Significance
The surface protein Pfs47 makes Plasmodium falciparum parasites “undetectable” to the mosquito immune system. We proposed the “lock and key” model in which a parasite expressing a compatible Pfs47 haplotype “the key,” is able to evade the mosquito immune system, by interacting with a mosquito Pfs47 receptor “the lock,” which differs between vectors from different continents. Here, we uncovered the identity of the mosquito midgut “Pfs47 receptor” (P47Rec). P47Rec has two natterin domains and structural homology to a cytoskeleton-interacting protein. It binds Pfs47 with high affinity, and silencing P47Rec reduced P. falciparum infection. The molecular interactions of P47Rec from different mosquito species with Pfs47 haplotypes from Africa and South America give further support to the lock and key model.
Keywords: Pfs47 receptor, mosquito, immune evasion, Anopheles, Plasmodium
Abstract
The surface protein Pfs47 allows Plasmodium falciparum parasites to survive and be transmitted by making them “undetectable” to the mosquito immune system. P. falciparum parasites express Pfs47 haplotypes compatible with their sympatric vectors, while those with incompatible haplotypes are eliminated by the mosquito. We proposed that Pfs47 serves as a “key” that mediates immune evasion by interacting with a mosquito receptor “the lock,” which differs in evolutionarily divergent anopheline mosquitoes. Recombinant Pfs47 (rPfs47) was used to identify the mosquito Pfs47 receptor protein (P47Rec) using far-Western analysis. rPfs47 bound to a single 31-kDa band and the identity of this protein was determined by mass spectrometry. The mosquito P47Rec has two natterin-like domains and binds to Pfs47 with high affinity (17 to 32 nM). P47Rec is a highly conserved protein with submicrovillar localization in midgut cells. It has structural homology to a cytoskeleton-interacting protein and accumulates at the site of ookinete invasion. Silencing P47Rec expression reduced P. falciparum infection, indicating that the interaction of Pfs47 with the receptor is critical for parasite survival. The binding specificity of P47Rec from distant anophelines (Anopheles gambiae, Anopheles dirus, and Anopheles albimanus) with Pfs47-Africa (GB4) and Pfs47-South America (7G8) haplotypes was evaluated, and it is in agreement with the previously documented compatibility between P. falciparum parasites expressing different Pfs47 haplotypes and these three anopheline species. Our findings give further support to the role of Pfs47 in the adaptation of P. falciparum to different vectors.
Malaria remains the most devastating human parasitic disease. It is caused by protozoan plasmodia parasites and is transmitted to humans by the bite of infected anopheline mosquitoes. Most malaria morbidity (92%) and mortality (93%) is caused by Plasmodium falciparum infections in Africa, with 200 million infections worldwide and 400,000 deaths per year, mostly of young African children (1). P. falciparum malaria originated in Africa (2) and was spread around the world by infected humans, as they migrated to regions that harbored different anopheline species. P. falciparum successfully adapted to more than 70 anopheline mosquitoes worldwide, some of them evolutionarily distant to the major African vectors (3, 4).
Plasmodium undergoes a complex development in the mosquito vector, including sexual reproduction (5–7). Mosquitoes become infected when they ingest blood from an infected host that contains both male and female Plasmodium gametocytes, which mature into gametes in a few minutes after they reach the lumen of the mosquito gut, where fertilization takes place. The resulting zygotes mature into motile ookinetes that invade and traverse the mosquito midgut epithelium. Studies with Plasmodium berghei (mouse malaria model) have shown that Anopheles gambiae, the main malaria vector in Africa, has an innate immune system capable of eliminating the majority of invading ookinetes through activation of the mosquito complement-like pathway. Effective deployment of this defense response requires the coordinated activation of epithelial, cellular, and humoral components of the mosquito immune system.
P. berghei ookinete midgut invasion causes irreversible cell damage and triggers a caspase-mediated apoptotic pathway involving activation of a strong epithelial nitration response by the JNK signaling pathway (8, 9). Hemocytes are constantly patrolling the basal surface of the midgut and are attracted to the invasion site by the release of midgut prostaglandins (10). Furthermore, we have previously shown that when hemocytes come in contact with an area of the midgut basal lamina that has been nitrated, they undergo apoptosis and release hemocyte-derived microvesicles (HdMvs), which traverse the basal lamina and reach the basal labyrinth. HdMv release is critical for effective activation of the complement-like system (11). The thioester containing protein 1 (TEP1), a homolog of the vertebrate C3 complement factor, is a key effector of the mosquito complement, which covers the ookinete surface and forms a complex that ultimately lyses the parasite (12–14). The component(s) released by HdMvs required for TEP1-mediated parasite lysis remains to be defined.
In contrast, the A. gambiae immune system cannot mount an efficient immune response to the African P. falciparum lines NF54 and GB4 (15–17), as well as others, presumably because these parasites are well adapted to their natural vector. We have shown that this lack of response is in part due to the parasite’s ability to evade the mosquito immune system, that is mediated by Pfs47. This protein is present on the surface of female gametes, zygotes, and ookinetes (18). Pfs47 disrupts JNK/caspase-mediated apoptosis in the invaded midgut cell, preventing epithelial nitration through an unknown mechanism (19).
Pfs47 is a polymorphic protein with multiple haplotypes that exhibit a striking geographic population structure (20, 21). Pfs47 is one of the genes in the P. falciparum genome with the highest fixation indexes (FST) between populations from different continents (22). The strong geographic population structure of Pfs47 is consistent with natural selection of specific haplotypes during the adaptation of P. falciparum to different anopheline species around the world. We have shown that P. falciparum lines from different continents infect sympatric vectors at much higher levels, and that the haplotype of Pfs47 present in a parasite is a major determinant of its compatibility with a given anopheline vector (21). When a vector is infected with an incompatible P. falciparum line, such as infection of New World Anopheles albimanus mosquitoes with P. falciparum NF54 from Africa, the parasites are eliminated (21). Moreover, the elimination of incompatible combinations is mediated by the mosquito immune system, because disruption of the mosquito complement-like system allows incompatible parasites to survive. We have proposed the “lock and key” model in which a parasite expressing a compatible Pfs47 haplotype “the key,” is able to evade the mosquito immune system, survives, and is selected by the vector through interaction with a mosquito Pfs47 receptor “the lock,” which differs between malaria vector species from different continents, such as A. gambiae (Africa), Anopheles dirus (South East Asia), and A. albimanus (Central and South America) (21). Here we investigated the nature of the “Pfs47 receptor” in anopheline mosquitoes and uncovered its identity. We found that the molecular interaction of different Pfs47 haplotypes with mosquito receptors from different vector species is consistent with the proposed lock and key model.
Results
Molecular Interactions of Pfs47 with the Mosquito Midgut.
