Significance
The persistent Cas9 activity following on-target editing and the lack of spatial precision have severely constrained the current CRISPR-Cas9 systems from complicated and diverse genome-editing contexts. Herein, we develop an optogenetically activatable CRISPR-Cas9 nanosystem (termed nanoCRISPR), and demonstrate that programmable and inducible genome editing can be simply manipulated through photothermal regulation of nanoCRISPR in the second near-infrared optical window. The genome-editing activity and spatial specificity can be precisely programmed by photothermal activation of nanoCRISPR in vitro and in vivo, and the optogenetic regulation minimizes the off-target mutations of CRISPR-Cas9 in potential off-target sites. The photothermal nanoCRISPR offers a useful tool to expand the current applications of CRISPR-Cas9 toward high precision and spatial specificity.
Keywords: CRISPR-Cas9, gene delivery, spatiotemporal specificity, photoregulation, off target
Abstract
We herein report an optogenetically activatable CRISPR-Cas9 nanosystem for programmable genome editing in the second near-infrared (NIR-II) optical window. The nanosystem, termed nanoCRISPR, is composed of a cationic polymer-coated Au nanorod (APC) and Cas9 plasmid driven by a heat-inducible promoter. The APC not only serves as a carrier for intracellular plasmid delivery but also can harvest external NIR-II photonic energy and convert it into local heat to induce the gene expression of the Cas9 endonuclease. Due to high transfection activity, the APC shows strong ability to induce a significant level of disruption in different genomic loci upon optogenetic activation. Moreover, the precise control of genome-editing activity can be simply programmed by finely tuning exposure time and irradiation time in vitro and in vivo and also enables editing at multiple time points, thus proving the sensitivity and inducibility of such an editing modality. The NIR-II optical feature of nanoCRISPR enables therapeutic genome editing at deep tissue, by which treatment of deep tumor and rescue of fulminant hepatic failure are demonstrated as proof-of-concept therapeutic examples. Importantly, this modality of optogenetic genome editing can significantly minimize the off-target effect of CRISPR-Cas9 in most potential off-target sites. The optogenetically activatable CRISPR-Cas9 nanosystem we have developed offers a useful tool to expand the current applications of CRISPR-Cas9, and also defines a programmable genome-editing strategy toward high precision and spatial specificity.
The RNA-guided clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated nuclease protein 9 (Cas9) was originally an adaptive immune defense system, which many bacteria exploit to protect themselves from invading genetic elements (1). It has been recently harnessed as an efficient tool for genome editing in both single cells and whole organisms for a wide range of biomedical applications in biology, genetics, medicine, and so forth (2, 3). In principle, CRISPR-Cas9 is composed of a single-guide RNA (sgRNA) for the identification of DNA targets and a Cas9 endonuclease that can bind and process the recognized DNA targets (4). CRISPR-Cas9–based genome-editing technology offers a powerful and reliable strategy for targeted modifications of the genome, enabling the precise perturbation of virtually any genomic sequence in living cells (2–5). Due to its genome-wide specificity and multiplexing capability, Cas9 and its variants have shown great potential in the generation of loss-of-function animals (6), the correction of genetic disorders (7), functional genome screening (8, 9), and the treatment of infectious diseases (10). Despite these exciting possibilities, the lack of temporal and spatial precision during the editing process has severely constrained current CRISPR-Cas9 systems from complicated and diverse genome-editing scenarios. Furthermore, off-target activity has now become a major concern when the CRISPR-Cas9 system is exploited for therapeutic purposes.
To improve the spatiotemporal specificity of Cas9-mediated genomic manipulation, recent efforts have been dedicated to the development of inducible CRISPR-Cas9 architectures to enable the conditional control of Cas9 activity through either chemical (11–14) or optical (15, 16) means. By precisely limiting the time of Cas9 function, off-target activity is also expected to be controlled by minimizing unwanted prolonged Cas9 activity (12, 16). Chemical methods mainly refer to the regulation of endonuclease activity of Cas9 through small molecule-triggered Cas9 binding and self-splicing inteins (12, 13). Although a few examples have been illustrated for the temporal control of Cas9 activity (17–19), this strategy generally lacks spatial specificity and reversibility, making it difficult to be explored for in vivo studies. Furthermore, commonly used small molecules for chemical activation, such as rapamycin (14) and doxycycline (12, 13), may induce potential cytotoxicity toward both edited and nonedited cells. As opposed to chemical strategies, optical regulation of Cas9 function is more favorable in terms of its noninvasiveness, spatial specificity, and reversibility. In the past few years, several different photoactivatable systems have been adopted for the optical regulation of CRISPR-Cas9–based genome editing and transcriptional activation (20, 21). For example, a photoactivatable Cas9 consisting of two split, deactivated Cas9 (dCas9) fragments and photoinducible dimerization domains (magnets) was engineered to enable optogenetic control of CRISPR-Cas9 activity in human cells (16). Upon blue-light irradiation, the split Cas9 was fused to magnet domains to recover its genome-editing activity, which could be simply switched off by extinguishing the irradiation. More recently, optogenetic anti-CRISPR variants comprising a powerful Cas9 inhibitor (hybrids of AcrIIA4) and a LOV2 photosensor were engineered for the photoregulation of CRISPR-Cas9 activity (22). As photoirradiation enabled the release of dCas9 from the optogenetic variant of AcrIIA4, the inhibited Cas9 activity could be rapidly recovered to enable genome and epigenome editing. Nevertheless, most optically controlled CRISPR-Cas9 systems respond to photoactivation by blue light. This suggests these blue light-mediated activatable CRISPR-Cas9 systems are not only difficult for deep-tissue penetration through turbid human tissues but also are potentially phototoxic in realistic genome-editing applications. To address these issues, a far-red light-mediated CRISPR-dCas9 device, which is built based on the bacterial photoactivatable cyclic diguanylate monophosphate (c-di-GMP) synthase BphS and the c-di-GMP–responsive hybrid transactivator, has been recently developed for targeted epigenetic modulation both in vitro and in vivo (23). The optical activation is based on the light-emitting diode array (400 to 730 nm), which affords moderate tissue penetration up to 5 mm (24). Most recently, a near-infrared upconversion-activated CRISPR-Cas9 nanoparticle system has been proposed for the optical control of therapeutic gene editing toward cancer treatment (25). While the above studies revealed that infrared light is critical for the regulation of genome editing and epigenome editing in vivo, precise CRISPR-Cas9 genome editing in a programmable, inducible manner has not been demonstrated yet, not to mention those for in vivo applications. In addition, off-target activity induced by light-controlled editing modalities still remains elusive to date.
We herein report a photoactivatable CRISPR-Cas9 nanosystem for the optogenetic control of genome editing at the second near-infrared (NIR-II) optical window (1,000 to 1,700 nm). As shown in Fig. 1, this CRISPR-Cas9 nanosystem is typically composed of a cationic polymer-coated Au nanorod (APC) and the Cas9 plasmid driven by a heat-inducible promoter, HSP70 (HSP-Cas9). Whereas the cationic polymer is able to carry and deliver the plasmid into the targeted cells, the Au nanorod serves as a photothermal transducer to transform the harvested external light into intracellular local heat. As such, APC not only acts as the delivery carrier for the plasmid delivery but also serves as an intracellular photothermal converter to trigger the transcription of Cas9 and sgRNA. By incorporating the expression vector with the Cas9 gene cloned downstream of the heat-inducible HSP70 promoter, the elevated local temperature subsequently offers a cue to promote the gene expression of Cas9. Thus, Cas9 activity can be regulated by heat-induced gene expression and activated by photothermal signals. APC–plasmid is first internalized by the targeted cell through charge-mediated internalization, followed by the formation of endosomes. After the endosomal escape, whereas the plasmid released from APC enters into the nucleus, APC is still retained in the cytoplasm. Upon light irradiation at 1,064 nm, APC quickly generates localized heat in the intracellular microenvironment to induce the transformation of the heat-shock factor (HSF) from inactive monomers to active trimers, which are capable of translocating into the nucleus. Then, the binding of the intranuclear trimers to the heat-shock element (HSE) of the HSP70 promoter results in the activation of transcription (26). However, once the light irradiation is switched off, the decreased temperature releases the bound trimer from the HSE, triggering the retransformation of trimers back to monomers to inactivate the transcription process (27). Thus, APC acts as an optogenetic switch to regulate Cas9 expression and activity with high spatial specificity. As NIR-II light shows stronger tissue-penetration ability as compared with first NIR (NIR-I) light (650 to 950 nm), the regulation of genome editing in vivo is also afforded by APC through the optogenetic control in the NIR-II optical window. As we found in our study, APC-mediated optogenetic activation and spatial control of gene expression are demonstrated to direct Cas9 activity in a precise and programmable manner and significantly reduce off-target effects, thereby paving a safe way for in vivo therapeutic genome editing and the spatial control of CRISPR-Cas9 in vitro and in vivo.
