Significance
By computing genotype data from six major world populations, we aimed at calculating how many individuals are affected with an autosomal recessive (AR) form of inherited retinal disease (IRD) or carry a mutation that can be transmitted to future generations. By analyzing variants in 187 IRD-associated genes, we detected 10,044 mutations and estimated that 2.7 billion individuals worldwide are carriers of an IRD disease-causing mutation, whereas 5.5 million are expected to be affected. This study will assist clinicians in their decision about the need to perform relevant genetic tests when diagnosing patients with IRDs. Similar studies can take advantage of our approach to calculate the expected number of affected or carrier individuals for any genetic disease with known molecular etiology.
Keywords: carrier frequency, disease-causing mutation, human genome, inherited retinal diseases, genetic prevalence
Abstract
One of the major questions in human genetics is what percentage of individuals in the general population carry a disease-causing mutation. Based on publicly available information on genotypes from six main world populations, we created a database including data on 276,921 sequence variants, present within 187 genes associated with autosomal recessive (AR) inherited retinal diseases (IRDs). Assessment of these variants revealed that 10,044 were categorized as disease-causing mutations. We developed an algorithm to compute the gene-specific prevalence of disease, as well as the mutational burden in healthy subjects. We found that the genetic prevalence of AR-IRDs corresponds approximately to 1 case in 1,380 individuals, with 5.5 million people expected to be affected worldwide. In addition, we calculated that unaffected carriers of mutations are numerous, ranging from 1 in 2.26 individuals in Europeans to 1 in 3.50 individuals in the Finnish population. Our analysis indicates that about 2.7 billion people worldwide (36% of the population) are healthy carriers of at least one mutation that can cause AR-IRD, a value that is probably the highest across any group of Mendelian conditions in humans.
Following the completion of the human genome project (1, 2), one of the major focuses of genetic research has been to characterize benign genomic variations vs. variants that are associated with multifactorial and monogenic diseases (3). To this end, many databases containing sequencing information of a large number of individuals have been created, allowing a broader and more precise view on genetic diversity. It is therefore now possible to gather information from many genomes at once and extract valuable measures of human variation. Allele frequency, for example, is an extremely important parameter that can aid in the identification of the genetic cause of Mendelian diseases. In medical genetics, next generation sequencing (NGS) techniques often output extremely large numbers of variants, most of which are unrelated to the condition that is investigated. It is therefore crucial to distinguish between true disease-causing mutations and the remaining variants, including the precise assessment of carrier frequency (CF) for a collective set of mutations causing a given disorder.
Inherited retinal diseases (IRDs) are a group of heterogeneous conditions leading to vision loss, due to the progressive degeneration of the retina, and are mainly caused by Mendelian mutations in 1 out of at least 300 genes (RETNET; https://sph.uth.edu/retnet/). Clinical symptoms vary across different IRD subtypes and different disease genes. For example, in retinitis pigmentosa (RP), the most common form of IRD, retinal degeneration initially causes night blindness due to the loss of rod photoreceptor cells. On the other hand, in Stargardt disease (STGD), degeneration of a specific retinal region, the macula, causes a drastic change in central visual acuity. It is currently unknown how many genes are involved in IRDs, and even by using NGS techniques, including whole-exome sequencing (WES) and whole-genome sequencing (WGS), mutations are identified only in 50 to 75% of patients (4, 5). Therefore, genomic databases such as the Genome Aggregation Database (gnomAD) (6) (https://gnomad.broadinstitute.org/) can be very valuable for studying the genetic landscape of genetically heterogeneous Mendelian diseases, including, for instance, IRDs and intellectual disabilities.
A major question in human genetics is what percentage of individuals in the general population carry a disease-causing mutation for a specific disease and/or in a given gene. This information is population-specific and is highly important for genetic counseling. A few studies attempted to calculate CF for IRD-causing mutations in the general population. By analyzing 46 WGS data of control individuals, autosomal recessive (AR)-IRD CF was estimated to be about one out of four to five individuals, for loss-of function mutations only (7). We recently used NGS and online databases to assess the CF of AR-IRD–causing mutations in different Israeli subpopulations by applying statistical analyses and assessed CF to be one out of three to four individuals (8).
In the current work, we developed an analysis scheme allowing the research community to use genetic information from large-scale genomic databases to identify potential prevalent pathogenic mutations (some of which were not published before) in each of six major worldwide subpopulations and calculate CF and genetic prevalence (GP; the proportion of individuals in the population who are expected to be affected based on their genotype) for each mutation, gene, or phenotype of interest. The analysis allowed us to calculate both total CF and GP values for AR-IRD genes and estimate the expected worldwide values for each mutation, gene, and phenotype in various ethnicities.
Results
The AR-IRD Mutation Database.
