Significance
Green seaweeds are important primary producers along coastlines worldwide. Their diversification played a key role in the evolution of animals. To understand their origin and diversification, we resolve key relationships among extant green algae using a phylotranscriptomic approach. A time-calibrated tree, inferred from available fossil data, reconstructs important evolutionary events, such as transitions to benthic environments and the evolution of macroscopic growth in the late Tonian/Cryogenian periods, followed by a marked Ordovician diversification of macroscopic forms. This ancient proliferation of green seaweeds likely modified shallow marine ecosystems, which set off an evolutionary arms race between ever larger seaweeds and grazers.
Keywords: green algae, Chlorophyta, phylogenomics, phylogeny, Ulvophyceae
Abstract
The Neoproterozoic Era records the transition from a largely bacterial to a predominantly eukaryotic phototrophic world, creating the foundation for the complex benthic ecosystems that have sustained Metazoa from the Ediacaran Period onward. This study focuses on the evolutionary origins of green seaweeds, which play an important ecological role in the benthos of modern sunlit oceans and likely played a crucial part in the evolution of early animals by structuring benthic habitats and providing novel niches. By applying a phylogenomic approach, we resolve deep relationships of the core Chlorophyta (Ulvophyceae or green seaweeds, and freshwater or terrestrial Chlorophyceae and Trebouxiophyceae) and unveil a rapid radiation of Chlorophyceae and the principal lineages of the Ulvophyceae late in the Neoproterozoic Era. Our time-calibrated tree points to an origin and early diversification of green seaweeds in the late Tonian and Cryogenian periods, an interval marked by two global glaciations with strong consequent changes in the amount of available marine benthic habitat. We hypothesize that unicellular and simple multicellular ancestors of green seaweeds survived these extreme climate events in isolated refugia, and diversified in benthic environments that became increasingly available as ice retreated. An increased supply of nutrients and biotic interactions, such as grazing pressure, likely triggered the independent evolution of macroscopic growth via different strategies, including true multicellularity, and multiple types of giant-celled forms.
Marine macroalgae or “seaweeds,” ecologically important primary producers in marine benthic ecosystems worldwide, have played a prominent role in the global biosphere for many millions of years. Seaweeds comprise red, green, and brown lineages, which evolved independently from unicellular algal ancestors. The red algae (Rhodophyta) are ancient. Bangiomorpha and Raffatazmia, both reasonably interpreted as red algal fossils, point to an origin of multicellular red algae well into the Mesoproterozoic, 1.0 to 1.6 billion y ago (1, 2). In contrast, brown algae (Phaeophyceae) emerged much more recently, with molecular clocks pointing to an Early Jurassic origin (3, 4). As sister to red algae, the green lineage (Viridiplantae), possibly along with the unicellular glaucophytes (5), likely originated in Neo- or Mesoproterozoic environments (6, 7). The green lineage comprises microscopic and macroscopic forms and displays the richest repertoire of cytological organizations within all seaweeds, ranging from uninucleate or multinucleate multicellular organisms to siphonous (acellular) seaweeds, organisms constituted by a single giant cell with thousands to millions of nuclei that can undergo subcellular morphological and functional differentiation. Despite the intriguing variety of cytological organization, the lack of a well-supported phylogenetic framework has so far impeded clear interpretation of how many times and when green seaweeds emerged from unicellular ancestors (8).
There is general consensus that an early split in the evolution of the green lineage gave rise to two discrete clades. One, the Streptophyta, contains a wide morphological diversity of green algae, also known as charophytes, which occur in freshwater, damp terrestrial, and—in a few cases—inland saline aquatic habitats, as well as the land plants that evolved from ancestral charophytes during the Ordovician Period (9, 10). The second clade, the Chlorophyta, diversified as planktonic unicellular organisms, likely in both freshwater and marine habitats during the late Mesoproterozoic and early Neoproterozoic (11, 12). These ancestral chlorophyte green algae gave rise to several extant lineages of unicellular planktonic marine and freshwater algae, known as the prasinophytes, as well as the core Chlorophyta, which radiated in freshwater, terrestrial, and coastal environments and evolved a wide diversity of forms, ranging from microscopic unicellular and multicellular algae to macroscopic forms (8). Two large core chlorophytan classes, Chlorophyceae and Trebouxiophyceae, are almost entirely restricted to freshwater and terrestrial environments. In contrast, the Ulvophyceae contains the main green seaweed lineages in addition to some smaller microscopic clades, some freshwater species, and the terrestrial Trentepohliales (13, 14). Only two other groups of green seaweeds are known to have phylogenetic affinities outside the Ulvophyceae: The Prasiolales, which belong to the Trebouxiophyceae, and the Palmophyllales, which are allied to prasinophytes (8, 15). These groups are much less diverse compared to the ulvophycean lineages, and clearly evolved independently.
The diversification of Ulvophyceae in marine benthic environments involved the evolution of an astonishing diversity of forms, most strikingly the evolution of macroscopic, benthic growth-forms from small, planktonic unicellular ancestors. Macroscopic growth in green seaweeds presents itself in various forms, ranging from multicellular thalli to different types of giant-celled algae with highly specialized cellular and physiological characteristics (16). About 10 extant ulvophycean orders are currently recognized, each characterized by a distinctive set of morphological and cytological features (14). Some orders (e.g., Ulvales and Ulotrichales) evolved multicellularity with coupled mitosis and cytokinesis, resulting in uninucleate cells. The Cladophorales evolved siphonocladous multicellular algae, in which mitosis is uncoupled from cytokinesis, resulting in large multinucleate cells with nuclei organized in fixed cytoplasmic domains. The Dasycladales and Bryopsidales evolved siphonous (acellular) macroscopic forms composed of a single giant tubular cell containing thousands to millions of nuclei, or a single macronucleus, and exhibiting cytoplasmic streaming, which enables transport of RNA transcripts, organelles, and nutrients throughout the thallus. Some siphonous algae, including species of Caulerpa, reach meters in size, thus qualifying as the largest known cells. Although the available data are limited, it seems that acellular algae are similar to comparable multicellular algae with regard to maximum photosynthetic and nutrient acquisition rates, light absorbance, and ability to growth at low irradiances (17), and it has been speculated that the acellular morphology may be an adaptation to shading caused by epiphytes because it facilitates the rapid formation of new photosynthetic surfaces (18). The smaller, nonseaweed orders (e.g., Ignatiales, Scotinosphaerales, and Oltmannsiellopsidales), are morphologically less complex, and they grow as microscopic unicellular forms with uninucleate cells.
Understanding the origin and ecological diversification of green seaweeds requires a well-resolved phylogeny of the core Chlorophyta, and reliable estimates of the timing of inferred diversification events. Early studies based on ultrastructural features, such as the fine structure of the flagellar apparatus, cytokinesis, and mitosis have been instrumental in defining higher-level groupings of green algae, but have been inconclusive in determining the relationships among these groups (8) or estimating the timing of their diversification. In addition, monophyly of the Ulvophyceae has been questioned because of the absence of shared derived characters (8). Early molecular phylogenetic studies based on nuclear ribosomal DNA sequences were not able to resolve relationships among the main core chlorophytan lineages (8). A 10-gene phylogenetic analysis of Cocquyt et al. (14) was the first to recover the Ulvophyceae as a well-supported monophyletic group and to resolve relationships among its main clades, providing a framework for interpreting the cytological and morphological evolution in the green seaweeds. Macroscopic growth was hypothesized to have originated at least four times independently within the Ulvophyceae from marine unicellular ancestors, by different mechanisms: By developing multicellularity with coupled mitosis and cytokinesis, by developing multicellularity with uncoupled mitosis and cytokinesis, or by developing a siphonous architecture (14). Conversely, chloroplast phylogenomic analyses generally did not support monophyly of the Ulvophyceae (19–22), indicating multiple independent origins of green seaweeds within the core Chlorophyta. These conflicting studies make it clear that resolving relationships within the core Chlorophyta is a difficult task, which can be attributed to the antiquity of the clade and possibly further confounded by the rapidity of the early evolutionary radiations (14, 21). An accurate phylogenetic reconstruction will thus require more elaborate sampling, both in terms of species and genes.