Fig. 1.
Illustration of the optogenetic regulation of genome editing mediated by the photoactivatable CRISPR-Cas9 nanosystem. (A) Process of preparation of the APC–HSP complex. (B) Illustration of deep-tissue penetration by NIR-II light. (C) Intracellular delivery of APC–HSP-Cas9 complexes. (D) Mechanism of inducible optogenetic regulation of Cas9 expression and genome editing.
Results
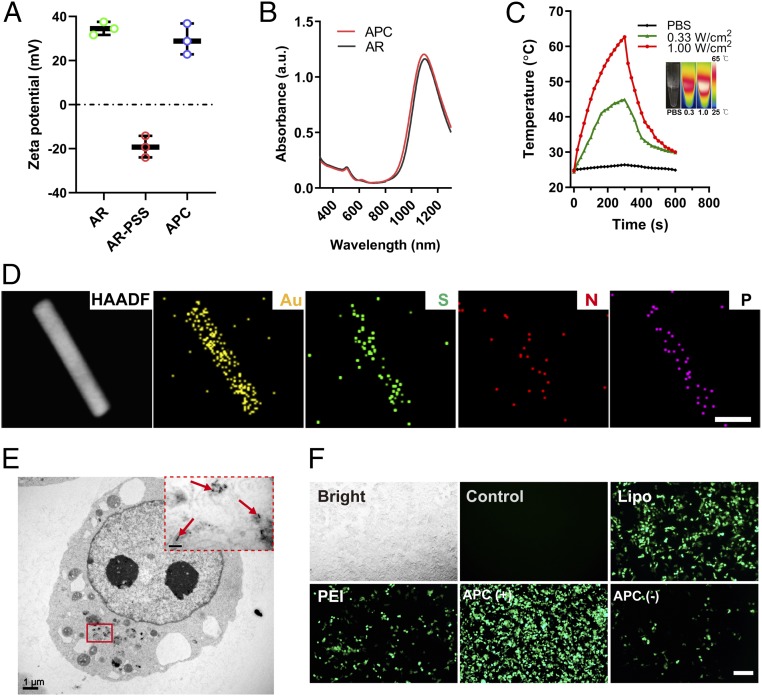
In our study, the classic cetyltrimethylammonium bromide-mediated synthesis approach was used for the preparation of Au nanorods (ARs) (28, 29), and uniform ARs with an aspect ratio of 7.1 (length 106.4 ± 14.1 nm; width 15.2 ± 3.3 nm) were obtained (SI Appendix, Fig. S1). Afterward, biocompatible polystyrene sulfonate (PSS), which acts as an interconnecting layer, was then coated on the AR surface through electrostatic force to form PSS-coated ARs. Subsequently, β-cyclodextrin-polyethyleneimine, a cationic polymer that has been well-demonstrated for the efficient transfection of plasmids both in vitro and in vivo (30), was assembled on top of the PSS layer. The layer-by-layer assembly process to prepare APC was verified by zeta-potential analysis (Fig. 2A), where the final product APC showed a positive surface charge (+29.5 mV). ARs displayed a strong absorption in the NIR-II region, with an absorption peak at ca. 1,070 nm (Fig. 2B). Noticeably, the assembly of polyelectrolytes on ARs barely affected the wavelength of maximum absorption. Such an optical feature of NIR-II is crucial for in vivo investigations. Upon continuous laser irradiation at 1,064 nm for 5 min, the temperature of the APC solution quickly increased and achieved a plateau of 42 °C under a power density at 0.33 W/cm2, as recorded by the infrared thermal camera (Fig. 2C). The maximum temperature generated by APC could be further adjusted to 65 °C at a power density of 1.00 W/cm2. The repeated heating and cooling of three cycles resulted in a similar temperature fluctuation (SI Appendix, Fig. S2) and laser irradiation merely changed the morphology of APC (SI Appendix, Fig. S3), thus demonstrating its good photothermal stability. As the optimal temperature for the activation of the HSP70 promoter was ∼42 °C (31), we also explored the irradiation mode that could stabilize the temperature at this degree. By discontinuous irradiation, the temperature could be finely tuned to a narrow range from 39.0 to 42.0 °C (SI Appendix, Fig. S4). Given the temperature elevation would start from body temperature for in vivo activation, we explored fine temperature control starting from 37 °C by discontinuous irradiation and found this irradiation approach could likewise control the temperature in an ideal range (41.5 to 42.0 °C) by slightly adjusting the discontinuous irradiation time (SI Appendix, Fig. S5). In the meantime, high-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM) and energy-dispersive X-ray spectroscopy (EDS) mapping were performed to verify the layer-by-layer (LBL) structure of APC–HSP-Cas9 (Fig. 2D). The distribution of S, N, or P elements overlapped well with the Au element. The LBL structure of APC–HSP-Cas9 complexes was also confirmed by X-ray photoelectron spectroscopy (SI Appendix, Figs. S6 and S7). To demonstrate whether APC was able to encapsulate the plasmid encoding Cas9, a gel electrophoresis assay was carried out. APC could completely inhibit plasmid DNA migration at an APC/plasmid weight ratio of 0.15, proving its excellent capability to condense and carry plasmid DNA for gene transfection (SI Appendix, Fig. S8). In the meantime, bio-TEM images indicated that APC was primarily located in the cytoplasm after GFP expression (Fig. 2E).
Fig. 2.
Characterization of APC and evaluation of transfection activity. (A) Zeta-potential analysis of Au nanorods, PSS-coated AR, and APC. Mean ± SD; n = 3. (B) Absorption spectrum of AR and APC. (C) Solution temperature of APC as a function of laser irradiation time. The laser wavelength was 1,064 nm. (C, Inset) Thermal image of a PBS solution at a laser power density of 0.33 W/cm2 (Left) and APC solution at a power density of 0.33 W/cm2 (Middle) and 1.00 W/cm2 (Right) at their respective maximum temperatures. (D) HAADF-STEM and EDS mapping of APC–HSP-Cas9. (Scale bar, 25 nm.) (E) Bio-TEM image of 293T cells after the transfection of APC–HSP-Cas9 complexes. The arrows show the presence of APC in the cytoplasm. (F) GFP expression mediated by APC–HSP-Cas9 with (+) or without (−) laser irradiation at 1,064 nm. Lipo- and PEI-mediated transfections at 42 °C were used as positive controls, whereas cells without any treatment were used as a negative control. (Scale bar, 200 µm.)