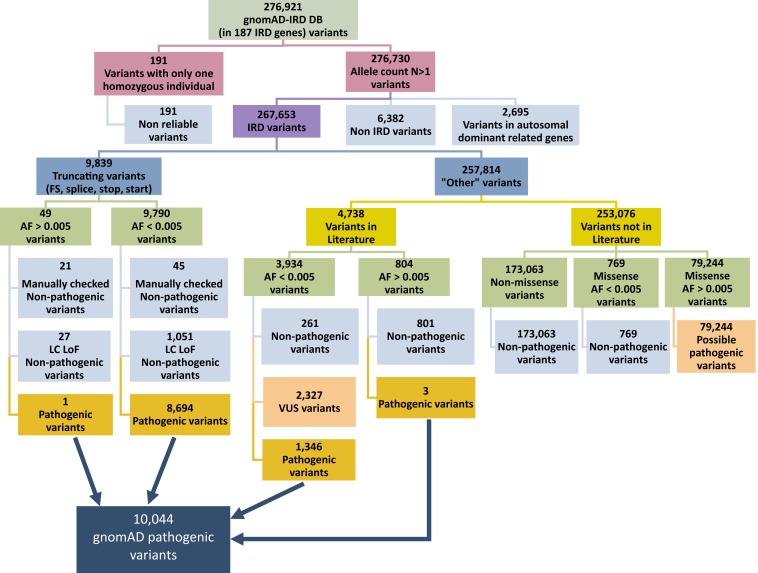
Aiming to assess worldwide CF and GP of AR-IRD mutations, we created a Structured Query Language (SQL) database containing 276,921 DNA variants within 187 known IRD genes (SI Appendix, Table S1). These variants were stratified according to the ethnical group of the individuals carrying them (Africans [AFR], East Asians [EAS], South Asians [SAS], Latinos [LAT], Europeans [non-Finnish] [EUR], and Finnish [FIN]), based on the information provided in the gnomAD database (version 2; all data are based on GRCh37/hg19). The SQL database also included information on variants and mutations that were published in the scientific literature and were extracted from the Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). It should be noted that multiple studies showed that not all variants that appear as pathogenic in HGMD and ClinVar are indeed disease-causing (9, 10). In order to identify true pathogenic mutations, we used a number of filtering steps (Fig. 1). First, we eliminated 191 variants that were each found in one homozygous individual only but were not found at all in heterozygotes, and therefore are likely to represent nonreliable reads. We subsequently eliminated 6,382 variants that were reported to cause a disease with no retinal involvement, as well as 2,695 variants identified in genes that were reported previously to cause IRD with an autosomal dominant inheritance pattern. The remaining 267,653 variants were divided into two subgroups: 9,839 probable truncating variants [frameshift, splice-site, nonsense variants, and start-loss variants, based on the American College of Medical Genetics classification for truncating variants (11)] and 257,814 “other” variants. We then validated the categorization information provided by gnomAD regarding truncating variants and stratified the truncating variants by allele frequency, using 0.005 as a cutoff threshold. Most of the truncating variants (9,790) had an allele frequency lower than the threshold and were considered more likely to be pathogenic. However, taking into account the low confidence loss of function flags in gnomAD, we discarded 1,051 variants. In addition, a manual analysis of these variants, considering the relevant published information (including cosegregation data, genotype in affected and control individuals, and biochemical data if available), led to the exclusion of 45 additional variants. The remaining 8,694 truncating variants were considered pathogenic. A similar analysis of 49 truncating variants with allele frequency >0.005 revealed that 1 of them was pathogenic, outputting a total of 8,695 likely pathogenic truncating mutations.
Fig. 1.
Flowchart showing the analytical scheme for the variants downloaded from gnomAD.
A different analysis was applied to study the 257,814 variants from the “other” group (Fig. 1). We first divided these variants into two subgroups based on their presence in HGMD and/or ClinVar. Most of the variants (253,076) did not appear in the literature and were divided into three groups: nonmissense variants, missense variants with an allele frequency higher than 0.005, and missense variants with an allele frequency lower than 0.005 (Fig. 1). The remaining 4,738 variants (appearing in HGMD and/or ClinVar) were subsequently divided into two subgroups according to allele frequency, followed by an analysis based on the reported interpretation. Variants interpreted as pathogenic in both ClinVar and HGMD were considered pathogenic in the current analysis, whereas variants that received contradicting interpretations (e.g., pathogenic in one database and benign in the other database) were manually examined by studying the relevant publications aiming to reach the more appropriate conclusion. This analysis resulted in 1,349 pathogenic variants, bringing the total number of likely pathogenic variants to 10,044.
Based on these data, we calculated CF and GP values as detailed below. A special attention was given to ABCA4 since 26 variants in this gene were reported as hypomorphic and therefore were treated differently in our analysis (SI Appendix, Supplementary Note).
Total CF and GP Analyses.
