Significance
The protein homeostasis system prevents proteins from misfolding and aggregation. It is unclear, however, how this system is organized to fulfil this role under different insults. To investigate this problem, we examined how six different stresses affected proteome solubility in a mouse neuroblastoma cell line. We found that, although different stresses substantially changed the solubility of many proteins, no proteins substantially aggregated in common. Instead, each stress distinctly remodeled protein–ligand networks involved in key cellular processes, notably SG formation and metabolism. We conclude that cells are endowed with a wide range of regulatory responses, which are tailored for distinct stresses and buffer against widespread protein aggregation.
Keywords: protein homeostasis, protein aggregation, protein misfolding, Huntington's disease, molecular chaperones
Abstract
The accumulation of protein deposits in neurodegenerative diseases has been hypothesized to depend on a metastable subproteome vulnerable to aggregation. To investigate this phenomenon and the mechanisms that regulate it, we measured the solubility of the proteome in the mouse Neuro2a cell line under six different protein homeostasis stresses: 1) Huntington’s disease proteotoxicity, 2) Hsp70, 3) Hsp90, 4) proteasome, 5) endoplasmic reticulum (ER)-mediated folding inhibition, and 6) oxidative stress. Overall, we found that about one-fifth of the proteome changed solubility with almost all of the increases in insolubility were counteracted by increases in solubility of other proteins. Each stress directed a highly specific pattern of change, which reflected the remodeling of protein complexes involved in adaptation to perturbation, most notably, stress granule (SG) proteins, which responded differently to different stresses. These results indicate that the protein homeostasis system is organized in a modular manner and aggregation patterns were not correlated with protein folding stability (ΔG). Instead, distinct cellular mechanisms regulate assembly patterns of multiple classes of protein complexes under different stress conditions.
The protein homeostasis system is vital for cell function as it ensures that proteins are properly translated, folded, trafficked to their correct cellular locations, and eventually degraded in a tightly controlled and timely manner. As a major task for this system is to prevent damaged and misfolded proteins from accumulating, it has been hypothesized that, when this system becomes unbalanced, proteins become prone to misfolding, which results in their mislocalization and deposition as aggregates (1, 2). Indeed, dysfunctional protein aggregation and protein homeostasis imbalance are central pathological features of common neurodegenerative diseases including Alzheimer’s (AD), Parkinson’s (PD), Huntington's, and motor neuron diseases (3, 4).
Different neurodegenerative diseases are characterized by the presence of signature proteins in characteristic deposits formed in the brains of affected patients. These proteins include TDP-43 and FUS in motor neuron disease, τ and Aβ in AD, α-synuclein in PD, and huntingtin (Htt) in Huntington's disease. It has been suggested that these aggregation-prone proteins, which may be affected by mutations (e.g., Htt), posttranslational modifications (e.g., τ phosphorylation), or environmental changes, lead to oversubscription of quality control resources that overloads the protein homeostasis system (5). This imbalance then triggers a cascade of protein folding defects that broadly impairs proteome function. In cell models expressing mutant Htt exon 1 (Httex1), for example, key molecular chaperone systems are sequestered into inclusions formed by Httex1, which depletes the resources of the protein homeostasis system (6, 7). Studies employing Caenorhabditis elegans models have shown that the aggregation of polyQ proteins can cause temperature-sensitive mutant proteins, which are metastable in their native states, to aggregate (7), which is consistent with a reduced capacity of the quality control system. Similarly, in aging, different genetic backgrounds and environmental stresses can alter the efficiency of the protein homeostasis system (8, 9).
Here, we sought to address three outstanding questions related to the factors capable of compromising protein homeostasis. The first question is how does aggregation of mutant Httex1, which previously has been suggested to unbalance protein homeostasis, impact the aggregation state of the wider proteome? The second is which proteins in the proteome are metastable to aggregation under different triggers of protein homeostasis stress? And the third is whether there is a subproteome that consistently requires a functional protein homeostasis machinery to remain folded and soluble, and if so, how is this subproteome regulated? Our results indicate that a substantial proportion of proteome undergoes large changes, both upward and downward, in solubility in response to stress, but also that each stress is associated with an articulated stress response that affects a different part of the metastable subproteome. Our data further suggest that the majority of the changes arises from the functional remodeling of protein–ligand complexes in adaptation (or response) to the stress and that the changes are highly specific to the different stress factors. Based on these results, we conclude that the resilience of the protein homeostasis system against widespread aggregation is based on the absence of a large common set of metastable proteins that aggregate under multiple stresses. Furthermore, the proteins that change aggregation do not correlate with the protein folding stability (ΔG). In this view, the proteins that change solubility most are functionally responsive to protein homeostasis regulators that are organized into different modules that respond specifically to different perturbations.
Results
Httex1 Mutation and Subsequent Aggregation Distinctly Remodels Proteome Solubility.
