å (His atom Nδ to NB of PCB) and 3.5 Å (His atom Nε to NC of PCB). The side chain of Asp498 comprises the central counterion interacting through its carboxylate group with the A-, B-, and C-pyrrolic nitrogen atoms (2.6, 3.0, and 2.6 Å to nitrogen NA, NB, and NC, respectively; Fig. 2A). This Asp counterion constellation is identical to AnPixJg2 and is different to phytochromes, e.g., Cph1, where the central position between pyrrole nitrogens A, B, and C is occupied by the backbone carbonyl group of an aspartate together with a water molecule. In Slr1393g3-Pr state, the propionate group at ring B is backfolded, making contact to Thr526 (2.8 Å) and to His529 (2.9 Å). The propionate group of ring C instead is pointing away from the pyrrole rings and interacts with Arg508 through the side chain as well as through the backbone carbonyl unit in a range of 2.8 to 3.2 Å; the carboxylic group of this propionate makes additional contacts to two closely located water molecules (Fig. 2A). The carbonyl substituents at rings A and D are in hydrogen-bonding distance to Asn532 and Trp496 (A-ring carbonyl group), and Thr543 and Tyr559 (D-ring carbonyl group). In addition, a water molecule is part of a hydrogen-bonding network between His529 and the D-ring nitrogen (Fig. 2A and SI Appendix, Fig. S4A and Table S2). For a detailed comparison of chromophore–protein interactions of Slr1393g3, see the two-dimensional (2D) ligand plots in SI Appendix, Fig. S4 A–F (AnPixJg2, Cph1, and TePixJg).
A tryptophan residue (here, Trp496) is proposed to stabilize the Pr state of CBCRs. In Slr1393g3, the tryptophan side chain is nearly parallel and in close π-stacking distance to ring D with distances in the range 3.49 to 3.87 Å (Fig. 2 A and C and SI Appendix, Fig. S4A). This residue undergoes the largest displacement upon Z–E isomerization (see below).
Structure of the Green-Absorbing Photoproduct State (Pg State).
The thermal stability of the photoproduct (Pg state) allowed crystallization and structure determination of Slr1393g3 in this green-absorbing state. Crystals of the photoproduct state of the in vivo-assembled Slr1393g3 diffracted to 1.86 Å with R values after refinement of Rwork 19.3% and Rfree 23.0%, respectively, and contain one monomer per ASU. The good quality of the electron density clearly depicts the PCB chromophore in the trans- (Z,Z,E,s,s,a) conformation.
In total, the secondary-structure elements and the overall protein folding of the Pr state remain the same in the photoproduct structure (rmsd 2.43 Å over 155 Cα atoms). The most significant change is the rearrangement of a long unstructured loop yielding an outward movement of Trp496 (being in π-stacking distance to ring D of the PCB molecule in the parental state) and a displacement by ∼14 Å with respect to the side-chain atom CZ2 (Fig. 2 B and C). This Trp outward movement opens the chromophore binding pocket allowing water molecules to enter (Figs. 2 and 3) (24). Part of this loop is rearranged into a short (two-turn) additional α-helix (α2′, Leu486 to Asn492; Fig. 1D). This new α-helical element adds several hydrogen bonds to this part of the protein and increases the stability of protein conformation and chromophore in the photoproduct.
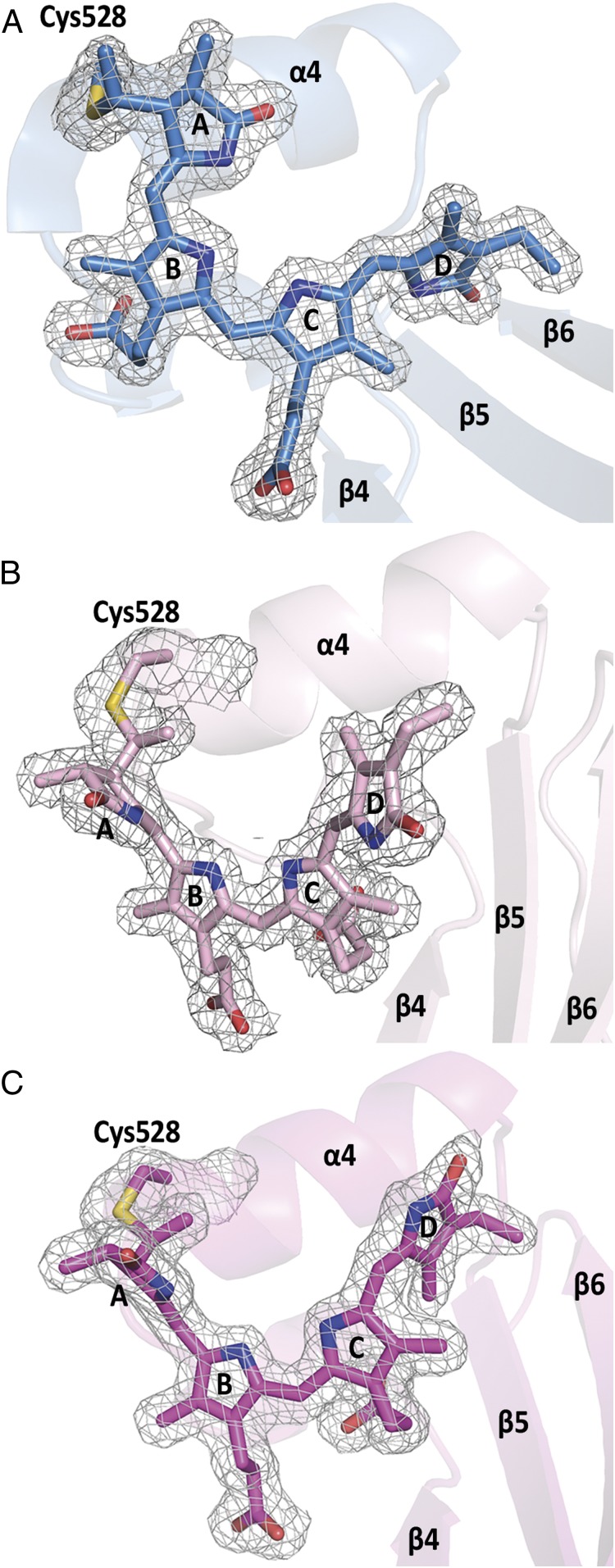
Fig. 3.