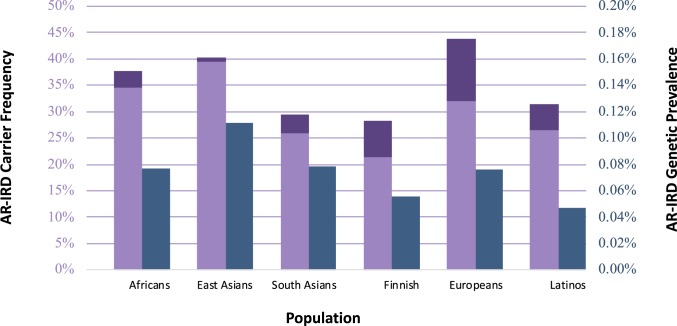
To determine the overall CF for all AR-IRD–causing mutations in different subpopulations, we initially calculated CF for each of the 10,044 likely pathogenic variants in each subpopulation (SI Appendix, Tables S2 and S3). We subsequently calculated global CF (taking into account the possibility that unaffected individuals might be heterozygous for multiple AR-IRD–causing mutations; Materials and Methods) using two different methods (based on the inclusion–exclusion principle [SI Appendix, Fig. S1] and the independence probability theory) yielding highly concordant results. Total CF for AR-IRD–causing mutations (Fig. 2, purple bars) varies from 28% of individuals (in the Finnish population) to 44% (in the European population). On average, we predict that 36% of the human population (i.e., 2.7 billion individuals) are unaffected carriers for at least one AR-IRD–causing mutation. Since hypomorphic ABCA4 variants are relatively common (and therefore might have a substantial effect on total CF) but contribute to prevalence only when combined with a severe variant, we calculated CF both with (Fig. 2, dark purple bars) and without these variants (Fig. 2, light purple bars). It has to be noted that hypomorphic variants show the largest effect in the European population, which is concordant with the fact that this ethnic group is studied more comprehensively than others.
Fig. 2.
AR-IRD CF and GP per subpopulation worldwide. CF is depicted by the purple bars and measured by the left vertical axis. The dark purple area of each bar represents the contribution of ABCA4 hypomorphic variants. GP is depicted by the dark blue bars (right vertical axis).
To compute the total GP of AR-IRDs, we initially calculated GP for each gene in each of the subpopulations, using an algorithm that is based on the product of individual CF values (Materials and Methods and SI Appendix, Table S2). To this end, we created a matrix for each combination of gene and subpopulation that includes all possible mutation combinations. In each cell of the matrix, we calculated the likelihood for affected individuals based on the CF value of each pathogenic mutation. Homozygous mutations are represented by the diagonal (blue cells in SI Appendix, Fig. S2), while the remaining cells represent compound heterozygous states (green cells). For example, we identified 10 RDH12 mutations in gnomAD carried by African individuals with CF ranging from 1.9 × 10−3 to 8.3 × 10−5. The product analysis calculated the number of affected individuals among Africans to be 3,442, i.e., individuals who are expected to carry biallelic RDH12 mutations and therefore to suffer from early-onset autosomal recessive retinitis pigmentosa (ARRP). A similar analysis was performed for each subpopulation in each of the 185 reported IRD genes (Materials and Methods). In two genes (COL2A1 and NEUROD1) that were included in the original analysis and cause mainly dominant phenotypes, the reported recessive mutations were not found in gnomAD. Therefore, there are no AR disease-causing mutations for these two genes in the final results, and data of the remaining 185 genes are presented (SI Appendix, Table S3). Following the product analysis for each gene, we summed the expected total GP of AR-IRDs (that include the number of individuals who are predicted to be affected by their genotype) in each subpopulation (Fig. 2, blue bars). The total GP of AR-IRDs ranges from 1:2,214 in the Latino subgroup (corresponding to 0.05%) to 1:1,003 in the East Asia group, with an average worldwide GP of 1:1,378 individuals.
Distribution of AR-IRD Mutations and Corresponding Genes Among Subpopulations.
The analysis of 185 IRD-causing genes highlighted three (ABCA4, USH2A, and EYS), for which mutations are highly prevalent in at least five of the six studied subpopulations. On the other hand, 20 of them (including PRCD, DHDDS, and IDH3A) harbor only extremely rare mutations in most subpopulations (SI Appendix, Table S2). A tabulation of CF values for each gene in each subpopulation (SI Appendix, Table S2) shows that ABCA4 has the highest number of mutations (225) in the European subpopulation, with a cumulative European CF value of 2.5% (not including hypomorphic variants). Based on the values presented in SI Appendix, Table S2, one can calculate the likelihood for a specific individual to be a carrier for a mutation in a gene of interest. Interestingly, some genes showed a noneven distribution of CF, with extremely high values in a specific subpopulation and much lower values in the remaining populations (SI Appendix, Fig. S3). For example, mutations in four genes (C8orf37, GPR125, MTTP, and PDE6G) are much more prevalent in Africans compared to other populations, with C8orf37 mutations showing an average ratio of 80 in GP in Africans vs. other groups. Interestingly, while in some populations (Africans, East Asians, Finnish, and Latinos) many genes are unique and contribute to GP mainly in a single subpopulation, only a single gene was found to be relatively prevalent in the remaining two populations (ZNF513 in South Asians and INPP5E in Europeans; SI Appendix, Fig. S3).
GP of AR-IRDs.