In the current work, we constructed a Cas9-encoding plasmid driven by an HSP70 promoter. The plasmid consists of a Cas9 gene driven by the HSP70 promoter (SI Appendix, Table S1), an enhanced green fluorescent protein (EGFP) reporter, and a luciferase reporter downstream of Cas9, all of which are separated by self-cleaving peptide P2A, followed by a segment of independent sgRNA sequence driven by the U6 promoter downstream of the luciferase reporter (SI Appendix, Fig. S9). Therefore, we first checked GFP expression after the intracellular delivery of APC–HSP-Cas9 complexes. As shown in Fig. 2F, very weak fluorescence generated from GFP was observed in the 293T cells without laser treatment, implying low background activity. In sharp contrast, strong green fluorescence was observed after the light irradiation on the cells. Flow cytometry analysis indicated that after APC-mediated transfection and photothermal activation, the percentage of GFP-positive cells reached more than 90% under laser irradiation, which is much higher than that from transfection supported by Lipofectamine 2000 (Lipo; 27.4% GFP-positive cells) or 25-kDa polyethyleneimine (PEI; 12.2% GFP-positive cells) at 42 °C (SI Appendix, Figs. S10 and S11). The high level of gene expression was further corroborated by a luciferase reporter assay, where strong luciferase expression was detected when the transfection was mediated by APC with laser irradiation (SI Appendix, Fig. S12). The incorporation of sgRNA cloned downstream in the plasmid merely affected the transfection activity of APC (SI Appendix, Fig. S13). In the meantime, we found that the level of luciferase expression could be modulated by laser intensity (SI Appendix, Fig. S14) and irradiation time (SI Appendix, Fig. S15), implying the transgene expression level is precisely tunable. In order to elucidate the role of specific internalization pathways, different inhibitors were added to the cell-culture medium before transfection in 293T cells (SI Appendix, Figs. S16 and S17). It was evident that the addition of methyl-β-cyclodextrin significantly reduced GFP expression, suggesting the internalization of APC–plasmid complexes primarily follows caveolae-dependent endocytosis. Additionally, the inhibition of transfection activity by bafilomycin A1 suggested the strong buffering capacity of APC, which is critical to facilitate the endosomal escape of the delivered plasmids. APC also showed high transfection activity toward different types of cell lines upon photothermal activation (SI Appendix, Fig. S18), and did not exhibit obvious cytotoxicity on 293T cells up to a concentration of 1.35 µg/mL (SI Appendix, Fig. S19).
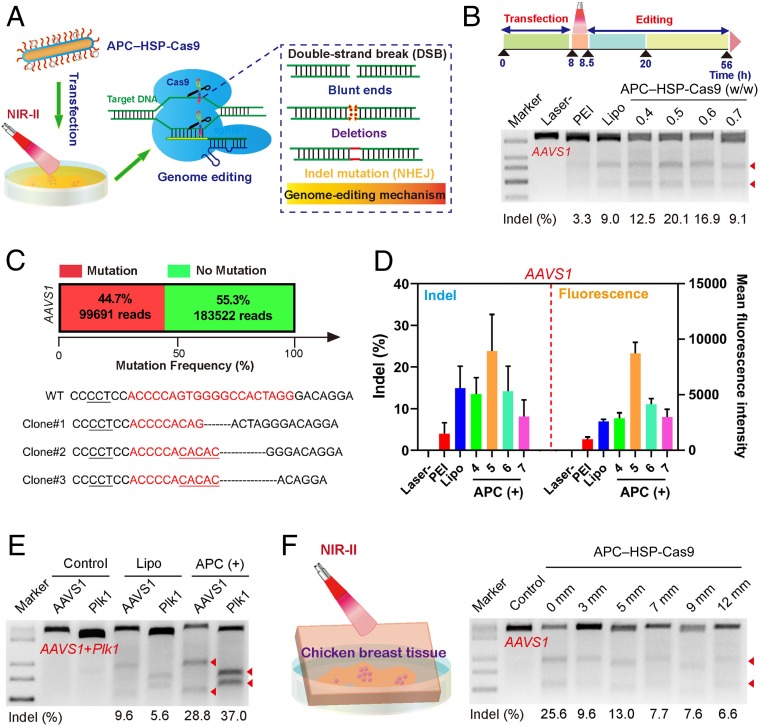
Based on the above optimized results, we subsequently investigated whether optogenetic control of CRISPR-Cas9 activity could be manipulated through efficient transfection and photothermal conversion by APC (Fig. 3A). We tested whether APC was capable of disrupting the EGFP gene in 293T cells that stably expressed EGFP (SI Appendix, Fig. S20). Upon the intracellular delivery of APC–HSP-Cas9 targeting EGFP, the intensity of GFP in 293T-EGFP cells decreased significantly with the laser irradiation, suggesting the strong ability of APC to mediate the disruption of the EGFP gene. Nevertheless, the treatment with APC–HSP-Cas9-sgEGFP without laser irradiation had negligible knockout effects. We also noticed that the temperature elevation to 42 °C by photothermal irradiation neither adversely altered the secondary structure of Cas9 protein (SI Appendix, Fig. S21A) nor affected its nuclease activities (SI Appendix, Fig. S21B). Additionally, we found Cas9 expression became strongest at 42 °C (SI Appendix, Fig. S22 A and B) and the photothermal activation at this optimal temperature induced a minimum degree of cell death (SI Appendix, Fig. S22C). To further validate the genome-editing efficiency, we studied the intracellular delivery of HSP-Cas9 plasmid targeting different genomic loci in the 293T cell line. Indels (insertions and deletions) detected by T7 endonuclease I (T7E1) digestion assays were carried out to evaluate the efficiency of genome editing at the targeted genome sites. After the transfection and photothermal activation, the bands from the digestion products of T7E1 distinguishing indels in the double-stranded DNA were clearly detected from the uncut bands at the genomic locus of adeno-associated virus integration site 1 (AAVS1). We noted that the editing efficiency is slightly dependent on the APC concentration, with the highest indel rate of 20.1% at the APC/plasmid weight ratio of 1:2. As expected, AAVS1 genome editing by Lipo and PEI resulted in indel rates of 8.9 and 3.3%, respectively, both of which were lower than that of APC-mediated genome editing (20.1% at the optimal weight ratio of 0.5; Fig. 3B). Sanger sequencing confirmed the mutations at the targeted loci, including base deletion, insertion, and substitution around the protospacer adjacent motif (PAM) (Fig. 3C and SI Appendix, Figs. S24 and S53), and deep-sequencing analysis showed that the mutation frequency was up to 44.6% when the transfection was mediated by APC. In the meantime, we further investigated whether the level of GPF expression is synchronized with the Cas9-mediated genome disruption. As expected, the level of GFP expression was well-correlated with the indel rate, suggesting the level of GFP expression could reflect and estimate the indel rate (Fig. 3D). Similarly, by screening different sequences of sgRNA (SI Appendix, Tables S2 and S3), the optimized genome editing at the rhomboid family member 1 (RHBDF1) locus mediated by APC showed an indel rate of 14.8%, which is more efficient than that of Lipo (6.5%) (SI Appendix, Fig. S23A) and was confirmed by Sanger sequencing (SI Appendix, Fig. S24). Deep-sequencing analysis indicates that sgRNA sequence design is also critical in affecting genome-editing activity, and the highest indel rate was 15% when the optimal sgRNA sequence (sg4) was used (SI Appendix, Figs. S23B and S56). Furthermore, we examined whether the optogenetic control could likewise activate the multiplex genome editing. To this end, we delivered two plasmids, both of which encoded a single but different sgRNA construct targeting AAVS1 and Plk1 (polo-like kinase 1), respectively. Both genomic loci showed an evident degree of editing, with indel rates of 28.8% (AAVS1) and 37.0% (Plk1) (Fig. 3E and SI Appendix, Fig. S25). The transfection of the HSP-Cas9 plasmid with Lipofectamine followed by irradiation was investigated as a control (SI Appendix, Fig. S26). Indel mutations were hardly detected in two genomic loci (AAVS1 and Plk1; SI Appendix, Fig. S26), suggesting the poor utility of Lipofectamine for photothermal conversion. In sharp contrast, APC-mediated transfection followed by irradiation induces significant mutations at both genomic loci. Since APC can well absorb NIR-II light that can afford deep-tissue penetration, we covered the cell-culture plate with breast chicken tissue of different thicknesses and investigated whether the irradiation could still activate the genome editing in the transfected cells in the presence of tissue (Fig. 3F). Though the increase of the tissue thickness reduced the genome-editing activity, the indel rate (6.6%) could still be detected in the presence of 12-mm breast chicken tissue. This suggested that the increase in the tissue thickness impaired the penetration ability of NIR-II light, thereby affecting the photothermal conversion efficiency, as we have demonstrated (SI Appendix, Fig. S27). Furthermore, the luciferase expression was temperature-dependent and became evident when the temperature reached 39 °C, reaching the highest at 42 °C (SI Appendix, Fig. S28). Interestingly, such optogenetic activation also works well for dCas9-mediated transcriptional activation of exogenous genes. For example, when three plasmids (HSP-dCas9-SPH, U6-sgRNA, and miniCMV-mCherry) were cotransfected in 293T cells, only very low basal fluorescence was observed before optogenetic activation, due to the weak ability of miniCMV to induce transcription. In sharp contrast, mCherry expression became very strong after the transcriptional activation of the heat-shock promoter, suggesting its potential for heat-inducible transcriptional activation (SI Appendix, Fig. S29). In the meantime, we investigated whether such a heat-shock approach affected cell cycles and induced potential apoptosis (SI Appendix, Fig. S30). As expected, cells treated with APC–HSP-Cas9 complexes with or without laser irradiation showed similar cell-cycle patterns as those treated with phosphate-buffered saline (PBS). In the meantime, cells transfected with APC–HSP-Cas9 complexes merely induced any apoptosis, suggesting the biocompatibility of APC and the safety of heat-shock optogenetic modality.