Chromophore conformations and electron densities in Slr1393g3. (A) Pr state, (B) hybrid state (back isomerized chromophore) showing a highly distorted Z,Z,Z,s,s,a configuration, and (C) Pg photoproduct showing the chromophore in Z,Z,E,s,s,a configuration. Note the changed relative orientation of the PCB molecule in B and C with respect to helix α4 and β-sheets β5, β6. Electron densities are shown for the chromophore and the covalently bound Cys528, contoured at 1σ.
As a further consequence of the ring D rotation, the chromophore is drawn closer to helix α4 (Fig. 2D). It should be mentioned that classical molecular dynamics and also NMR experiments performed at ambient temperature on the Pr state of AnPixJg2 revealed a subpopulation of Trp residues in a “nonstacking” conformation (20, 25), suggesting that the unstructured loop carrying this Trp is prone to move already in the parental state; however, the extent of this change might vary.
The Pg state of Slr1393g3 changes the propionate side chains’ counterion due to the chromophore movement (Fig. 2B): Arg508 now forms a salt bridge with the propionate side chain of ring B (photoproduct state) instead of that from ring C (parental state) (SI Appendix, Table S2 and Fig. 2 A and B). The released propionate group from ring C folds toward the chromophore and interacts with His529 (2.7 Å). This change of interaction between Arg508 and either of the two propionate side chains is caused by the slight rotation of chromophore around its vertical axis. Interestingly, His529 engages both imidazole nitrogen atoms: Interactions are in hydrogen-bonding distance to B-ring propionate in Pr through Nδ, whereas in Pg the interaction to C-ring propionate now directs to Nε.
As a result of this spatial rearrangement, Thr526, Asn532, Thr543, and Tyr559 no longer interact with the chromophore; instead, Thr499 stabilizes the A-ring (3.1 Å), together with Asp498 (2.74 Å), as also observed in Slr1393g3-Pr state (Fig. 2B and SI Appendix, Figs. S4B, S5A, and S6). An N-cyclohexyl-2-hydroxyl-3-aminopropanesulfonic acid (CAPSO) molecule from the precipitant is bound between two monomers close to the propionate group of ring B (SI Appendix, Fig. S6 and Table S2) interacting with Gln497 of one chain and Arg508 of the other and further stabilizing the network (Fig. 2B and SI Appendix, Fig. S4B). Four water molecules (2.64, 2.68, 2.71, and 2.76 Å) are in hydrogen bonding distance to the oxygen atoms of the propionate groups of ring B and C, whereas only three are present in the Pr state (Fig. 2 A and B). The D-ring experiences more changes in the protein interaction: its carbonyl group has contact to sodium 2 (Na2, 2.48 Å), and a water molecule formerly present in the parental state is now removed (Fig. 2B and SI Appendix, Fig. S4B).
The 15–16 double-bond isomerization of the PCB chromophore is associated with distortions along the single bonds between the four pyrrole rings that are larger than observed in the parental state. The neighboring 14–15 single bond is highly twisted (NC=14–15=16 = −109°) leading to an almost perpendicular D-ring with respect to the plane of the B and C rings. Also the A-ring is more twisted away from the B–C plane (4=5–6-NB = −61°). Hence, the twist of the single and double bond contributes to the overall rotation of the peripheral rings relative to the central rings. The tilting angle between rings A and B changes from 30° in the α-facial disposition to 67° in the β-facial disposition when going from dark state to the photoproduct. The angle between rings C and D changes from 48° to 62°, both in the α-facial disposition. Hence, the change of the tilt is more pronounced for the A ring than for the D ring even though the 15–16 double has isomerized from Z to E conformation. This observation raises the question which of the peripheral rings contributes more to the spectral tuning.
Structure of a Hybrid Form of Slr1393g3.