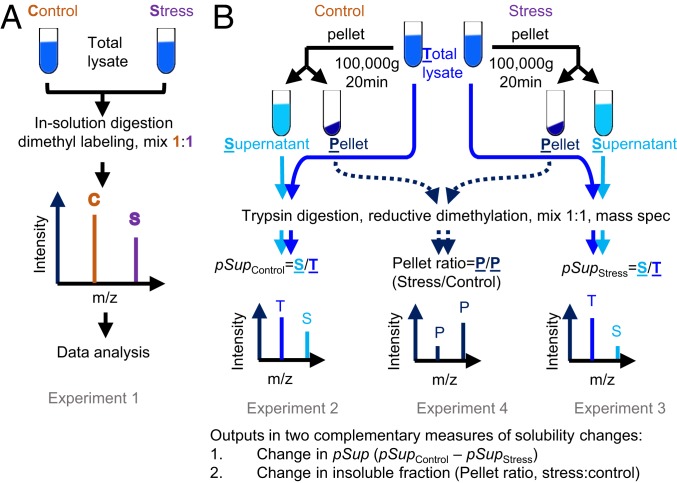
To investigate how protein homeostasis imbalance alters the aggregation state of the proteome, we employed a neuronal-like cell model system (mouse Neuro2a cells) and a quantitative proteomic workflow inspired by the work of Wallace et al. (10). In essence, the approach involved a fractionation strategy based on centrifugation of cell lysates prepared using a mild nonionic detergent based lysis condition (IGEPAL CA-630) with subsequent quantitative proteome analysis to monitor changes in the abundances of individual proteins between the supernatant versus the pellet, resulting from each stress (Fig. 1). Quantitation was performed using a reductive dimethylation labeling approach with n = 4 biological replicates and detection by nano-reversed-phase liquid chromatography coupled with tandem mass spectrometry (MS/MS). We measured the changes in the abundance of proteins in the total starting material (Experiment 1 in Fig. 1A) and applied two reported methods to measure changes in solubility that provide complementary information (10). Δ pSup was defined as the change in proportion of the protein in the supernatant by subtracting the proportion of the protein in the stress (pSupStress) from the supernatant of control (pSupControl) (Experiments 2 and 3 in Fig. 1B). We also measured the changes in the pellet fraction directly as the ratio of proteins in the stress:control treatments (Experiment 4 in Fig. 1B). The measure of pSup is expected to be the best estimate of change in the absolute yield of soluble protein and for proteins that show large proportional changes in solubility. On the other hand, pellet ratios are expected to be selectively sensitive for proteins that have very small fractions of aggregate or proteins that are less abundant in the proteome. Hereon, we use the term solubility to indicate the changes in protein mass measured by this specific experimental framework.
Fig. 1.
Proteomic workflow for quantitative measurements of stress-related proteome solubility changes. (A) Strategy to measure changes in total proteome abundances due to stress (Experiment 1). (B) Strategies to measure changes in solubility by a combination of experiments (Experiments 2–4).
Our first protein homeostasis stress model examined the effect of Httex1 mutation and the aggregation state on the overall proteome solubility. Huntington's disease mutations lead to the expansion of a polyglutamine (polyQ) sequence in Httex1 to lengths longer than 36Q, whereas the wild-type protein is typically less than 25Q (11). PolyQ expansion causes Httex1 to become highly aggregation prone, which manifests as intracellular inclusion bodies as the disease progresses (12). We and others have used transient expression constructs of Httex1 fused to fluorescent proteins as models for replicating essential features of the disease, including protein homeostasis stress (13, 14). Mutant Httex1-fluorescent protein constructs progressively form large cytosolic inclusions in cell culture over time.
We employed the flow cytometry method of pulse shape analysis to purify cells expressing mutant Httex1-mCherry into those with inclusions (i) from those without inclusions ([ni]; dispersed uniformly in the cytosol) at matched median expression levels (15). This strategy enabled us to assess how the aggregation state of mutant Httex1 (97Q ni and i) affected proteome solubility compared to a wild-type state (25Q ni—note that 25Q does not form aggregates) (Fig. 2A and SI Appendix, Fig. S1).
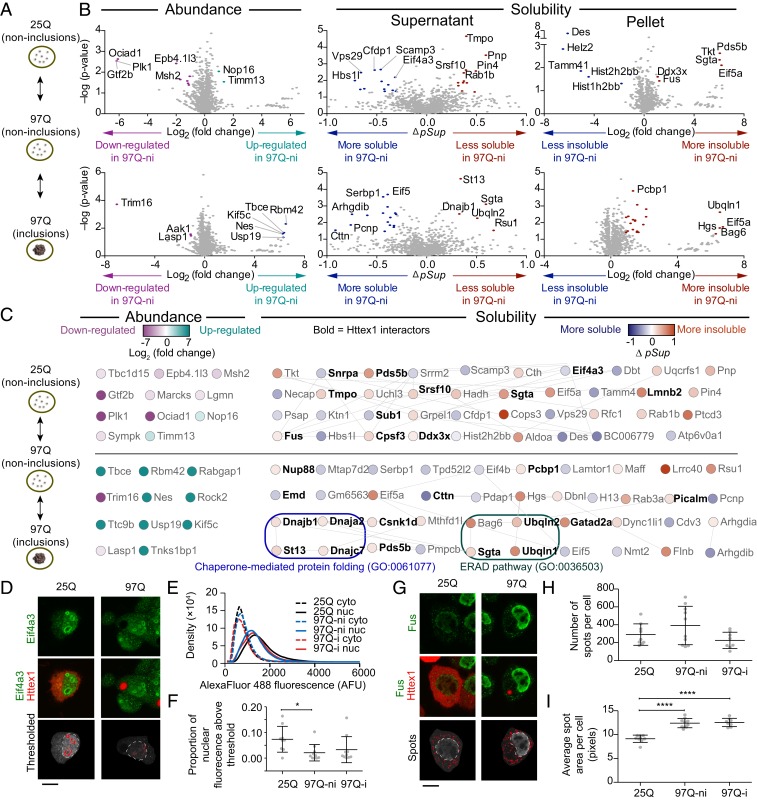
Fig. 2.