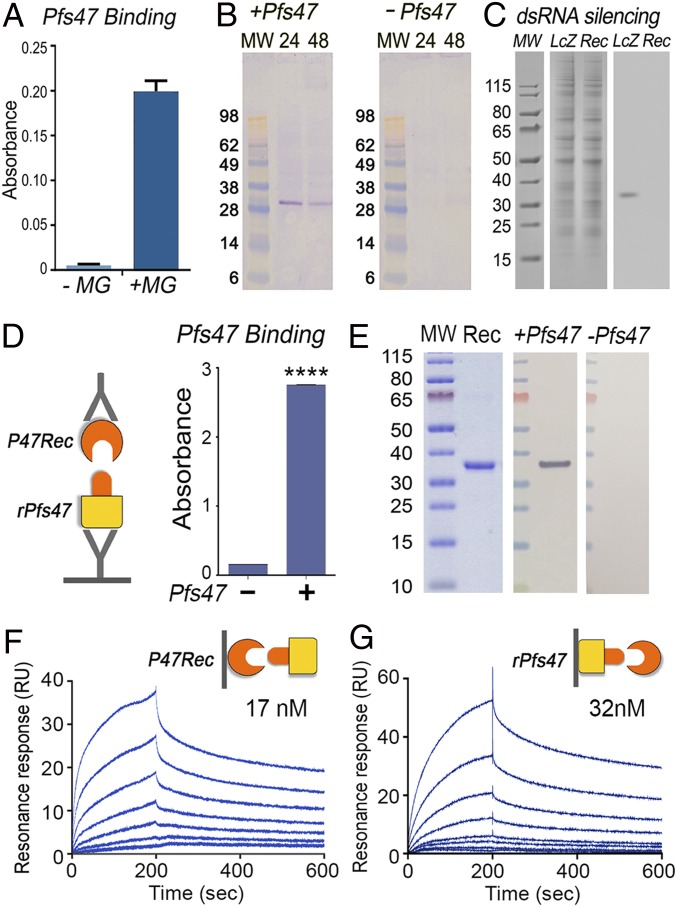
The observed differences in midgut epithelia cellular responses to invasion by wild-type (WT) and Pfs47 knockout (KO) parasites (19) suggests that Pfs47 interacts specifically with some cellular component(s) of the mosquito midgut. The ability of recombinant Pfs47 to interact with protein extracts from the mosquito midgut was therefore explored. Purified recombinant Pfs47 (rPfs47) of the GB4 haplotype, the most frequent Pfs47 haplotype in Africa, had strong binding to immobilized A. gambiae midgut homogenates in ELISA (Fig. 1A). Furthermore, rPfs47 also bound to a single band of about 30 kDa in far-Western assays, in which A. gambiae midgut homogenates collected from females 24 h or 48 h postfeeding (PF) on human serum were separated by SDS/PAGE under nonreducing conditions and transferred to a nitrocellulose membrane (Fig. 1B and SI Appendix, Fig. S1). rPfs47 bound to a protein band present in the pellet fraction (containing membrane- and cytoskeleton-associated proteins) that was not present in the cytosol (SI Appendix, Fig. S1). This band was not present in control membranes that were not incubated with rPfs47 (Fig. 1B). rPfs47 also bound to a single protein spot of the same size (∼31 kDa) present in the pellet fraction of midgut homogenates subjected to two-dimensional (2D) nonreducing SDS gel electrophoresis, followed by far-Western blot analysis (SI Appendix, Fig. S2A). The interacting protein spot and surrounding gel areas (negative controls) were excised from a corresponding Coomassie blue stained 2D gel (SI Appendix, Fig. S2B) and subjected to mass spectrometry analysis. Bioinformatic analysis of the detected peptides identified A. gambiae protein AGAP006398 as the most abundant protein in the area interacting with rPfs47 (Dataset S1, spot A2). Surrounding areas of the rPfs47 interacting spot also presented AGAP006398 as the most abundant mosquito protein (Dataset S1, spots A1 and 3 to 8). Silencing AGAP006398 expression by systemic injection of dsRNA disrupted the interaction of rPfs47 with the 31-kDa band present in the midgut homogenates (Fig. 1C). Thus, we will refer to AGAP006398 as A. gambiae P47 receptor (AgP47Rec). Recombinant AgP47Rec protein was expressed in Escherichia coli, purified, and used to establish the affinity of the interaction with rPfs47 (GB4 haplotype). rPfs47 binds to AgP47Rec in ELISA (Fig. 1D) and far-Western assays (Fig. 1E). Furthermore, binding kinetics analysis using surface plasmon resonance revealed a high-affinity interaction between rPfs47 and AgP47Rec. The affinity constant (Kd) of soluble rPfs47 binding to AgP47Rec immobilized in the sensor was 17 nM (Fig. 1F), while the Kd of soluble AgP47Rec binding to immobilized rPfs47 was 32 nM (Fig. 1G), both in the nanomolar range. Altogether, this evidence indicates that Pfs47 binds with high affinity to a specific midgut protein.
Fig. 1.
Binding of Pfs47 to a specific protein in A. gambiae midgut. (A) Binding of rPfs47 haplotype GB4 (Pfs47) to midgut homogenate (MG) in an ELISA. (B) Binding of rPfs47 to midgut proteins in a far-Western of midgut membrane/cytoskeleton fractions collected 24 or 48 h postfeeding and separated by SDS/PAGE. A control far-Western was run without incubation with rPfs47 (−Pfs47). (C) Effect of dsRNA-mediated silencing of the AgP47Rec (Rec) in binding of rPfs47 in a far-Western of midguts separated by SDS/PAGE. Control mosquitoes were injected with dsLacZ (LcZ). Coomassie blue staining of the midgut protein is shown on the Left, far-Western on the Right. (D) Binding of AgP47Rec to immobilized rPfs47 in a sandwich ELISA. t test, ****P < 0.0001. (E) Coomassie blue staining of AgP47Rec run in an SDS/PAGE (Left), binding of rPfs47 to AgP47Rec in a far-Western (Middle), and negative control not incubated with rPfs47 (Right). (F) Binding kinetics of rPfs47 at different concentrations to immobilized AgP47Rec by plasmon resonance (Kd = 17 nM). (G) Binding kinetics of AgP47Rec at different concentrations to immobilized rPfs47 by surface plasmon resonance (Kd = 32 nM). MW, molecular weight protein markers.
AgP47Rec Gene Structure, Expression, and Effect on Plasmodium Infection.
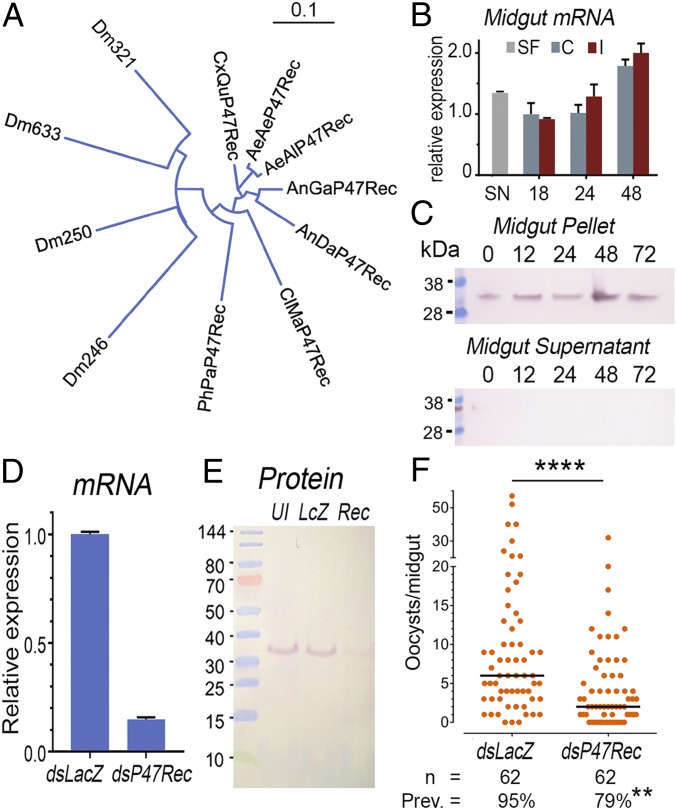
The AgP47Rec gene has four exons that encode a 290-amino acid (aa) protein (SI Appendix, Fig. S3) with a predicted molecular weight (MW) of 31.28 kDa and an isoelectric point of 6.51, in close agreement with the spot detected in the 2D far-Western. The AgP47Rec protein has four DM9 repeats, a motif described in several Drosophila proteins of unknown function (23), organized as two natterin-like domains (SI Appendix, Fig. S3). Natterins are a family of secreted proteins with two DM9 repeats that are abundant in fish venom (24). In A. gambiae, P47Rec is most similar in sequence to plasmodium responsive salivary gland 1 (PRS1) (AGAP006102), and AGAP006103, two smaller proteins with a single natterin-like domain, which share 51% and 56% aa identity, respectively, with AgP47Rec. Interestingly, PRS1 is an agonist for ookinete invasion of the midgut and for sporozoite salivary gland invasion in mosquitoes infected with either P. berghei or P. falciparum parasites (25).
AgP47Rec is the only protein in A. gambiae with two natterin-like domains, and has a clear one-to-one ortholog in other mosquito species, including anophelines (e.g., Anopheles darlingi) and culicines (e.g., Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus) with 83 to 84% amino acid identity (Fig. 2A and SI Appendix, Table S1). Putative orthologs are also present in more distant diptera of the same suborder (Nematocera), such as Phlebotomus papatasi (sandfly) and Clunio marinus (a nonbiting midge), which share 68% identity with AgP47Rec (Fig. 2A, SI Appendix, Fig. S4 and Table S1, and Dataset S2). In Drosophila (suborder Brachycera), there are several proteins with two natterin-like domains, but they are more divergent (45 to 58% identity) (Fig. 2A and SI Appendix, Table S1). Therefore, AgP47Rec is a protein of ancient origin in dipteran evolution. Proteins with two natterin domains are also present in other nondipteran insects. For example, the protein from the emerald ash borer (Agrilus planipennis), a coleoptera, has 54% identity to AgP47Rec, while those from two lepidopteran species, the cabbage looper (Trichoplusia ni) and the cabbage moth (Plutella xylostella), both share 50% identity. However, it is not clear whether these proteins are true functional orthologs of AgP47Rec.
Fig. 2.