Fig. 3.
Optogenetic activation of CRISPR-Cas9 genome editing by APC–HSP-Cas9. (A) Illustration of optogenetic activation mediated by APC. (B) Indel mutations of the AAVS1 locus of 293T cells transfected with APC–HSP-Cas9 complexes with or without laser irradiation. APC was complexed with HSP-Cas9 at different weight ratios. Lipo and PEI were used as positive controls. (C) Deep-sequencing analysis of mutation frequency at the AAVS1 locus (optimal weight ratio of 0.5) and Sanger sequencing results of T–A cloning from 293T cells (AAVS1) after APC-mediated transfection, followed by optogenetic activation. The target sequences are marked in red. The PAM is underlined (black). Substitutions, insertions, and deletions are marked by red base sequences, underlining (red), and dotted lines, respectively. A library of genomic DNA pooled from the sample in triplicate was subjected to deep-sequencing analysis. (D) Analysis of indel rate, as revealed by the grayscale density of cut bands from T7E1 results, after the transfection of APC–HSP-Cas9 targeting AAVS1 with or without laser treatment (Left). The corresponding GFP expression was evaluated; 4, 5, 6, and 7 refer to APC/HSP-Cas9 weight ratios 0.4, 0.5, 0.6, and 0.7, respectively. Mean ± SD; n = 3. (E) Indel mutation from multiplex genome editing by HSP-Cas9 targeting Plk1 and AAVS1 with laser irradiation. (F) Illustration of cultured cells exposed to irradiation in the presence of a piece of chicken breast tissue (Left). Indel mutations of the AAVS1 locus from 293T cells transfected with APC–HSP-Cas9, followed by irradiation in the presence of breast chicken tissue of different thicknesses (Right). The arrowheads in B, E, and F show the cleaved DNA fragments of the target genome.
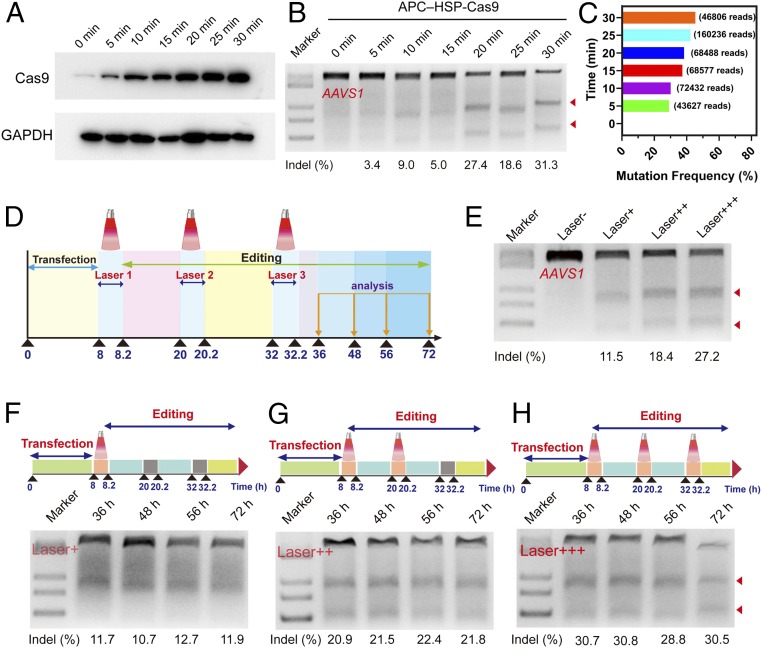
While successfully establishing the above strong evidence of optogenetic genome editing, we were curious about whether such a modality could precisely control the degree of editing. We first monitored the continuous bioluminescence (BL) intensity to reflect the amount of Cas9 expression upon optogenetic activation, and the level of luciferase expression was studied as a function of time. As reflected by SI Appendix, Fig. S31, after transfection for 24 h, the BL intensity increased quickly following irradiation for 5 min, suggesting a fast gene expression by optogenetic activation. Luciferase expression further increased and reached 3.3 × 105 relative light unit (RLU) per mg after optogenetic activation for 30 min. The level of luciferase expression remained stable over a period of 24.5 h after the removal of irradiation. However, once the laser was switched on again, BL intensity rapidly increased up to 4.4 × 105 RLU per mg, and became stable upon the removal of irradiation. The above information suggested photothermal control of gene expression by APC is inducible and APC may serve as an optogenetic switch to regulate Cas9 expression and activity. Then, Western blot analysis demonstrated that the Cas9 expression by nanoCRISPR was inducible, and the expression of Cas9 protein can be exactly controlled by time of irradiation (Fig. 4A and SI Appendix, Fig. S32). By controlling the time length of irradiation from 5 to 30 min, we found the indel range could be precisely tuned from 3.4 to 31.4% through the T7E1 assay (Fig. 4B and SI Appendix, Fig. S33). Deep sequencing further indicated that mutation frequency ranged from 29.1 to 45.4% (Fig. 4C and SI Appendix, Fig. S54), which was also reflected by GFP expression with different irradiation times (SI Appendix, Fig. S34). Furthermore, the temporal control of genome-editing activity could also be simply realized by adjusting the number of irradiation episodes (Fig. 4D). For example, when the irradiation was conducted for only one time length (10 min), the resulted indel rate was 11.5%; however, the indel rate could be improved simply by increasing the number of irradiation times to reach the expected ones (Fig. 4E and SI Appendix, Fig. S35). It is worthy to mention that such a stepwise, optogenetic activation modality is also stable. By analyzing the indel rate at different time points after genome editing, we found the degree of editing was generally stable, irrespective of the irradiation times and harvesting time points (Fig. 4 F–H and SI Appendix, Fig. S36). Western blot assays were carried out to examine the time course of Cas9 expression level after transfection (SI Appendix, Fig. S37). In general, the expression level dropped with time from 24 to 72 h following the photothermal induction, implying the decreased Cas9 activity following the on-target genome editing by nanoCRISPR.
Fig. 4.