Impact of a Httex1 mutation and subsequent aggregation on solubility of endogenous proteins. (A) Neuro2a cells transiently expressing Httex1-GFP for wild-type polyQ (25Q) and mutant polyQ (97Q) were presorted into three groups by PulSA with matched expression levels as shown. ni denotes cells without inclusions, and i denotes cells with inclusions (B) Volcano plots of protein abundances and solubility, aligned to the schematic in A. Gray-colored points indicate proteins below the threshold of significance (twofold change and P value of 0.05 for abundance and Δ pSup value of 0.3, Pellet ratio log2 value of 1 and P value of 0.05). (C) Protein networks represented by STRING (v.11.0) analysis with medium confidence interactions. Selected significantly enriched gene ontology (GO) terms are annotated. All enriched GO terms are included in Dataset S2. (D) Confocal micrographs of Neuro2a cells transfected with Httex1-mCherry and immunostained with Eif4a3. Thresholds for nuclear ring structures (red pattern) and nucleus boundary (white dashed lines) are shown. (E) Histograms of Eif4a3 immunofluorescence intensity inside the nucleus and in the cytoplasm (at least, nine cells were analyzed for each histogram). (F) Proportion of nuclear Eif4a3 immunofluorescence above a defined brightness threshold that corresponds to the ring structures. (G) Confocal micrographs of Neuro2a cells transfected with Httex1-mCherry and immunostained with Fus. Thresholds annotated as described for D. (H) Number of spots per cell. (I) Average size of spots per cell. Data in F, H, and I all show datapoints as values from individual cells and with means ± SD shown. These panels also show results of one-way ANOVA and Tukey’s multiple comparisons test. *P < 0.05 and ****P < 0.0001. (Scale bars, 10 μm.)
First, we assessed the protein abundances using the Experiment 1 pipeline from Fig. 1A. A total of 2,013 proteins were identified (Dataset S1). Only a handful of them (22) significantly changed abundance among the 25Q-ni, 97Q-ni, and 97Q-i samples (Fig. 2B). None of these had known protein–protein interactions (Fig. 2C), and the dataset was enriched with only one GO term (cytoskeleton GO:0005856, Dataset S2). However, several of the proteins have reported roles in Huntington's disease-relevant mechanisms. Further discussion of the functional details of these proteins is in SI Appendix, Note 1.
For the assessment of protein solubility changes, we observed 2,519 proteins* (Fig. 2B). For the comparison between the 97Q-ni and the wild-type 25Q-ni, we observed a slightly higher proportion of proteins that decreased solubility in 97Q-ni (17 more soluble and 22 less soluble). Likewise, a slightly higher proportion of proteins became more insoluble once Httex1 formed inclusions (97Q-ni versus 97Q-i) (16 more soluble and 25 less soluble). None of these differences were statistically significant (χ2 test P = 0.89; Dataset S3—test 1).
Of the proteins that significantly changed solubility due to dispersed or inclusion-localized 97Q Httex1, almost one-third (24 out of 78) were previously reported as interactors of Httex1, five of which became more soluble (16) and 19 of which becaame more insoluble (16–20) (Fig. 2C, shown in bold). The enrichment of known Httex1 interactors was not significant (Fisher's exact test P = 0.36; Dataset S3—test 2). One interesting feature was that 11 of the Httex1-interacting proteins were reported to localize to SGs including Cttn, Pds5b, Cpsf3, Ddx3x, Dnajc7, Eif4a3, Ubqln2, Nup88, Pcbp1, Fus, and Srsf10 (21–26). Eight other SG proteins were also observed to change in solubility, including Helz2, Mthfd1, Serbp1, Eif5a, Eif4b, Cdv3, Pdap1, and Flnb. The enrichment of SG proteins to the proteins that changed solubility was statistically significant overall and within the Httex1 interactors (Fisher's exact test P = 0.0009 and P = 0.02; Dataset S3—tests 3 and 4). Further examination of two of these SG proteins (Eif4a3 and Fus) by immunofluorescence (Fig. 2 D–I) indicated subtle changes in their localization. Eif4a3 resided predominately in the nucleus and was enriched in punctate structures within the nucleus similar to what has been previously described (27) (Fig. 2D). The 97Q treatments led to reduced nuclear staining of Eif4a3 (Fig. 2E) and, in particular, within the ringlike structures (Fig. 2F). Fus also resided mostly in the nucleus but also formed cytoplasmic puncta as anticipated for SGs (Fig. 2G). The 97Q treatment did not appear to change the number of Fus puncta (Fig. 2H), but it did increase the size of the puncta (Fig. 2I).
Of note is the broader role for SG abnormalities involved with misfolded proteins and neurodegenerative diseases (28–30). A GO analysis revealed 37 terms enriched in the proteins that changed their solubility when mutant Httex1 97Q formed inclusions. This set of terms included chaperone-mediated protein folding (GO:0061077) and ER-associated protein degradation pathway (GO:0036503) (full list of enriched GO terms in Dataset S2). Collectively, these data suggested that mutant Httex1 causes two major effects on the proteome. The first is a substantial remodeling of SG proteins into different cellular locations both before and after aggregation into inclusions, which includes some elements becoming more soluble and some less soluble. The second is that the quality control systems involved in ER stress and protein misfolding appear to be selectively remodeled to become less soluble as Httex1 inclusions form, which is consistent with the inclusions recruiting molecular chaperones and other quality control machinery in attempts to clear them (6, 16).
Different Triggers of Protein Homeostasis Stress Invoke Distinct Functional Remodelings of Proteome Solubility.