Dating divergence times in the phylogeny of the Chlorophyta has been challenging because of difficulties in interpreting fossils with respect to extant taxa. Microfossils in Paleo- to Neoproterozoic rocks have sometimes been assigned to green algae (23–25), but these interpretations are uncertain as they rely on comparisons of simple morphologies (26). Similarly, the assignment of the middle Neoproterozoic filamentous fossil Proterocladus (ca. 750 Mya) to the Cladophorales (27, 28) is questioned (3, 29). Reliable chlorophytan fossils include resistant outer walls of prasinophyte cysts known as phycomata in Ediacaran and Paleozoic deposits (30, 31) and fossils of siphonous seaweeds (Bryopsidales, Dasycladales) from the Cambro-Ordovician onward (32–36). Although reliable green algal fossils from the Neoproterozoic are scarce, organic biomarkers (steroids) indicate that green algae were present and persisted through the Cryogenian Period, and rose to ecological prominence between the Sturtian and Marinoan glaciations (659 to 645 Mya) (37–39).
The principal goals of our study were to examine the monophyly of, and resolve evolutionary relationships among the main lineages of core Chlorophyta, and to reconstruct key evolutionary events, such as transitions to benthic marine environments and the evolution of macroscopic growth. We use a rigorous phylotranscriptomic approach, thereby increasing nuclear gene sampling by an order-of-magnitude, and produce a tree calibrated in geological time with available fossil data. The phylogenetic results enable us to formulate hypotheses regarding the ecological and evolutionary context of green seaweed origins.
Results
Transcriptome Data.
We collected and analyzed nuclear encoded protein-coding genes from 55 species mined from 15 genomes and 40 transcriptomes (SI Appendix, Tables S1 and S2). Thirteen transcriptomes were generated during this study (40, 41). Our dataset includes representatives from the major lineages of Streptophyta and Chlorophyta. A denser taxon sampling of the core Chlorophyta, including all main orders of Ulvophyceae (Bryopsidales, Cladophorales, Dasycladales, Ignatiales, Oltmannsiellopsidales, Scotinosphaerales, Trentepohliales, Ulotrichales and Ulvales), representative of all main morphological and cytological types found in this class, was obtained to consolidate evolutionary relationships in this group and advance our understanding of the origin and diversification of green seaweeds.
Eight sequence alignments were assembled for phylogenetic analyses (41) (SI Appendix, Table S3). The largest alignment consisted of 539 high-confidence single-copy genes (hereafter referred to as coreGF) (SI Appendix, Material and Methods). From this dataset a subset of 355 genes was selected, with at least one sequence in each ulvophycean order (hereafter referred to as ulvoGF). Alignments were manually curated in an orthology-guided approach to retain only orthologous single-copy genes for downstream phylogenetic analyses (Materials and Methods). Residual overlapping and nonoverlapping partial sequences from the transcriptomes, corresponding to genes not containing a complete open-reading frame, were checked for concordant phylogenetic signal. Sequences with concordant signal were either scaffolded (scaffolded dataset) or removed (unscaffolded dataset), resulting in a more comprehensive and a more conservative version of the coreGF and ulvoGF datasets (SI Appendix, Fig. S1). Finally, ambiguously aligned regions were removed to obtain the corresponding trimmed datasets. The eight datasets were analyzed with a range of phylogenetic methods, including supermatrix and the coalescence-based analyses. The significance of conflicting topologies was tested to assess the robustness of our findings.
Green Algal Phylogeny.
Trees estimated by the various methods and datasets were well-supported and congruent, although different topologies were recovered for a few specific relationships. Most differences were due to analysis method (i.e., supermatrix versus coalescence-based) rather than the dataset used (SI Appendix, Fig. S4). Because both methods have their analytical advantages and limitations (e.g., supermatrix analyses are based on large datasets and can use complex models that can account for the heterogeneity of the substitution process, but cannot account for gene-tree species-tree incongruence, while coalescence-based approaches accommodate incomplete lineage sorting, but rely on topologies inferred from small datasets using simpler models that make them more sensitive to stochastic noise) (42, 43), the topologies from the two analyses are displayed in Figs. 1 and 2A and SI Appendix, Fig. S3. Chlorodendrophyceae and Pedinophyceae were recovered as the two earliest diverging lineages of the core Chlorophyta, although their relative position differed in the different analyses (Fig. 1 and SI Appendix, Figs. S4 and S5 and Table S4). The Trebouxiophyceae and Chlorophyceae were recovered as monophyletic groups with high support, with the Trebouxiophyceae consisting of two distinct clades, the Chlorellales and the core Trebouxiophyceae. The Trebouxiophyceae were recovered sister to the clade containing the Chlorophyceae and Ulvophyceae in all analyses.
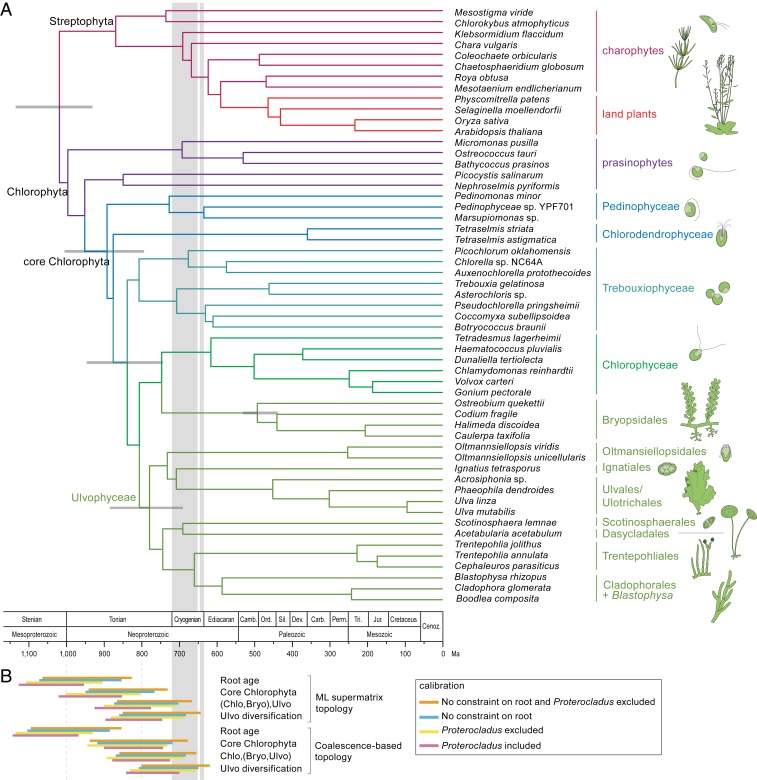
Fig. 1.