Under extended duration of X-ray radiation at standard data collection conditions of the photoproduct crystals described above, the chromophore, crystallized as Pg under red light, partly converted from its 15-E state (photoproduct conformation) into the 15-Z state of the parental form. The surrounding protein, however, remained in the photoproduct conformation (rmsd 0.45 Å over 157 Cα atoms; SI Appendix, Fig. S4), with the features indicative for the photoproduct as described in the former paragraph. To exclude that this hybrid conformation (henceforth Ph) is artificially resulting from radiation damage, we carefully processed the data collection images in packages of 100 each and compared the data statistics within these packages and with the whole processed dataset. However, neither from data statistics nor from the electron density (Fig. 3 and SI Appendix, Fig. S8) we observed radiation damage [as was proven as a potentially occurring event in structurally similar proteins by Li et al. (26)]. A comparison between the chromophore configurations in all three structurally solved states—Slr1393g3-Pr, Slr1393g3-Ph, and Slr1393g3-Pg—is presented on the basis of their electron density in Fig. 3 and SI Appendix, Fig. S5 to demonstrate the very similar protein configuration. This comparison also highlights the different relative orientation of the chromophore with respect to the adjacent secondary-structure elements, e.g., α4 and β5. This change in absorption and conformation is not of generic nature as would occur from unintended exposure of the crystal to light. Instead, it is clearly limited to the location of the X-ray beam, as is apparent by the absorbance of the crystal at the position of the X-ray beam exposure recorded before and after the data collection conditions (SI Appendix, Fig. S7) that reveal an increase in absorbance at the long-wavelength flank of the photoproduct spectrum. For both recording conditions, the detection device is fully saturated, and therefore generation of a difference spectrum and the determination of the absorption maximum are greatly impeded. We identify the increase in absorbance in the range between 630 and 650 nm (shoulder in SI Appendix, Fig. S7) with the dominant absorbance still documenting the photoproduct form with λmax around 550 nm. With all precaution due to the experimental challenges, the maximum of the difference spectrum is found around 640 nm.
Crystals of this hybrid form of Slr1393g3 diffracted to 2.1 Å and contain one monomer per ASU. The entire sequence of Slr1393g3 could be modeled into the electron density (residues 441 to 596) with three C-terminal residues of the expression tag (Glu597, Leu598, Glu599; R values after refinement Rwork 17.0% and Rfree 22.2%, respectively; SI Appendix, Table S1). The protein fold of this hybrid form is nearly identical to the Pg state (rmsd 0.45 Å over 157 Cα atoms). The PCB chromophore is further shifted into the protein cleft and positioned closer to helix α4 compared to the Pr state, but shows a somewhat smaller lateral rotation. The electron density around the chromophore clearly identifies the PCB conformation as 15–16 Z (Z,Z,Z,s,s,a configuration, Fig. 3B).
Despite the different C15–C16 isomeric state, there are only few changes in the hydrogen bonding pattern between the PCB chromophore and the Slr1393g3-Ph protein compared to the Pg state (SI Appendix, Table S2 and Fig. S4). Trp496 remains oriented outward and in close proximity to Cys528 (more than 10 Å apart from the D ring) being out of reach for any π-stacking. The positionally conserved tryptophan in the ortholog protein AnPixJg2 [Trp289, assigned as Trp90 by Narikawa et al. (19)] had been suggested to move upon red-to-green conversion based on molecular-dynamics simulations (24). These authors proposed a sideways movement of Trp289, causing an inflow of water molecules into the binding pocket of the photoproduct state as a potential explanation for the hypsochromic shift of the absorption peak.
Also, the chromophore geometry in the Ph state shows barely any changes in comparison to the Pg state despite the full isomerization of the C15–C16 double bond. The most noticeable modification in comparison to the Pg form is the change of the D ring from an α-facial to a β-facial orientation. This is well reflected in the dihedral angle of the single bond NC=14–15=16 = 142° (pdb labels: NA=C4A-CHB=C1B) and visualized in Fig. 4. Besides that, the twist of the A ring as well as the orientation of both propionates are nearly coinciding with those in the Pg state. Therefore, this hybrid form suits well to study the molecular mechanism that determines the spectral tuning that cannot be deduced unambiguously from the structures of the Pr and Pg forms. In the following, we apply quantum-chemical calculations also for this structure along with the parental state and the photoproduct.
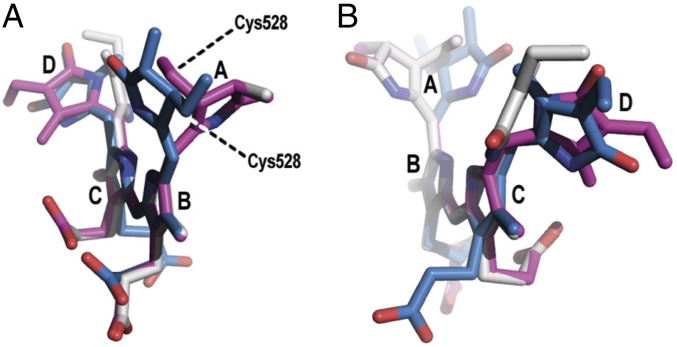
Fig. 4.