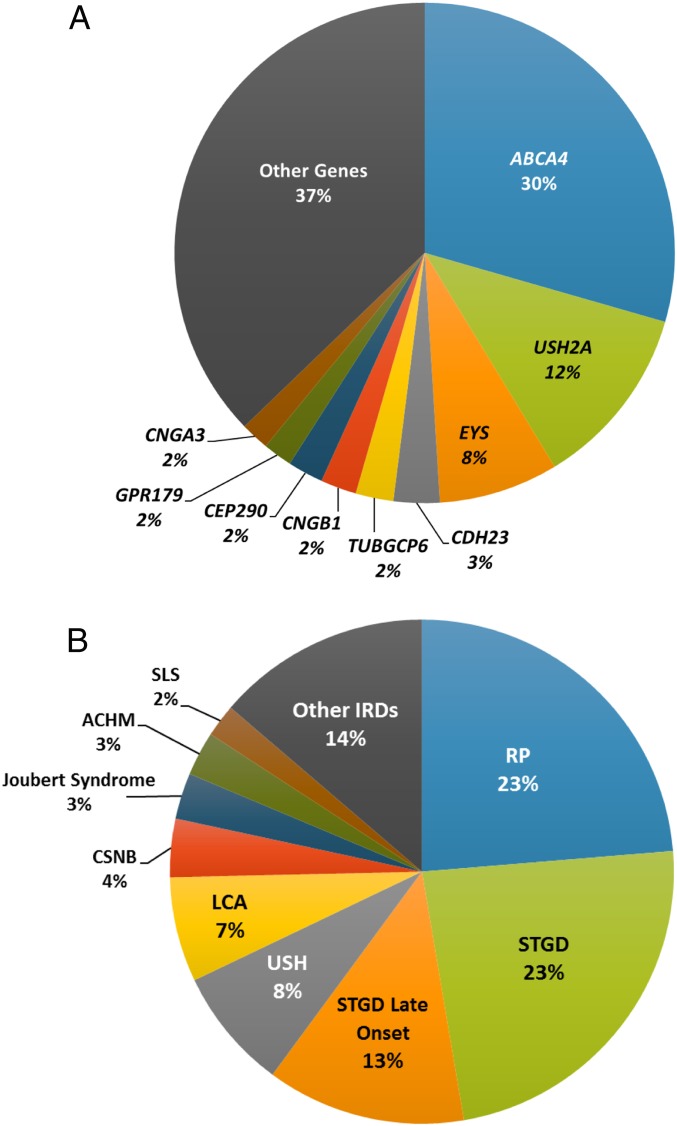
Based on the values we obtained from the product analysis, we calculated GP of AR-IRD per causing gene (Fig. 3A and SI Appendix, Table S2) as well as the expected GP of various IRD phenotypes (Fig. 3B), allowing us to predict the number of individuals who are expected to be affected worldwide by biallelic mutations in each gene. For example, the analysis predicts a total of 15,620 biallelic RPE65 patients worldwide, most of whom (9,484 individuals—over 60% of all affected individuals worldwide) are from the African population, while only 9% are Europeans. It should be noted here that for congenital diseases such as Leber congenital amaurosis (LCA) caused by RPE65 mutations, GP is likely to be highly similar to the actual disease prevalence. The most prevalent AR-IRD gene is ABCA4 (Fig. 3A), mutations in which are responsible for about 30% of all IRD cases, followed by USH2A (12%), and EYS (8%). The cumulative GP of ABCA4-associated IRDs is predicted to correspond to 1.4 million affected individuals worldwide (SI Appendix, Table S2). This value is much higher with respect to USH2A (with 557,723 expected affected individuals) and EYS (with 363,411 expected affected individuals). Recent studies have reported that ABCA4 hypomorphic variants cause disease only in a compound heterozygous state, in trans with a severe ABCA4 mutation (frameshift, nonsense, splice-site, etc.). We therefore considered 26 ABCA4 variants as hypomorphic variants based on these reports (12–16), as well as on the LOVD database (http://www.lovd.nl/3.0/home). When calculating the GP for ABCA4, we paired these hypomorphic changes only with truncating mutations and discarded the values resulting from homozygous genotypes or compound heterozygous states with missense or intronic mutations. We subsequently used the known information regarding the range of diseases caused by each mutation in each of the IRD genes to calculate the GP of each condition. While for some IRD genes this calculation was straightforward (e.g., all FAM161A mutations cause only one phenotype, RP, in AR pattern), for other genes each mutation was paired with a disease based on previous publications. For example, mutations in USH2A can cause either Usher syndrome type 2 (USH2) or nonsyndromic RP with a clear genotype–phenotype correlation: while biallelic truncating mutations were reported to cause USH2, specific missense mutations [e.g., p.(Cys759Phe) (17)] on at least one of the two alleles cause ARRP. Similar analyses were performed for the remaining IRD genes (SI Appendix, Table S4), revealing that STGD is expected to be the most common AR-IRD phenotype (23% as well as 13% late-onset STGD), followed by RP (23%), USH (8%), LCA (7%), and congenital stationary night blindness (CSNB - 4%) (Fig. 3B). Accordingly, ARRP and STGD show the highest expected GP (1 affected in 6,562 individuals and 1 in 6,578, respectively) followed by AR-USH (1 in 19,890 individuals). Alone, these three conditions are expected to affect almost 2.7 million individuals worldwide, out of an estimated total number of AR-IRD expected affected individuals of 5.5 million. It should be noted that the abovementioned values also include relatively young individuals who are currently healthy but are expected to develop an IRD later in life, based on their genotype.