Time-calibrated phylogeny of the green algae. (A) The topology of the tree is based on the ML analysis inferred from a concatenated amino acid alignment of 539 nuclear genes (supermatrix analysis of the coreGF scaffolded untrimmed dataset) (SI Appendix, Fig. S2). Branch lengths are based on a relaxed molecular clock analysis of the 10 most clock-like genes from the scaffold trimmed dataset, and excluding Proterocladus as calibration point. Error bars are indicated for a number of key nodes. (B) Divergence time confidence intervals of key nodes inferred from different analyses (SI Appendix, Fig. S9 and Table S7). Vertical gray bands indicate Sturtian (716 to 659 Mya) and Marinoan (645 to 635 Mya) glaciations. Bryo, Bryopsidales; Chlo, Chlorophyceae; Ulvo, Ulvophyceae (excluding Bryopsidales).
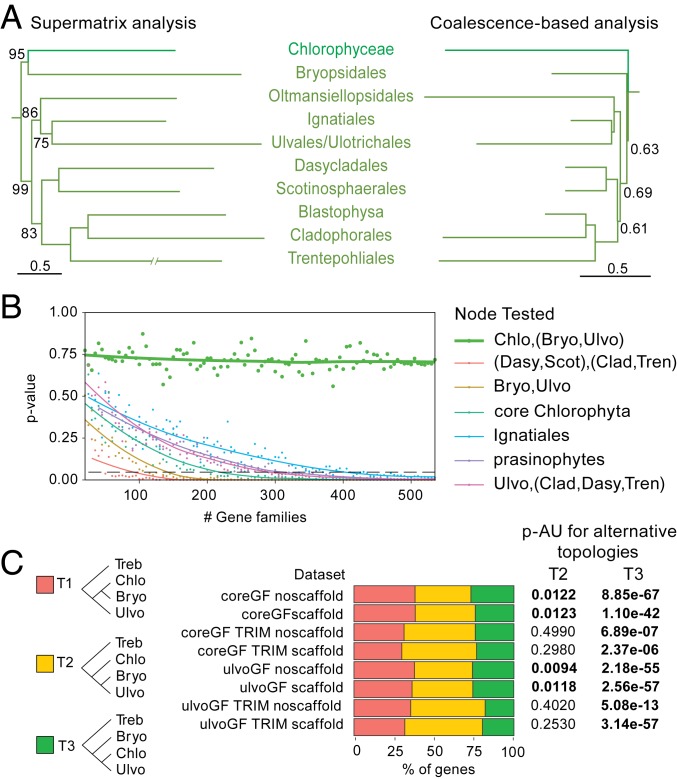
Fig. 2.
Relationships between Bryopsidales, remaining Ulvophyceae, and Chlorophyceae. (A) Phylogeny as recovered by the supermatrix (SI Appendix, Fig. S2) and coalescence-based analysis. Only the Chlorophyceae-Ulvophyceae clade is shown, and the tree is pruned down to one taxon per lineage. (B) Test for polytomy null-hypothesis for selected relationships based on the coreGF scaffold trimmed dataset (see Materials and Methods and SI Appendix, Fig. S7 for details). For the Chlorophyceae, Bryopsidales, and remaining Ulvophyceae (thick green line), increasing gene numbers did not reduce the P value of rejection. (C) Proportion of genes supporting a sister relationship between Bryopsidales and Chlorophyceae (T1), a sister relationship between the Bryopsidales and remaining Ulvophyceae (T2), and a sister relationship between Chlorophyceae and Ulvophyceae (Bryopsidales excluded) (T3); and summary of the AU test for the constrained topologies (T2 and T3) (see SI Appendix, Table S4 for details). Bryo, Bryopsidales; Chlo, Chlorophyceae; Clad, Cladophorales+Blastophysa; Dasy, Dasycladales; Scot, Scotinosphaerales; Treb, Trebouxiophyceae; Tren, Trentepohliales; Ulvo, Ulvophyceae (excluding Bryopsidales).
The siphonous seaweed order Bryopsidales was resolved as the sister clade of the Chlorophyceae, rendering the Ulvophyceae nonmonophyletic, in the supermatrix analyses (Figs. 1 and 2A and SI Appendix, Fig. S2). Conversely, in the coalescent-based analyses, the Bryopsidales was sister to the remaining Ulvophyceae, with very short branches separating the Bryopsidales, remaining Ulvophyceae, and Chlorophyceae (Fig. 2A and SI Appendix, Fig. S3). A polytomy test could not reject the null hypothesis that the length of the branches in question equals zero, indicating a hard polytomy (Fig. 2B and SI Appendix, Fig. S7). In addition, approximately unbiased (AU) tests for the trimmed datasets could not reject the topology constrained to conform the coalescence-based analyses (Ulvophyceae monophyletic). For the untrimmed datasets, however, Ulvophyceae monophyly was significantly rejected (SI Appendix, Table S4). Analysis of the gene-wise log-likelihood showed that the majority of the genes supported the monophyly of Ulvophyceae for the trimmed datasets (Fig. 2C). A third topology in which Chlorophyceae are sister to Ulvophyceae excluding Bryopsidales, which would be compatible with a pure coalescent model, was not recovered in any of our phylogenetic trees. AU tests consistently rejected this topology, although it was supported by a relatively small proportion of the genes (Fig. 2C and SI Appendix, Table S4).
The supermatrix and coalescence-based analyses supported the same overall relationships among the remaining orders of Ulvophyceae. Two major clades were recovered, both containing seaweed and unicellular lineages. The position of the Ignatiales was not well-supported in any of the phylogenetic analyses, and similar numbers of genes supported five different relationships (SI Appendix, Fig. S5 and Table S4). The inclusion of Ignatius was also found to cause instability in the phylogenetic analyses, and removing it led to an overall higher support for the relationships among the main ulvophycean clades (SI Appendix, Fig. S6).
Time-Calibrated Phylogeny.
To estimate the timeframe of diversification, we inferred chronograms based on two sets of the 10 most clock-like genes, as assessed against the supermatrix and the coalescence-based topologies, using different molecular clock models, and constraining the analyses with supermatrix- and coalescence-based topologies (41). The choice of calibration points largely follows that of Jackson et al. (7). A series of analyses was performed in order to evaluate the effect of calibration nodes (SI Appendix, Table S6 and Fig. S9). Results indicate that the core Chlorophyta emerged during the Neoproterozoic Era, approximately 1,000 to 700 Mya (Fig. 1B and SI Appendix, Table S7). Conditional on the calibration points, diversification of the main ulvophycean lineages took place just before or during the Cryogenian.
Transition to Marine Benthic Habitats and Evolution of Macroscopic Growth.
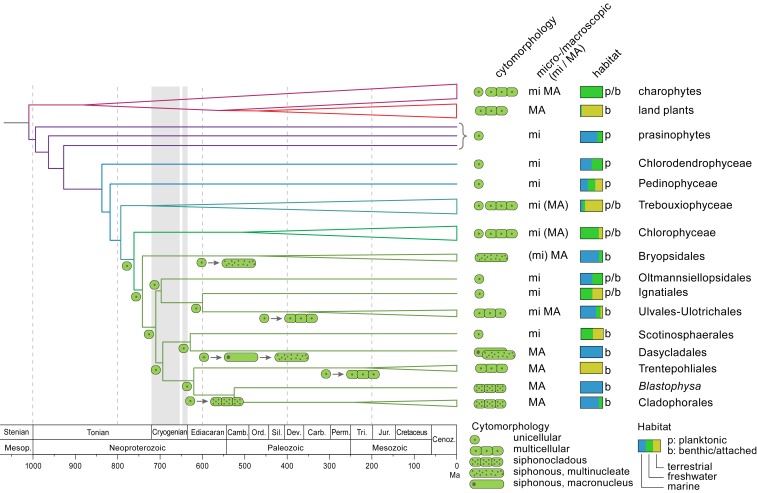
Our statistical inference of the ancestral states of key ecological and cyto-morphological traits (SI Appendix, Fig. S10) indicates that the Trebouxiophyceae and Chlorophyceae diversified mainly in freshwater environments. The radiation event at the base of the diversification of Chlorophyceae, Bryopsidales, and Ulvophyceae is associated with a major switch back to marine environments early in the evolution of Bryopsidales and Ulvophyceae, populating coastal environments with marine benthic green seaweeds.