Characterization of AgP47Rec in A. gambiae and effect of dsRNA-mediated silencing on P. falciparum infection. (A) Neighbor-joining phylogenetic tree (without distance correction) of AgP47Rec orthologs and related proteins in other dipterans, including Drosophila melanogaster (Dm), P. papatasi (PhPa), C. marinus (CIMa), A. darlingi (AnDa), A. gambiae (Anga), Aedes albopictus (AeAl); Aedes aegypti (AeAe); and C. quinquefasciatus (CxQu). (B) Expression of AgP47Rec in A. gambiae midguts of sugar fed (SF), control blood fed (C), or P. falciparum-infected blood fed (I) mosquitoes at different times. (C) Western blot of AgP47Rec protein expression in A. gambiae midgut pellet (Upper) and midgut cytosol (midgut supernatant, Lower) from blood-fed mosquitoes 12, 24, 48, 72 h PF, or sugar fed (0 h). (D) qPCR quantitation of AgP47Rec mRNA in whole mosquitoes after injection of AgP47Rec dsRNA (dsP47Rec) or control LacZ dsRNA (dsLacZ). (E) Western blot of AgP47Rec in midguts from mosquitoes after injection of AgP47Rec dsRNA (Rec) or control LacZ dsRNA (LcZ). (F) P. falciparum oocyst infection levels in midguts from mosquitoes injected with AgP47Rec dsRNA (dsP47Rec) or control LacZ dsRNA (dsLacZ). n, number of mosquitoes analyzed; Pev., prevalence of infection. ****P < 0.0001 Mann–Whitney U test, **P < 0.01 χ-squared test.
AgP47Rec is expressed in larval, pupae, and adult stages (SI Appendix, Fig. S5A). In adult females, it is expressed in both the midgut and carcass (Fig. 2B and SI Appendix, Fig. S5B). AgP47Rec is constitutively expressed in the midgut of adult females, increasing by 48 h PF, but with no significant difference between noninfected and P. falciparum-infected midguts (Fig. 2B). However, expression increased significantly in the carcass of P. falciparum-infected females relative to blood-fed uninfected controls 48 h PF (P < 0.001, t test) (SI Appendix, Fig. S5B).
Recombinant AgP47Rec protein was also used to immunize mice and generate antisera. The antibodies detected a protein of the expected size, which was constitutively expressed in the midgut of sugar-fed and blood-fed females dissected 12, 24, 48 and 72 h PF. The protein was readily detected in the pellet fraction of midgut homogenates, but was absent from the soluble supernatant (Fig. 2C and SI Appendix, Fig. S6). This agrees with the positive rPfs47 binding in the midgut pellet fraction observed in the initial far-Western assays. Systemic injection of dsAgP47Rec reduced mRNA expression by 85% (Fig. 2D) and also greatly reduced protein expression of AgP47Rec (Fig. 2E). Anti-AgP47Rec polyclonal antibodies were specific, as they detected a single band present in the pellet fraction of the midgut homogenate (Fig. 2E). The effect of silencing AgP47Rec on P. falciparum infection was evaluated. Reducing expression of AgP47Rec by dsRNA injection 3 d before feeding significantly reduced the number of P. falciparum oocysts present 10 d post infection (PI) (Mann–Whitney U test, P < 0.0001) and the prevalence of infection (χ2, P < 0.01) (Fig. 2F). We confirmed that P47Rec silencing had no effect on mosquitoes infected with NF54-Pfs47KO parasites (SI Appendix, Fig. S7).
Subcellular Localization of AgP47Rec.
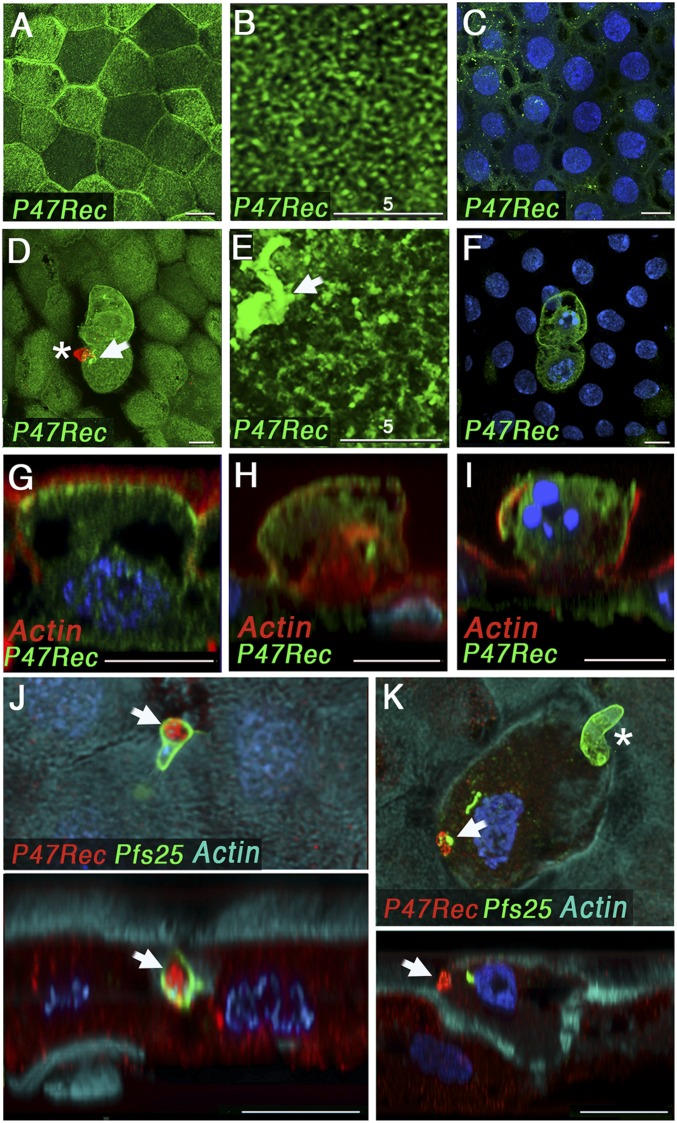
Immunofluorescence staining of mosquitoes fed uninfected blood showed that AgP47Rec protein is expressed in midgut epithelial cells, with higher protein expression levels in some cells (Fig. 3A). At a subcellular level, the receptor is mostly localized in the apical side (Fig. 3 A–C, G) and exhibits a reticular pattern at higher magnification (Fig. 3B). Expression in the middle plane of the cell, at the level of the nuclei, is low (Fig. 3C). A side view revealed that the apical staining is localized immediately below the microvilli (submicrovillar region) (Fig. 3G). In cells damaged by P. falciparum invasion, the submicrovillar localization is less prominent, and more protein localizes throughout the cell (Fig. 3 F and H). In some cases, a total loss of the apical surface of the cells is observed in invaded cells (Fig. 3I). We were able to capture one ookinete as it invaded the epithelial cell and we documented direct interaction of AgP47Rec protein with the surface of the parasite (Fig. 3J). AgP47Rec protein accumulates at the site of ookinete invasion (Fig. 3 D, E, and K) and colocalizes with Pfs25 (Fig. 3 K and J), a glycophosphatidylinositol (GPI)-anchored protein present on the surface of P. falciparum ookinetes that is shed during cell traversal (26). This appears to be a general response, as it is also observed in epithelial cells invaded by NF54-Pfs47KO or P. berghei (ANKA 2.34) ookinetes (SI Appendix, Fig. S8). Taken together, these data indicate that the parasite’s surface comes in close contact with AgP47Rec and triggers local aggregation of the receptor as ookinetes penetrate the apical end of midgut epithelial cells.
Fig. 3.
Subcellular localization of the AgP47Rec in the A. gambiae midgut cells (26 to 28 h postfeeding) by immunostaining and confocal microscopy. AgP47Rec (green) subcellular localization after a control blood meal: (A) apical section, (B) magnified apical section, (C) mid cell section, and (G) side view. AgP47Rec (green) subcellular localization in P. falciparum-invaded cells: (D) apical section*, (E) magnified apical section, (F) mid cell section of two invaded cells, and (H and I) cross-sections of invaded cells. Apical and cross-section views during ookinete traversal of the apical surface of the cell (J) and after cell traversal was completed* (K) with AgP47Rec in red. Midgut actin staining (cyan) and P. falciparum ookinete surface protein Pfs25 immunostaining (green) are also shown. White arrows indicate the accumulation of AgP47Rec at the site of invasion of the apical end of the cell. *In D and K, ookinete staining was overlaid with the surface AgP47Rec staining to indicate the position of the parasite relative to the invaded cell. In both cases the ookinetes had traversed the midgut and reached the basal plane. This is indicated by the asterisk in the figures. (Scale bars, 10 µm; magnified images in B and E, 5 µm.)