Fig. 3.
Pie charts showing (A) the expected disease prevalence per gene worldwide and (B) the fraction of expected affected cases per diagnostic class out of all AR-IRD cases. ACHM, achromatopsia; SLS, Senior-Loken syndrome.
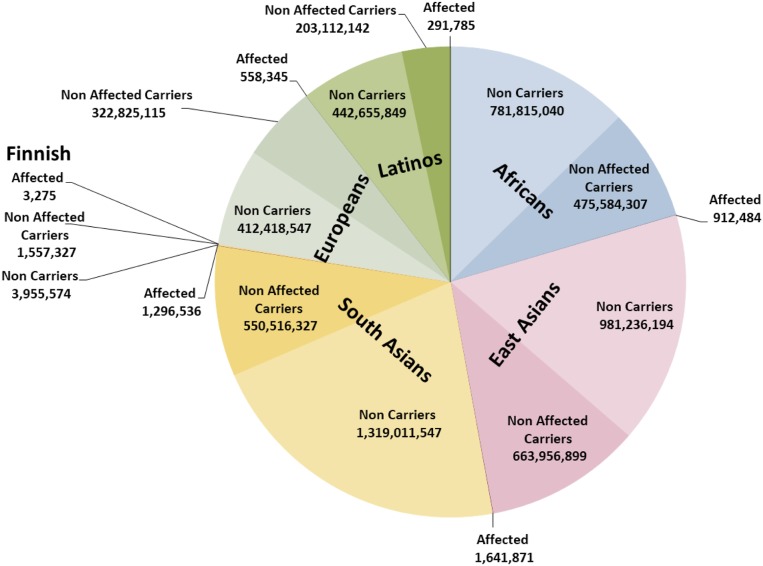
In the Asian populations, GP is the highest, i.e., 1 in 1,003 to 1,443, with ∼3 million expected affected individuals. Worldwide CF is expected to be 1 in 2.68 individuals, the highest value being from the European population (1 in 2.26 individuals). The distribution of IRD genotypes (Fig. 4, noncarriers in light color and carriers and affected in dark colors) in the different subpopulations was also calculated, and since more than half of the human population are of Asian origin, their contribution to the total number of individuals is the most significant.
Fig. 4.
Worldwide distribution of healthy WT individuals, healthy AR-IRD carriers, and expected affected individuals. The numbers displayed are the number of individuals in each group.
Finally, we inquired whether gnomAD contains individuals who are likely to be affected by AR-IRDs but are still asymptomatic or for whom their condition was not immediately recognized. We identified 56 gnomAD individuals who are homozygous for an AR-IRD–causing mutation (SI Appendix, Table S5), including congenital forms of IRDs (LCA and achromatopsia). The expected GP of homozygous AR-IRD in gnomAD is much lower, and only 7.9 homozygous individuals are expected in this group of individuals. A similar analysis on gnomAD individuals with possible compound heterozygous mutations is not possible, and therefore, we predict that the number of IRD affected individuals in gnomAD is even higher.
Discussion
The clinical and genetic heterogeneity of IRDs allows a unique opportunity to study various aspects of clinical genetics, including the assessment of the total CF and GP in different subpopulations. For example, RP is the most common reported IRD with an AR prevalence of 1:8,000 individuals (18–20) and 65 reported genes. If only a single mutation in a single gene had to cause RP, based on the Hardy–Weinberg equation, the total CF for ARRP can be calculated as 1:45 individuals. However, due to genetic heterogeneity, a much higher CF can be maintained in the human population. In 2002, we calculated a theoretical value for frequency of unaffected carriers for ARRP, based uniquely on disease prevalence and the predicted number of causing genes (21), leading to an estimated value of 0.18 (1:6 individuals). This oversimplified analysis was based on the assumption that every gene contributes equally to disease prevalence and that every gene would carry only one mutation. A subsequent analysis of 46 WGS control samples revealed that 22% (1:4.5 individuals) were heterozygous for truncating AR-IRD mutations alone (7). In the current study, we analyzed a much larger set of individuals (about 138,000) for truncating as well as other variants reported as pathogenic and a larger set of genes (187 genes), leading to a higher global CF value of 36% (or 1 out of 2.8 individuals). Since disease-causing mutations are identified in 50 to 75% of IRD patients (4, 5), this value is likely to rise as additional AR-IRD–causing mutations and genes are identified, and therefore we expect that about half of the human population worldwide might carry at least one AR-IRD mutation. Although global CF of other inherited diseases is still unknown, once similar studies are conducted on other phenotypes, the scientific community will be able to assess more precisely the number of pathogenic disease-causing mutations in each human genome for representative subpopulations.