Discussion
We present a phylogenetic reconstruction of the core Chlorophyta based on the most comprehensive multigene dataset to date, and contextualize the relationships in light of transitions to marine benthic environments and evolution of multicellularity and macroscopic growth (Fig. 3). Although this study focused on the evolution of the Chlorophyta, the relationships among the main lineages of Streptophyta are consistent with current phylogenetic consensus, validating the strength of our phylotranscriptomic approach in confidently resolving difficult phylogenetic relationships, such as identifying the closest living relative of the land plants (44, 45). Our analyses mark a significant change to current views on chlorophyte evolution, and resolve some long-standing phylogenetic questions within the Chlorophyta, including the evolutionary placement of the unicellular Pedinophyceae and Chlorodendrophyceae as the earliest diverging lineages of the core Chlorophyta, and monophyly of the Trebouxiophyceae (SI Appendix, Phylogenetic relationships within the core Chlorophyta). Molecular phylogenetic studies based on nuclear and chloroplast gene data have yielded ambivalent or contradictory results regarding relationships among the main core chlorophytan lineages, Trebouxiophyceae, Chlorophyceae, and Ulvophyceae (reviewed in refs. 46 and 47). Our analyses suggest a hard polytomy between the Chlorophyceae, siphonous seaweeds of the Bryopsidales, and the remaining Ulvophyceae. Roughly equal numbers of genes supported alternative topologies, indicative of ancient incomplete lineage sorting or a nonbifurcating evolutionary history (43, 48). Although coalescence-based analyses have the ability to detect and incorporate conflicting signals in gene-trees in phylogenetic reconstruction, and supermatrix analyses may converge on a wrong species-tree when incomplete lineage sorting is high (43, 49), we hypothesize that these lineages represent a rapid ancient radiation.
Fig. 3.
Hypothesis for the evolution of multicellularity and macroscopic growth, and transition to benthic marine habitats in the Ulvophyceae. The topology of the tree is based on the coalescence-based analysis of the coreGF scaffolded untrimmed dataset. Branch lengths are based on a relaxed molecular clock analysis of the 10 most clock-like genes from the coreGF scaffolded trimmed dataset, and excluding Proterocladus as calibration point. Vertical gray bands indicate Sturtian (716 to 659 Mya) and Marinoan (645 to 635 Mya) glaciations.
Phylogenetic uncertainty in the core Chlorophyta has hampered the reconstruction of key evolutionary events, such as transitions to benthic marine environments and the evolution of macroscopic growth. Our phylogenetic results allow us to propose a scenario for the evolutionary history of green seaweeds. Given that several early-branching ulvophycean lineages consist of unicellular algae (including the Ignatiales, Oltmansiellopsidales, Scotinosphaerales, and some lineages of Ulotrichales and Ulvales), the ancestral ulvophycean was likely a small unicellular organism with a single nucleus. Our phylogeny affirms the hypothesis by Cocquyt et al. (14) that macroscopic growth evolved independently in various lineages of ulvophyceans from ancestral unicellular green algae, and by different mechanisms: Multicellularity with coupled mitosis and cytokinesis (independently in the Ulvales-Ulotrichales and Trentepohliales), multicellularity with uncoupled mitosis and cytokinesis (siphonocladous organization in the Cladophorales and Blastophysa), and a siphonous architecture (independently in the Bryopsidales and Dasycladales). The sister relationship of mainly marine Cladophorales (plus Blastophysa) and strictly terrestrial Trentepohliales may indicate a single origin of multicellularity at the base of this clade, followed by the evolution of multinuclear cells in the Cladophorales. However, the phragmoplast-mediated cell division in Trentepohliales, producing plasmodesmata, differs strongly from cell division in the Cladophorales, which takes place by ingrowth of a diaphragm-like cross wall (8). It may therefore be more reasonable to think that multicellularity evolved independently in the Trentepohliales and the Cladophorales in response to different environmental pressures in terrestrial and marine/freshwater habitats. Contrary to previous phylogenetic studies, which indicated a sister relationship between Bryopsidales and Dasycladales (14, 19), the separate positions of these orders point toward an independent evolution of siphonous organization in Bryopsidales and Dasycladales from ancestral unicellular green algae. The distinct position of the Bryopsidales is unexpected from a morphological view point, but is supported by ultrastructural features, as well as by independent molecular data, such as the presence of an alternative nuclear genetic code (SI Appendix, Phylogenetic relationships within the core Chlorophyta). The Trentepohliales, Cladophorales, Dasycladales, and Scotinosphaerales are characterized by a noncanonical nuclear genetic code while the Bryopsidales and all other green algae possess the canonical code (50), pointing toward a single origin of a noncanonical nuclear code in the green algae.
Our relaxed molecular clock analyses indicate that the early diversification of core Chlorophyta took place in the Tonian Period, with several green algal lineages persisting throughout the Cryogenian ice ages. The early diversification of ulvophycean lineages took place during this interval as well. Diversification before and survival during the Cryogenian glacial interval has been inferred for other groups of eukaryotes based on fossil evidence, including complex forms with blade-stipe-holdfast differentiation that have been interpreted as benthic macroalgae (51–53). It is important to note, however, that the scarce fossil record and the uncertain nature of ancient green algal fossils, along with methodological bias, impose a limit on the precision of ancient divergence time estimates (54). These unavoidable uncertainties in node age assignments have to be accommodated in our interpretations, and therefore one cannot dismiss the hypothesis that green seaweeds began to diversify earlier, in the middle or even early Tonian Period. One concrete, well-dated fossil that has been put forward as important to estimate the age of the Ulvophyceae is Proterocladus, from approximately 780-Mya shales of the upper Svanbergfjellet Formation in Svalbard, originally interpreted as a Cladophora-like alga (28). Proterocladus has typical branches that are in cytoplasmic contact with the parent cell, which is as a derived feature in the Cladophorales (55). Assignment of Proterocladus to the Cladophorales would imply that the order would be older than 780 My, and would push the divergences of the main ulvophycean lineages even further back in time, inconsistent with our time-calibrated phylogeny, which suggests an Ediacaran or Cambrian origin of the Cladophorales. While Proterocladus is certainly eukaryotic, and appears to be coenocytic, we prefer to consider its taxonomic affinity as uncertain based on the late appearance of Cladophorales in our molecular clock analyses.