P47Rec and the Lock and Key Model.
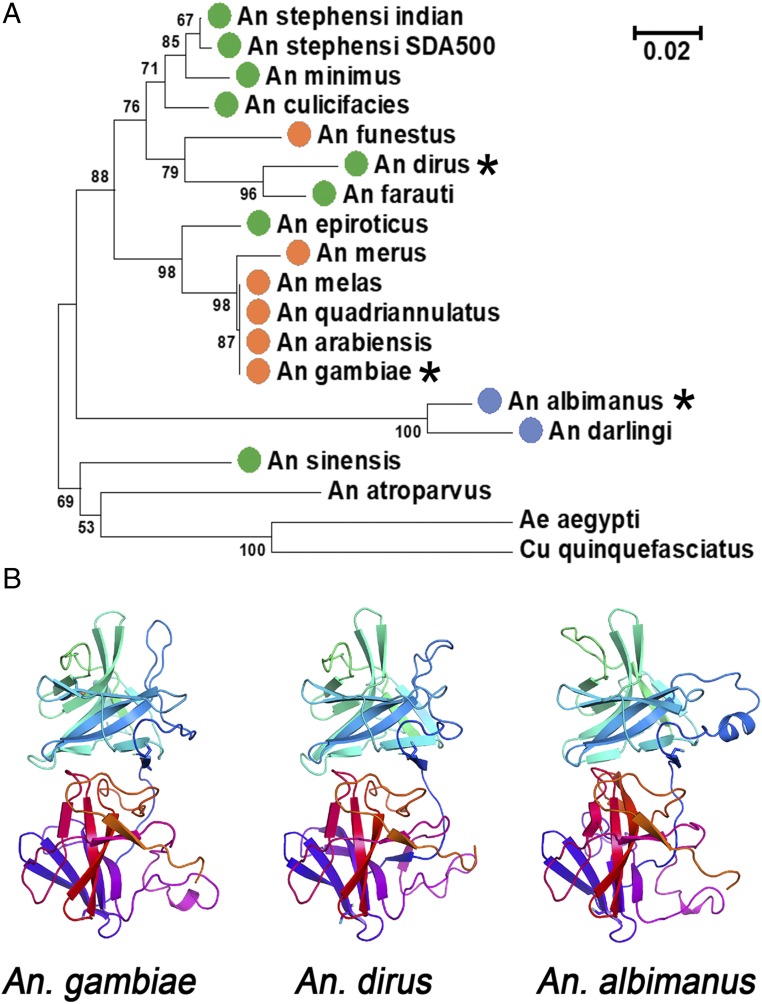
We previously proposed that ookinetes need to express a haplotype of Pfs47 compatible with the P47Rec present in the midgut of a given species of anopheline mosquito in order to effectively infect it. A detailed phylogenetic analysis of P47Rec sequences available from anophelines indicates that all of them have a clear ortholog, and that sequence divergence follows the speciation of anophelines (Fig. 4A and Dataset S3). This, and the clear presence of orthologs in other mosquito species, including culicines, indicates that AgP47Rec has an ancestral basic function in mosquitoes and other dipterans, besides its role in parasite evasion of antiplasmodial immunity. AgP47Rec is identical in most anophelines of the A. gambiae complex, and AgP47Rec has 91% amino acid identity with its ortholog in Anopheles funestus, another major African malaria vector. The amino acid identity of AgP47Rec with its orthologs in the evolutionarily distant Anopheles dirus (Asia) and A. albimanus (New World) are 93% and 85%, respectively.
Fig. 4.
Phylogenetic relationship and predicted structure of AgP47Rec orthologs. (A) Neighbor-joining tree of P47Rec amino acid sequences from orthologs in different anophelines present in Africa (orange dots), Asia (green dots), Central and South America (blue dots), and Europe (Anopheles atroparvus). Aedes aegypti and Culex quinquefasciatus orthologs were included as outgroups. *Indicates the receptors in the phylogenetic tree from vectors that are compared below (A. gambiae, A. dirus, and A. albimanus). Bootstrap values higher than 50% (1,000 replications) are shown. Scale indicates the number of amino acid substitutions per site. (B) Predicted protein structure of AgP47Rec and orthologs in A. dirus and A. albimanus by Phyre2. Each receptor has two natterin domains (one shown in blue-cyan-green and a second in orange-red-purple).
In silico three-dimensional (3D) modeling of the AgP47Rec protein using the Phyre2 software (27) revealed a high confidence match (confidence scores greater than 98%) to nematode sperm cell motility protein 2 (MFP2) (28) (SI Appendix, Fig. S9), a protein with two natterin-like domains (Protein Data Bank [PDB] accession no. 2BJQ). Ascaris sperm motility is mediated by the polymerization of major sperm protein (MSP), a 14-kDa protein that forms dense meshwork of filaments that pack the lamellipod. MFP2 is required for sperm amoeboid motility by promoting the polymerization of MSP (28). Structural modeling based on the MFP2 protein (20 to 22% amino acid identity) shows the presence of a highly conserved core region consisting of two domains having the same β-sheet fold in A. gambiae, A. dirus, and A. albimanus P47Rec (Fig. 4B and SI Appendix, Fig. S10). The three mosquito proteins contain the same number of amino acids and high homology, suggesting that the positions of main chain atoms in this structure are likely to be very similar through the length of the protein, including the loop regions connecting the β-strands. As a first step in identifying determinants of ligand selectivity, we used molecular models and sequence alignments to identify solvent-accessible polymorphic sequence positions that are not conserved between A. gambiae and A. albimanus P47Rec. A total of 24 sites meeting these criteria were identified; they are potential Pfs47 interaction points and may determine differences in binding (SI Appendix, Fig. S10 A and B).
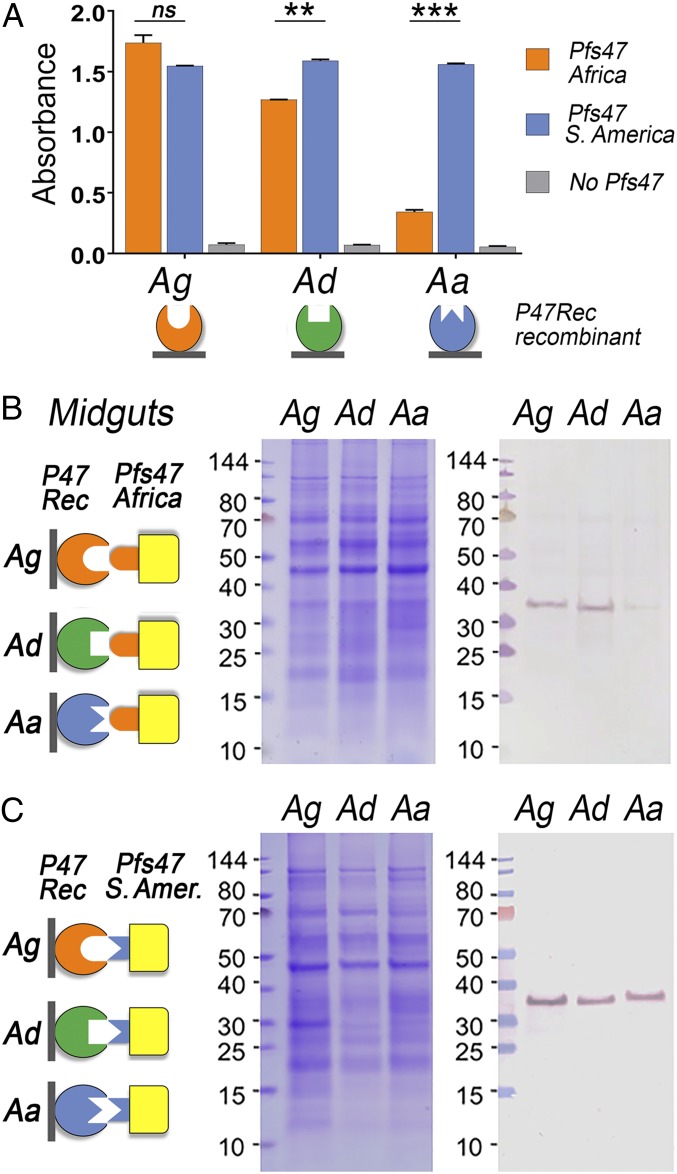
The ability of rPfs47-Africa (GB4 haplotype) and rPfs47-South America (S. America, 7G8 haplotype) to bind to recombinant P47Rec from these three evolutionarily distant mosquito species, which differ in their susceptibility to infection with parasites carrying different Pfs47 haplotypes, was compared. Recombinant P47Rec proteins from the three vectors, A. gambiae (Ag), A. dirus, (Ad), and A. albimanus (Aa) were immobilized on ELISA plates, and the binding of soluble rPfs47-Africa and rPfs47-S. America were evaluated. rPfs47-S. America bound to all receptors with similar strength, while rPfs47-Africa was more selective and bound less to the A. dirus receptor and much less to the one from A. albimanus, than to the P47Rec from A. gambiae (Fig. 5A). There was no significant difference between rPfs47-Africa and rPfs47-S. America binding to A. gambiae P47Rec. However, rPfs47-Africa binding to A. dirus P47Rec was lower (17%) than the binding of rPfs47-S. America (P < 0.01, ANOVA), and rPfs47-Africa binding to A. albimanus P47Rec was much weaker (4.3 times less) than rPfs47-S. America (P < 0.001, ANOVA) (Fig. 5A).