To investigate whether the proteins that changed solubility upon Httex1 aggregation are relevant to protein homeostasis stress more generally, we expanded our analysis to examine proteome solubility changes associated with five other triggers of protein homeostasis stress that have previously reported roles leading to protein misfolding and aggregation. These stresses included three specific inhibitors of key protein homeostasis hubs (Hsp70, Hsp90, and the proteasome) whereby defects are reported in models of Huntington's disease, protein aggregation, degradation of misfolded proteins, and/or other markers of protein homeostasis stress (6, 16, 31) and two exogenous stress states that reflect pathology observed in neurodegenerative disease settings and protein aggregation in cell models (namely, oxidative stress and ER stress) (32–35). We chose approaches that could be readily and relatively specifically targeted pharmacologically and that have been well studied previously to cause protein homeostasis stress.
The Hsp70 chaperone system was targeted by the small molecule inhibitor Ver-155008, which binds to the ATPase domain of Hsp70 family proteins (Kd of 0.3 µM and IC50 of 0.5–2.6 µM) (36, 37) and can promote the aggregation of a reporter metastable protein in transfected cells (38). Hsp90 was targeted with the ATP binding competitor novobiocin, which can unbalance the protein homeostasis system without activating a compensatory heat shock response and induce the aggregation of a metastable bait protein (38). Proteasome activity was targeted with the inhibitor MG132 (39). ER stress was triggered using the N-linked glycosylation inhibitor tunicamycin, which impairs flux of folding via the calnexin–calreticulin folding pipeline (40). Oxidative stress was induced with arsenite (41, 42). Our experimental design followed the dosages and timings as performed in prior studies as indicated above.
The changes in protein abundance from these treatments are shown in SI Appendix, Fig. S2A (the full proteomics datasets are summarized in Datasets S4–S9). The treatments led to changes consistent with their function based on GO assignment as well as protein–protein interactions (Fig. 3A; complete list of GO terms in Dataset S2). Of note was that many more proteins were observed to have changed solubility (upwards and downward) than had changed abundance, which suggests that protein solubility change, rather than changes in protein expression, is a particularly substantial response to stress (volcano plots in SI Appendix, Fig. S2B). Similar to what was observed for protein abundance changes, the GO terms indicated functional groupings anticipated from protein functions related to the treatment (Fig. 3B for three examples of Hsp90 inhibition by novobiocin treatment, proteasome inhibition, and oxidative stress; SI Appendix, Fig. S3 for the other two of Hsp70 inhibition and ER stress). This result, therefore, indicates that the protein assembly state, not just abundance, is key to understanding the function of the protein homeostasis function (Fig. 3B). For example, MG132 treatment indicated enrichment for proteolysis (GO: 0006508) as anticipated. An effect on proteasome activity was also indicated by MG132 increasing the abundance of ubiquitin and proteasome subunits (SI Appendix, Fig. S4A). Almost all of the proteolysis GO terms were associated with proteins becoming more insoluble, suggesting that the proteasome-degradation machinery forms larger molecular weight complexes when the proteasome is inhibited, which is consistent with the prior knowledge that proteasome inhibition induces the formation of ubiquitin- and proteasome-enriched cellular aggregates (43) (Fig. 3B). Further insight into this functional remodeling of the proteasome machinery was provided by the substantial decrease in solubility of total cellular ubiquitin, even though an overall increase in total ubiquitin abundance was observed (SI Appendix, Fig. S4B).
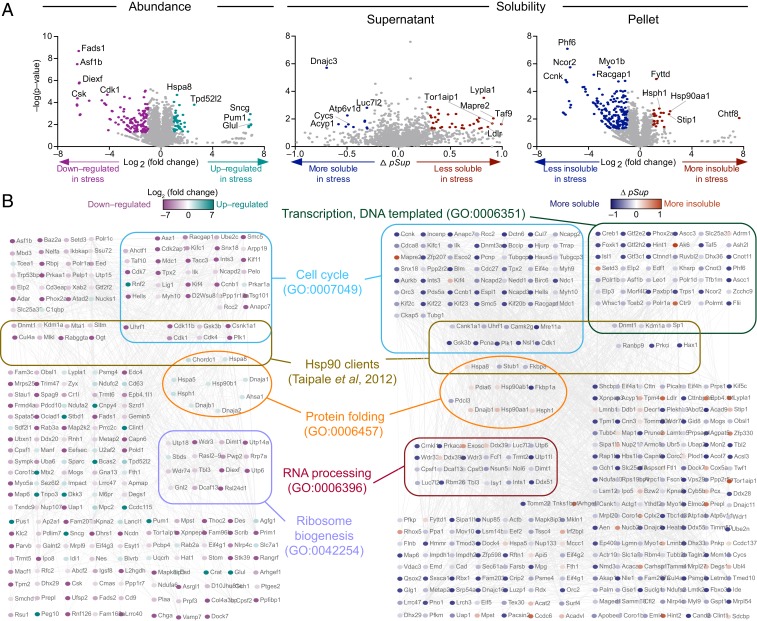
Fig. 3.
Impact of three protein homeostasis stresses on proteome abundance and solubility. Protein–protein interaction analysis of proteins with significant changes in abundance (A) or solubility (B) due to protein homeostasis stresses (Hsp90 inhibition [with novobiocin], proteasome inhibition [with MG132], and oxidative stress [with arsenite], top view) was performed with built-in String (v.11) in Cytoscape (v3.7.1) at the medium confidence. Selected significantly enriched GO terms are annotated. All enriched GO terms are included in Dataset S2. Hsp90 interactions were manually added based on String and shown with thicker black connectors. SG curated list from Markmiller et al. (22).