Ulvophycean diversification could have been promoted by the highly dynamic climatic conditions and habitat variability of the Cryogenian periods (56, 57). Little is known about the paleontological conditions of these periods that would have allowed lineage diversification and survival, but a number of different scenarios can be put forward. The later Tonian Period was a time of evolutionary innovation among both photosynthetic and heterotrophic eukaryotes in a number of major eukaryotic clades (57); thus, inferred divergence of green seaweeds would appear to be part of a broader pattern of Tonian diversification. This has sometimes been related to an increase in atmospheric oxygen levels (e.g., ref. 57), but evidence in support of this hypothesis is mixed, suggesting that any redox change was small relative to later Ediacaran oxygenation (e.g., ref. 58). Alternatively, evolutionary drivers may have included the expansion of eukaryovorous protists in the oceans, supported by both fossils and molecular clock estimates for major eukaryote-eating protistan lineages (59, 60). Experiments show that predation can select for multicellularity in originally unicellular prey populations (61, 62), consistent with the observed and inferred Tonian diversification of multicellular and coenocytic forms, as well as scales and other protistan armor (57, 60).
As already noted, the ensuing Cryogenian Period was characterized by two widespread glaciations, the older, protracted Sturtian ice age and the younger and shorter Marinoan glaciation (56). Among the most extreme climatic events in recorded Earth history, these ice ages involved the freezing of large parts of the ocean surface for millions of years and dramatic fluctuations in biogeochemical cycling. Snowball Earth conditions were certainly unfavorable for planktonic eukaryotes, resulting in greatly diminished photosynthesis in the marine realm. The fossil record is consistent with this notion, showing low overall eukaryotic diversity during this interval (63). An important key to the success and diversification of ulvophycean green algae during the Cryogenian, thus may have been an evolutionary transition from a planktonic to benthic lifestyle, a shared feature of all extant ulvophycean lineages. During glaciated conditions, benthic environments may have been among the few suitable habitats left for green algal lineages to persist, and diversification may have been promoted in different ways. First, the Sturtian glaciation (716 to 659 Mya) lasted a long time, but shows evidence of glacial waxing and waning, resulting in secular variation in suitable habitat availability (56). During this period, when colonizable benthic substrates would have oscillated between rare and uncommon, ulvophycean populations likely experienced prolonged isolation and demographic declines that would amplify the effects of genetic drift. We hypothesize that, in concert with local adaptation, this has promoted early diversification of ulvophycean lineages. Macroscopic compressions of probable algae in shales within the Marinoan Nantuo Tillite demonstrate that macroalgae could and did survive at least locally during times when ice was widespread (53). As ice sheets decayed, habitat space would have increased dramatically, perhaps helped along by a transient increase in nutrients (especially P) associated with high postglacial weathering fluxes. Arguably, the emergence of new ulvophycean lineages could have been rapid in this new permissive ecology, where competition for newly available habitats would have been low, allowing a broader suite of mutations to persist (64).
We hypothesize that following the transition to a greenhouse world at the end of the Marinoan glaciation, surviving lineages would have been able to diversify, and new morphological experiments ensued, resulting in the different types of macroscopic growth, such as uniseriate and branched filaments, sheets, and different morphologies of siphonous forms. The rapid diversification of eukaryotic life in the early Ediacaran Period is evidenced by molecular biomarkers, which document the rise to global ecological prominence of green algal phytoplankton (39), as well as the diversification of macroscopic seaweeds [both florideophyte reds and greens (65, 66)] and animals (67–69). The transition from predominantly cyanobacteria to green phytoplankton has been hypothesized to reflect increasing nutrient availability, a conjecture supported by a state change in the abundance of phosphorite deposits during the Ediacaran Period (70). Concomitant increase in oxygen availability, recorded by multiple geochemical signatures (58) fulfills another prediction of increased productivity.
The combination of increasing oxygen and food supply likely facilitated the radiation of animals (37, 71), and metazoan evolution, in turn, fed back onto seaweed diversification. Indeed, the Paleozoic evolution of multicellularity and macroscopic growth in ulvophyceans may have been triggered by the introduction of novel grazing pressures by animals in the late Cambrian and Ordovician (33, 69). This may have started an evolutionary arms race between grazers and algae, which would have resulted in increasingly complex feeding and defense strategies, such as larger thalli, larger cells, and mineralized skeletons (52, 62). The relatively late appearance of large grazing animals in the late Cambrian and Ordovician, such as jawed polychaetes (33, 69), would explain the considerable time lag between the origin of ulvophyceans, and further diversification and origin of crown group ulvophyceans. This scenario is consistent with the predation hypothesis of Stanley (72), who postulated that grazing by benthic animals resulted in a mutual feedback system that drove increased diversity of both macroalgae and animals. The spread of calcium carbonate skeletons in both red and green algae is also consistent with the predation hypothesis. Macroscopic growth may additionally have been facilitated by other factors, including competition for space in benthic habitats, pressure for upright growth to overshadow benthic unicellular algae, a means of increasing nutrient uptake from the water column, or a means of spreading reproductive propagules more widely. In turn, the proliferation of green macroalgae may have resulted in more efficient energy transfer, richer food webs, and the modified shallow marine ecosystems, which may have allowed the evolution of larger and more complex animals (33, 73).
The long-term isolation of ulvophycean lineages in Cryogenian refugia could explain the independent evolution of macroscopic growth in the different clades using radically different mechanisms, as inferred from our phylogeny. Our phylogenetic results, combined with the fossil record indicates that there may have been a time lag of 100 My or more after the early diversification of ulvophyceans where these microbial ulvophyceans persisted, but never became dominant, followed by transitions to macroscopic growth, which may have occurred in different periods in the different lineages. Our analyses indicate that macroscopic crown group ulvophyceans may have originated between the early Paleozoic (e.g., Bryopsidales) and early Mesozoic (e.g., Cladophorales). This hypothesis is supported by the fossil record, which shows that seaweed morphogroups of the Ediacaran remained largely unchanged during the Cambrian, but that there was a major replacement of this early seaweed flora concomitant with the Great Ordovician Biodiversification Event (33). These observations are also consistent with the fact that the earliest fossils that can be reliably linked to extant ulvophycean clades (Bryopsidales, Dasycladales, and Ulotrichales) are only found from the Ordovician onward (but see discussioin of Proterocladus above); most (but not all) have calcium carbonate skeletons.
In conclusion, our phylogenetic analysis of the green algae, based on a comprehensive nuclear gene dataset, provides a general picture on the evolution of green seaweeds in which the origin and early diversification of the ulvophyceans likely took place late in the Tonian and Cryogenian, followed by a marked Ordovician diversification of macroscopic crown group taxa, including those that produce calcium carbonate skeletons and are well preserved in the fossil record. That this trajectory is similar to that of animals reminds us not to forget these organisms when we think about the environmental circumstances of early green algal evolution. Finally, our analyses provide a phylogenetic framework to study the evolution of the genetic toolkit for multicellularity and macroscopic growth in green seaweeds, a group that has been largely neglected in studies of large-scale gene discovery (74).
Materials and Methods
Dataset Retrieval, RNA Extraction, and Sequencing.
DNA sequence data were mined from 15 genomes and 40 transcriptomes; 13 transcriptomes were generated for this study based on cultured strains or freshly collected specimens (SI Appendix, Material and Methods), while the remaining data were retrieved from publicly available repositories (SI Appendix, Table S1). RNA extractions follow Palmer (75). RNA quality and quantity were assessed with Qubit and Nanodrop spectrophotometer, and integrity was assessed with a Bioanalyzer 2100. RNA-sequencing libraries were sequenced as reported in SI Appendix, Table S2.
Transcriptome Assembly, Frameshift Error Correction, and ORF Detection.