APC-mediated optogenetic control of programmable genome editing in vitro. (A) Western blot analysis of Cas9 protein expression after the transfection followed by the thermal induction at different irradiation times. (B) Indel mutations of the AAVS1 locus detected by T7E1 assay. 293T cells were first transfected with APC–HSP-Cas9 complexes, and then exposed to the laser irradiation from 5 to 30 min. The indel mutations were evaluated 72 h after the irradiation. (C) Deep-sequencing analysis of mutation frequency at the AAVS1 locus. The experimental conditions were the same as those described in Fig. 3B. (D) Illustration of the transfection, irradiation, and genome-editing processes. The irradiations were conducted at 8, 20, and 32 h after the transfection, and irradiation time was 10 min each time. The indel analysis was conducted at 36, 48, 56, and 72 h. (E) Indel mutations of the AAVS1 locus detected by T7E1 assay. The irradiation was conducted at 8 h (Laser+), 8 and 20 h (Laser++), and 8, 20, and 32 h (Laser+++) after the transfection. The indel mutations were analyzed at 72 h after the transfection. (F–H) Indel mutations of the AAVS1 locus detected by T7E1 assay after exposure to irradiation for different times. The irradiation was conducted at 8 h (Laser+), 8 and 20 h (Laser++), and 8, 20, and 32 h (Laser+++) after the transfection, and the cells were harvested at 36, 48, 56, and 72 h for indel analysis. The arrowheads in B, E, and H show the cleaved DNA fragments of the target genome.
To validate the potential of such an optogenetic control modality in vivo, A549 cells transfected with APC–HSP-Cas9 complexes were subcutaneously transplanted into the back of BALB/c mice ex vivo. Following laser irradiation on the subcutaneously transfected cells, BL intensity gradually became strong along the increased irradiation time (Fig. 5A). We next harvested these transplanted cells and checked whether the optogenetic activation worked for in vivo genome editing. As shown from T7E1 assay results (Fig. 5 B and C), indel rates ranging from 4.7 to 20.1% could also be well-manipulated by tuning the irradiation time, implying the controllable activation of Cas9 expression and programmable regulation of genome-editing activity in vivo. Moreover, we explored whether direct in vivo transfection and optogenetic activation are possible. To this end, APC–HSP-Cas9 complexes were first delivered into the hind limb of BALB/c mice via intramuscular injection, and optogenetic activation was conducted after 8 h (Fig. 5D). Strikingly, strong in vivo bioluminescence was clearly detected in the hind limb 40 h after optogenetic activation. Based on these findings, we further harvested and lysed the tissue from the muscle of the hind limb to evaluate the indel mutation of the edited cells. The indel rate reached 18.1% with laser irradiation; however, the indel was hardly detectable after the in vivo transfection of APC–HSP-Cas9 complexes without laser irradiation (Fig. 5 E and F). This further motivated us to explore whether optogenetic genome editing could be manipulated in the deep tissue of local lesions. For this purpose, BALB/c nude mice bearing A549 xenograft tumors were first injected with APC–HSP-Cas9 complexes through peritumoral injection, and irradiation was then carried out in the presence of a piece of breast chicken tissue (5-mm thickness) covering the tumor position to simulate the deep-tissue condition (Fig. 5G). The presence of tissue slightly decreased the tumor temperature to 40.0 to 41.4 °C. Excitingly, strong BL intensity was clearly observed over the tumor position, suggesting that moderate hyperthermia could well activate gene expression. In the meantime, a significant level of genome editing was detected from both the surface and deep layer of the tumor tissues, with an indel rate of 16.0 and 14.9%, respectively (Fig. 5 H and I). These results strongly implied that such optogenetic control may be suitable for regulating genome-editing activity in a deep-tissue environment. Given that the elevated temperature improved the Cas9 activity (32), such a heat-shock approach may also improve the editing capacity of the photothermal nanoCRISPR. We also found that the spatial specificity could be well-manipulated as well through optogenetic regulation. The systemic administration of APC–HSP-Cas9 to BALB/c mice by tail-vein injection resulted in strong luciferase expression in the liver that was exposed to irradiation, whereas the same treatment of mice with APC–CMV-Cas9 resulted in luciferase expression primarily in the lung (Fig. 5J and SI Appendix, Fig. S38). Indel mutation (9.9%) was clearly detected in the liver when the transfection was mediated by photothermal nanoCRISPR (Fig. 5K), and such mutations were hardly found in any other organs including heart, spleen, lung, and kidney. As a control, indel mutations were also barely detected in the liver when the transfection of photothermal nanoCRISPR was conducted in the absence of NIR-II irradiation (SI Appendix, Fig. S39A). In sharp contrast, indel mutations were found in liver (3.8%), spleen (1.7%), and lung (0.7%) when the transfection was mediated by APC–CMV-Cas9 (SI Appendix, Fig. S39B). In agreement with the results from transcriptional activation of exogenous genes in vitro, the transcriptional activation of mCherry expression was also verified in vivo, when APC–plasmid complexes were codelivered into the hind limb either through ex vivo transfection or direct in vivo tissue transfection, followed by optogenetic activation (Fig. 5L and SI Appendix, Figs. S40 and S41). Importantly, we demonstrated that the optogenetic activation through different administration approaches, including ex vivo transfection, direct in vivo intramuscular administration, and systemic administration, merely induced toxicity in the major organs (heart, liver, spleen, lung, and kidney) after the irradiation (SI Appendix, Figs. S42–S44). The above results demonstrated that spatial and programmable genome editing could also be safely achievable in vivo as well.
Fig. 5.
APC-mediated optogenetic control of programmable genome editing in vivo. (A) Schematic illustration (Left). Transfection with APC–HSP-Cas9 complexes, and then subcutaneous implantation. Whereas the right implanted position was exposed to irradiation from 5 to 30 min, the left position (nonirradiation) was used as the control. In vivo luciferase expression (Right). (B) Indel mutations detected by T7E1 assay. (C) Quantitative analysis of indel mutations. Mean ± SD; n = 3. (D) Schematic illustration (Left). APC–HSP-Cas9 complexes were subcutaneously injected into the muscle of the hind limb of BALB/c mice, followed by irradiation at 1,064 nm. In vivo luciferase expression (Right). (E) Indel mutations detected by T7E1 assay. (F) Quantitative analysis of indel mutations. Mean ± SD; n = 3. (G) Schematic illustration (Left). Tumor-bearing mice were administered APC–HSP-Cas9 complexes through peritumoral injection, and the tumor (Right) was then exposed to irradiation for 30 min. In vivo luciferase expression (Middle). (H) Indel mutations detected by T7E1 assay. (I) Quantitative analysis of indel mutations. Mean ± SD; n = 3. (J) Evaluation of in vivo luciferase expression through intravenous injection, followed by optogenetic activation in the liver. Schematic illustration (Left). BALB/c mice were injected with APC–HSP-Cas9 complexes via tail-vein injection, and the liver position was exposed to irradiation for 30 min after the transfection. (K) Indel mutations from different tissues after the systemic administration of nanoCRISPR through intravenous injection, followed by injection with APC–HSP-Cas9 complexes via tail-vein injection, and the liver position was exposed to the irradiation for 30 min after the transfection. (L) Optogenetic regulation of transcriptional activation of mCherry expression through in vivo transfection and optogenetic activation. The arrowheads in B, E, H, and K show the cleaved DNA fragments of the target genome.