Another notable finding from this analysis was the ability to extract novel information on the effect of novobiocin treatment on assembly states of macromolecular machines. Novobiocin did not change the levels of Hsp90 or proteins involved in the heat shock response as anticipated but decreased known Hsp90 client proteins, in accordance with previous studies (Fig. 3A) (44, 45). GO and network analysis of the changes in solubility identified many more Hsp90 clients than those detected from expression level analysis as well as large changes in the solubility of proteins in diverse complexes including those that form the proteasome, mitochondrial ribosome, DNA repair machinery, RNA splicing machinery, RNA transport machinery, and respiratory chain complexes (Fig. 3B). Despite these substantial changes in proteome solubility, the enrichment of Hsp90 clients was not significant (Fisher's exact test P = 0.32 and P = 0.75; Dataset S3—tests 5 and 6), which may be a consequence of off-target impacts driving much of the changes in the proteome since novobiocin has an IC50 value around 700 μM (46). To explore this idea further, we tested a different Hsp90 inhibitor, 17-allylamino-17-demethoxy-geldanamycin (17-AAG) (EC50 of 7.2 μM) that operates through a distinct mechanism by inhibiting ATPase function through binding to its amino-terminal domain (47). Unlike novobiocin, inhibition by this mechanism is known to activate the heat shock response (48). In accordance with this effect, 17-AAG increased heat shock protein Hspa8 and other proteins in the protein folding GO term (GO: GO:0006457) (Fig. 4). Like that for the novobiocin treatment, we observed a substantial change in solubility (upward and downward) of a wide range of proteins, including a statistical enrichment of Hsp90 client proteins (Fisher's exact test P = 0.02; Dataset S3—test 7) and proteins in the protein folding GO term. There was a limited overlap in proteins that changed solubility with novobiocin (shown in Dataset S5). The most notable difference between the treatments was that novobiocin appeared to impair some complexes from properly assembling into large molecular weight machines, including the mitochondrial respiratory chain, which contains five multimeric membrane-anchored complexes (49). We observed more than half of identified subunits of mitochondrial respiratory complexes I, III, and IV becoming more soluble after novobiocin treatment, suggesting a failure of these complexes to assemble into their mature states as part of large membrane-anchored complexes, which are anticipated to partition into the insoluble fraction under our pelleting regime.
Fig. 4.
Impact of Hsp90 inhibition by 17-AAG on proteome abundance and solubility. (A) Shown are effects of 17-AGG treatment on abundance and solubility. (B) Protein:protein interaction analysis of proteins with significant changes in abundance (Left) or solubility (Right) due to indicated protein homeostasis stresses was performed using built-in String (v.11.0) in Cytoscape (v3.7.1) at the medium confidence. Selected significantly enriched GO terms are annotated. All enriched GO terms are included in Dataset S2.
Consensus Features of Metastable Subproteomes Changing Solubility under Stress.
To address the fundamental question of whether there is a common metastable subproteome that is more aggregation prone under any condition of stress due to protein misfolding, we next sought to assess whether stress increased the net insolubility of the proteome by measuring the protein mass concentration in the supernatants before versus after pelleting†. There was a small but significant decrease in solubility of 3.6% (P = 0.0047 by two-way ANOVA; Dataset S3) (Fig. 5A). These results suggest that aggregation arising from misfolded proteins increases marginally under stress but does not reflect a dramatic accumulation of misfolded protein states that aggregate.
Fig. 5.
Dynamic remodeling of proteome solubility involving a core enrichment of proteins involved in nucleocytoplasmic transport and SGs. (A) Overall proteome solubility changes due to treatments. Comparison of the proportion of protein amount in the supernatant fraction out of the total lysate measured by bicinchoninic acid assay between the control and the treatment groups. Error bars represent SD. N = 3 or 4. (B) List of proteins found in common as shown. (C) Venn diagram presents the overlaps of more insoluble or soluble proteins across six stresses. The area enclosed by the thick black line represents the overlap regions with, at least, three stresses. Protein–protein interaction of common proteins that became more insoluble (red) and more soluble (blue) in, at least, three stresses are shown. Purple bold represents known SG proteins.
At the individual protein level, there were no proteins that consistently decreased or increased in solubility across six stresses using our criteria for a significant change. Of the proteins detected in all experiments, 408 proteins significantly decreased solubility by any one or more of the stress treatments, which represent 10.5% of the proteins detected (4,278). In addition, 315 proteins had significantly increased solubility (8.2%). A further 183 proteins increased or decreased solubility depending on the stress (4.9%). Altogether, the proteins that changed solubility was just over one-fifth of the proteome (21.2%) with a significant bias to proteins becoming more insoluble (based on 95% confidence intervals [CIs] on proportions; Dataset S3). Collectively, these data indicate that significant remodeling in proteome solubility occurs under stress but that most of the increase in insoluble protein load is counterbalanced by other proteins increasing in solubility.
Despite this widespread change in solubility, the changes were largely specific to the type of stress invoked. For example, we observed that only 26 proteins became more insoluble in, at least, three stresses, which represent just 0.6% of the proteome detected (Fig. 5B) while 139 proteins (2.6% of the proteins) became more insoluble in, at least, two stresses. Only three proteins, Pcbp1, Bag3, and Sqstm, were more insoluble in four stresses, and only one protein, asparagine synthetase (Asns), was more insoluble in five of the stresses (Dataset S9). Likewise, for proteins that became more soluble, we observed only a low number of proteins (8) that became more soluble in, at least, three of the stresses (0.2% of the proteome detected) (Fig. 5B and Dataset S9). No protein was found to be more soluble in more than three stresses. Expanding the analysis to two stresses yielded 73 proteins (1.7% of the proteome).