Fig. 5.
Binding of rPfs47 haplotypes from Africa (GB4) and South America (7G8) with P47Rec from vectors from Africa, A. gambiae (Ag, orange); Asia, A. dirus (Ad, green); and South America, A. albimanus (Aa, blue). (A) Binding of rPfs47-Africa and S. America haplotypes to recombinant AgP47Rec from Aa, Ad, and Aa by ELISA. Column and error bars represent mean titer ± SE of two biological replicate assays. ns, nonsignificant, P > 0.05; **P ≤ 0.01; ***P ≤ 0.001 t test. (B) Binding of rPfs47-Africa haplotype in a far-Western of Ag, Ad, and Aa midgut homogenate pellets separated by SDS/PAGE. (C) Binding of rPfs47-S. America haplotype in a far-Western of Ag, Ad, and Aa midgut homogenate pellets separated by SDS/PAGE. In B and C, the diagrams on the Left illustrate the immobilized P47Recs from the midgut homogenate pellets of different mosquito species and the two rPfs47 haplotypes tested; the Middle shows the Coomassie blue stained SDS/PAGE gels of the midgut homogenate pellets; and the Right shows the far-Western blots.
The binding of these two recombinant Pfs47 proteins to midgut homogenates from these three mosquito species was also compared using far-Western assays (Fig. 5 B and C and SI Appendix, Fig. S11). Similar binding patterns were observed, with rPfs47-Africa having a stronger binding to midgut homogenate pellets from A. gambiae and A. dirus than to A. albimanus (Fig. 5B), while rPfs47- S. America bound equally well to the P47Recs from these three mosquito species (Fig. 5C). No signal was observed in the negative controls that were not incubated with rPfs47 (SI Appendix, Fig. S11).
Discussion
Several lines of evidence indicate that Pfs47 is critical for P. falciparum to survive in the mosquito vector. Parasites expressing a compatible Pfs47 haplotype disrupt caspase-mediated apoptosis in invaded midgut cells and epithelial nitration and survive by preventing complement activation (19). The Pfs47 haplotype expressed by the parasite is a major determinant of compatibility with a specific mosquito species (21). For example, P. falciparum parasites carrying the African Pfs47 NF54 haplotype are able to infect the genetically selected A. gambiae refractory L3-5 strain, while replacement with the South American Pfs47 7G8 haplotype (with the NF54 genetic background) leads to elimination and melanization of the parasite (18). The differences in compatibility between parasites expressing the African (GB4) and New World (7G8) Pfs47 haplotypes in the A. gambiae L3-5 refractory strain are determined by four amino acids in a region of domain 2 (Pfs47-D2) flanked by two cysteines (18, 29). These two cysteines are predicted to form a loop structure and are likely to be involved in binding to AgP47Rec. If any of these four amino acids in the Pfs47-D2 cysteine loop of NF54 parasites are substituted by the one present in 7G8, the parasite becomes visible to the mosquito immune system and is eliminated, indicating that a strong and specific interaction is necessary to evade immunity in A. gambiae L3-5 mosquitoes (29). Taken together, these observations predicted specific interactions of Pfs47 with a molecule(s) present in the mosquito and are consistent with the observed high-affinity binding of Pfs47 with the AgP47Rec protein (Fig. 1 A–G).
Most AgP47Rec protein localized at the apical end of midgut epithelial cells, in the submicrovillar region (Fig. 3G), and the staining had a “mesh-like” reticular pattern (Fig. 3B). Furthermore, AgP47Rec was recovered in the pellet faction of midgut homogenates containing membrane/cytoskeleton-associated proteins (Fig. 1B). These observations, in conjunction with the predicted protein structure similarity to MFP2, a protein that promotes polymerization of the cytoskeletal protein MSP in Ascaris sperm (28), suggest a role of AgP47Rec in the organization of the cytoskeletal structure of the cell. The apical end of the cell is damaged when the ookinete penetrates and traverses the midgut cell. The observed accumulation of AgP47Rec at the site of ookinete invasion may reflect an attempt by the cell to rearrange the cytoskeleton to contain the damage and stabilize the cell (Fig. 3 D, E, and K). The parasite presumably comes into contact with AgP47Rec upon entering the midgut cell, and Pfs47 binding disrupts the normal function of AgP47Rec. Reducing expression of AgP47Rec decreased P. falciparum wt infection (Fig. 2F) but had no effect on P. falciparum Pfs47 KO, indicating that the interaction of Pfs47 with the mosquito receptor is critical for parasite survival. We have previously shown that parasites expressing a compatible Pfs47 haplotype can invade midgut epithelial cells without activating the JNK pathway (19). Presumably, the interaction of Pfs47 with the mosquito P47Rec may prevent activation of JNK signaling and caspase-mediated apoptosis. However, further studies are necessary to understand how Pfs47 affects the response of epithelial cells to parasite invasion and the role of AgP47Rec in defining the mechanism of cell death.
The phylogenetic analysis of the P47Rec sequences in anophelines (Fig. 4A) follows the known phylogenetic relationships between these mosquito species based on whole genome sequence analysis (30). It is worth noting that A. funestus, a dominant malaria vector in Africa, is evolutionarily closer to Asian vectors (30). As expected, A. funestus P47Rec is also phylogenetically closer to that of Asian vectors such as A. dirus and Anopheles farauti, than to African vectors of the A. gambiae complex (Fig. 4). This raises the interesting possibility that A. funestus could be one of the drivers of genetic diversity of Pfs47 in Africa, and may have served as a bridge vector by selecting P. falciparum parasites with Pfs47 haplotypes compatible with Asia vectors. Furthermore, P. falciparum originated from a horizontal transfer of Plasmodium praefalciparum parasites from gorillas to humans (2). Interestingly, the reported gorilla malaria vectors (Anopheles vinckei, Anopheles marshallii, and Anopheles moucheti) (31) are phylogenetically closer to A. dirus and A. funestus than to A. gambiae s.l (32). This suggests that the ancestral P. falciparum Pfs47 haplotype may have been more compatible with A. funestus than with A. gambiae. This could have facilitated adaptation of the parasite to other Asian vectors phylogenetically close to A. funestus, leading to the spread of malaria in Asia. In Africa, adaptation of the ancestral P. falciparum parasites to highly anthropophilic mosquitoes from the A. gambiae complex, the increase in the human population size, and migration across the continent resulted in a dramatic and rapid expansion and diversification of the parasite population.
The genetic background of the mosquito vector is also a major determinant of P. falciparum’s ability to evade immunity. For example, the A. gambiae refractory L3-5 has the same AgP47Rec amino acid sequence as the susceptible A. gambiae G3 strain (33), but P. falciparum 7G8 (Brazilian) parasites are eliminated by the refractory L3-5 strain, while they survive in the G3 strain. The mosquito immune response is complex, involving multiple genes and cell types, and both the speed and the intensity of the response can result in very different outcomes. For example, the A. gambiae L3-5 is in a chronic state of oxidative stress (34) and the JNK pathway is constitutively overactivated (8). These two mechanisms are predicted to enhance epithelial nitration (8, 34). NF54 parasites that express Pfs47 are able to suppress epithelial nitration, while NF54 parasites in which the Pfs47 gene has been disrupted trigger strong epithelial nitration and activate the complement system both in G3 and L3-5 A. gambiae mosquitoes (18). The L3-5 strain also expresses a haplotype of Tep1 (Tep1-R), which is more effective at eliminating P. berghei parasites than the haplotype present in the susceptible G3 strain (Tep1-S) (12), and this may also affect P. falciparum survival. These observations indicate that having a Pfs47 compatible with the P47Rec of a given vector is necessary, but may not be sufficient for P. falciparum to evade immunity and survive.