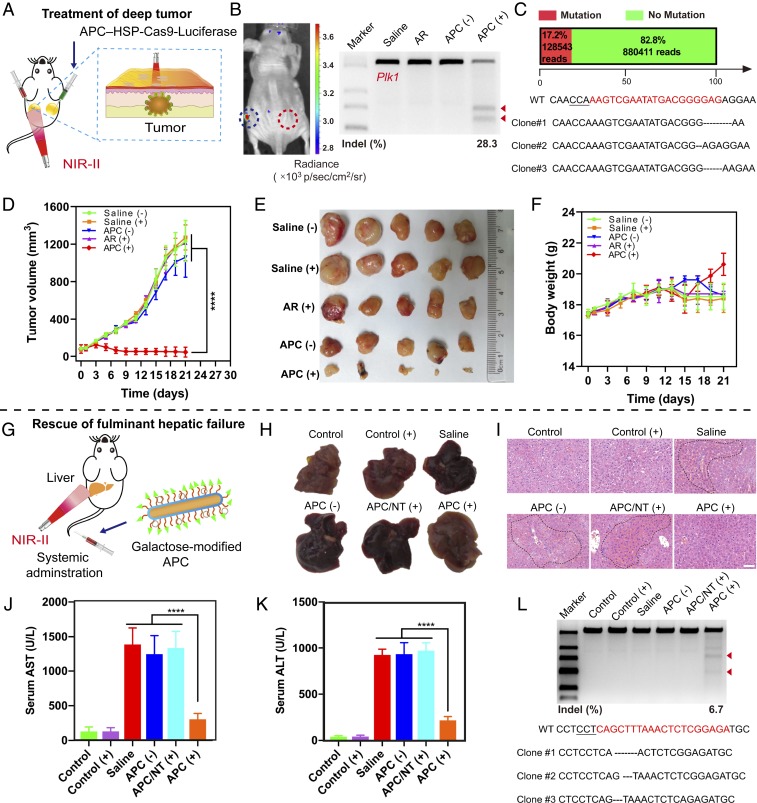
We further investigated the optogenetic activation of photothermal nanoCRISPR in vivo as proof-of-concept examples for therapeutic genome editing. To this end, we first delivered Cas9 plasmid with sgRNA targeting Plk1, a master regulator of mitosis (33), and activated expression after transfection in A549 cells. Indel analysis indicated that significant mutation was detected in the targeted genomic locus, with an indel rate up to 41.5% when the optimized sgRNA targeting Plk1 was used (SI Appendix, Fig. S45A). The editing-induced mutation was also confirmed by Sanger sequencing results, where significant deletions and insertions were detected at the targeted loci around the PAM (SI Appendix, Fig. S24). Deep-sequencing analysis indicated that the mutation frequency was up to 51.4% (SI Appendix, Figs. S45B and S55). Western blot analysis indicated that the level of Plk1 expression was remarkably reduced after the transfection and activation process (SI Appendix, Fig. S45C). These results in vitro well establish the fact that optogenetically regulated genome editing enables the efficient knockout of the target Plk1 gene. Based on these results, we next investigated whether tumor growth could be effectively inhibited on BALB/c nude mice bearing A549 xenograft tumors by this therapeutic modality (Fig. 6A). After the peritumoral injection of APC–HSP-Cas9 complexes and incubation for 48 h, we noticed that the BL was still visible but became weak after irradiation activation (Fig. 6B). In the meantime, T7E1 assay results suggested significant genome disruption in the Plk1 site in the tumor tissue (Fig. 6B and SI Appendix, Fig. S47), and deep-sequencing analysis of a single library prepared from genomic DNA pooled from five mice also indicated significant mutation in the Plk1 locus (17.2%) (Fig. 6C and SI Appendix, Fig. S57). Therefore, the reduced expression was probably attributable to the presence of large amounts of apoptotic cells induced by Plk1 disruption, leading to the poor ability to express luciferase. In fact, the above speculation was verified by the in vivo tumor-inhibition assay, where the tumor-bearing mice injected with APC–HSP-Cas9 targeting Plk1 exhibited significant tumor regression after irradiation treatment. The tumor size also became much smaller in comparison with the initial size before treatment (43.9 mm3 on average). In sharp contrast, mice treated with the same formulation but without laser treatment exhibited rapid tumor progression, reaching a final tumor volume of 1,270 mm3 at 21 d (Fig. 6 D and E). As an indicator of systemic toxicity, we monitored body weight through a therapy session and noticed that a slight increase in body weight was observed at the end of the treatment (20.6 g on average; Fig. 6F). In the meantime, toxicity to major organs was investigated by hematoxylin and eosin (H&E) staining (SI Appendix, Fig. S46), and blood biochemistry was also evaluated to reflect liver and kidney index (SI Appendix, Fig. S48). As compared with the saline control group (without laser treatment), the tumor slice showed the fewest tumor cells combined with a significant degree of necrosis. Whereas H&E staining suggested that such a therapeutic modality was generally safe and biocompatible in the major organs, the function index of blood biochemistry further validated that such an optogenetic treatment merely caused any damage to the liver and kidney.
Fig. 6.
APC-mediated optogenetic activation for therapeutic genome editing in vivo. (A) Illustration of optogenetic activation for in vivo cancer therapy. APC–HSP-Cas9 complexes were administered through peritumoral injection, followed by the exposure of the tumor to irradiation. (B) In vivo luciferase expression in the tumor tissue. The left tumor was exposed to laser irradiation, and the right one, without laser exposure, was used as the control. T7E1 assay of indel mutations of Plk1 in the tumor tissue. (C) Analysis of in vivo Plk1 gene mutation efficiency through deep sequencing and Sanger sequencing. The mutation frequency was determined from a single deep-sequencing library prepared from genomic DNA pooled from five mice. (D) Tumor growth curve after the transfection of APC–HSP-Cas9 complexes in the tumor tissue, followed by exposure to irradiation. The treatment was carried out twice a week, and continued for 3 wk. Mean ± SD; n = 5 (two-way ANOVA with a Bonferroni post hoc test, ****P < 0.0001). (E) Images of dissected tumor tissues from tumor-bearing BALB/c mice with different treatments. (F) The body-weight change during the treatment. (G) Illustration of optogenetic activation of nanoCRISPR in vivo to rescue mice from fulminant hepatic failure. Mice were injected intravenous through the tail vein with APC–HSP-Cas9–Fas complexes, followed by irradiation over the liver position. (H) Images of dissected livers from mice with fulminant hepatic failure. (I) H&E staining of liver slices from mice 10 d after the treatment. The regions in the dotted lines denote accumulation of blood cells. (Scale bar, 50 µm.) (J and K) Serum AST and ALT from a mouse injected with saline, APC–HSP-Cas9-NT with irradiation (NT, nontargeted scramble sgRNA), and APC–HSP-Cas9–Fas (with or without irradiation) 10 d after treatment. Mean ± SD; n = 4 (one-way ANOVA with a Tukey post hoc test, ****P < 0.0001). (L) Indel mutation analysis of the Fas gene by T7E1 assay and mutation Sanger sequencing after the optogenetic activation of nanoCRISPR in the liver of mice with fulminant hepatic failure. The arrowheads in B and L show the cleaved DNA fragments of the target genome.
Last, we explored whether the optogenetic activation of photothermal nanoCRISPR could protect mice from fulminant hepatic failure. A wide range of liver diseases are associated with Fas-mediated apoptosis, and the inhibition of Fas gene expression by RNA interference was shown to protect mice from fulminant hepatic failure (34). Therefore, we speculate that Cas9-mediated genome editing of Fas may protect fulminant hepatic failure. Additionally, by controlling the irradiation of NIR-II light over the liver, we were able to improve the spatial specificity of genome editing exclusively in the liver. First, we found the optogenetic activation of photothermal nanoCRISPR with the HSP-Cas9 plasmid encoding sgRNA targeting Fas (HSP-Cas9–Fas) induced an 18.0% indel mutation rate in Hepa1-6 cells (SI Appendix, Fig. S49A), which was confirmed by Sanger sequencing (SI Appendix, Fig. S49B). Since galactose-decorated nanoparticles have been widely explored to improve liver-targeting ability by interacting with asialoglycoprotein receptors on the membrane of hepatocytes (35), we further delivered galactose-modified APC–HSP-Cas9–Fas complexes by tail-vein injection and examined whether such a prophylactic measure could protect mice from concanavalin A (Con A)-induced fulminant hepatic failure (Fig. 6G). The mice treated with galactose-modified APC–HSP-Cas9–Fas significantly reduced hyperemia (Fig. 6 H and I), in contrast with the ones without irradiation (APC [−]) or treated with scramble sgRNA (APC/NT [+]). In the meantime, blood biochemistry indicated the optogenetic activation of nanoCRISPR in the liver could effectively reduce AST (aspartate transaminase) and ALT (alanine aminotransferase) levels (303 and 217 U/L, respectively), which were close to that of mice without any treatment (Fig. 6 J and K). The in vivo editing was validated by the T7E1 and Sanger sequencing results, where significant genome disruption in the Fas site (6.7% indel mutation) was found in the liver tissue (Fig. 6L). Hematological indicators were also evaluated after the systemic administration of APC to reflect the liver and kidney index in vivo (SI Appendix, Fig. S50). We monitored the mice for up to 14 d, and found the systemic administration of APC at the therapeutic dose (12.5 µg/mL) merely induced any liver and kidney toxicity during the observation period.