Superposition of the PCB chromophores in different perspectives. Geometries of PCB in Slr1393g3 in Pr state (blue; PDB ID code 5DFY), hybrid state (gray; PDB ID code 5M85), and Pg state (magenta; PDB ID code 5M82). Ring B and C were used as reference points. A shows the chromophore conformation along rings B–C, (B) along rings C–B. Note the two different orientations of ring A in the Pr state versus the Pg and hybrid states. The D ring is oriented differently in the hybrid state, compared to the Pr and Pg states.
Quantum-Chemical Calculations.
The high-resolution structures of both parental and photoproduct state for one and the same protein encouraged us to investigate the spectral tuning mechanism of this protein using quantum-chemical calculations in a separate publication (27). Here, we also address the hybrid (Ph) structure by the ab initio quantum chemical method CC2 to calculate the excitation energies and thereby to identify the parameters that cause the spectral shift due to its peculiar chromophore and protein conformation as outlined above. Furthermore, the simulation allows us to determine the excitation energy of this hybrid state, which cannot be determined from the crystal. The three structures Pr, Pg, and Ph were subjected to the same simulation protocol that comprised calculations of the vertical excitation energies for three sets of geometries. The first set of geometries contained the chromophores extracted from the crystal structure and then fully relaxed in vacuo (these models are called “vacuo”). The other two sets were obtained from hybrid quantum-mechanics/molecular-mechanics (QM/MM) calculations in which the chromophore was optimized in the presence of a protein environment. These models are denoted as “strained” and “protein.” Both of them are based on the same chromophore geometry from the QM/MM energy minimization, where the quantum chemically described chromophore was optimized while the protein was kept at the crystallographic position (see Methods and SI Appendix for full details). However, the protein models include the electrostatic effect of the environment, while in the strained models the charges of the environment are set to zero. Hence, the systematic comparison between the vacuo, strained, and protein models allows to analyze the protein-induced effect on the excitation energy (Fig. 5 and SI Appendix, Tables S3 and S4). In particular, it allows us to decompose the effect in two contributions: the influence of the protein on the chromophore conformation and the direct electrostatic effect on the excitation energy. While we have addressed the latter for the Pr-to-Pg transition (27), the former will give us further insight into the steric effect of the protein.
Fig. 5.
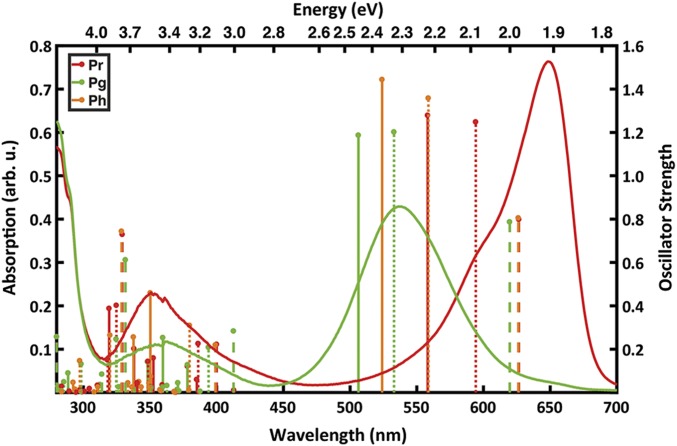
Experimental absorption spectra and calculated excitation energies of Slr1393g3. The experimental absorption spectra (9) are shown as lines for Pr (red) and Pg (green). The calculated excitation energies are represented as sticks for the Pr (red), Pg (green), and the hybrid Ph (orange) forms. The vacuo models are represented by dashed lines, strained models by dotted lines, and protein models as solid sticks. The height of each stick is proportional to the oscillator strength of the corresponding transition.
For Slr1393g3, the optimization of the vacuo models of the Pr and Ph forms leads to two enantiomeric geometries as they are mirror images of one another (SI Appendix, Figs. S9 and S10). This difference stems from the opposed pretwist of all three methine bridges in the crystal structures that serve as starting points for the geometry optimization. In the Pr form the A and the D rings are in the α-facial orientation, while in the hybrid form both rings are in the β-facial orientation (SI Appendix, Figs. S5 and S10). Hence, this leads to a negligible difference of 0.0013 eV (due to numerical accuracy) in the excitation energies between the vacuo models of Pr and Ph forms. The absorption of the Pg vacuo model shows a slight blue shift of 0.02 eV relative to the other two forms, despite the isomerized 15–16 bond (SI Appendix, Tables S3 and S4). The larger difference of the Pg form results from the steric interaction between the two methyl groups at the C and D rings, which prevents a more planar geometry (SI Appendix, Fig. S9C), in agreement with previous findings by Rockwell et al. (28). Hence, we conclude that the spectral change due to the double-bond isomerization is clearly below 0.1 eV.
The calculated excitation energies of the protein models are systematically higher than the experimental absorption maxima (Fig. 5 and SI Appendix, Table S3). For the Pr and Pg states, the results deviate by +0.31 and +0.14 eV from the maxima, respectively. However, the computed Pr-to-Pg spectral shift is 0.23 eV, while the experimental value is 0.40 eV. Comparison between the protein and strained models shows lower excitation energies (0.13 and 0.12 eV for Pr and Pg, respectively). Therefore, the calculated spectral shift remains nearly unaltered with 0.24 eV. This indicates that the protein environment does not have a direct electrostatic effect on the tuning of the photoproduct, and the spectral shift is mainly due to chromophore distortions in the protein environment. This finding is supported by the calculation of the excitation energy in the hybrid form. The difference between the protein and the strained model of the Ph form is 0.15 eV, similar to the other two forms.