GO analysis of the proteins that changed solubility in two or more stresses illuminated the key mechanisms involved (Fig. 6A). The most enriched molecular function terms for the more insoluble protein list were proteasome-activating ATPase activity (82.6-fold enrichment), proteasome binding (36.7-fold), structural constituent of nuclear pore (35.9-fold), ATPase activator activity, (9.9-fold), and heat shock protein binding (9.8-fold) ubiquitin protein ligase binding (6.0-fold). These functions are consistent with protein quality control mechanisms being engaged to respond to proteome folding stress. Other major biological process and cellular component terms of enrichment include lamin filament (99.0-fold), nuclear pore outer ring (66-fold), mRNA export from the nucleus (22.0-fold), and protein import into the nucleus (14.6-fold), which are consistent with previously reported findings that protein folding stress, more generally, impacts nucleocytoplasmic transport mechanisms (24, 50–55). For the proteins that become more soluble, we observed terms highly enriched for mitochondrial activity including ATPase activity, coupled to the transmembrane movement of ions, rotational mechanism (46.9-fold), proton-exporting ATPase activity (40.0-fold), proton transmembrane transporter activity (15.5-fold), ATP biosynthetic process (21.8-fold), and mitochondrial protein complex (9.7-fold).
Fig. 6.
Biological pathways involved in the functional remodeling of proteome solubility. (A) GO terms for proteins significantly changed in solubility in two or more stresses. Shown are the top tier GO terms tested with Fisher's exact test and Bonferroni post hoc testing with P < 0.05. (B) The proportion of detected SG proteins using the curated list of Markmiller et al. (22) for proteins in the indicated number of stresses. Black circle indicates background proteome. Colored circles indicate proteins that significantly changed solubility. Fisher's exact test results comparing stress to background are shown (*P < 0.05; **P < 0.01; ****P < 0.0001) (Dataset S3—test 9). Mean and 95% CIs shown from proportions analysis (Dataset S3) (C) Barcode graph of SG proteins from Markmiller et al. (22) and solubility.
Another striking observation was that 183 of the proteins that become more insoluble in one or more of the stresses also became more soluble in one or more of the other stresses (Dataset S9). This finding suggests that these proteins are functionally tailored to different roles in different stresses. Clues to the key pathways involved came from examination of the proteins that became more insoluble in, at least, three stresses, revealing GO enrichments for nucleocytoplasmic transport and cellular response to stress, which are consistent with stress and prior findings that nuclear-cytoplasmic transport is altered in neurodegenerative disease settings (Fig. 5C) (50–54). Another finding was the profound enrichment of SG proteins in both proteins that become more insoluble as well as those that become more soluble in, at least, three stresses (Fig. 5C). Indeed, there was a significant enrichment of proteins involved in SGs that become both more insoluble and more soluble across one or more stresses (Fig. 6B). Curiously, a majority of the SG proteins displayed differentially altered solubility depending on the stress (Fig. 6C). These observations indicate a great diversity, dynamism, and heterogeneity in the complexes that are formed by SG proteins and are suggestive of an elaborate tailoring of the functional responses of the SG structures to the stress.
Discussion
In this paper, we found that about one-fifth of the proteome of mouse neuroblastoma cells undergoes solubility changes in response to an array of stresses to the protein homeostasis system. We found that most of the proteins that decrease in solubility are counteracted by other proteins that increase in solubility. Different stresses, therefore, evoke a regulatory response that rebalances homeostasis that involves a large functional remodeling of protein–ligand interactions. It is also apparent that protein aggregation arising from the perturbation of protein quality control under these conditions accounts for only a minor fraction of the changes in the proteome solubility (probably less than 5%). Indeed, if marginally stable proteins mediate the shift in aggregation under protein homeostasis stress, we might predict there to be a correlation between the free energy of folding (ΔGF) and solubility. Analysis of the proteome ΔGF values from ref. 56 for the proteins in our dataset revealed no correlations for any individual stress or the pooled data (SI Appendix, Fig. S5 and Dataset S10). Our results, thus, reveal an important principle underlying the robustness of the protein homeostasis system, namely, that there is no bellwether set of proteins that are profoundly metastable due to being difficult to fold, including obvious candidate proteins, such as TDP-43 or τ which are commonly mislocalized and/or misfolded in neurodegenerative disease contexts (57, 58). Instead, changes are the assembly state of proteins that are dominated by functional responses to different stresses (an alternative analysis of the core proteins that change solubility to the data in Fig. 5C is shown in SI Appendix, Fig. S6).
These findings are informative to a large-scale proteomic study of mouse models of different neurodegenerative diseases, including AD, PD, and amyotrophic lateral sclerosis (59). In that study, 91 proteins were found to become more sodium dodecyl sulfate (SDS) insoluble in three transgenic mouse models. Our data show a significant enrichment for these proteins when we considered proteins that decreased solubility by, at least, one stress (Fisher's exact test P = < 0.0001; Datasets S9 and S3–test 10). These findings point to the general principles of proteome solubility remodeling operating as found in the mouse neuroblastoma cell context also in more chronic disease settings.