The pipeline we developed allowed us to calculate two measures: frequency of unaffected carriers and GP of each AR-IRD phenotype in different subpopulations worldwide. The GP is the proportion of individuals in the population who are expected to be affected by their genotype and therefore might include relatively young individuals who are too young to be diagnosed with the relevant disease (excluding congenital forms of IRDs, such as achromatopsia and LCA). The analysis allowed filtering for a large number of variants (almost 280,000), mainly via computer-based procedures and, in rare cases, manual analysis. This scheme may be used to perform CF analyses and GP prevalence calculations in other conditions, either based on large-scale online databases such as gnomAD, ClinVar, and HGMD or including data from extended cohorts of patients. In addition, machine learning techniques (22) should be developed to better classify variants for pathogenicity, without the need for manual analysis of research data as published in scientific manuscripts. This will allow a global and comprehensive analysis of a large number of genes and diseases in humans and might provide important information regarding CF and GP for every inherited disease in various subpopulations worldwide, information that is highly important for accurate and meaningful genetic counseling.
The product analysis we performed revealed GP that is mostly correlated with reported disease prevalence values. For example, our analysis revealed that ABCA4 mutations result in a GP of STGD cases corresponding to 1:6,578 individuals, while STGD prevalence was previously estimated empirically to be 1:10,000 (23). Taking into account that the vast majority of STGD cases show recessive inheritance and that in about 75 to 85% of cases ABCA4 mutations are identified (24–26), the computed ABCA4-related AR-STGD GP is in line with the epidemiological data detected for this condition. Similarly, for RP the total prevalence was reported to be 1:4,000 (20, 27–29) with 50 to 60% of those being recessive (19) and causing mutations being identified in ∼65% of cases (30, 31). These data would lead to an expected ARRP prevalence of 1:9,850, compared to a calculated GP of 1:6,562 in this current study.
Among the 187 AR-IRD genes studied here, a single gene, ABCA4, had to be treated differently. First, ABCA4 mutations were reported to cause variable AR-IRD phenotypes, and although truncating variants are considered to result in a more widespread retinal phenotype (i.e., RP/CRD versus STGD), no clear genotype–phenotype is currently established. Moreover, at least 26 of the 358 ABCA4 mutations were reported to be hypomorphic and will lead to a retinal phenotype only when in trans with a severe mutation. Recent reports indicated that these variants usually result in a late-onset macular phenotype, and therefore, ABCA4-related disease prevalence might be much higher than current estimates. It should be noted here that this information is based mainly on variants identified in Europeans and North Americans, and therefore, many other such common and hypomorphic variants might exist in other populations, possibly in much higher numbers than those detected so far.
As expected, we detected a large variability in CF of specific mutations and genes among different subpopulations. We expect that our mutation database is relatively enriched for pathogenic variants identified in the European and North American populations, while disease-causing mutations (mainly nontruncating) that are frequent in other populations (e.g., Africans) but are not reported in the literature will rather be cataloged as variants of unknown significance. In addition, our GP calculation does not take into account features that are not compatible with the Hardy–Weinberg equation, such as, for instance, high levels of consanguinity and intracommunity marriages. Therefore, in specific subpopulations [e.g., some countries in Africa and South Asia (32)], GP might be higher than the values calculated here.
Moreover, our analysis revealed a surprising result regarding the number of gnomAD individuals who are expected to be affected with AR-IRDs. We identified an excess (of approximately sevenfold) of people with homozygous AR-IRD mutations, compared to their expected proportion among gnomAD individuals. We therefore predict that the gnomAD database includes individuals or a cohort of individuals affected by IRDs and possibly other Mendelian disorders, including congenital ones. This might lead to an overestimation of the allele frequency of pathogenic mutations, although the number of homozygous gnomAD individuals is small (56 out of 138,000), and their effect on allele frequency is not expected to affect dramatically our analysis. However, it should be noted that the number of expected affected individuals in gnomAD is theoretically even higher since subjects with compound heterozygous mutations could not be detected and were not included in the analysis. We therefore recommend using gnomAD data cautiously when searching for disease-causing mutations in WES and WGS analyses, avoiding drastic filtering on variants with homozygous gnomAD individuals. We predict that once machine learning techniques reach proven efficacy and are applied systematically in similar analyses, a much broader view of all inherited diseases will be available for the human genome.
In summary, we present here a comprehensive analysis of AR-IRD CF and GP, worldwide, as well as in major world subpopulations. The data presented show that the number of unaffected carriers of IRD recessive mutations could very well correspond to half of the entire human population, providing important information for genetic counseling, assessment of disease prevalence that is adapted to specific subpopulations, and prediction of pathogenic mutations in different ethnic groups.
Materials and Methods
Statistical Data.