At the time of the experiment, preassembled transcriptomes were only available for Acrosiphonia sp., Blastophysa rhizopus, and Caulerpa taxifolia (SI Appendix, Table S1). All remaining assemblies were performed in house, starting from the raw reads using a custom semiautomated pipeline (SI Appendix, Material and Methods). For each of the 40 transcriptomes, transcripts were clustered with CD-HIT-EST v4.6.1 (76) with a similarity cutoff of 97.5%, and only the longest transcript was retained for downstream analysis as representative of the cluster. After taxonomic profiling of the transcripts (SI Appendix, Material and Methods), only eukaryotic transcripts were retained for downstream analysis, bacterial transcripts, and transcripts lacking sequence similarity to known proteins were discarded.
Transcripts with putative frameshift errors were identified after initial processing in TRAPID (77), and transcripts carrying a putative frameshift error were corrected with FrameDP 1.2.2 (78), using the Chlamydomonas proteome as reference to guide the frameshift correction step (SI Appendix, Material and Methods). Each transcript was translated into the corresponding amino acid sequences with the transeq algorithm from the EMBOSS package, using the appropriate translation table, and added to the proteome data of the 15 genomes, resulting in 1,228,821-amino acid sequences.
Gene Family Inference.
Sequences were used to build a custom PLAZA 4.0 instance (79), and single-copy families were selected by identifying the 620 picoPLAZA single-copy genes (80) (SI Appendix, Material and Methods). To remove potential contaminants in the transcriptomic data from the single-copy gene families, only sequences that were classified as “Viridiplantae” after an additional sequence similarity search with the GhostKOALA (81) webserver were retained for downstream analyses. To further reduce the residual redundancy of the transcriptome datasets in the single-copy gene families, for each gene family the nucleotide sequences of each species were collapsed with CAP3 (82), using stringent parameters to avoid artifactual creation of chimeras: Gap penalty 12 (-g) and overlap percent identity cutoff 98% (-p). This set of 620 quasi–single-copy genes was used for the downstream phylogenetic analyses.
Sequence Aligments and Filtering.
Amino acid sequences of the 620 single-copy genes were aligned with MAFFT v7.187 (83), using accuracy-oriented parameters (–localpair–maxiterate 1,000) and an offset value (–ep) of 0.075. To identify and trim eventual residual in-paralogs, we followed a phylogeny-guided approach, in which alignments were manually cured to retain only full length or fragments of orthologous single-copy genes (SI Appendix, Material and Methods). After filtering, 539 high-confidence single-copy genes of the 620 initial quasi–single-copy genes were retained (referred to as coreGF). The coreGF dataset formed the basis for the construction of seven derived datasets based on different filtering approaches (SI Appendix, Material and Methods and Table S3).
Phylogenetic Analysis.
The eight datasets were analyzed using a supermatrix and a coalescence-based phylogenetic approach. Maximum-likelihood (ML) supermatrix analyses were performed using IQtree with two settings: 1) A gene-wise partitioned analysis (84) was performed, assigning the best substitution model inferred to each partition; 2) an analysis using mixture models was performed using an LG+F+G plus a C20-profile mixture model of substitution rates (85). To estimate the best substitution model of each partition, ML trees were built with IQtree for each single-copy gene, inferring the best model and rate of heterogeneity across sites. All ML analyses were run with 1,000 μLtra-fast bootstrap and SH-aLRT branch test replicates.
Gene trees were used also for the coalescent-based analyses, using ASTRAL v5.6.1 (86). First, for each ML gene tree, low support branches (μLtra-fast bootstrap support < 10) were collapsed with Newick Utilities v1.6 (87). Branches contracted in the ML gene trees were removed as well from the pool of the corresponding 1,000 bootstrap trees generated during the ML reconstruction. Then, two independent runs were performed either using the ML tree for each gene (BestML), or using the multilocus bootstrap support (MLBS) approach. For the MLBS analysis, 100 replicates were run (-r) starting from the 1,000 contracted bootstrap trees for each gene, allowing gene and site resampling (–gene-resampling flag).
Statistical tests for rejecting the null hypothesis of polytomies were performed in ASTRAL (-t 10 flag), following Sayyari and Mirarab (88). Briefly, for each of the eight datasets, the coalescence-based MLBS tree was tested (-q flag) by random sampling subsets of ML gene trees representing 1 to 100% of the total gene families in the dataset, with a minimum of 20 ML gene trees per subset. For each dataset and for each subset, 10 independent replicates were generated and analyzed with ASTRAL on BestML mode with the –t 10 flag –q flag to score the MLBS trees. The support for a number of key relationships was analyzed in each subset of the datasets and the median of the P values for each subset of trees was calculated.
Different topologies for a number of key relationships were tested using AU tests (89) implemented in IQtree, with 100,000 RELL resamplings (90). In addition to the AU test, the gene-wise log-likelihood scores and the percentage of genes supporting alternative topologies were calculated as outlined by refs. 91 and 48.
Calibrated Phylogenetic Tree.
Due to the high computational cost, the molecular clock analysis was restricted to the 539 coreGF scaffold trim dataset. Clock-likeliness of each gene was assessed with the package SortDate (92) against the ML supermatrix and the coalescence-based topologies, scoring the trees on minimal conflict, low root-to-tip variance, and discernible amounts of molecular evolution (SI Appendix, Fig. S8 and Table S5). The 10 most clock-like genes for each topology were concatenated and subjected to relaxed clock analyses (2,806- and 2,857-amino acid residues for the ML supermatrix and the coalescence-based topology, respectively). Node calibrations were transferred from fossil information and from node age estimates from previous studies (SI Appendix, Table S6). All analyses were run with the same set of calibration nodes, except for the UB (Proterocladus) and the RT (root age, i.e.: Streptophyta-Chlorophyta split) nodes. The UB node was either constrained (UB1) or unconstrained in time (UB0). For the root age, three different priors were used (RT1 to RT3), or the root was left unconstrained (RT0) (SI Appendix, Table S6). Relaxed molecular clock analyses were run with PhyloBayes 4.1b (93). Two sets of analyses were run on two fixed topologies (the ML supermatrix and the coalescence-based topologies), using the set of clock-like genes for each topology. Both lognormal autocorrelated clock (-n flag) and uncorrelated γ-multiplier clock (-ugam flag) models were tested for each dataset, and the models were run with either LG+Γ4 or CATGTR+Γ4 models of amino acid substitutions. In total, 64 different analyses were run (SI Appendix, Table S7) to test the influence of different models, and of root and key prior ages on the age estimations. For each analysis, two distinct Markov chain Monte Carlo chains were run for at least 10,000 generations. The convergence of the log likelihoods and parameters estimates was tested in PhyloBayes. Chains were summarized after discarding the first 750 generations as burn-in.
The ultrametric trees were used to guide the ancestral state reconstruction of the ecological and cyto-morphological traits in Phytools (94). The posterior probabilities of the ancestral state of each node were calculated from summaries of 1,000 replicates of simulated stochastic character map (make.simmap), using empirical Bayes method under the ADR model, which permits backward and forward rates between states to have different values.
Data Availability.
Data associated with this paper are available to download from open-access repositories: https://zenodo.org/record/3242517 and https://figshare.com/articles/Green_algal_transcriptomes_for_phylogenetics_and_comparative_genomics/1604778. The uploaded data include transcriptome assemblies, gene and protein sequences, sequence alignments, and inferred phylogenies. Raw sequence reads have been deposited to the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA548654. Part of this work has been included and presented in the PhD dissertation of A.D.C. (95).