The in vitro binding of recombinant rPfs47 Africa (GB4) or South America (7G8) haplotype proteins with recombinant receptors, or to midgut homogenates from the different vectors (A. gambiae, A. dirus, and A. albimanus) (Fig. 5), are consistent with the ability of transgenic P. falciparum NF54 parasites carrying these two different Pfs47 haplotypes to infect these three mosquito species (21). The most striking difference in binding is the ability of rPfs47-Africa (GB4) to bind to recombinant AgP47Rec and AdP47Rec, or to midgut homogenates from A. gambiae and A. dirus, but not to those from A. albimanus (Fig. 5 A and B). This observed difference in binding is in accordance with the known absence of the most common Africa Pfs47 haplotypes (GB4 and NF54) in the Americas, and the very low infectivity of NF54 in A. albimanus (21). Although the binding of rPfs47-Africa to recombinant AgP47Rec, or to midgut homogenates, was similar for A. gambiae and A. dirus, the infectivity of P. falciparum NF54 parasites was much higher in A. gambiae than in A. dirus (21). Again, this suggests that the ability of Pfs47 to bind to the mosquito AgP47Rec is necessary, but may not be sufficient for NF54 parasites to efficiently infect A. dirus.
In contrast, recombinant rPfs47-7G8 bound equally well to the three different P47Rec recombinant proteins, and to all midgut homogenates (Fig. 5 A and C). Moreover, there was no significant difference in the infectivity of transgenic P. falciparum NF54 parasites expressing this haplotype between the three vectors (21). Nevertheless, the infectivity of the P. falciparum 7G8 line was significantly higher in A. albimanus than in A. dirus or A. gambiae (21). These differences in infectivity of the same Pfs47-7G8 haplotype in different genetic backgrounds (7G8 vs. NF54) indicate that there are other genes in P. falciparum, besides Pfs47, that are also major determinants of infectivity in A. albimanus. We conclude that the identification of the AgP47Rec provides further support for the “lock and key” model, which predicts natural selection of compatible Pfs47 haplotypes during the adaptation of P. falciparum to evolutionarily distant vectors. Our findings also show that although the interaction of Pfs47 with the mosquito receptor is critical for P. falciparum survival, the efficiency of malaria transmission is the result of a complex interplay between multiple parasite and mosquito genes, that may also be affected by environmental factors.
Materials and Methods
Anopheles Mosquitoes and Plasmodium Parasites.
The mosquitoes used were A. gambiae G3, A. dirus A s.s., and A. albimanus. Mosquitoes were reared at 27 °C and 80% humidity on a 12-h light/dark cycle under standard laboratory conditions as previously described (21). The P. falciparum NF54 strain was cultured in O+ human erythrocytes using RPMI medium 1640 supplemented with 25 mM Hepes, 10 mg/L hypoxanthine, 25 mM NaHCO3, and 10% (vol/vol) heat-inactivated type O+ human serum (Interstate Blood Bank, Inc.) at 37 °C and with a gas mixture of 5% O2, 5% CO2, and balance N2 (35).
Experimental Infection of Mosquitoes with P. falciparum.
P. falciparum gametocytogenesis was induced as previously described (36). Female mosquitoes were infected artificially by membrane feeding with P. falciparum gametocytes (stages IV and V at 14 to 16 d) at 37 °C for 30 min. Mosquitoes were transferred to an incubator at 22.5 °C 6 h later. After 36 h, the mosquitoes were transferred back to 26 °C. This temperature switch has been shown to increase sensitivity of dsRNA-mediated gene silencing (19). Midguts were dissected 8 to 10 d after feeding, and oocysts were stained with 0.1% (wt/vol) mercurochrome in water and counted by light microscopy (21). The distribution of parasite numbers in individual mosquitoes between control and experimental groups were compared using the nonparametric Mann–Whitney U test. All parasite phenotypes were confirmed in at least two independent experiments.
Pfs47 and P47Rec Recombinant Proteins.
Expression of recombinant Pfs47 protein was done as preciously described (37). Briefly, the mature full-length P. falciparum Pfs47 coding sequence, extending from the predicted signal peptide cleavage site to the GPI anchor site (Leu28 to Ala414; GenBank: ALQ44015.1), was codon optimized for insect cell expression and synthesized by GenScript. The Pfs47 construct for baculovirus expression, the BVPfs47 construct, was built by PCR amplification of the Pfs47 coding sequence from Thr32 to Tyr420, which was subcloned into a modified pAcSecG2T vector (Pharmingen) containing a tobacco etch virus (TEV) protease cleavable N-terminal hexahistidine/maltose binding protein (MBP) tag. Recombinant Pfs47 constructs were produced in insect cells using established protocols (38). High titer expression viruses were generated in Spodoptera frugiperda 9 (Sf9) cells and used to infect T. ni (Hi5) cells. Secreted proteins were harvested from the Hi5 cell culture supernatant after 65 h of infection and purified by nickel affinity chromatography. Using TEV protease, the hexahistidine/MBP tag was cleaved, and the tag was removed by ion exchange chromatography. Proteins were purified to homogeneity by size exclusion chromatography in Hepes-buffered saline (20 mM Hepes pH 7.5, 150 mM NaCl) containing 2% glycerol.
The mature full-length AGAP006398 (GenBank XP_316431.3) coding sequence was codon optimized for E. coli cell expression and synthesized by BioBasic. The AGAP006398 construct was built by PCR amplification and subcloned by In-Fusion (Clontech) into a pET17 vector (EMD Millipore) between NdeI and XhoI sites, with primers containing a N-terminus FLAG epitope (DYKDDDDK) and a C-terminus histidine tag. Orthologs of AGAP006398 in A. albimanus (VectorBase AALB004659-PA) and A. dirus (VectorBase ADIR015582-PA) were amplified from A. albimanus and A. dirus unfed midgut cDNA, respectively, and cloned by In-Fusion (Clontech) into a pET17 vector (EMD Millipore) between NdeI and XhoI sites, with primers containing an N-terminus FLAG epitope (DYKDDDDK) and a C-terminus histidine tag. All constructs were expressed in BL21 E. coli (Invitrogen). Hepes-buffered saline (20 mM Hepes pH 7.5, 150 mM NaCl) 8 M urea solution containing inclusion bodies of cultures induced for 3 h at 37 °C with 1 mM isopropyl β-d-thiogalactopyranoside (Sigma) was purified by nickel affinity chromatography and stored after being dialyzed in Hepes-buffered saline 2 M urea. All gel images derive from the same experiment and were processed in parallel. Sequences of A. gambiae, A. dirus, and A. albimanus P47Rec are included in Dataset S4.
Antibody Generation.
All animal procedures were performed according to protocols approved by the National Institute of Allergy and Infectious Diseases (NIAID) and the NIH Animal Care and Use Committee. Five- to 8-wk-old naïve, female BALB/c mice were purchased from Charles River and maintained at a facility at the NIH. Groups (n = 5) of female BALB/c mice were immunized subcutaneously (s.c.) in their ears with a 20-μL solution that contained 5 µg of Hepes-buffered saline-dialyzed recombinant AGAP006398 protein emulsified in Magic mouse adjuvant (Creative Diagnostics CDN-A001). The immunized mice were boosted twice at 2-wk intervals with the same quantity of protein. Blood was collected on day 0 (preimmune sera) and 2 wk after each subsequent immunization for analysis of antibody titer or terminal bleeding. Alternatively, rabbit antiserum was obtained from immunized rabbit with recombinant AGAP006398 (Noble Sciences, Inc). IgG from mouse and rabbit sera was purified as previously described (39).
Far-Western Blot.