In our dataset, there was only one protein, filamin C (Flnc), that appeared to stand out as a potential bona fide bellwether protein for substantial aggregation by misfolding. Flnc was observed to become more insoluble in three stresses (ER stress, oxidative stress, and proteasome inhibition). Mutations in Flnc cause myofibrillar myopathies, which are characterized by protein deposits, a defective ubiquitin–proteasome system, and oxidative stress (60). Flnc is a cytoskeletal protein that has been shown to aggregate in cells that lack activity of small heat shock protein HspB7 (61). We also observed a binding partner of Flnc, Desm, to be more soluble in four stresses (but, of note, was not seen in all of the stress datasets). This result would be expected if Flnc misfolds and aggregates and is unable to act as an appropriate scaffold to interacting partners. Further evidence is that other genes that cause myofibrillar myopathies when mutated are Desm and chaperones DnajB6, HspB8, Cryab, and Bag3 which suggest these chaperones are critical to stabilizing the folded state of Flnc (62). We also noted Bag3, which also colocalizes in SGs, as one of our proteins that become more insoluble in, at least, three stresses. Thus, it remains possible that the machinery involving Bag3 and other SGs is very competent at buffering against aggregation of misfolded proteins under stress (63) and that Flnc might be one of the first proteins vulnerable to aggregation under prolonged stress.
The signatures for the stresses were generally distinct, although we observed a strong association with SG proteins and folding stress as the major activator of quality control systems. In concert, these responses appear to robustly buffer misfolded proteins from actually aggregating and are reminiscent of findings from a prior study that found heat shock in yeast led to an adaptive autoregulatory assembly and disassembly of protein complexes and minimal aggregation from denatured endogenous proteins (10). Our results extend from this finding to show that such responses are generally applicable to stress and that each stress provides a unique pattern of response.
One of the intriguing findings was the heterogeneity and dynamism of SG proteins. There are now, at least, 238 proteins that have been curated to reside in SGs (22). It is apparent that there is compositional heterogeneity in the assembly of SG (22, 64) and our data further suggest an even more diverse level of heterogeneity and specificity to different forms of stress than currently understood.
The protein seen in five stresses, Asns, has been reported to form filaments in yeast under stress (65), suggesting it also is functionally remodeled in the stress response. When we consider the solubility changes in proteins found in, at least, three stresses in terms of KEGG pathways, we observed a clustering into core metabolic pathways of lipid metabolism, carbohydrate metabolism, nucleotide metabolism, amino acid metabolism, and energy metabolism (SI Appendix, Fig. S7). The clusters included proteins that increased and decreased solubility and suggest that the remodeling of the proteome solubility is functionally linked to core responses associated with the protein homeostasis stress induction. One last noteworthy point is that enzymes involved in metabolism pathways, including specifically those found in our paper as hotspots for changes in solubility, have been shown to form molecular condensates from phase separation in yeast and other cell models (66), which suggests that functional aggregation changes by phase separation could drive many of the functional changes seen in the data presented here, such as SGs. Overall, our findings suggest a link among metabolic responses, SG formation, and proteome solubility remodeling involving about a fifth of the proteome and reveal a modular organization of the protein homeostasis system that regulates metastable proteins against aggregation.
Methods
Expanded details are provided in SI Appendix, Supplementary Methods.
Stress Conditions.
Neuro2a cells were cultured in a medium with the following stress treatments: 20 µM Ver155008 (Sigma-Aldrich) for 18 h; 800 µM novobiocin (Sigma-Aldrich) for 6 h; 5 µM MG132 (Sigma-Aldrich) for 18 h; 500 µM sodium arsenite (Sigma-Aldrich) for 1 h; 5 µM tunicamycin (Sigma-Aldrich) for 18 h; 10 µM 17-AAG (Sigma-Aldrich) for 24 h. For Httex1 stress, Neuro2a cells were transiently transfected with the Httex1 fusions to mCherry.
Cell Fractionation by Ultracentrifugation.
Neuro2a cells were harvested, pelleted, and frozen −80 °C. The pellets were resuspended in buffer 1 (50 mM Tris⋅HCl, 150 mM NaCl, 1% [vol/vol] IGEPAL CA-630, 10 units/mL DNase I, and 1:1,000 protease inhibitor) and lysed by extrusion. The cell lysate was then split into two, one for total protein assessment and the other pelleted at 100,000 g for 20 min at 4 °C. The resulting supernatant was the collected sample.
Protein Sample Preparation for Mass Spectrometry.
Some 100 μL (∼100 µg) of proteins from each sample were reduced, alkylated, and treated to a standard quantitative proteomics workflow following trypsin digestion (details in SI Appendix, Supplementary Methods). Quantitation was reductive dimethyl labeling.
Statistical Analysis.
The details of the tests were indicated in the figure legends. Significant results were defined for P < 0.05 in the figures. Dataset S3 shows the statistical results for tests cited in the paper.
Data Availability.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers (accession nos. PXD014420 and PXD015573).
Supplementary Material
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.G. is a guest editor invited by the Editorial Board.
Data deposition: The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers (accession nos. PXD014420 and PXD015573).
*This includes the sum of proteins seen in the 25Q-ni versus 97Q-ni and 97Q-ni versus 97Qi treatments.
†Note we were unable to measure solubility for the Huntington’s disease cell model due to small yields of cells after flow cytometry sorting.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912897117/-/DCSupplemental.