Supplementary Material
Acknowledgments
We thank Diedrik Menzel for providing cultures of Acetabularia acetabulum; Endymion Cooper for generating transcriptomic data of charophyte taxa; and Denis Baurain for his comments on an earlier version of the manuscript. This work was supported by Ghent University (BOF/01J04813) with infrastructure funded by European Marine Biological Resource Centre Belgium/Research Foundation - Flanders Project GOH3817N (to O.D.C.); the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant agreement H2020-MSCA-ITN-2015-675752; the Australian Research Council Grant DP150100705 (to H.V.); the Maryland Agricultural Experiment Station; and the National Science Foundation GRAToL 10136495 (to C.F.D.). The University of Dundee is a registered Scottish charity, No. 050196.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject PRJNA548654); https://zenodo.org/record/3242517 https://figshare.com/articles/Green_algal_transcriptomes_for_phylogenetics_and_comparative_genomics/1604778.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910060117/-/DCSupplemental.
References
- 1.Butterfield N. J., Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26, 386–404 (2000). [Google Scholar]
- 2.Bengtson S., Sallstedt T., Belivanova V., Whitehouse M., Three-dimensional preservation of cellular and subcellular structures suggests 1.6 billion-year-old crown-group red algae. PLoS Biol. 15, e2000735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berney C., Pawlowski J., A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. Biol. Sci. 273, 1867–1872 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J. W., Sorhannus U., A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): Substantive underestimation of putative fossil ages. PLoS One 5, e12759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price D. C., Steiner J. M., Yoon H. S., Bhattacharya D., Löffelhardt W., “Glaucophyta” in Handbook of the Protists, Archibald J. M., Simpson A. G. B., Slamovits C. H., Margulis L., Eds. (Springer, Cham, 2017), pp. 23–87. [Google Scholar]
- 6.Sánchez-Baracaldo P., Raven J. A., Pisani D., Knoll A. H., Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl. Acad. Sci. U.S.A. 114, E7737–E7745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson C., Knoll A. H., Chan C. X., Verbruggen H., Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci. Rep. 8, 1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leliaert F., et al. , Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 31, 1–46 (2012). [Google Scholar]
- 9.McCourt R. M., Delwiche C. F., Karol K. G., Charophyte algae and land plant origins. Trends Ecol. Evol. 19, 661–666 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kenrick P., Crane P. R., The origin and early evolution of plants on land. Nature 389, 33–39 (1997). [Google Scholar]
- 11.Porter S. M., “The fossil record of early eukaryotic diversification” in The Paleontological Society Papers 10: Neoproterozoic-Cambrian Biological Revolutions, Lipps J. H., Waggoner B., Eds. (Paleontological Society, New Haven, 2004), pp. 35–50. [Google Scholar]
- 12.Knoll A. H., Javaux E. J., Hewitt D., Cohen P., Eukaryotic organisms in Proterozoic oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1023–1038 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Škaloud P., Rindi F., Boedeker C., Leliaert F., Freshwater Flora of Central Europe, Vol 13: Chlorophyta: Ulvophyceae, Süßwasserflora von Mitteleuropa (Springer Spektrum, Berlin, Heidelberg, 2018), vol. 13. [Google Scholar]
- 14.Cocquyt E., Verbruggen H., Leliaert F., De Clerck O., Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol. Biol. Evol. 27, 2052–2061 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Leliaert F., et al. , Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 6, 25367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mine I., Menzel D., Okuda K., Morphogenesis in giant-celled algae. Int. Rev. Cell Mol. Biol. 266, 37–83 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Raven J. A., Knight C. A., Beardall J., Genome and cell size variation across algal taxa. Perspect. Phycol. 6, 59–80 (2019). [Google Scholar]
- 18.Littler M. M., Littler D. S., Blade abandonment/proliferation: A novel mechanism for rapid epiphyte control in marine macrophytes. Ecology 80, 1736–1746 (1999). [Google Scholar]
- 19.Fučíková K., et al. , New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2, 63 (2014). [Google Scholar]
- 20.Leliaert F., Lopez-Bautista J. M., The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): Compact genomes and genes of bacterial origin. BMC Genomics 16, 204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turmel M., Otis C., Lemieux C., Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 7, 994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang L., et al. , Improving phylogenetic inference of core Chlorophyta using chloroplast sequences with strong phylogenetic signals and heterogeneous models. Mol. Phylogenet. Evol. 127, 248–255 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Tappan H., Palaeobiology of Plant Protists (Freeman, San Francisco, 1980). [Google Scholar]
- 24.Loron C., Moczydłowska M., Tonian (Neoproterozoic) eukaryotic and prokaryotic organic-walled microfossils from the upper Visingsö Group, Sweden. Palynology 42, 220–254 (2018). [Google Scholar]
- 25.Moczydłowska M., Algal affinities of Ediacaran and Cambrian organic-walled microfossils with internal reproductive bodies: Tanarium and other morphotypes. Palynology 40, 83–121 (2016). [Google Scholar]
- 26.Javaux E. J., Knoll A. H., Micropaleontology of the lower Mesoproterozoic Roper Group, Australia, and implications for early eukaryotic evolution. J. Paleontol. 91, 199–229 (2017). [Google Scholar]
- 27.Butterfield N. J., Modes of pre-Ediacaran multicellularity. Precambrian Res. 173, 201–211 (2009). [Google Scholar]
- 28.Butterfield N. J., Knoll A. H., Swett K., Paleobiology of the Neoproterozoic Svanbergfjellet Formation, Spitsbergen. Fossils and Strata 34, 1–84 (1994). [Google Scholar]
- 29.Graham L. E., Digging deeper: Why we need more Proterozoic algal fossils and how to get them. J. Phycol. 55, 1–6 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Colbath G. K., Grenfell H. R., Review of biological affinities of Paleozoic acid-resistant, organic-walled eukaryotic algal microfossils (including “acritarchs”). Rev. Palaeobot. Palynol. 86, 287–314 (1995). [Google Scholar]
- 31.Arouri K. R., Greenwood P. F., Walter M. R., Biological affinities of Neoproterozoic acritarchs from Australia: Microscopic and chemical characterisation. Org. Geochem. 31, 75–89 (2000). [Google Scholar]
- 32.LoDuca S. T., New Ordovician marine macroalgae from North America, with observations on Buthograptus, Callithamnopsis, and Chaetocladus. J. Paleontol. 93, 197–214 (2018). [Google Scholar]
- 33.LoDuca S. T., Bykova N., Wu M., Xiao S., Zhao Y., Seaweed morphology and ecology during the great animal diversification events of the early Paleozoic: A tale of two floras. Geobiology 15, 588–616 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Verbruggen H., et al. , A multi-locus time-calibrated phylogeny of the siphonous green algae. Mol. Phylogenet. Evol. 50, 642–653 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Satterthwait D. F., “Paleobiology and paleoecology of middle Cambrian algae from western North America,” PhD thesis, University of California, Los Angeles (1976).