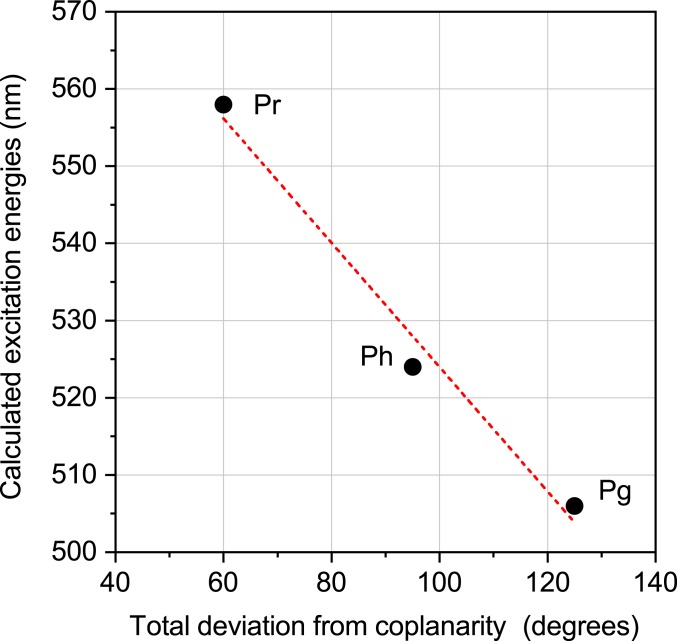
The comparison of the strained to the vacuo models shows that the excitation energies for the three vacuo geometries are within 0.02 eV; however, there is a large variation of 0.24 eV for the strained models. This spectral shift between the vacuo and the strained model is increasing with the extent of the distortion of the peripheral rings. For the Pr form, the smallest shift of 0.11 eV is associated with the A- and D-rings being 22° and 40° out of the plane (SI Appendix, Table S5). The maximum shift due to distortion from planarity is observed for the Pg form with 0.33 eV and corresponding A and D ring deviations of 63° and 61°. Interestingly, the Ph form lies in-between with 0.24 eV shift and out-of-plane distortions of 62° and 32°. A superposition of the chromophores from the Ph and Pg forms (Fig. 4) shows that the A-ring and the propionates are nearly superimposable and the difference originates mainly from the D-ring distortion from the B–C plane, i.e., this shift of 0.11 eV between the Ph and Pg form is mainly due to the D-ring rotation. In contrast, distortions of both peripheral rings from planarity are smaller in the Pr form, indicating that it is not possible to separate the effect of the A and D ring on the spectral tuning. However, the calculations confirm the distortion imposed by the protein environment on the chromophore as the mechanism of spectral tuning, supporting previous studies by Lagarias and coworkers (18, 20, 28–30). It is also in line with a correlation between the extent of coplanarity of the pyrrole rings and the absorption maxima in the PCB chromophores of phycobilisomes presented by Zhao and coworkers (31). This points to a more general mechanism of color regulation; when following their approach for Slr1393g3 (Fig. 6), our findings concur nicely with the proposal from Zhao and coworkers.
Fig. 6.
Relationship between the calculated QM/MM excitation energies for three structures and the coplanarity of the chromophore. Coplanarity is defined by summing the absolute values of the dihedrals between rings A/B, B/C, and C/D as defined in ref. 31.
Discussion
The crystallization of cyanobacterial bilin-binding GAF3 domain from Slr1393 in both parental and photoproduct states with high resolution allows a direct comparison of conformational changes induced by light in one and the same protein. On the basis of these structural data, we could study the origin of the red/green shift in Slr1393g3, which is in contrast to the red/far-red shift of canonical phytochromes. Hence, the understanding of the factors leading to the change of the absorption could pave the way for rational design of bilin-binding photoreceptors.
Despite the overall well-preserved tertiary structure of both Slr1393g3 states, significant conformational changes are precisely identified: Whereas the parental state shows a long unstructured loop extending over the chromophore, part of this loop is formed into a small α-helix in the photoproduct, thus adding structural stabilization to the chromophore conformation. Importantly, a tryptophan residue (Trp496), part of this loop and in π-stacking arrangement with ring D in the parental state, is moved away from the isomerized chromophore by ∼14 Å, opening the binding pocket to the external medium (Fig. 2). The chromophore isomerization causes also a rotation of the PCB molecule around its central orthogonal axis such that Arg508 (interacting amino acid to the propionate group of ring C in the parental state) now interacts with the propionate group of ring B, concomitant with a reduction of the distance between chromophore and helix α4.