Mosquito midguts were collected from human serum fed mosquitoes 24 to 72 h PF in solubilization buffer (SB) (15 mM Tris pH 8, 150 mM NaCl, 5 mM EDTA containing 1× fresh cOmplete ULTRA Protease Inhibitor, Roch). The mosquitoes were fed with human serum to facilitate the removal of the bolus from the midgut epithelia. Midguts were homogenized with an electric hand homogenizer at 0.5 midgut/μL, and then frozen at −80 °C. In order to fractionate cells, midguts were thawed, homogenized, and centrifuged at low speed (0.5 × g) for 10 min (4 °C), in order to separate and remove large pieces of tissue. The supernatant was recovered for ultracentrifugation at 100,000 × g for 20 min at 4 °C. The new supernatant was separated (cytosolic fraction) and kept on ice, while the pellet was resuspended in SB containing 0.5% Triton X-100 at 1 midgut/μL. Cytosol and pellet were subjected to NuPage SDS/PAGE with 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Invitrogen). This was done under denaturing nonreducing conditions, heating the cytosol and pellets at 95 °C for 10 min. Three SDS/PAGE gels were run side by side, one for staining (Coomassie or silver staining), one for far-Western blot with bait protein, and one for a no-bait control. The proteins separated by SDS/PAGE were transferred to a nitrocellulose membrane using the iBlot, following manufacturer’s instructions (Invitrogen). In the case of runs with A. gambiae, A. dirus, and A. albimanus midgut pellets, the membrane was treated to refold the proteins without including a reducing agent (40) to maximize binding. The nitrocellulose membrane was blocked overnight shaking at 4 °C with TBST with blocking solution (5% nonfat milk and 1× fresh protease inhibitor) containing 0.1 mM levamisole. The membrane was then incubated overnight shaking at 4 °C with blocking solution containing bait protein at 500 nM. The membranes were rinsed with Tris-buffered saline (TBS) 0.1% Tween 20 (TBST) five times (10 min each) with TBST and then incubated with primary antibody (anti-His or anti-Pfs47 monoclonal JH11) diluted 1 to 1,000. The membrane was rinsed again five times and incubated with secondary antibody, alkaline phosphase or peroxidase-conjugated anti-mouse antibody. The membrane was rinsed five times and developed with alkaline phosphatase reagent at room temperature (RT) or by chemiluminescence using Super Sigma West Femto (Thermo Scientific).
Two-Dimensional Far-Western Blot and Mass Spectrometry Analysis.
A. gambiae midgut membrane/cytoskeleton protein was prepared as described above. The protein pellet from 10 A. gambiae midguts was separated in a 3- to 10-pH precast gel (Invitrogen) and run in a second dimension in the NuPAGE SDS/PAGE with Mops buffer and MW marker proteins, SeeBLue Plus 2 (Invitrogen). The separated protein was transferred to a nitrocellulose membrane and probed with rPfs47 as described above. A second 2D SDS/PAGE was stained with Coomassie blue for protein visualization. Digital images of the far-Western blot and the Coomassie blue-stained gel were compared to identify the protein band labeled in the far-Western blot. The corresponding labeled protein band as well as surrounding bands were excised from the gel for mass spectrometry analysis.
ELISA of rPfs47 Binding to A. gambiae Midgut Homogenate or P47Rec in Different Anophelines.
Flat-bottom 96-well ELISA plates (Immunolon 4 VWR) were coated (50 μL/well) in triplicate with midgut homogenate in SB with 0.5% Triton X-100 (0.1 midgut/μL) diluted 1:10 in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) overnight at 4 °C. In the case of recombinant P47Rec proteins (AGAP006398, AALB004659, and ADIR015582) they were diluted to 1 μg/mL in coating buffer before being used to coat the plates overnight at 4 °C. The plates were washed three times with TBST and blocked with general block ELISA blocking buffer (ImmunoChemistry cat. no. 640) for 2 h at 37 °C. Plates were incubated with recombinant baculovirus-expressed P. falciparum Pfs47 NF54 for 16 h at 4 °C. After three washes with TBST, plates were incubated with 1 μg/mL mouse monoclonal anti-His in the case of A. gambiae homogenate or monoclonal antibody JH11 anti-Pfs47 in the case of P47Rec ELISA, for 2 h at 37 °C. The plates were then washed three times with TBST and incubated with goat anti-mouse IgG conjugated to alkaline phosphatase (Seracare cat. no. 5220-0303) secondary antibodies (0.2 μg/mL) for 2 h at 37 °C. The plates were washed again, and detection was performed using 100 µL/well of p-nitrophenyl disodium phosphate solution (Sigma 104 phosphatase substrate; 1 tablet per 5 mL of coating buffer). After a 20-min incubation, absorbance was read at 405 nm with a VersaMax ELISA plate reader.
dsRNA-Mediated Gene Knockdown and qPCR.
Individual female A. gambiae mosquitoes were injected 1 to 2 d postemergence as previously described (41). Briefly, mosquitoes were injected with 69 nL of a 3 μg/μL dsRNA solution 3 to 4 d before receiving a Plasmodium-infected blood meal. The control dsRNA (LacZ) and A. gambiae dsAGAP006398 were produced as previously described (41, 42). The dsRNA was produced using the MEGAscript RNAi Kit (Ambion), using DNA templates obtained by nested PCR using cDNA from whole female mosquitoes. The primers used to obtain the DNA template for AGAP006398 for external PCR were: 6398AgEx_F 5′-CGCACGGATGTCTGTACATT-3′ and 6398AgEx_R 5′-TGTTCGATCAGCACCTCGTA-3′. PCR conditions were as follows: 94 °C for 3 min, 25 cycles of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min with final extension 72 °C for 5 min. For the nested PCR, with primers that contained the T7 promoter, the primer sequences were: AGAP6398dsT7_InF 5′-TAATACGACTCACTATAGGGACGAGGTGCTAGTGGGACAG-3′ and AGAP6398dsT7_InR 5′-TAATACGACTCACTATAGGG GGGATGTAGAGCGTCTGGTG-3′. PCR conditions were 94 °C for 3 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min with final extension 72 °C for 5 min, using 1 μL of the external primer PCR. Silencing was assessed in whole sugar-fed mosquitoes by quantitative real-time PCR (qPCR) using the S7 ribosomal protein gene as internal reference. The primers used for qPCR were AGAP_6398_qP2_F 5′-GTGTACGGTGGAAACGATCA-3′, AGAP_6398_qP2_R 5′-CCTGCTTGGATGGGATCA-3′, S7F 5′-AGAACCAGCAGACCACCATC-3′, and S7R 5′-GCTGCAAACTTCGGCTATTC-3′. qPCRs were performed under standard conditions using 0.5 μM of each primer with an initial denaturation step of 15 min at 95 °C and then 45 cycles of 10 s at 94 °C, 20 s at 55 °C, and 30 s at 72 °C, with a final extension of 5 min at 72 °C. The silencing efficiency in dsRNA-injected mosquitoes was 81 to 84%, relative to dsLacZ-injected controls.
Western Blot.
Recombinant proteins were boiled under reducing conditions in SDS/PAGE loading buffer, separated using 12% SDS/PAGE gels, and transferred on nitrocellulose membranes. The membrane was blocked for 2 h with 5% nonfat dry milk in TBST. The proteins were detected using 6×-His epitope tag primary antibodies (Thermo Fisher cat. no. MA1-21315) or animal sera at 1:1,000 dilutions. The membranes were then washed three times with TBST and incubated with goat anti-mouse or anti-rabbit secondary antibodies conjugated to alkaline phosphatase at 1:5,000 dilution. The proteins were then detected using Western Blue Stabilized Substrate for alkaline phosphatase (Promega cat. no. S3841) after a 5-min incubation.
Binding Analysis by Surface Plasmon Resonance.
Binding affinity of recombinant mosquito receptor (P47Rec) and P. falciparum surface protein (rPfs47) were analyzed at 25 °C by surface plasmon resonance (SPR) spectrometry using a BIAcore T100 instrument (GE Healthcare Life Sciences). Contact time, flow rate, dissociation, and regeneration cycles are described for each condition below. Recombinant AgP47Rec (AGAP006398), at 30 µg/mL in acetate buffer with a pH of 4.5, was covalently immobilized on the surface of a CM5 sensor chip using the amine coupling kit supplied by Biacore, which resulted in a final immobilization level of 1683.8 RU. Blank flow cells were used to subtract the buffer effect on the sensorgrams. For kinetic experiments, recombinant Pfs47, in concentrations ranging from 0.4 to 400 nM, was flowed for 200 s at 30 mL/min in HBS-P buffer (10 mM Hepes, 150 mM NaCl, 0.005% surfactant P20, pH 7.4). The complex dissociation was monitored for 600 s, and the active cell regenerated with 45-s pulse of 10 mM HCl 40 µL/min. Additionally, rPfs47 (30 µg/mL in acetate buffer, pH 4.5) was immobilized on a CM5 sensor at 1549.2 RU and AgP47Rec at concentrations ranging from 4.4 to 282 nM was used as analyte. The Biacore T100 Evaluation software 2.0.4 was utilized for kinetic evaluation. Kinetic parameters (kon, koff, and KD) were determined by global fitting of the sensorgrams and application of the 1:1 binding model (Langmuir binding model). These experiments were carried out in duplicates.
Midgut Immunostaining and Confocal Microscopy.