References
- 1.Schneider K., Bertolotti A., Surviving protein quality control catastrophes–From cells to organisms. J. Cell Sci. 128, 3861–3869 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Knowles T. P., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Bertram L., Tanzi R. E., The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 115, 1449–1457 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hipp M. S., Park S. H., Hartl F. U., Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Cox D., Raeburn C., Sui X., Hatters D. M., Protein aggregation in cell biology: An aggregomics perspective of health and disease. Semin. Cell Dev. Biol., 10.1016/j.semcdb.2018.05.003 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Park S. H., et al. , PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I., Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Gidalevitz T., Krupinski T., Garcia S., Morimoto R. I., Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 5, e1000399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther D. M., et al. , Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace E. W., et al. , Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duyao M., et al. , Trinucleotide repeat length instability and age of onset in huntington’s disease. Nat. Genet. 4, 387–392 (1993). [DOI] [PubMed] [Google Scholar]
- 12.DiFiglia M., et al. , Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S., Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Ramdzan Y. M., et al. , Huntingtin inclusions trigger cellular quiescence, deactivate apoptosis, and lead to delayed necrosis. Cell Rep. 19, 919–927 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Ramdzan Y. M., et al. , Tracking protein aggregation and mislocalization in cells with flow cytometry. Nat. Methods 9, 467–470 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kim Y. E., et al. , Soluble oligomers of PolyQ-expanded huntingtin target a multiplicity of key cellular factors. Mol. Cell 63, 951–964 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Kino Y., et al. , FUS/TLS acts as an aggregation-dependent modifier of polyglutamine disease model mice. Sci. Rep. 6, 35236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratovitski T., et al. , Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirasaki D. I., et al. , Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcombe E. A., et al. , Tadpole-like conformations of huntingtin exon 1 are characterized by conformational heterogeneity that persists regardless of polyglutamine length. J. Mol. Biol. 430, 1442–1458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markmiller S., et al. , Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain S., et al. , ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K., et al. , Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dao T. P., et al. , Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C. H., Ohn T., Ivanov P., Tisdale S., Anderson P., eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One 5, e9942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daguenet E., et al. , Perispeckles are major assembly sites for the exon junction core complex. Mol. Biol. Cell 23, 1765–1782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boeynaems S., et al. , Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackenzie I. R., et al. , TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateju D., et al. , An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36, 1669–1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosp F., et al. , Spatiotemporal proteomic profiling of huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hetz C., Saxena S., ER stress and the unfolded protein response in neurodegeneration. Nat. Rev. Neurol. 13, 477–491 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Tamás M. J., Sharma S. K., Ibstedt S., Jacobson T., Christen P., Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules 4, 252–267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson T., et al. , Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J. Cell Sci. 125, 5073–5083 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Medicherla B., Goldberg A. L., Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 182, 663–673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson D. S., et al. , Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 52, 1510–1513 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Schlecht R., et al. , Functional analysis of Hsp70 inhibitors. PLoS One 8, e78443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood R. J., et al. , A biosensor-based framework to measure latent proteostasis capacity. Nat. Commun. 9, 287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg A. L., Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ron D., Harding H. P., Protein-folding homeostasis in the endoplasmic reticulum and nutritional regulation. Cold Spring Harb. Perspect. Biol. 4, a013177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamann I., Klotz L. O., Arsenite-induced stress signaling: Modulation of the phosphoinositide 3′-kinase/Akt/FoxO signaling cascade. Redox Biol. 1, 104–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flora S. J., Arsenic-induced oxidative stress and its reversibility. Free Radic. Biol. Med. 51, 257–281 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Kopito R. R., Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Taipale M., et al. , Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma K., et al. , Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol Cell Proteomics 11, M111.014654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donnelly A., Blagg B. S., Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr. Med. Chem. 15, 2702–2717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte T. W., Neckers L. M., The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother. Pharmacol. 42, 273–279 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R., Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Mimaki M., Wang X., McKenzie M., Thorburn D. R., Ryan M. T., Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta 1817, 851–862 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Woerner A. C., et al. , Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351, 173–176 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Grima J. C., et al. , Mutant huntingtin disrupts the nuclear pore complex. Neuron 94, 93–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eftekharzadeh B., et al. , Tau protein disrupts nucleocytoplasmic transport in alzheimer’s disease. Neuron 99, 925–940 (2018). Erratum in: Neuron 101, 349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freibaum B. D., et al. , GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K., et al. , The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreira P. A., The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration. Cell. Mol. Life Sci. 76, 2247–2273 (2019). Erratum: Cell. Mol. Life Sci. 76, 2275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker E. J., Bettinger J. Q., Welle K. A., Hryhorenko J. R., Ghaemmaghami S., Global analysis of methionine oxidation provides a census of folding stabilities for the human proteome. Proc. Natl. Acad. Sci. U.S.A. 116, 6081–6090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumann M., et al. , Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Wang Y., Mandelkow E., Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Pace M. C., et al. , Changes in proteome solubility indicate widespread proteostatic disruption in mouse models of neurodegenerative disease. Acta Neuropathol. 136, 919–938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrer I., Olivé M., Molecular pathology of myofibrillar myopathies. Expert Rev. Mol. Med. 10, e25 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Juo L. Y., et al. , HSPB7 interacts with dimerized FLNC and its absence results in progressive myopathy in skeletal muscles. J. Cell Sci. 129, 1661–1670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kley R. A., Olivé M., Schröder R., New aspects of myofibrillar myopathies. Curr. Opin. Neurol. 29, 628–634 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Fu H., et al. , A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 22, 47–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aulas A., et al. , Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927–937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S., Ding K., Shen Q.-J., Zhao S., Liu J.-L., Filamentation of asparagine synthetase in Saccharomyces cerevisiae. PLoS Genet. 14, e1007737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prouteau M., Loewith R., Regulation of cellular metabolism through phase separation of enzymes. Biomolecules 8, E160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers (accession nos. PXD014420 and PXD015573).