In addition to the direct comparison of two states of Slr1393g3, important information was provided by the structural determination of a hybrid form with the protein in the photoproduct conformation and the chromophore in the parental-state–like 15-Z conformation. This structure unveils key factors affecting the absorption and challenges previously proposed mechanisms that regulate the absorbance of the various states. In the X-ray beam-exposed parts of the crystal, the chromophore conformation is clearly assigned as 15-Z configuration (Fig. 3B), irrespective of the presence (parental state) or absence (photoproduct) of the π-stacking Trp496, and unaffected by an altered protein surrounding, including interactions with amino acids and the water molecules. It demonstrates that a change in absorption does not originate from the rearrangement of the binding pocket as proposed earlier on the basis of classical molecular-dynamics simulations (24), but the absorption properties are intrinsic to the distortion of the chromophore itself. Both 15-Z and 15-E isomers of Slr1393g3 show high out-of-plane distortions of A- and D-rings, although the angles are different for each state (Fig. 4). Strong distortions in the A- and D-ring dihedral angles are also found in various phytochrome structures (SI Appendix, Table S6); however, so far, no attention has been given to the relation of distortional angles and absorption. This additional information along with our quantum-chemical calculations strongly support the proposed mechanism of spectral tuning where the chromophore conformation is mainly responsible for the absorption in agreement with recent calculations (27).
Although being instrumental for the identification of factors regulating the absorption maxima, the mechanisms for formation of the hybrid form remain unsolved. As commented above, we can exclude radiation damage as a potential effect and also unintended background illumination, as the “color change” is clearly focused on the X-ray beam exposed position. One might speculate that the X-ray exposure generates a global increase of protein dynamics, potentially sufficient for the chromophore to overcome a very shallow barrier and to commence isomerization. It is tempting to assign this hybrid structure to an early intermediate of the Pg-to-Pr conversion, as it proceeds into the “correct” direction (15-E to 15-Z). However, experiments by time-resolved vibrational spectroscopy documenting a concomitant change of the protein conformation would be essential to propose an involvement of this hybrid form in the Pg-to-Pr conversion, and thus, a speculation of its biological role appears to be reaching out much too far.
Methods
Protein Preparation.
GAF3 domain of Slr1393 from the cyanobacterium Synechocystis PCC6803 was prepared and purified as recently described, following two protocols for chromophore assembly: 1) a two-plasmid expression protocol yields protein and chromophore during expression (in vivo) and allows formation of the holoprotein during biosynthesis (9, 17), and 2) an in vitro assembly by expressing the apoprotein and adding the PCB chromophore after the IMAC purification (details in SI Appendix).
Crystallization Conditions and Structure Determination.
Slr1393g3-Pr state, in vivo.
Initial crystals were optimized using sitting-drop vapor diffusion method with drops containing of 1 µL of protein solution (10 mg/mL in 50 mM Tris⋅HCl, pH 7.5, 100 mM NaCl) mixed with 1 µL of reservoir solution (0.02 M Tris⋅HCl, pH 7.0, 0.1 M sodium chloride, 7.7% [wt/vol] PEG 4000), equilibrated over 300-µL reservoir at 12 °C. Crystallization experiments were set up under dim green light, and crystallization plates were covered with aluminum foil. Blue rhomboid-shaped crystals grew to a final size of 100 × 100 × 70 µm3 within 8 wk. All crystal-containing drops (see also following) were overlaid with mineral oil for cryoprotection before harvesting the crystals, and crystals were shock-frozen with liquid nitrogen.
Slr1393g3-Pr state, in vitro.
Crystallization trials and crystal harvesting were performed as described above but under different crystallization conditions and in hanging drops. The optimized precipitant contained 0.02 M sodium citrate, pH 5.6, 0.1 M sodium chloride, and 11% (wt/vol) PEG 3350. Blue rhomboid-shaped crystals grew to a final size of 120 × 120 × 100 µm3 within 8 wk.
Slr1393g3-Pg state and -hybrid state.
Prior to crystallization experiments, the protein solution was irradiated with red light-emitting diodes (LEDs) to generate the photoproduct state. All crystallization experiments were set up under dim red light. Initial crystals were optimized using sitting-drop vapor diffusion method with drops containing 1 µL of protein solution (8–10 mg/mL in 50 mM Tris⋅HCl, pH 7.2, 100 mM NaCl) mixed with 1-µL reservoir solution (0.1 M CAPSO, pH 9.5, 0.1 M sodium chloride, 100 to 175 mM lithium sulfate, 12 to 14% [wt/vol] PEG 4000), equilibrated over 300-µL reservoir at 4 °C. The crystallization plates were kept under red light. Pink rhomboid-shaped crystals grew to a final size of 90 × 90 × 60 µm3 within 2 wk.
Data Collection.
Slr1393g3-Pr state (in vivo).
Data were collected at 100 K from a single crystal at ID29, European Synchrotron Radiation Facility (ESRF) (Grenoble, France). Data were processed by XDS and XSCALE (32, 33). The structure was solved by molecular replacement (MR) using the autorickshaw pipeline (34) providing the protein sequence and processed data as input files. The structural models of this protein, its in vitro-assembled counterpart, the photoproduct, and the hybrid form were further built and refined using iterative cycles of manual building in coot (35) and refinement cycles using REFMAC5 (36) from the ccp4 suite (37). The structure was deposited in the PDB under ID code 5DFX.
Slr1393g3-Pr state (in vitro).