To analyze the off-target effects generated by this editing modality, we used an off-target searching tool, Cas-OFFinder (36), to estimate the potential off-target sites, and further carried out both Sanger sequencing and deep sequencing to evaluate whether genomic mutations could be detected in the estimated off-target sites (SI Appendix, Tables S4 and S5). First, Sanger sequencing analysis proved that the sequence that was suspected to off-target disruption displayed the same intact sequence as the wild-type one without any treatment (SI Appendix, Fig. S51). We further carried out deep sequencing to quantitatively analyze the off-target mutation at different time points following photothermal transfection of nanoCRISPR. APC-mediated transfection of Cas9 plasmid driven by the cytomegalovirus (CMV) promoter, a constitutive promoter, was used as a control. As revealed by deep-sequencing analysis, the transfection of APC–CMV-Cas9 complexes only resulted in moderate gene disruption at the on-target site of AAVS1 2 d after the transfection, with an indel mutation frequency of 16.6%. The degree of on-target mutation increased a further 4 d following the transfection, reaching an indel mutation frequency of 27.6%. After 6 d of transfection, the on-target mutation rate decreased to 15.2% (SI Appendix, Fig. S52A and Table S6). In comparison with CMV-Cas9, the mutation frequency induced by photothermal transfection of HSP-Cas9 was at least two times higher 2 d after the transfection, suggesting high on-target genome-editing activity. Note that on-target mutation frequency dropped significantly 4 d following the photothermal transfection of HSP-Cas9, and further decreased to 20.0% 6 d after the transfection. The decreased on-target mutation might be partially due to the delayed cell division in the mutated daughter population, where 53BP1 nuclear bodies function as a key regulator to restrain the replication of disrupted genomic loci until late S phase (37). In addition, detectable off-target mutations were found throughout the whole experimental window, which were more frequent and significant over the photothermal transfection of HSP-Cas9 in general, though the latter also resulted in traceable off-target editing. The mean specificity ratio [defined as the ratio of on-target activity to off-target activity (38)] of HSP-Cas9–induced genome editing was remarkably higher than that of CMV-Cas9–induced editing during this time window (SI Appendix, Fig. S52B). These results collectively demonstrated that optogenetically activatable nanoCRISPR may minimize off-target effects through optogenetic control of Cas9 expression to reduce prolonged Cas9 activity.
Discussion
Remote activation with noninvasive NIR light has been extensively exploited in a wide range of biomedical applications, such as microRNA detection (39), brain stimulation (40), modulation of gene expression (31), and immunomodulation (41), largely owing to the low photocytotoxicity and deep-tissue penetration capability of NIR light. In comparison with NIR-I light, NIR-II light has been validated to afford deeper tissue penetration, a key challenge preventing many optogenetic control strategies from in vivo investigation. Although a few types of organic (42, 43) and inorganic nanomaterials (28) that absorb NIR-II light have been developed, AR was selected as a building block for APC largely due to its high photothermal conversion rate and photothermal stability (44). Furthermore, the AR not only converts the external photonic energy into intracellular local heat but also serves as a template where the cationic polymers are assembled for the subsequent encapsulation of large plasmids. As an unconventional finding, the assembly of PC over AR surprisingly resulted in far more efficient condensation of the Cas9 plasmid in comparison with PC alone (30), probably owing to the higher aspect ratio of gold nanostructure being more favorable for entangling the plasmid. This feature of AR may also facilitate APC to enter the nucleus by passive fusion, as high-aspect ratio nanoparticles, such as nanorods and nanoworms, were previously demonstrated to be superior to spherical ones with identical surface chemistries in terms of nuclear entry (45). It is also noteworthy that the incorporation of the Cas9 plasmid and photothermal transducer into the same carrier ensures the delivery of two payloads into the same cell population, thereby maximizing the sensitivity and efficiency of optogenetic activation of the Cas9 transcription. As opposed to other light-activated editing or transcriptional regulation systems that require the cotransfection of multiple plasmids by commercial transfection agents (16, 22, 23), nanoCRISPR is a relatively straightforward and simple system that only contains an “all-in-one” plasmid and a photothermal nanocarrier, both of which are easy to construct. The efficient transfection and photothermal conversion enabled by APC ensure the successful induction of Cas9 expression as well as the precise control of Cas9 nuclease activity.
Heat-induced transcription of genes encoding a major heat-shock protein (HSP70) is a cytoprotective mechanism which a wide variety of cells exploit to protect themselves from heat shock and other deleterious stresses (46). HSP70 promoters are regulated by cytosolic HSF, which becomes active in response to moderate hyperthermia (41.5 to 42.0 °C) to induce the expression of downstream heat-shock proteins that are critical for cellular defense (47). The heat-responsive HSP70 promoters have been previously explored for the spatial and temporal control of gene expression through photothermal effects (48, 49). In the current study, we constructed a Cas9-encoding plasmid driven by an HSP70 promoter, which indeed serves as a photothermal switch to regulate Cas9 transcription by sensing the surrounding temperature. As the temperature could be finely tuned by controlling the irradiation time length and is closely correlated with Cas9 expression and activity, we are therefore capable of programming the degree of editing simply by adjusting exposure time and irradiation time. Hence, this editing modality may be applicable in the context where CRISPR-Cas9 activity is required to fulfill editing missions at multiple time points. For example, the optogenetically activatable CRISPR-Cas9 nanosystem may serve as an ideal platform for inducible editing at multiple time points that is required for CRISPR-Cas9 barcode editing to trace lineage information of different cells during development and disease (50). For many other applications, CRISPR-Cas9 activity should be inhibited following the on-target editing, and prolonged activity may otherwise cause undesired side effects. For instance, restriction of Cas9 activity to a narrow temporal window is critical in germ-line editing, as the persistent Cas9 activity following the initial rounds of mitosis contributes to mosaicism (51). Furthermore, nanoCRISPR may serve as an inducible genome-editing platform to precisely control the dose of Cas9, as a number of side effects, such as genotoxicity, immunogenic response, and chromosomal translocations, are associated with elevated levels of Cas9. Collectively, our system also provides a robust method to diminish Cas9 activity after certain editing events simply by switching off the light.
Spatial specificity of CRISPR-Cas9 is essential for many potential therapeutic purposes in that Cas9 activity in ancillary tissues may give rise to safety risks. As a proof-of-concept study, we demonstrated that the spatial control of CRISPR-Cas9 activity can well be manipulated in the liver exclusively through optogenetic activation after the systematic administration. Thanks to the NIR-II–absorbing feature of APC, spatial optogenetic control is validated to be realizable in deep tissue, which opens an avenue for broader in vivo investigations. In addition, the current findings also suggest that precise control of Cas9 activity by light is important to diminish off-target effects and other potential genotoxicities (52). In addition to 53BP1-mediated delayed cell division, the decreased off-target effects may also be attributed to the shortened exposure time of the genome to Cas9/sgRNA through optogenetic control, which minimized the tolerable mismatches between sgRNA and genomic loci bearing similar sequences (53). Our future efforts will be dedicated to the intensive investigation of off-target effects at the whole-genome level, in order to understand the safe use of this genome-editing modality.