The immune stainings were performed as previously described (26, 43). Briefly, the midguts were dissected 26 to 28 h postinfection with P. falciparum. The blood bolus was carefully removed by longitudinally opening the midguts on ice-cold phosphate-buffered saline (PBS). After fixation with 4% paraformaldehyde in PBS for 1 h at RT, the midguts were washed two times with 0.1% Triton-X 100 (Sigma) in PBS (PBST), followed by blocking for at least 1 h with 5% BSA (Sigma), 0.1% gelatin (Sigma) in PBST. Then, the midguts were incubated overnight with rabbit IgG purified anti-6398 (SI Appendix, Fig. S12) at 5 µg/mL and mAb anti-Psf25 at 1:500 primary antibodies, followed by incubation at room temperature with secondary antibodies anti-rabbit Alexa-fluor 488 (Thermo Fisher) and anti-mouse Alexa-fluor 594 (Thermo Fisher)-conjugated antibody diluted 1:1,000. After washing, the midguts were counterstained with Alexa-fluor 647 phalloidin (1:40) (Thermo Fisher), and 2 µM Hoechst (Thermo Fisher) in PBST for 30 min at room temperature. The midguts were mounted with Prolong gold (Thermo Fisher) and analyzed by confocal microscopy. Images were obtained using a Leica TCS SP8 confocal microscope (Leica Microsystems) 63× oil immersion objective. Images were taken using sequential acquisition and variable z-steps. Images were processed using Imaris 9.0.0 (Bitplane AG) and Adobe Photoshop CC (Adobe Systems).
P47Rec Phylogenetic Analysis.
The sequences of P47Rec orthologs were obtained from OrthoDB (https://www.orthodb.org). Alignment of sequences was done with ClustalW (www.clustal.org). Phylogenetic analysis of sequences was done with Mega6 (44).
In Silico Modeling of AgP47Rec.
Molecular modeling of AgP47Rec orthologs was performed using the Phyre2 server where sequence alignments and secondary structure predictions are generated using sequence databases representing known protein folds (27). Alignment and secondary structure information was used to search the library of protein structures, in order to identify homologs that allow modeling of the protein core. Loops and insertions were modeled using fragment libraries, and side chains were added in the last step. The quality of fold identification was assessed using a confidence score that indicated the probability of the query sequence matching the identified fold. In the cases of A. gambiae, A. dirus, and A. albimanus P47Rec orthologs, the confidence scores were greater than 98 (on a scale of 100) for matches with the crystal structure of the nematode sperm motility protein MFP2 (PDB accession no. 2BJQ) (28). Sequence identities for pairwise alignments of MFP2 with the mosquito proteins ranged from 20 to 22%.
Data Availability.
All of the data described are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research Z01AI000947. We thank Andre Laughinghouse and Kevin Lee for insectary support. We acknowledge the assistance of the Research Technology Branch NIAID NIH, specifically the help of Ming Zhao, Carl Hammer, Lisa Olano, and Raynaldo Martin for 2D gel protein and mass spectrometry analysis.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917042117/-/DCSupplemental.
References
- 1.WHO , World Malaria Report 2018 (World Health Organization, Geneva, 2018), p. 166. [Google Scholar]
- 2.Loy D. E., et al. , Out of Africa: Origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int. J. Parasitol. 47, 87–97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina-Cruz A., Zilversmit M. M., Neafsey D. E., Hartl D. L., Barillas-Mury C., Mosquito vectors and the globalization of Plasmodium falciparum malaria. Annu. Rev. Genet. 50, 447–465 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Sinka M. E., et al. , A global map of dominant malaria vectors. Parasit. Vectors 5, 69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennink S., Kiesow M. J., Pradel G., The development of malaria parasites in the mosquito midgut. Cell. Microbiol. 18, 905–918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinden R. E., The cell biology of malaria infection of mosquito: Advances and opportunities. Cell. Microbiol. 17, 451–466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachou D., Schlegelmilch T., Runn E., Mendes A., Kafatos F. C., The developmental migration of Plasmodium in mosquitoes. Curr. Opin. Genet. Dev. 16, 384–391 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Garver L. S., de Almeida Oliveira G., Barillas-Mury C., The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS Pathog. 9, e1003622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira Gde. A., Lieberman J., Barillas-Mury C., Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barletta A. B. F., Trisnadi N., Ramirez J. L., Barillas-Mury C., Mosquito midgut prostaglandin release establishes systemic immune priming. iScience 19, 54–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo J. C., Ferreira A. B. B., Trisnadi N., Barillas-Mury C., Activation of mosquito complement antiplasmodial response requires cellular immunity. Sci. Immunol. 2, eaal1505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blandin S., et al. , Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Fraiture M., et al. , Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 5, 273–284 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Povelones M., Waterhouse R. M., Kafatos F. C., Christophides G. K., Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324, 258–261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohuet A., et al. , Anopheles and Plasmodium: From laboratory models to natural systems in the field. EMBO Rep. 7, 1285–1289 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins F. H., et al. , Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science 234, 607–610 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Molina-Cruz A., et al. , Some strains of Plasmodium falciparum, a human malaria parasite, evade the complement-like system of Anopheles gambiae mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 109, E1957–E1962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina-Cruz A., et al. , The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science 340, 984–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramphul U. N., Garver L. S., Molina-Cruz A., Canepa G. E., Barillas-Mury C., Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl. Acad. Sci. U.S.A. 112, 1273–1280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthony T. G., Polley S. D., Vogler A. P., Conway D. J., Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol. Biochem. Parasitol. 156, 117–123 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Molina-Cruz A., et al. , Plasmodium evasion of mosquito immunity and global malaria transmission: The lock-and-key theory. Proc. Natl. Acad. Sci. U.S.A. 112, 15178–15183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manske M., et al. , Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponting C. P., Mott R., Bork P., Copley R. R., Novel protein domains and repeats in Drosophila melanogaster: Insights into structure, function, and evolution. Genome Res. 11, 1996–2008 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Magalhães G. S., et al. , Natterins, a new class of proteins with kininogenase activity characterized from Thalassophryne nattereri fish venom. Biochimie 87, 687–699 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Chertemps T., et al. , Anopheles gambiae PRS1 modulates Plasmodium development at both midgut and salivary gland steps. PLoS One 5, e11538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y. S., Thompson J., Kafatos F. C., Barillas-Mury C., Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: The time bomb theory of ookinete invasion of mosquitoes. EMBO J. 19, 6030–6040 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant R. P., Buttery S. M., Ekman G. C., Roberts T. M., Stewart M., Structure of MFP2 and its function in enhancing MSP polymerization in Ascaris sperm amoeboid motility. J. Mol. Biol. 347, 583–595 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Canepa G. E., Molina-Cruz A., Barillas-Mury C., Molecular analysis of Pfs47-mediated Plasmodium evasion of mosquito immunity. PLoS One 11, e0168279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neafsey D. E., et al. , Mosquito genomics. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makanga B., et al. , Ape malaria transmission and potential for ape-to-human transfers in Africa. Proc. Natl. Acad. Sci. U.S.A. 113, 5329–5334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbach R. E., “The phylogeny and classification of Anopheles” Anopheles Mosquitoes–New Insights into Malaria Vectors, Manguin S. (InTech, London, UK, 2013). [Google Scholar]
- 33.Padrón A., et al. , In depth annotation of the Anopheles gambiae mosquito midgut transcriptome. BMC Genomics 15, 636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S., et al. , The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 100, 14139–14144 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolg J. W., MacLeod A. J., Dickson I. H., Scaife J. G., Plasmodium falciparum: Modifications of the in vitro culture conditions improving parasitic yields. J. Parasitol. 68, 1072–1080 (1982). [PubMed] [Google Scholar]
- 36.Ifediba T., Vanderberg J. P., Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature 294, 364–366 (1981). [DOI] [PubMed] [Google Scholar]
- 37.Canepa G. E., et al. , Antibody targeting of a specific region of Pfs47 blocks Plasmodium falciparum malaria transmission. NPJ Vaccines 3, 26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonkin M. L., et al. , Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41. J. Biol. Chem. 288, 12805–12817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolaeva D., et al. , Functional characterization and comparison of Plasmodium falciparum proteins as targets of transmission-blocking antibodies. Mol. Cell. Proteomics. mcp.RA117.000036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y., Li Q., Chen X.-Z., Detecting protein-protein interactions by Far western blotting. Nat. Protoc. 2, 3278–3284 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Jaramillo-Gutierrez G., et al. , Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol. 9, 154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molina-Cruz A., et al. , Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J. Biol. Chem. 283, 3217–3223 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Kumar S., Gupta L., Han Y. S., Barillas-Mury C., Inducible peroxidases mediate nitration of anopheles midgut cells undergoing apoptosis in response to Plasmodium invasion. J. Biol. Chem. 279, 53475–53482 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data described are included in the manuscript and SI Appendix.