Data were collected at 100 K from a single crystal at ID23-1, ESRF (Grenoble, France). Data were processed by XDS (32, 33). The structure was solved by MR using 5DFX as search model. For structural model building and refinement, see above (Pr form).
Slr1393g3-hybrid state.
Data were collected at 100 K from a single crystal at P13, DESY, EMBL, Hamburg, Germany (38). During data collection the crystal area exposed to X-rays immediately turned from pink to dark blue indicating a conformational change in the chromophore arrangement. Data were processed by XDS (32). The structure was solved by MR using 5DFX as search model. Structural model building and refinement see above.
Slr1393g3-Pg state.
Data were collected at 100 K from a single crystal at P13, German Synchrotron Radiation Facility (DESY), European Molecular Biology Laboratory (EMBL) (Hamburg, Germany) (38). Experienced from using the pink (photoproduct) crystals for determination of the hybrid state, we knew the crystal area exposed to X-rays was highly susceptible to turn from pink to blue; we here used a helical data collection strategy with minimum exposure time and minimum total oscillation to reduce the effect of the X-rays to the chromophore arrangement (39). Data were processed by XDS (32, 33). The structure was solved by MR using 5DFX as search model. For structural model building and refinement, see above.
Detailed information about all data collection and refinement statistics are given in SI Appendix, Fig. S1.
Images of the models were prepared using PyMOL (the PyMOL Molecular Graphics System). The 2D ligand plots of interactions were generated using the webservice “PDBsum Generate” (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html).
Absorption Spectra of Slr1393g3 Crystals.
For recording the absorption spectra, two crystals of Slr1393g3 in photoproduct state were used (mounted in loops): one remained unexposed to X-rays (pink crystal), and the other one was measured after collection of a complete dataset (areas exposed to the X-ray beam turned to dark blue, whereas the unexposed parts of the crystal remained pink). Absorption was measured at 100 K at ID29S-Cryobench (ESRF, Grenoble, France) (40).
QM/MM Calculations.
The crystal structures of Slr1393g3 [PDB ID codes 5DFX (Pr), 5M82 (Pg), and 5M85 (hybrid form)] that are reported in this study were used as a starting point for hybrid QM/MM calculations. The full details of the computational setup of the protein and the subsequent QM/MM calculations are given in SI Appendix. The models obtained at the RI-CC2/cc-pVDZ/Amber level of theory of the chromophore inside the frozen protein are denoted “protein.” In these models, all atoms of the environment within a distance of 12 Å to any of the QM atoms were taken into account via point charges in the excited state calculations. In the strained models, the environment represented by point charges was discarded, but the QM region is kept in the protein-induced geometry. A full relaxation of the strained models in the gas phase resulted in the vacuo models.
We have used the ab initio RI-CC2 method to go beyond the semiempirical DFTB2+D-based optimizations in our previous study. The excited-state calculations were realized consistently to the ground state description by using the same method. The RI-CC2 calculations were done in Turbomole (10), which is interfaced with ChemShell (11) for QM/MM calculations. Details of the optimizations and excited-state calculations are given in SI Appendix.
Data Availability.
The crystal structure of Slr1393g3 has been deposited in the PDB, https://www.wwpdb.org/ (under ID codes 5DFY and 5DFX [in vitro and in vivo assembled] in the dark state, under ID code 5M82 in the light state, and under ID code 5M85 in the hybrid state).
Note Added in Proof.
During preparation of this manuscript, a manuscript on the same protein had been accepted for publication: D. Buhrke et al., Red, orange, green: Light- and temperature-dependent color tuning in a cyanobacteriochrome. Biochemistry; https://doi.org/10.1021/acs.biochem.9b00931 (2019).
Supplementary Material
Acknowledgments
The expert technical help from Stefanie Kobus, Center for Structural Studies (CSS), Heinrich-Heine-Universität Düsseldorf, is gratefully acknowledged. We thank Sander Smits (Institute of Biochemistry and CSS) for helpful discussion and advice during development of data collection strategies. We acknowledge help from the European Synchrotron Radiation Facility, and we thank Anton Royant and Ulrich Zander for assistance in using beamlines ID23-1 and ID29, and Guillaume Gotthard for measuring absorption at beamline ID29S. We also thank Isabel Bento and Guillaume Pompidor at P13, DESY (EMBL, Hamburg, Germany), for support during data collection. Quantum chemical calculations were performed on resources from the Paderborn Center for Parallel Computing. C.W. acknowledges support by Deutsche Forschungsgemeinschaft (DFG) (WI 4853/1-1). This project has received funding from the European Research Council (ERC), European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement 678169, ERC Starting Grant “PhotoMutant”). I.S. is grateful for the Mercator Fellowship and support from the DFG (Grant SFB 1078 “Protonation Dynamics in Protein Function”). K.-H.Z. gratefully acknowledges support by the National Natural Science Foundation of China (Projects 31861143029 and 31770822). W.G. gratefully acknowledges support from the Max Planck Society. The CSS is funded by the DFG (417919780; INST 208/740-1 FUGG).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structure of Slr1393g3 has been deposited in the Protein Data Bank (PDB), https://www.wwpdb.org/ (accession nos. 5DFY and 5DFX [in vitro and in vivo assembled] in the dark state, 5M82 in the light state, and 5M85 in the hybrid state).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910208117/-/DCSupplemental.