Materials and Methods
Experimental details and methods can be found in SI Appendix, including the synthesis and characterizations of the APC, heat-inducible Cas9/dCas9 plasmid construction, target design and sgRNA plasmid construction, in vitro photothermal studies, in vitro and in vivo APC-mediated optogenetic control of programmable genome editing, T7E1 assay and deep-sequencing analysis, off-target analysis, temperature control by irradiation, in vivo optogenetic genome editing in varying animal models, hematological and histological analysis, and statistical analysis. All animal treatments and procedures were approved by the Laboratory Animal Welfare and Ethics Committee of Zhejiang University.
Data Availability Statement.
The authors declare that all data supporting the findings of this study are available within the paper and SI Appendix. The deep-sequencing data generated in this paper are available in the NCBI Sequence Read Archive (Bioproject ID PRJNA599254).
Supplementary Material
Acknowledgments
We thank National Key Research and Development Program of China (2018YFA0901800), National Natural Science Foundation of China (81872807), Fundamental Research Funds for the Central Universities (2018XZZX001-14), and Thousand Talents Plan (Y.P.) for the financial support of this work. We also acknowledge Dr. Di Wu for helpful discussions on the synthesis of gold nanorods, and Shuaishuai Zhang for help with the construction of plasmids. We also appreciate Prof. Xing Chang from Westlake University for the helpful discussions of deep-sequencing analysis, Prof. Xue Gao from Rice University for the useful discussions of off-target analysis, and Prof. Yunxian Yu from Zhejiang University for the helpful advice on statistical analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.Z. is a guest editor invited by the Editorial Board.
Data Deposition: The datasets generated in this paper are available in the NCBI Sequence Read Archive (Bioproject ID PRJNA599254).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912220117/-/DCSupplemental.
References
- 1.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox D. B., Platt R. J., Zhang F., Therapeutic genome editing: Prospects and challenges. Nat. Med. 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan T., et al. , Material solutions for delivery of CRISPR/Cas-based genome editing tools: Current status and future outlook. Mater. Today 26, 40–66 (2019). [Google Scholar]
- 4.Nishimasu H., et al. , Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H. X., et al. , CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 117, 9874–9906 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Yang L., et al. , Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350, 1101–1104 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Min Y. L., Bassel-Duby R., Olson E. N., CRISPR correction of Duchenne muscular dystrophy. Annu. Rev. Med. 70, 239–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalem O., Sanjana N. E., Zhang F., High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manguso R. T., et al. , In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soppe J. A., Lebbink R. J., Antiviral goes viral: Harnessing CRISPR/Cas9 to combat viruses in humans. Trends Microbiol. 25, 833–850 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Liu K. I., et al. , A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 12, 980–987 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Aubrey B. J., et al. , An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 10, 1422–1432 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Dow L. E., et al. , Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33, 390–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen D. P., et al. , Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat. Commun. 7, 12009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemphill J., Borchardt E. K., Brown K., Asokan A., Deiters A., Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 137, 5642–5645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nihongaki Y., Kawano F., Nakajima T., Sato M., Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zetsche B., Volz S. E., Zhang F., A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maji B., et al. , Multidimensional chemical control of CRISPR-Cas9. Nat. Chem. Biol. 13, 9–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis K. M., Pattanayak V., Thompson D. B., Zuris J. A., Liu D. R., Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 11, 316–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain P. K., et al. , Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. Engl. 55, 12440–12444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polstein L. R., Gersbach C. A., A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 11, 198–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bubeck F., et al. , Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat. Methods 15, 924–927 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Shao J., et al. , Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, E6722–E6730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ash C., Dubec M., Donne K., Bashford T., Effect of wavelength and beam width on penetration in light-tissue interaction using computational methods. Lasers Med. Sci. 32, 1909–1918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y., et al. , Near-infrared upconversion–activated CRISPR-Cas9 system: A remote-controlled gene editing platform. Sci. Adv. 5, eaav7199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosser D. D., Duchaine J., Massie B., The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol. Cell. Biol. 13, 5427–5438 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abravaya K., Myers M. P., Murphy S. P., Morimoto R. I., The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 6, 1153–1164 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Li X., et al. , In vitro and in vivo photothermal cancer therapeutic effects of gold nanorods modified with mushroom β-glucan. J. Agric. Food Chem. 66, 4091–4098 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Chen Y. S., Zhao Y., Yoon S. J., Gambhir S. S., Emelianov S., Miniature gold nanorods for photoacoustic molecular imaging in the second near-infrared optical window. Nat. Nanotechnol. 14, 465–472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., et al. , Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 40, e1800068 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Andersson H. A., Kim Y. S., O’Neill B. E., Shi Z. Z., Serda R. E., HSP70 promoter-driven activation of gene expression for immunotherapy using gold nanorods and near infrared light. Vaccines (Basel) 2, 216–227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang G., Zhang X., An C., Cheng C., Wang H., Temperature effect on CRISPR-Cas9 mediated genome editing. J. Genet. Genomics 44, 199–205 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Wang P., et al. , Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. Engl. 57, 1491–1496 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Song E., et al. , RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9, 347–351 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Lee K., et al. , In vivo delivery of transcription factors with multifunctional oligonucleotides. Nat. Mater. 14, 701–706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae S., Park J., Kim J. S., Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spies J., et al. , 53BP1 nuclear bodies enforce replication timing at under-replicated DNA to limit heritable DNA damage. Nat. Cell Biol. 21, 487–497 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Hendel A., Fine E. J., Bao G., Porteus M. H., Quantifying on- and off-target genome editing. Trends Biotechnol. 33, 132–140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W., et al. , Dual quantification of microRNAs and telomerase in living cells. J. Am. Chem. Soc. 139, 11752–11759 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Chen S., et al. , Near-infrared deep brain stimulation via upconversion nanoparticle-mediated optogenetics. Science 359, 679–684 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Tan P., He L., Han G., Zhou Y., Optogenetic immunomodulation: Shedding light on antitumor immunity. Trends Biotechnol. 35, 215–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., et al. , Metabolizable semiconducting polymer nanoparticles for second near-infrared photoacoustic imaging. Adv. Mater. 31, e1808166 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Zhu S., Tian R., Antaris A. L., Chen X., Dai H., Near-infrared-II molecular dyes for cancer imaging and surgery. Adv. Mater. 31, e1900321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conde J., Oliva N., Zhang Y., Artzi N., Local triple-combination therapy results in tumour regression and prevents recurrence in a colon cancer model. Nat. Mater. 15, 1128–1138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinde E., et al. , Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 12, 81–89 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Morimoto R. I., Cells in stress: Transcriptional activation of heat shock genes. Science 259, 1409–1410 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Abravaya K., Phillips B., Morimoto R. I., Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperatures. Genes Dev. 5, 2117–2127 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Lyu Y., et al. , Dendronized semiconducting polymer as photothermal nanocarrier for remote activation of gene expression. Angew. Chem. Int. Ed. Engl. 56, 9155–9159 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Miyako E., et al. , Photothermic regulation of gene expression triggered by laser-induced carbon nanohorns. Proc. Natl. Acad. Sci. U.S.A. 109, 7523–7528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raj B., et al. , Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol. 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen S. T., et al. , Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 393, 3–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gangopadhyay S. A., et al. , Precision control of CRISPR/Cas9 using small molecules and light. Biochemistry 58, 234–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao J., et al. , An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 44, e149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and SI Appendix. The deep-sequencing data generated in this paper are available in the NCBI Sequence Read Archive (Bioproject ID PRJNA599254).