- 36.Conway Morris S., Robison R. A., More soft-bodied animals and algae from the middle Cambrian of Utah and British Columbia. Kansas Univ. Paleont. Contr. 122, 1–48 (1988). [Google Scholar]
- 37.Brocks J. J., et al. , The rise of algae in Cryogenian oceans and the emergence of animals. Nature 548, 578–581 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Love G. D., et al. , Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457, 718–721 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Hoshino Y., et al. , Cryogenian evolution of stigmasteroid biosynthesis. Sci. Adv. 3, e1700887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper E., Delwiche C., Green algal transcriptomes for phylogenetics and comparative genomics. Figshare. 10.6084/m9.figshare.1604778. Accessed 24 December 2019. [DOI]
- 41.Del Cortona A., et al. , Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds. Zenodo. https://zenodo.org/record/3242517. Deposited 11 June 2019. [DOI] [PMC free article] [PubMed]
- 42.Philippe H., et al. , Pitfalls in supermatrix phylogenomics. Eur. J. Taxon. 283, 1–25 (2017). [Google Scholar]
- 43.Bravo G. A., et al. , Embracing heterogeneity: Coalescing the tree of life and the future of phylogenomics. PeerJ 7, e6399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B., Liu L., Yan Z., Penny D., Origin of land plants using the multispecies coalescent model. Trends Plant Sci. 18, 492–495 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Wickett N. J., et al. , Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Cortona A., Leliaert F., Molecular evolution and morphological diversification of ulvophytes (Chlorophyta). Perspect. Phycol. 5, 27–43 (2018). [Google Scholar]
- 47.Fang L., Leliaert F., Zhang Z. H., Penny D., Zhong B. J., Evolution of the Chlorophyta: Insights from chloroplast phylogenomic analyses. J. Syst. Evol. 55, 322–332 (2017). [Google Scholar]
- 48.Shen X.-X., Hittinger C. T., Rokas A., Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1, 0126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirarab S., Bayzid M. S., Warnow T., Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Syst. Biol. 65, 366–380 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Cocquyt E., et al. , Complex phylogenetic distribution of a non-canonical genetic code in green algae. BMC Evol. Biol. 10, 327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knoll A. H., The multiple origins of complex multicellularity. Annu. Rev. Earth Planet Sci. 39, 217–239 (2011). [Google Scholar]
- 52.Knoll A. H., Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb. Perspect. Biol. 6, a016121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye Q., et al. , The survival of benthic macroscopic phototrophs on a Neoproterozoic snowball Earth. Geology 43, 507–510 (2015). [Google Scholar]
- 54.dos Reis M., et al. , Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 25, 2939–2950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leliaert F., De Clerck O., Verbruggen H., Boedeker C., Coppejans E., Molecular phylogeny of the Siphonocladales (Chlorophyta: Cladophorophyceae). Mol. Phylogenet. Evol. 44, 1237–1256 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Hoffman P. F., et al. , Snowball Earth climate dynamics and Cryogenian geology-geobiology. Sci. Adv. 3, e1600983 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao S., Tang Q., After the boring billion and before the freezing millions: Evolutionary patterns and innovations in the Tonian Period. Emerg. Top. Life Sci. 2, 161–171 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Lyons T. W., Reinhard C. T., Planavsky N. J., The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Knoll A. H., Lahr D. J., “Fossils, feeding, and the evolution of complex multicellularity” in Multicellularity, Origins and Evolution, Niklas K. J., Newman S. A., Eds. (The Vienna Series in Theoretical Biology, MIT Press, Cambridge, MA, 2016), pp. 1–16. [Google Scholar]
- 60.Cohen P. A., Riedman L. A., It’s a protist-eat-protist world: Recalcitrance, predation, and evolution in the Tonian–Cryogenian ocean. Emerg. Top. Life Sci. 2, 173–180 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Boraas M. E., Seale D. B., Boxhorn J. E., Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol. Ecol. 12, 153–164 (1998). [Google Scholar]
- 62.Coates J. C., Umm-E-Aiman, Charrier B., Understanding “green” multicellularity: Do seaweeds hold the key? Front. Plant Sci. 5, 737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohen P. A., Macdonald F. A., The Proterozoic record of eukaryotes. Paleobiology 41, 610–632 (2015). [Google Scholar]
- 64.Knoll A. H., Life on a Young Planet: The First Three Billion Years of Evolution on Earth (Princeton University Press, Princeton, NJ, 2003). [Google Scholar]
- 65.Xiao S., Yuan X., Steiner M., Knoll A. H., Macroscopic carbonaceous compressions in a terminal Proterozoic shale: A systematic reassessment of the Miaohe biota, South China. J. Paleontol. 76, 347–376 (2002). [Google Scholar]
- 66.Xiao S., Knoll A. H., Yuan X., Pueschel C. M., Phosphatized multicellular algae in the Neoproterozoic Doushantuo Formation, China, and the early evolution of florideophyte red algae. Am. J. Bot. 91, 214–227 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Yuan X., Chen Z., Xiao S., Zhou C., Hua H., An early Ediacaran assemblage of macroscopic and morphologically differentiated eukaryotes. Nature 470, 390–393 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Shen B., Dong L., Xiao S., Kowalewski M., The Avalon explosion: Evolution of Ediacara morphospace. Science 319, 81–84 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Wood R., et al. , Integrated records of environmental change and evolution challenge the Cambrian Explosion. Nat. Ecol. Evol. 3, 528–538 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Cook P. J., Shergold J. H., Phosphate Deposits of the World: Volume 1: Proterozoic and Cambrian Phosphorites (Cambridge University Press, Cambridge, UK, 1986). [Google Scholar]
- 71.Knoll A. H., Biogeochemistry: Food for early animal evolution. Nature 548, 528–530 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Stanley S. M., An ecological theory for the sudden origin of multicellular life in the late Precambrian. Proc. Natl. Acad. Sci. U.S.A. 70, 1486–1489 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brocks J. J., The transition from a cyanobacterial to algal world and the emergence of animals. Emerg. Top. Life Sci. 2, 181–190 (2018). [DOI] [PubMed] [Google Scholar]
- 74.De Clerck O., et al. , Insights into the evolution of multicellularity from the sea lettuce genome. Curr. Biol. 28, 2921–2933.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Palmer J. D., Physical and gene mapping of chloroplast DNA from Atriplex triangularis and Cucumis sativa. Nucleic Acids Res. 10, 1593–1605 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Van Bel M., et al. , TRAPID: An efficient online tool for the functional and comparative analysis of de novo RNA-seq transcriptomes. Genome Biol. 14, R134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gouzy J., Carrere S., Schiex T., Frame D. P., FrameDP: Sensitive peptide detection on noisy matured sequences. Bioinformatics 25, 670–671 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Bel M., et al. , PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vandepoele K., et al. , pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ. Microbiol. 15, 2147–2153 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Huang X., Madan A., CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chernomor O., von Haeseler A., Minh B. Q., Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quang S., Gascuel O., Lartillot N., Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24, 2317–2323 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 19 (suppl. 6), 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Junier T., Zdobnov E. M., The Newick utilities: High-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sayyari E., Mirarab S., Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes (Basel) 9, E132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimodaira H., An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Kishino H., Miyata T., Hasegawa M., Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 31, 151–160 (1990). [Google Scholar]
- 91.Chiari Y., Cahais V., Galtier N., Delsuc F., Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10, 65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith S. A., Brown J. W., Walker J. F., So many genes, so little time: A practical approach to divergence-time estimation in the genomic era. PLoS One 13, e0197433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lartillot N., Lepage T., Blanquart S., PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 95.Del Cortona A., “The rise of algae: Molecular evolution of macroscopic growth in green algae” PhD Thesis, Ghent University, Ghent, Belgium (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this paper are available to download from open-access repositories: https://zenodo.org/record/3242517 and https://figshare.com/articles/Green_algal_transcriptomes_for_phylogenetics_and_comparative_genomics/1604778. The uploaded data include transcriptome assemblies, gene and protein sequences, sequence alignments, and inferred phylogenies. Raw sequence reads have been deposited to the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA548654. Part of this work has been included and presented in the PhD dissertation of A.D.C. (95).