References
- 1.Ikeuchi M., Ishizuka T., Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Rockwell N. C., Lagarias J. C., A brief history of phytochromes. ChemPhysChem 11, 1172–1180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders K., Essen L. O., The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Montgomery B. L., Lagarias J. C., Phytochrome ancestry: Sensors of bilins and light. Trends Plant Sci. 7, 357–366 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Müller M. G., Lindner I., Martin I., Gärtner W., Holzwarth A. R., Femtosecond kinetics of photoconversion of the higher plant photoreceptor phytochrome carrying native and modified chromophores. Biophys. J. 94, 4370–4382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slavov C., Xu X., Zhao K. H., Gärtner W., Wachtveitl J., Detailed insight into the ultrafast photoconversion of the cyanobacteriochrome Slr1393 from Synechocystis sp. Biochim. Biophys. Acta 1847, 1335–1344 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Kim P. W., et al. , Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 1. Forward dynamics. Biochemistry 51, 608–618 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Kim P. W., et al. , Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 2. Reverse dynamics. Biochemistry 51, 619–630 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Xu X. L., et al. , Combined mutagenesis and kinetics characterization of the bilin-binding GAF domain of the protein Slr1393 from the cyanobacterium Synechocystis PCC6803. ChemBioChem 15, 1190–1199 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Fukushima Y., et al. , Photoconversion mechanism of a green/red photosensory cyanobacteriochrome AnPixJ: Time-resolved optical spectroscopy and FTIR analysis of the AnPixJ-GAF2 domain. Biochemistry 50, 6328–6339 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Narikawa R., Fukushima Y., Ishizuka T., Itoh S., Ikeuchi M., A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 380, 844–855 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., et al. , Fused-gene approach to photoswitchable and fluorescent biliproteins. Angew. Chem. Int. Ed. Engl. 49, 5456–5458 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Rockwell N. C., et al. , A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry 47, 7304–7316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockwell N. C., Martin S. S., Lagarias J. C., Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry 51, 9667–9677 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Rockwell N. C., Martin S. S., Feoktistova K., Lagarias J. C., Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U.S.A. 108, 11854–11859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon J., Losi A., Zhao K. H., Gärtner W., FRET in a synthetic flavin- and bilin-binding protein. Photochem. Photobiol. 93, 1057–1062 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Chen Y., et al. , Photophysical diversity of two novel cyanobacteriochromes with phycocyanobilin chromophores: Photochemistry and dark reversion kinetics. FEBS J. 279, 40–54 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Rockwell N. C., Martin S. S., Gulevich A. G., Lagarias J. C., Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 53, 3118–3130 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Narikawa R., et al. , Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim S., et al. , Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. U.S.A. 115, 4387–4392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgie E. S., et al. , Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Burgie E. S., Walker J. M., Phillips G. N. Jr, Vierstra R. D., A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Velazquez Escobar F., et al. , Photoconversion mechanism of the second GAF domain of cyanobacteriochrome AnPixJ and the cofactor structure of its green-absorbing state. Biochemistry 52, 4871–4880 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Scarbath-Evers L. K., et al. , Structural heterogeneity in a parent ground-state structure of AnPixJg2 revealed by theory and spectroscopy. Phys. Chem. Chem. Phys. 19, 13882–13894 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Li F., et al. , X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome. J. Am. Chem. Soc. 137, 2792–2795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiebeler C., Rao A. G., Gärtner W., Schapiro I., The effective conjugation length is responsible for the red/green spectral tuning in the cyanobacteriochrome Slr1393g3. Angew. Chem. Int. Ed. Engl. 58, 1934–1938 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Rockwell N. C., Martin S. S., Lim S., Lagarias J. C., Ames J. B., Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A protonated bilin ring system in both photostates. Biochemistry 54, 2581–2600 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Rockwell N. C., Martin S. S., Lim S., Lagarias J. C., Ames J. B., Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A hydrophobic pocket for the C15-E, anti chromophore in the photoproduct. Biochemistry 54, 3772–3783 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Rockwell N. C., Martin S. S., Gan F., Bryant D. A., Lagarias J. C., NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 14, 258–269 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Tang K., et al. , The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore. Proc. Natl. Acad. Sci. U.S.A. 112, 15880–15885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A., Auto-rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collaborative Computational Project , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 38.Cianci M., et al. , P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat. 24, 323–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flot D., et al. , The ID23-2 structural biology microfocus beamline at the ESRF. J. Synchrotron Radiat. 17, 107–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Stetten D., et al. , In crystallo optical spectroscopy (icOS) as a complementary tool on the macromolecular crystallography beamlines of the ESRF. Acta Crystallogr. D Biol. Crystallogr. 71, 15–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure of Slr1393g3 has been deposited in the PDB, https://www.wwpdb.org/ (under ID codes 5DFY and 5DFX [in vitro and in vivo assembled] in the dark state, under ID code 5M82 in the light state, and under ID code 5M85 in the hybrid state).