Demographic data on the worldwide populations (in terms of the number of individuals per each subpopulation) are based on the Department of Economic and Social Affairs of the United Nations Secretariat 2017 Revision published on June 2017 (https://www.un.org/development/desa/publications/world-population-prospects-the-2017-revision.html) as follows: total worldwide population size, 7.6 billion; Africans, 1,258,311,831; East Asians, 1,646,834,964; South Asians, 1,870,824,410; Finnish, 5,516,175; European (non-Finnish), 735,802,007; Latino, 646,059,776; and other populations that are not represented in the gnomAD database (Oceania, 40,722,400; Northern America, 360,963,0043; West Asia, 268,093,436; Southeastern Asia, 649,200,488; and South-Central Asia, 1,941,780,322).
Assessment of Pathogenicity of Sequence Variants by Manual Analysis.
Although most of the filtering decisions regarding the pathogenicity of sequence variants was automated, for a small group of variants (Fig. 1) we applied a manual decision process to improve the assessment of pathogenicity for each sequence variant. The following groups were included in the manual analysis: truncating variants with allele frequency >0.005, truncating variants affecting the beginning or the end of the ORF, and nontruncating variants with contradicting predictions in HGMD versus ClinVar. We collected information about each of the variants from scientific literature, ClinVar, and HGMD regarding the following parameters (in order of importance): segregation analysis (was it performed? was the family size large enough?), availability of biochemical analyses (protein localization assays in cell cultures or biochemical assays) supporting pathogenicity, presence of the variant in patients vs. controls, and biallelically vs. monoallelically.
CF Analysis.
The gnomAD database provides the following values for each subpopulation: allele count (representing the number of detected alleles in a given subpopulation), allele number (total number of genotyped alleles at the genomic position of the variant considered), and homozygote count (total number of homozygous individuals for that specific allele). Based on these values, we calculated, as previously described in ref. 8, the following parameters: allele frequency, total number of individuals, number of heterozygous individuals, number of wild-type (WT) individuals, and CF. When CF was calculated for a specific mutation or gene, only individuals who are heterozygous for a single mutation in the studied gene were included, whereas homozygotes were excluded. It should be noted, however, that this CF value might include patients who are biallelic for mutations in another AR-IRD gene that causes the same phenotype as the studied gene.
Since unaffected carriers might harbor a heterozygous pathogenic mutation in more than one gene, we corrected the total CF for each subpopulation aiming to eliminate counting individuals more than once. To this end, we used two different calculation methods. The first method is based on the inclusion–exclusion principle, and the second is based on the independence probability theory as detailed below. Both calculation methods resulted in highly concordant values, and extremely mild differences were attributed to Python’s use of a limited number of decimal digits.
The inclusion–exclusion principle allows one to deduce overlapping elements within a given number of datasets. This principle is based on assessing the fraction of all carriers for any mutation in a specific gene and then calculating the fraction of individuals who are also carriers for a mutation in another gene; this value was excluded from the total CF value aiming to prevent a situation in which a particular carrier is considered twice. As described in SI Appendix, Fig. S1, every circle represents a group of carriers for a heterozygous mutation in a specific gene, and the overlapping areas represent carriers for mutations in more than one gene. However, while only 3 genes are represented in this example, the total number of AR-IRD genes in which mutations were found in the current analysis is 185, and therefore, a large number of possible overlapping areas is possible, intersecting at least 2 genes. By simply summing the carrier probabilities in all genes, carriers of multiple mutations are counted more than once, therefore increasing CF values. We therefore calculated the probabilities for double carriers among all 185 genes (by multiplying the probabilities of being a carrier in each gene) and exclude them from the total sum. This calculation takes all of the different possibilities of carriers in more than 1 gene, from carriers in 2 genes and carriers in the 185 genes. The following formula is used in this calculation:
The independence probability theory is based on the independent probability of individuals as carriers of mutations in different genes. The independence probability theory was calculated using the following formula:
where represents CF of gene i; represents the fraction of individuals who are not healthy carriers of mutations in gene i, and therefore, 1 minus the product of these values represents the cumulative frequency of heterozygous carriers for any IRD mutation, in any gene.
GP Calculations.
We calculated the GP of affected individuals for AR-IRDs in the worldwide subpopulations using a product-based algorithm for allele matrices. For each gene, we calculated the likelihood of two individuals who are carriers of an AR-IRD–causing mutation in the same gene to have an affected offspring. In order to calculate AR-IRD prevalence, we created a matrix of all of the possible genetic combinations of different mutations for each gene and multiplied the CF of the two mutations in each pair, including both homozygous and compound heterozygous combinations. We aggregated all of the sums of each multiplication based on the following equation:
The only exceptions were hypomorphic alleles (SI Appendix).
Data Availability.
All data are available in the manuscript.
Supplementary Material
Acknowledgments
We thank Segev Meyer for excellent help in cataloging IRD-related genes; Eyal Banin for fruitful discussions; and Liam Hanany, Yaniv Sadeh, and Adar Sharon for mathematical inputs. This study was funded by the Israeli Ministry of Health (grant number 3-12583), the Foundation Fighting Blindness (grant number BR-GR-0518-0734), and the Swiss National Science Foundation (grant number 176097).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.P.D. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913179117/-/DCSupplemental.
References
- 1.Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Erratum in: Nature412, 565 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Venter J. C., et al. , The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Tabor H. K., et al. ; NHLBI Exome Sequencing Project , Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: Implications for the return of incidental results. Am. J. Hum. Genet. 95, 183–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beryozkin A., et al. , Whole exome sequencing reveals mutations in known retinal disease genes in 33 out of 68 Israeli families with inherited retinopathies. Sci. Rep. 5, 13187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carss K. J., et al. ; NIHR-BioResource Rare Diseases Consortium , Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 100, 75–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karczewski K. J., et al. , Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv:10.1101/531210 (30 January 2019).
- 7.Nishiguchi K. M., Rivolta C., Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS One 7, e41902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanany M., et al. , Carrier frequency analysis of mutations causing autosomal-recessive-inherited retinal diseases in the Israeli population. Eur. J. Hum. Genet. 26, 1159–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu H.-Q., Wang X., Tian L., Hakonarson H., Application of ACMG criteria to classify variants in the human gene mutation database. J. Hum. Genet. 64, 1091–1095 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Rim J. H., et al. , Systematic evaluation of gene variants linked to hearing loss based on allele frequency threshold and filtering allele frequency. Sci. Rep. 9, 4583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S., et al. ; ACMG Laboratory Quality Assurance Committee , Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collison F. T., et al. , Clinical characterization of Stargardt disease patients with the p. N1868I ABCA4 mutation. Retina 39, 2311–2325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis S. S., et al. , In silico functional meta-analysis of 5,962 ABCA4 variants in 3,928 retinal dystrophy cases. Hum. Mutat. 38, 400–408 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zernant J., et al. , Extremely hypomorphic and severe deep intronic variants in the ABCA4 locus result in varying Stargardt disease phenotypes. Cold Spring Harb. Mol. Case Stud. 4, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaway C. S., Adams M. K. M., Jenkins K. S., Layton C. J., A novel ABCA4 mutation associated with a late-onset Stargardt disease phenotype: A hypomorphic allele? Case Rep. Ophthalmol. 8, 180–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zernant J., et al. , Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J. Med. Genet. 54, 404–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivolta C., Sweklo E. A., Berson E. L., Dryja T. P., Missense mutation in the USH2A gene: Association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet. 66, 1975–1978 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boughman J. A., Conneally P. M., Nance W. E., Population genetic studies of retinitis pigmentosa. Am. J. Hum. Genet. 32 223–235 (1980). [PMC free article] [PubMed] [Google Scholar]
- 19.Hartong D. T., Berson E. L., Dryja T. P., Retinitis pigmentosa. Lancet 368, 1795–1809 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Bunker C. H., Berson E. L., Bromley W. C., Hayes R. P., Roderick T. H., Prevalence of retinitis pigmentosa in Maine. Am. J. Ophthalmol. 97, 357–365 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Rivolta C., Sharon D., DeAngelis M. M., Dryja T. P., Retinitis pigmentosa and allied diseases: Numerous diseases, genes and inheritance patterns. Hum. Mol. Genet. 11, 1219–1227 (2002). Erratum in: Hum. Mol. Genet.12, 583–584 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Sundaram L., et al. , Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 50, 1161–1170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Hollander A. I., Black A., Bennett J., Cremers F. P. M., Lighting a candle in the dark: Advances in genetics and gene therapy of recessive retinal dystrophies. J. Clin. Invest. 120, 3042–3053 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassisi M., et al. , Expanding the mutation spectrum in ABCA4: Sixty novel disease causing variants and their associated phenotype in a large French Stargardt cohort. Int. J. Mol. Sci. 19, 1–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz H. L., et al. , Mutation spectrum of the ABCA4 gene in 335 Stargardt disease patients from a multicenter German cohort—Impact of selected deep intronic variants and common SNPs. Invest. Ophthalmol. Vis. Sci. 58, 394–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riveiro-Alvarez R., et al. , Outcome of ABCA4 disease-associated alleles in autosomal recessive retinal dystrophies: Retrospective analysis in 420 Spanish families. Ophthalmology 120, 2332–2337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bundey S., Crews S. J., A study of retinitis pigmentosa in the City of Birmingham. II Clinical and genetic heterogeneity. J. Med. Genet. 21, 421–428 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haim M., Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol. Scand. Suppl., 1–34 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Sharon D., Banin E., Nonsyndromic retinitis pigmentosa is highly prevalent in the Jerusalem region with a high frequency of founder mutations. Mol. Vis. 21, 783–792 (2015). [PMC free article] [PubMed] [Google Scholar]
- 30.Stone E. M., et al. , Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology 124, 1314–1331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H., et al. , Systematic evaluation of a targeted gene capture sequencing panel for molecular diagnosis of retinitis pigmentosa. PLoS One 13, e0185237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamamy H., Consanguineous marriages: Preconception consultation in primary health care settings. J. Community Genet. 3, 185–192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript.