Abstract
Purpose:
Increased glycolysis and glucose dependence is a hallmark of malignancy that enables tumors to maximize cell proliferation. In HER2+ cancers, an increase in glycolytic capacity is associated with trastuzumab resistance. IGF-1R activation and t-Darpp over-expression both confer trastuzumab resistance in breast cancer. We therefore investigated a role for IGF-1R and t-Darpp in regulating glycolytic capacity in HER2+ breast cancers.
Experimental design:
We examined the relationship between t-Darpp and IGF-1R expression in breast tumors and their respective relationships with patient survival. To assess t-Darpp’s metabolic effects, we used the Seahorse flux analyzer to measure glucose metabolism in trastuzumab-resistant SK-BR-3 cells (SK.HerR) that have high endogenous t-Darpp levels and SK.tDrp cells that stably over-express exogenous t-Darpp. To investigate t-Darpp’s mechanism of action, we evaluated t-Darpp:IGF-1R complexes by co-immunoprecipitation and proximity ligation assays. We used pathway-specific inhibitors to study the dependence of t-Darpp effects on IGF-1R signaling. We used siRNA knockdown to determine if glucose reliance in SK.HerR cells was mediated by t-Darpp.
Results:
In breast tumors, PPP1R1B mRNA levels were inversely correlated with IGF-1R mRNA levels and directly associated with shorter overall survival. t-Darpp over-expression was sufficient to increase glucose metabolism in SK.tDrp cells and essential for the glycolytic phenotype of SK.HerR cells. Recombinant t-Darpp stimulated glucose uptake, glycolysis and IGF-1R-Akt signaling in SK-BR-3 cells. Finally, t-Darpp stimulated IGF-1R heterodimerization with ErbB receptors and required IGF-1R signaling to confer its metabolic effects.
Conclusions:
t-Darpp activates IGF-1R signaling through heterodimerization with EGFR and HER2 to stimulate glycolysis and confer trastuzumab resistance.
Keywords: PPP1R1B, IGF-1R, Breast cancer, Glycolysis, Metabolism
INTRODUCTION
Breast cancer is the most common malignancy worldwide and the second leading cause of cancer death in women in the developed world. Breast cancers in which the human epidermal growth factor receptor 2 (HER2) is amplified account for about 20% of all U.S. cases of the disease. HER2-positive (HER2+) breast cancers are characterized by rapid growth and lower survival rates, but they are also responsive to therapies that target the HER2 receptor itself, most notably the humanized monoclonal antibody trastuzumab (1,2).
We and others identified transcripts of the PPP1R1B gene, which encodes the dopamine- and cAMP-regulated neuronal phosphoprotein 32 (Darpp-32) and a truncated isoform (t-Darpp), as being up-regulated in trastuzumab-resistant HER2+ breast cancer. High levels of t-Darpp, in particular, are observed in HER2+ breast cancer cell lines selected for trastuzumab resistance and t-Darpp over-expression is sufficient to confer trastuzumab resistance, promote cell growth, and inhibit apoptosis (3–7). Elevated t-Darpp levels have also been demonstrated in human prostate cancers, as well as in gastric and esophageal cancer samples, where t-Darpp is also associated with trastuzumab resistance (8–10). Moreover, t-Darpp over-expression can confer resistance to a variety of anti-proliferative agents besides trastuzumab (11), suggesting that it promotes a growth advantage through HER2-independent mechanisms that have not yet been identified. Cells over-expressing t-Darpp are able to maintain activation of the PI3K/Akt signaling pathway in the presence of cytostatic agents, such as trastuzumab, and they are more resistant to apoptosis induced by cytotoxic drugs (4,6,7,9,11–13).
Most evidence to date suggests that t-Darpp activates alternative signaling pathways to compensate for direct inhibition of HER2 signaling by trastuzumab. These include the protein kinase A (PKA) and the epidermal growth factor receptor (EGFR) pathways, acting mostly via sustained PI3K/Akt signaling (11–14). Signaling through the type 1 insulin-like growth factor receptor (IGF-1R) has also been associated with tumor cell proliferation and trastuzumab resistance (15–17). Activation of IGF-1R and its downstream mitogen-activated protein kinase (MAPK) and PI3K/Akt signaling pathways contributes to increased proliferation, migration and invasion in several types of cancer including breast, prostate, pancreatic and colon cancer (18–20). In cells selected for trastuzumab resistance, IGF-1R was shown to dimerize with the HER2 receptor (17). Although both IGF-1R and t-Darpp have been independently associated with trastuzumab resistance, a possible connection between t-Darpp and IGF-1R has not previously been reported. Moreover, although dysregulated IGF-1R signaling has been implicated in acquired chemoresistance, the mechanisms by which IGF-1R signaling is activated in chemoresistant cancers are not completely understood (21).
Activation of the IGF-1R signaling pathway results in increased glucose uptake and glycolysis that is subject to feedback regulation under normal growth factor-dependent cell growth. However, uncontrolled glycolysis is a metabolic signature of cancer cells first described by Otto Warburg (22) that supports the malignant phenotype (23). Most highly proliferative cells, including cancer cells, take up more glucose than non-proliferating cells, but only a small fraction of the glucose undergoes complete catabolism in the mitochondria despite adequate oxygen required for oxidative phosphorylation (24). Instead, the breakdown of glucose via aerobic glycolysis allows for rapid ATP synthesis and generates metabolic intermediates required for biosynthesis of nucleotides, lipids and protein to support cell proliferation (25,26). Although the molecular mechanisms underlying the Warburg effect are not completely clear, this metabolic reprogramming confers a growth advantage to tumor cells and is associated with acquired drug resistance in some forms of cancer including HER2+ breast and gastric cancers (27,28).
In this study, we provide evidence for a novel link between t-Darpp, IGF-1R stimulation and increased glycolysis in the mechanism of trastuzumab resistance. We show that PPP1R1B expression in breast tumors is inversely correlated with IGF-1R expression and that the magnitude of PPP1R1B over-expression is associated with reduced overall survival of patients. We also demonstrate that t-Darpp over-expression is sufficient to confer a glycolytic phenotype mimicking that seen in cells selected for trastuzumab resistance. Conversely, knocking down t-Darpp expression in trastuzumab-resistant cells reverses their glucose reliance, indicating t-Darpp is essential for the glycolytic phenotype of these cells. Additionally, pharmacological inhibition and genetic IGF-1R knockdown phenocopies the effects of t-Darpp inhibition in trastuzumab resistant cells. We describe a molecular mechanism in which t-Darpp interacts directly with IGF-1R to promote heterodimerization with EGFR and HER2, resulting in activation of the downstream signaling pathway and increased glycolysis.
MATERIALS AND METHODS
Cell Culture.
SK-BR-3 cells were maintained in McCoy’s medium containing 10% fetal bovine serum (FBS) supplemented with Glutamax (Gibco). For SK-BR-3 cells expressing a control vector (SK.empty) and SK-BR-3 cell over-expressing t-Darpp (SK.tDrp) , G418 at a concentration of 0.5mg/ml was added to maintain expression of the transfected vectors. Trastuzumab resistant SK-Br-3 cells (SK.HerR) were maintained in McCoy’s supplemented with 4ug/ml trastuzumab in addition to Glutamax and 10% FBS. Expression knockdown was performed using validated siRNA: IGF-1R (Ambion IGF1R Silencer validated AM51331, ID:74), INSR (AM51331, ID:29) and PPP1R1B (Santa Cruz Biotechnology, sc-35173) and transfected using RNAiMAX (Invitrogen, Lipofectamine RNAiMAX, 13778–075), according to the manufacturer’s protocol.
Metabolic phenotype analysis.
All experiments were performed using the XFe24 Flux analyzer (Seahorse Bioscience). SK-BR-3 and derived cell lines were plated at 20,000 cell/well in McCoy’s medium supplemented with 10% FBS and Glutamax. The following day, the cells were washed three times in DMEM base medium supplemented with 12mM glucose, 2mM glutamine and 2mM pyruvate, then incubated with the same medium for 1 hour in a CO2-free incubator. Cellular metabolism was analyzed in the SeahorseXFe24 flux analyzer using Mitostress and Glycolysis stress kits (Seahorse Bioscience). Briefly, basal oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured for three cycles followed by successive delivery via port injection of recombinant t-Darpp protein (or medium), the ATP synthase inhibitor oligomycin (2µM) (ATP-coupled OCR), the mitochondrial uncoupler FCCP (4µM) (maximal OCR), and then a combination of rotenone and antimycin A (1µM) (non-mitochondrial oxidation). For glycolysis assays base media was supplemented with 2mM L-glutamine and basal ECAR was measured prior to injection of 16mM glucose. Maximal glucose was assessed after injection of 2µM oligomycin and non-glycolytic acidification was assessed by measuring ECAR in the presence of 100mM 2-Deoxy-D-glucose (2-DG).
Cellular ATP content.
Cells were plated in clear-bottom 96-well plates (Costar 3610) at 5,000 cells/well. The following day the cells were washed with PBS and fresh medium containing treatment (2.5µM trastuzumab, 50 mM 2-DG or t-Darpp) was added to the plate. Cells were incubated at 37°C with 5% CO2 for the desired time. Cellular ATP levels were than measured using Luminescent ATP Detection Assay Kit (Abcam), as described in the instruction manual. Briefly, 50µl cell lysis buffer was added to 100µl of medium and incubated for 5 minutes at room temperature with agitation. 50µl of reaction buffer was then added to each well and incubated for 5 minutes at room temperature with agitation, in the dark. Luminescence was measured using a Wallac 1420 multilabel counter (Perkin Elmer).
Purification of recombinant t-Darpp.
t-Darpp cDNA was cloned into the pET28a bacterial expression vector (Novagen) with an in-frame 6X-His tag at the C-terminus and transformed into ClearColi (BL21 DE3). The detailed purification protocols used to prepare recombinant t-Darpp for use in experiments are provided in supplementary methods.
Co-immunoprecipitation.
Cell lines were grown to 80% confluence in T75 cell culture flasks, cells were washed with PBS, treated with 0.5% trypsin and pelleted at 500xg for 3 minutes. 500µl of modified RIPA buffer (50mM Tris, 1%NP-40, 150mM NaCl, 0.5mM EDTA, pH 8.0) was added to the pellet, incubated at 4°C for 30 minutes and centrifuged at 8000xg for 15 minutes to remove cell debris. Lysates (500µg) were pre-cleared by incubation with 50µl protein A agarose (Cell Signaling Technology) for 1 hour and spun for 1 minute at 1000xg. The supernatant was transferred to a fresh tube and incubated with the appropriate antibody overnight at 4°C (Cell Signaling Technology #2306 for t-Darpp and #3027 for IGF-1R). Protein A agarose (50µl) of was added and the binding reactions were incubated for 1 hour. The tubes were centrifuged at 1000xg for 1 minute and supernatant was collected as unbound proteins. Pellets were washed three times with modified RIPA and bound proteins were eluted by boiling in Laemmli sample buffer. Eluted proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane and blocked for 1 hour in 5% skim-milk. The membranes were incubated overnight with primary antibody diluted 1:1000 in skim milk at 4°C (Cell Signaling Technology #2306 for t-Darpp and #3027 for IGF-1R). The following day membranes incubated for 1 hour with secondary antibody at room temperature and developed using Pierce ECL Plus (Thermo Scientific).
Proximity ligation assay.
The assay was performed as described by the manufacturer for 96-well Duolink (Sigma-Aldrich) assay with modifications for per-cell data normalization. Briefly, SK-BR-3 and derived cells were seeded on a black clear-bottom 96-well plate (Corning) at 10,000 cells/well. In parallel, triplicates of serial dilutions of the same cell type were seeded at 2,500–20,000 cells/well. The cells were washed, fixed with 4% paraformaldehyde prior to performing the Duolink assay. The cells were then incubated for 5 minutes with 1µg/ml DAPI in PBS, and washed 3 times for 5 minutes in PBS. The fluorescent signal was quantified using a fluorescent plate reader (Tecan infinite M1000) for both the Duolink and the DAPI channels and the seeded serial dilutions were used to generate a standard curve that transformed the fluorescence value to cell number. The Duolink fluorescence was then divided by the number of cells measured in each well to produce a normalized value of interaction per cell.
Cell proliferation assay.
Cells were seeded on 6 well plates at 100,000 cells/well. The following day media was then replaced with serum free media (SFM) supplemented with trastuzumab (2.5uM), NVP-AEW (2uM) or the combination of both. The cells were incubated for 3 days with the drugs, then the media was replaced with SFM supplemented with 1X WST-1 and incubated for 1 hour. Absorbance was measured at 440nM with a reference wavelength at 690nM. absorbance measurements in treated conditions were normalized to the mean absorbance of cells treated with SFM alone.
Clinical data analysis.
All data sets used for analysis were obtained from cBioPortal (29) and KM-Plot (30) and includes patients with all cancer stages and treatments. Mutual exclusivity analysis was performed using the cBioPortal server. Overall survival (OS) analysis was performed using Prism (Prism 7 for Windows, GraphPad software) using anonymized patient ID, gene expression and OS data downloaded from cBioPortal and KM-Plot.
Statistical analysis.
All statistical analysis was performed in Prism V6.01 (GraphPad). p-values were calculated using unpaired, two-sided t-tests or two-way ANOVA. A Kaplan-Meier plot was created to calculate hazard ratios (HR) and 95% confidence interval ranges from breast cancer patient data acquired from CBioPortal.
RESULTS
2.1. PPP1R1B expression inversely correlates with IGF-1R expression in breast tumors and PPP1R1B over-expression is associated with reduced survival
Both IGF-1R activation and t-Darpp over-expression have been shown to play important roles in the development of trastuzumab resistance in HER2+ tumors. To determine the possible connection between t-Darpp and IGF-1R in clinical breast cancers, we examined their relative expression in the METABRIC dataset (n=2369 patients) using cBioPortal (29). The major mRNA species transcribed by the PPP1R1B gene, Darpp-32 and t-Darpp, are not distinguished in the dataset, so our analysis necessarily reflects both transcripts. First, we looked at tumors that over-express and under-express the PPP1R1B and IGF1R genes (+/− two S.D. away from their respective means). A co-occurrence analysis showed that PPP1R1B and IGF1R genes are rarely over-expressed at the same time in breast tumors (p<0.001; log odds ratio <−1.75). Moreover, an inverse correlation in expression levels exists between PPP1R1B and IGF-1R transcripts. Increased PPP1R1B expression is significantly more likely in tumors with low IGF-1R transcript levels (p<0.001; log odds ratio=1.63) and high IGF-1R expression is more frequently observed in tumors with low PPP1R1B transcript levels (p=0.01; log odds ratio=1.016) (Table S1). At the protein level, an analysis of the Breast Invasive Carcinoma dataset showed a similar inverse relationship between Darpp-32/t-Darpp and IGF-1R expression (Figure S1).
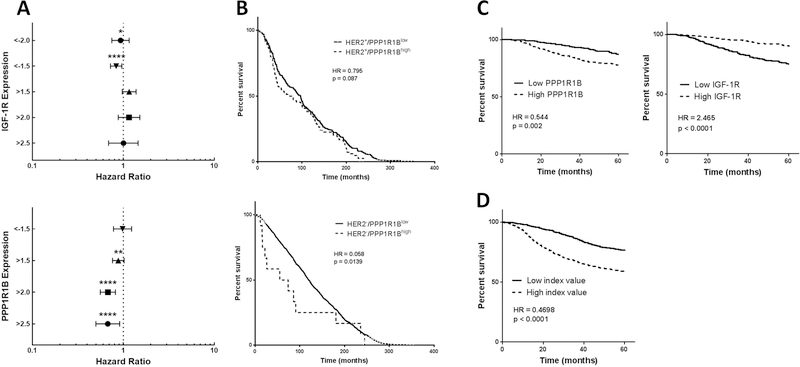
We next examined the relationship between PPP1R1B and IGF-1R and 25 years overall survival (OS) in breast cancer patients, using the METABRIC database. We observed a significant direct relationship between low IGF-1R levels and shorter OS (HR=0.8352; p<0.001) and a inverse relationship between high PPP1R1B levels and shorter OS. The effect of PPP1R1B was more pronounced with PPP1R1B over-expression ≥1.5 S.D. above the mean (HR=0.675–0.881; p<0.001) (Figure 1A and Table S2).
Figure 1. Clinical effect of PPP1R1B and IGF-1R expression.
(A) Forest plots of the hazard ratios for 20 year OS, associated with expression of IGF-1R (top) and PPP1R1B (bottom). (B) Kaplan-Meier survival curves for patients with HER2+ (top) and HER2− (bottom) tumors, comparing tumors with PPP1R1B over-expression (PPP1R1Bhigh) to tumors with normal PPP1R1B (PPP1R1Blow) levels. (C) Kaplan-Meier survival curves for the effect of PPP1R1B (left) and IGF-1R (right) on 5-year OS in breast cancer patients. (D) Kaplan-Meier survival curve using an index value comprised of the mean expression of PPP1R1B and the inverse expression of IGF-1R (PPP1R1B expression + (−1) * IGF-1R expression). HR=hazard ratio; all p-values displayed are log rank values, *p<0.05, **p<0.01, ****p<0.0001.
Since PPP1R1B is on the same amplicon as the ErbB2 (HER2) gene, we examined whether the relationship between PPP1R1B transcript over-expression and OS was related to HER2 expression status. We performed Kaplan-Meier analysis for OS of patients with HER2+ and HER2-minus (HER2−) tumors, with and without over-expression of PPP1R1B, and compared the hazard ratios (HR) of the groups. Within the HER2+ group (n=278), PPP1R1B over-expression was associated with shorter survival but the result did not achieve statistical significance (HR=0.7959; Log rank p=0.087 Gehan-Breslow-Wilcoxon p=0.1681). In the HER2− group (n=1701), PPP1R1B over-expression was significantly associated with shorter survival (HR=0.5805; Log rank p=0.130 Gehan-Breslow-Wilcoxon p=0.0139) (Figure 1B). To examine further the relationship between PPP1R1B and IGF-1R expression and patient prognosis we used the dataset created by Gyroffy et al. (30) and determined the relationship between PPP1R1B and IGF-1R expression and 5-year overall survival (OS) in breast cancer patients (n=1764). We found PPP1R1B to be significantly associated with shorter OS (HR=0.544; Log rank p=0.0017 Gehan-Breslow-Wilcoxon p=0.0007) and IGF-1R to be associated with longer OS (HR=2.465; p<0.0001 for both tests) (Figure 1C). In light of the significant but inverse prognostic value of each gene, we examined the prognostic value of an index comprised of the mean PPP1R1B expression level and the mean inverse of IGF-1R expression. We found this index to be strongly predictive of disease free survival (HR=0.4698 p<0.0001; Figure 1D). Finally, we used multivariate analysis of PPP1R1B and IGF-1R and found both to contribute significantly to 5-year OS, independent of HER2 status (Table S3).
2.2. Cells over-expressing t-Darpp have increased glycolytic capacity
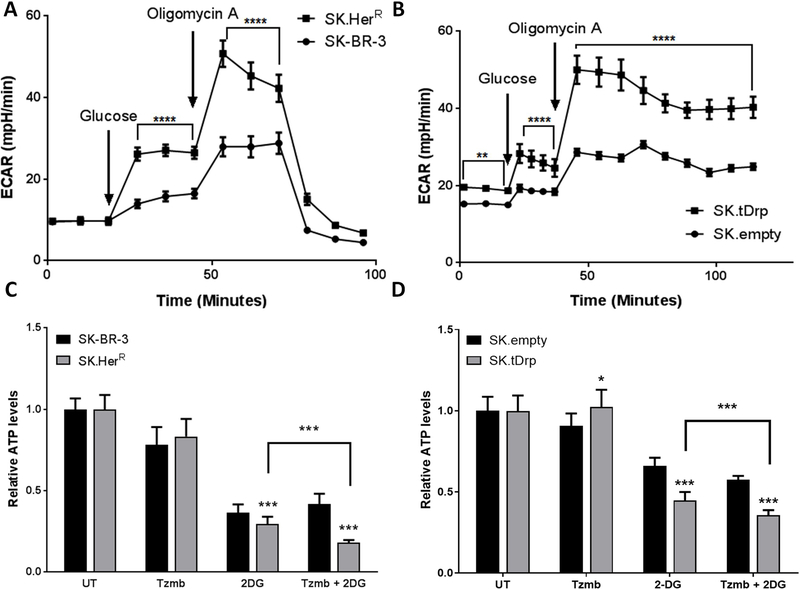
We and others have shown that t-Darpp over-expression confers trastuzumab resistance and that trastuzumab resistance is associated with increased glycolytic capacity (4–6,9,14,27,28). In light of the genetic interaction described between the expression of PPP1R1B and IGF1R genes we set out to examine whether t-Darpp might act through IGF-1R to confer resistance to trastuzumab by promoting a metabolic switch towards glycolysis. To begin to examine the role of t-Darpp in the metabolic phenotype of trastuzumab-resistant cells, we used the XFe24 flux analyzer to compare the metabolic characteristics of HER2+ SK-BR-3 cells selected for trastuzumab resistance (SK.HerR), which are known to over-express endogenous t-Darpp (4–6), and SK-BR-3 cells that stably over-express exogenous t-Darpp (SK.tDrp), which confers resistance to trastuzumab (4,5). The parental SK-BR-3 cells or SK.empty cells, transfected with an empty expression vector, were used as controls, respectively. The basal glycolysis rate was measured as the extracellular acidification rate (ECAR) following addition of glucose, and mitochondrial oxygen consumption rate (OCR) was analyzed in parallel. We observed no significant differences in basal OCR or respiration coupled to ATP synthesis (+oligomycin) among the SK-BR-3, SK.HerR, SK.empty and SK.tDrp cells (Figure S2). However, both SK.HerR and SK.tDrp cells exhibited increased basal glycolysis and maximal glycolytic capacity (+oligomycin) compared to SK-BR-3 and SK.empty cells (Figure 2A and 2B).
Figure 2. SK.tDrp and SK.HerR cells share similar metabolic characteristics.
(A and B) Seahorse analysis of the metabolic phenotype of SK-BR-3 cells compared to SK.HerR cells (A) and SK.empty cells compared to SK.tDrp cells (B). Assays were performed on the XFe24 flux analyzer. A change in ECAR following injection of glucose indicates glycolysis and a change in ECAR following oligomycin A injection indicates maximal glycolytic capacity. (C and D) Cellular ATP levels were measured in untreated (UT) cells and in cells following 30 min exposure to 2.5µM trastuzumab, 50mM 2-DG, or the combination. Mean values (±S.D.) normalized to UT (=1.0) are reported for SK-BR-3 compared to SK.HerR cells (C) and SK.empty compared to SK.tDrp cells (D). *p<0.05, ***p<0.001, ****p<0.0001
To assess whether the increased glycolytic characteristics of SK.HerR and SK.tDrp cells would result in increased dependence on glycolysis to meet energy needs, we measured changes in ATP levels following inhibition of glycolysis with 2-deoxy-D-glucose (2-DG). Since 2-DG and trastuzumab have a synergistic effect on breast cancer cell growth (28), we also examined the effect of trastuzumab + 2-DG on the energetic state of our cells. ATP levels were significantly lower in both SK.HerR and SK.tDrp cells, compared to their respective controls, after exposure to 2-DG (Figure 2C and 2D). In addition, trastuzumab appeared to sensitize SK.HerR and SK.tDrp cells to 2-DG. This effect was not observed in SK-BR-3 and SK.empty cells (Figure 2C and 2D). These results suggest that t-Darpp over-expression in SK.tDrp cells is sufficient to increase reliance on glycolysis, similar to the metabolic switch observed in trastuzumab-resistant SK.HerR cells.
2.3. t-Darpp and IGF-1R are required for the glycolytic phenotype of SK.HerR cells
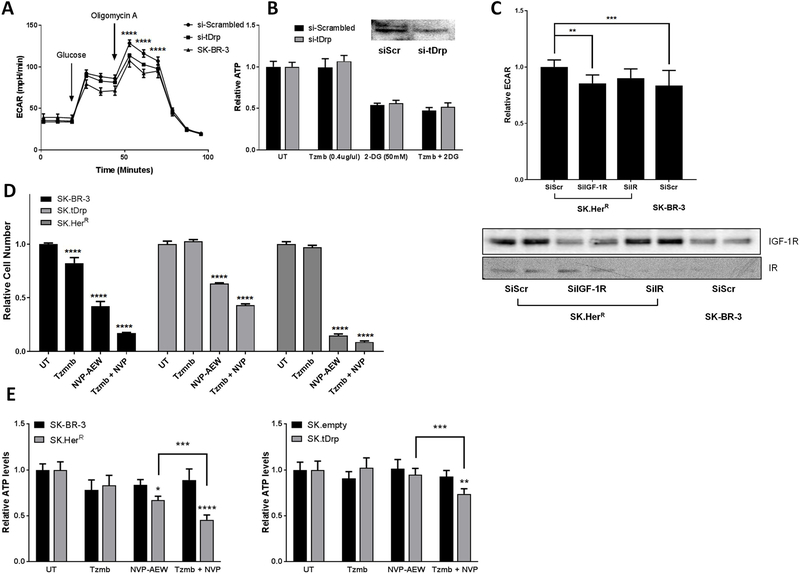
Based on our observation that t-Darpp over-expression is sufficient to increase glycolytic capacity and dependency in SK.tDrp cells, we tested whether t-Darpp is essential for the enhanced glycolytic phenotype observed in SK.HerR cells. We knocked down t-Darpp expression using siRNA and analyzed glycolytic capacity and dependency. Following t-Darpp knockdown, the glycolysis rate in SK.HerR cells was significantly reduced to levels equivalent to SK-BR-3 cells (Figure 3A). However, the sensitivity to inhibition of glycolysis by 2-DG alone, or in combination with trastuzumab, was essentially unchanged in response to t-Darpp knockdown (Figure 3B). Taken together, these data suggest that t-Darpp is required for the increased glycolytic capacity but not the increased glycolytic dependency in SK.HerR cells.
Figure 3. t-Darpp knockdown reverses the glycolytic phenotype of SK.HerR cells.
(A) Seahorse flux analysis comparing glycolysis and glycolytic capacity between cells transfected with t-Darpp siRNA (si-tDrp) and control siRNA (si-Scrambled). (B) ATP levels in untreated (UT) cells and in cells following 30 min exposure to2.5µM trastuzumab, 50mM 2-DG, or both, in cells transfected with t-Darpp/PPP1R1B (si-tDrp) siRNA or control siRNA (si-Scrambled). (C) Fold change in glycolysis in SK.HerR cells following siRNA knock-down of IGF-1R or IR compared to a scrambled siRNA control, with corresponding Western blots of the receptors. (D) Growth inhibition of SK-BR-3 derived cell lines by trastuzumab, NVP-AEW or combined treatment. SK-BR-3, SK.tDrp and SK.HerR were incubated for 3 days in SFM only (untreated, UT) or in the presence of either trastuzumab, NVP-AEW or both. Cell number was measured by WST-1 and normalized to untreated wells. (E) Cellular ATP levels were measured in untreated (UT) cells and in cells following 30 min exposure to 2.5µM trastuzumab, 2µM NVP-AEW, or the combination. Mean values (±S.D.) normalized to UT (=1.0) are reported for SK-BR-3 versus SK.HerR cells (left) and SK.empty versus SK.tDrp cells (right). ***p<0.001 ****p<0.0001
In light of the observed relationship between PPP1R1B and IGF-1R in clinical samples and the role for IGF-1R and Insulin receptor (IR) in the regulation of glycolysis, we examined the effect of IGF-1R and IR knock-down on the glycolytic phenotype of SK.HerR cells (Figure 3C and Figure S3). We observed a significant decrease in both glycolysis and glycolytic capacity of SK.HerR cells following knock-down of IGF-1R. Knock-down of IR resulted in a more modest decrease in glycolysis that was not statistically significant (Figure 3C). Since IR expression is lower than IGF-1R expression in our cell lines, the extent of siRNA mediated knockdown and the modest effects on glycolysis following IR knockdown might be expected but does not completely exclude a role for IR. Nevertheless, the data support that IGF-1R is an important mediator of the glycolytic phenotype in trastuzumab resistant cells.
To further evaluate the role for IGF-1R in t-Darpp’s ability to confer resistance to trastuzumab, we incubated SK-BR-3, SK.tDrp and SK.HerR cell lines with serum free media (SFM) complemented with either trastuzumab (2.5µM), NVP-AEW (2 µM), a potent IGF-1R/IR small molecule inhibitor, or the combination of both. We then examined the relative cell number of treated wells compared to controls incubated with SFM alone. We found that trastuzumab reduced cell number in SK-BR-3 cells by ~20% compared to SFM alone (p<0.0001) while SK.tDrp and SK.HerR were unaffected. Treatment with NVP-AEW alone inhibited growth to varying degree with SK.HerR showing the greatest (~85% inhibition p<0.0001) and SK.tDrp the least sensitivity (~45% inhibition p<0.0001). The combination treatment resulted in a significant growth inhibition in all 3 cell lines, compared to untreated and to either compound alone (Figure 3D). In agreement, acute NVP-AEW treatment significantly reduced intracellular ATP levels only in SK.HerR cells (−33%, p<0.05) but not in parental SK-BR-3 (−16%, n.s.); whereas combined treatment of NVP-AEW and trastuzumab reduced cellular ATP content in both SK.tDrp and SK.HerR cells (Figure 3E).
2.4. t-Darpp mediates glycolytic effects through IGF-1R
One of the mechanisms involved in acquired trastuzumab resistance is enhanced signaling through IGF-1R and the heterodimerization of IGF-1R with EGFR and HER2, which activates signaling downstream of the receptors (17,18,21,31–34). We therefore asked if t-Darpp’s mechanism of action in promoting glycolysis might be mediated through interaction with IGF-1R or activation of IGF-1R signaling.
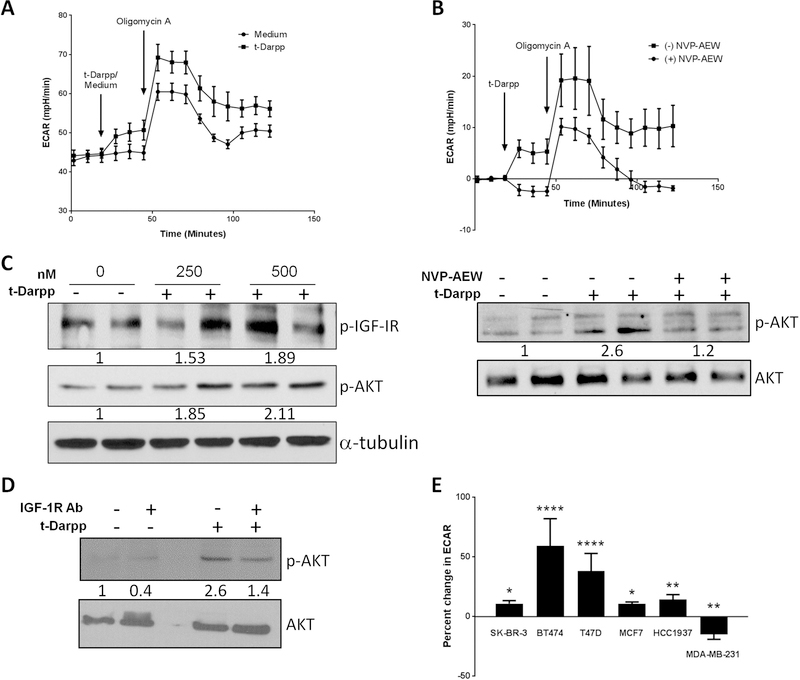
To determine if t-Darpp regulates IGF-1R signaling directly via the receptor complex, we tested the effects of purified, recombinant t-Darpp on glycolysis in SK-BR-3 cells. Similar to cells with t-Darpp over-expression, the addition of recombinant t-Darpp to SK-BR-3 cells led to an acute increase in glycolysis and glycolytic capacity (Figure 4A), with no effect on OCR (Figure S4). Pre-treatment with NVP-AEW for 1 hour abolished the increase in ECAR (p<0.01; Figure 4B). Next, we examined whether t-Darpp activated the IGF-1R pathway by detecting phosphorylation of IGF-1R and the downstream Akt kinase in SK-BR-3 cells. We found that treatment with recombinant t-Darpp stimulated a dose dependent increase in IGF-1R and Akt phosphorylation that was abolished by pre-treatment with IGF-1R inhibitor (Figure 4C).
Figure 4. Recombinant t-Darpp confers an IGF-1R-dependent increase in glycolysis.
(A) Seahorse analysis of ECAR at baseline in 1mM glutamine, 1mM pyruvate without glucose and following injection of 16mM glucose (Medium) or glucose supplemented with 500nM recombinant t-Darpp (t-Darpp). (B) SK-BR-3 cells were pre-treated in media minus or plus 5µM NVP-AEW for one hour. ECAR was analyzed at baseline in 1mM glutamine, 1mM pyruvate and 16mM glucose and following injection of medium supplemented with 500nM recombinant t-Darpp, the data is presented as net change in ECAR following injection. (C) Western blot analysis of IGF-1R and Akt phosphorylation following treatment with recombinant t-Darpp protein (left) and the effect of IGF-1R inhibition with NVP-AEW prior to treatment with recombinant t-Darpp (right) Numbers indicate fold change compared to untreated samples after normalization to total protein (D) Western analysis comparing Akt phosphorylation following treatment with recombinant t-Darpp (400nM), an inhibitory IGF-1R monoclonal antibody (Ab) or both. (E) Percent change in ECAR following treatment with 500nM recombinant t-Darpp of a panel of breast cancer cell lines (SK-BR-3, BT474, T47D, MCF7, HCC1937 and MDA-MB-231). Shown is the mean change in ECAR normalized to untreated controls (n=3–6). *p<0.05, **p<0.01 ***p<0.001 ****p<0.0001
To validate the role for IGF-1R in t-Darpp-mediated Akt activation, we treated SK-BR-3 cells with t-Darpp (400nM) along with an inhibitory monoclonal antibody targeting IGF-1R. In the absence of the inhibitory antibody, recombinant t-Darpp resulted in nearly3-fold increase in p-AKT, while co-treatment with the inhibitory antibody mostly abolished the effect (Figure 4D).
To examine the generality of metabolic regulation by t-Darpp, we tested a panel of breast cancer cell lines representing the main subtypes of the disease (triple-negative: MD-MBA-231 and HCC1143; HER2+: SK-BR-3 and BT474; luminal: MCF7 and T47D). The addition of recombinant t-Darpp to the culture medium increased glycolysis in all but one of the examined cell lines (Figure 4E). The increase in ECAR ranged from 10% (MCF7 and SK-BR-3) to 60% (BT474), whereas MDA-MB-231 cells showed no significant response to t-Darpp. Notably, MD-MBA-231 cells have very low levels of IGF-1R cell surface expression (33). In addition, we examined the effects of acute treatment with recombinant t-Darpp on IGF-1R and Akt activity in L6 myotubes, a non-tumorigenic IGF-1-responsive cell type (Figure S5). Phosphorylation of both Akt and IGF-1R was increased in a dose and time dependent manner in the myotubes. Similar to the effects observed in SK-BR-3 cells, treatment with NVP-AEW abolished IGF-1R pathway activation by t-Darpp.
2.5. t-Darpp promotes heterodimerization of IGF-1R with ErbB receptors
IGF-1R and HER2 have been reported to heterodimerize in trastuzumab-resistant cells (17). Based on our findings that t-Darpp enhances the glycolytic phenotype in SK.HerR and SK.tDrp cells via IGF-1R, we wanted to determine more directly if t-Darpp was also involved in heterodimerization between IGF-1R and HER2. First, we used a proximity ligation assay (PLA) to confirm the observation that IGF-1R forms dimers with ErbB receptors in cells selected for trastuzumab resistance. We examined the proportions of IGF-1R/EGFR, IGF-1R/HER2 or IGF-1R/Her3 complexes in SK-BR-3 parental cells and SK.HerR cells. We observed 2.0- to 2.5-fold more of each heterodimeric interaction per cell in SK.HerR cells than in parental SK-BR-3 cells (Figure 5A). We also detected significantly more IGF-1R/EGFR and IGF-1R/HER2 heterodimers, but not IGF-1R/Her3 heterodimers, in SK.tDrp cells than in SK.empty cells (Figure 5B).
Figure 5. Heterodimerization of IGF-1R and ErbB receptors.
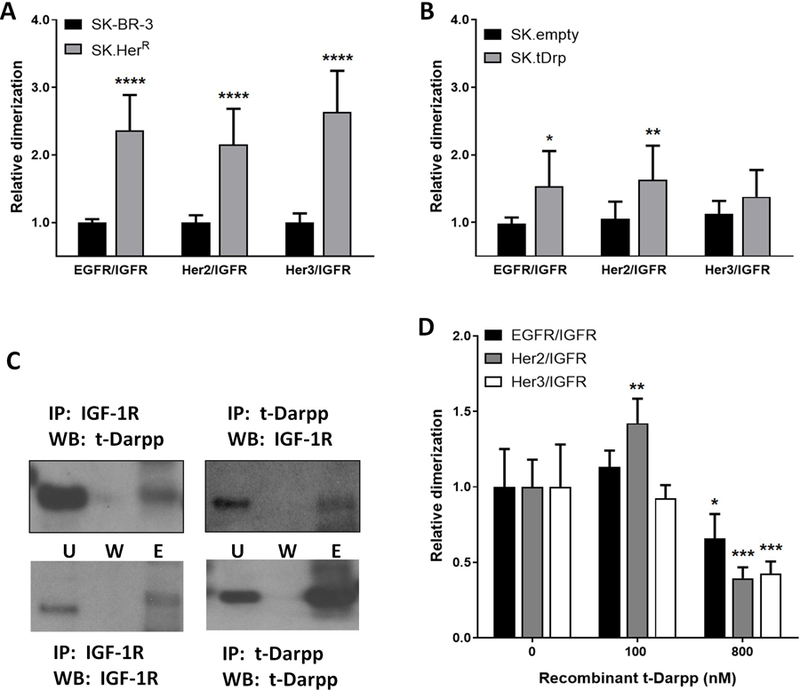
Quantitative proximity ligation assay analysis of the interaction between IGF-1R and ErbB receptors (EGFR, HER2 and Her3), in SK-BR-3 versus SK.HerR cells (A) and SK.empty versus SK.tDrp cells (B). (C) Co-immunoprecipitation (IP) assays using antibody to IGF-1R or t-Darpp for the IP, followed by Western analyses (WB) with t-Darpp or IGF-1R antibody, as indicated. (U=Unbound, W=Wash, E=Elution). (D) Proximity ligation assay to detect interaction between IGF-1R and EGFR, HER2 or Her3 following the addition of recombinant t-Darpp at the indicated concentrations. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
Next, based on the ability of recombinant t-Darpp to turn on IGF-1R signaling, we asked if t-Darpp might directly bind to IGF-1R to confer its effects. Using a co-immunoprecipitation approach, we found that immunoprecipitation with anti-IGF-1R antibody pulled down t-Darpp and the reciprocal immunoprecipitation with anti-Darpp antibody pulled down IGF-1R (Figure 5C). We then incubated immobilized recombinant t-Darpp with lysates from SK-BR-3 and BT474 cell lines and used MS/MS to examine protein-protein interactions. Both IGF-1R and HER2 were seen to interact with t-Darpp in both cell lines (Table S4). Moreover, addition of recombinant t-Darpp to the growth medium of parental SK-BR-3 cells stimulated heterodimerization of IGF-1R with ErbB receptors. Specifically the interaction between IGF-1R and HER2 was significantly increased following the addition of 100nM t-Darpp (Figure 5D). IGF-1R/EGFR complexes were modestly increased in response to t-Darpp, but the effect was not statistically significant, and the IGF-1R/Her3 interaction was not affected by recombinant t-Darpp. A higher concentration of t-Darpp (800nM) resulted in decreased dimerization of IGF-1R with all three ErbB receptors (Figure 5D). These data suggest that t-Darpp can promote IGF-1R dimerization with HER2, consistent with earlier reports of IGF-1R/HER2 complexes in trastuzumab-resistant cells (17) . At higher concentrations, t-Darpp appears to be inhibitory, possibly due to receptor saturation as characterized for ligands binding to IGF-1R (35,36).
DISCUSSION
We provide evidence that suggests for the first time that t-Darpp, a protein over-expressed by several cancer types, confers a metabolic advantage not only when over-expressed in transfected or selected cells but also when added to cells extracellularly as a recombinant protein. We also show that reversing the over-expression of t-Darpp in cells selected for trastuzumab resistance abolishes most of their glycolytic phenotype. This effect appears to be mediated by binding of t-Darpp to IGF-1R to promote heterodimerization with HER2 and possibly EGFR. This represents a new function for t-Darpp and a previously unknown mechanism for activation of IGF-1R signaling, which is commonly up-regulated in breast cancer. It could also be a key mechanism by which resistance develops to drugs that target specific tyrosine kinase receptors (e.g. trastuzumab) (15–17,37). Moreover, recent studies have shown that metabolic reprogramming, which causes cells to become more reliant on glucose, is a component of the chemoresistance phenotype in many cancers (28,38,39). Given that t-Darpp over-expression is capable of conferring trastuzumab resistance (4–6,11), the activation of glycolysis characterized herein may relate to the mechanism by which t-Darpp promotes resistance. Thus, this work has relevance to well-described phenotypes of several cancer types, including the HER2+ breast cancer model used in this study.
t-Darpp confers a metabolic advantage in trastuzumab-resistant cells.
We have shown that t-Darpp is both sufficient and necessary to confer a shift in the metabolic phenotype of SK-BR-3 cells in culture. There are striking similarities in maximal ECAR and 2-DG sensitivity between cells that over-express exogenous t-Darpp (SK.tDrp) and cells selected for trastuzumab resistance (SK.HerR) in which endogenous t-Darpp expression is up-regulated. The reversal of the glycolytic capacity back to wild-type levels after t-Darpp knockdown in SK.HerR cells indicates that t-Darpp is required for this phenotype. The glycolytic phenotype driven by t-Darpp is sufficient to confer sensitivity to inhibitors of glycolysis, resulting in reduced ATP and growth restriction, similar to SK.HerR cells, suggesting that the magnitude of t-Darpp effects are biologically relevant. Trastuzumab has been shown to inhibit glycolysis in breast cancer (28), and increased glycolysis and glycolytic capacity have been associated with reduced sensitivity to anti-cancer therapies (for a review see (40). It is therefore reasonable to conclude that trastuzumab resistance mediated by t-Darpp over-expression can be attributed, at least in part, to the increase in glycolytic capacity described in this report.
A novel mechanism of IGF-1R activation by t-Darpp.
Tumors with acquired trastuzumab resistance commonly exhibit activation of IGF-1R signaling and heterodimerization of IGF-1R with ErbB receptors (17,21,32,34). Since ErbB receptors are not natural dimerization partners for IGF-1R and since IGF-1R is a ligand-dependent receptor, the heterodimerization patterns described here and elsewhere cannot be attributed solely to increased expression of IGF-1R. In fact, although IGF-1R/ErbB dimerization levels in SK.HerR cells are increased, relative to SK-BR-3 cells, the total IGF-1R, EGFR, HER2 and HER3 protein levels are similar in the two cell lines (Figure S6). Our experiments with recombinant t-Darpp and the co-immunoprecipitation data suggest a mechanism in which t-Darpp interacts directly with IGF-1R and facilitates heterodimerization with one or more ErbB receptor. The differential response to recombinant t-Darpp exhibited by the different breast cancer cell lines (Figure 4E) indicates that IGF-1R expression levels alone are not sufficient to predict the magnitude response to t-Darpp. High levels of IGF-1R have been reported for both MCF-7 and T47D, yet the response of T47D to treatment with t-Darpp was considerably greater than that observed in MCF-7 cells. Moreover, while MDA-MB-231 cells have been reported to express similar IGF-1R levels as SK-BR-3 and BT474 cells, they were not responsive to t-Darpp (41,42). It is possible that a combination of cell surface availability and post-translational modifications of IGF-1R influence t-Darpp’s ability to activate glycolysis in various cell types. More likely, as suggested by our data, t-Darpp may promote IGF-1R dimerization with HER2 and HER3 and mediate its signaling and metabolic effects via these hybrid receptors (Figure 6). t-Darpp has no known enzymatic activity and is mostly an unstructured protein, suggesting that it might act as a scaffold for IGF-1R/ErbB dimerization. Further clarification of the molecular nature of the interaction of t-Darpp with IGF-1R and the mechanism behind t-Darpp stimulated IGF-1R/ErbB heterodimerization could help shed light on receptor dynamics and choice of dimerization partners. One possible model would be that t-Darpp physically interacts with both dimerization partners, thus bringing the two receptors together. Alternatively, the interaction between t-Darpp and IGF-1R may confer a conformational change that allows IGF-1R to dimerize with non-natural partners such as the ErbB receptors.
Figure 6. A schematic model of IGF-1R activation by t-Darpp.
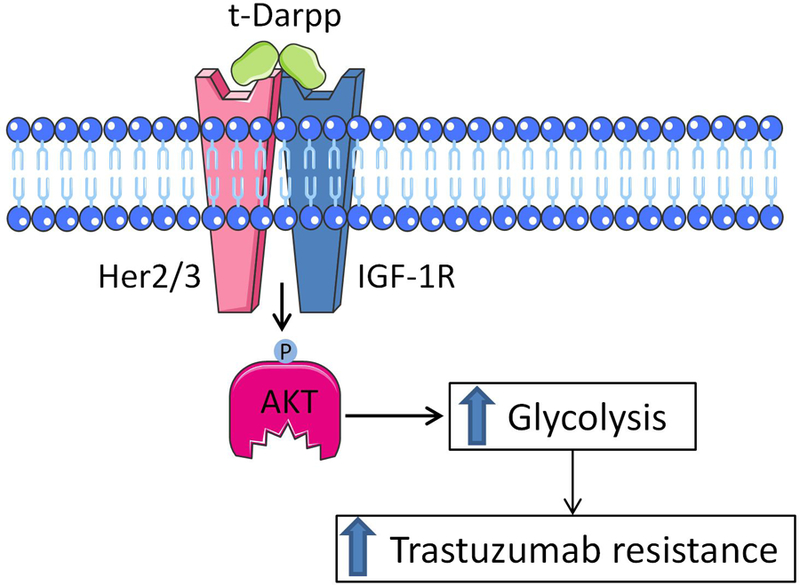
t-Darpp interacts with IGF-1R and HER2 to promote heterodimerization and activation of IGF-1R signaling. This results in enhanced glycolysis and trastuzumab resistance.
Clinical association between expression of IGF1R and PPP1R1B genes.
We have determined that in all sub-types of breast cancer, the over-expression of PPP1R1B transcript is mutually exclusive from the over-expression of IGF-1R and PPP1R1B is enriched in tumors that under-express IGF-1R. However, this observation is not unique to breast cancers. In the TGCA prostate cancer dataset of 491 tumors, there are 20 tumors that over-express PPP1R1B and 20 tumors that over-express IGF-1R (≥2 S.D. above the mean) with no overlap between the groups. Indeed, regardless of the cancer type or data acquisition method, the mutually exclusive nature of PPP1R1B and IGF-1R over-expression holds true (Table S5). This intriguing observation could be attributed to a shared regulatory network in which the factors responsible for the up-regulation of t-Darpp in malignancy may have an inhibitory effect on the expression of IGF-1R. In vitro selection for trastuzumab resistance results in over-expression of t-Darpp in BT474 and SK-BR-3 cell lines, yet changes in the IGF-1R levels were observed only in BT474 cells ((17,43) and Figure S6), indicating that the rise in t-Darpp expression under these conditions was not necessarily linked to suppressed IGF-1R expression. An alternative explanation for the inverse correlation between IGF-1R and PPP1R1B in tumors could be functional. We have shown that t-Darpp confers its glycolytic advantage through activation of IGF-1R. It is therefore likely that in tumors that over-express IGF-1R, the positive impact of PPP1R1B expression on tumor fitness would be less advantageous or completely redundant. This hypothesis could also explain the enrichment for PPP1R1B over-expression in tumors with low IGF-1R expression, since activation of the receptor in these tumors would be expected to have a greater impact on proliferation and drug resistance. We have also shown that PPP1R1B over-expression is significantly associated with shorter survival time in breast cancer patients and that the effect is more pronounced with higher levels of expression. Interestingly, the relationship between PPP1R1B expression and OS is independent of HER2 status of the tumors. IGF-1R expression levels have had inconsistent prognostic value in previous studies (44,45). Our results show that while the correlation of IGF-1R expression with increased OS in breast cancer patients is weak, an index value comprised of PPP1R1B expression levels and inverse IGF-1R expression levels has powerful predictive value of overall survival and may have valuable prognostic implications .
Summary of findings.
In this study we provide a molecular mechanism of action through which t-Darpp can directly bind and promote the dimerization and activation of IGF-1R. IGF-1R activation by t-Darpp results in increased glycolysis and glycolytic capacity that may contribute to drug resistance (28,38,39). We also show that the exogenous over-expression of t-Darpp, known to confer trastuzumab resistance (3–5), results in a metabolic shift towards increased dependency on glycolysis for energetic homeostasis. The same metabolic characteristics are observed in cells selected for trastuzumab resistance, which express high levels of endogenous t-Darpp. siRNA-mediated knockdown of t-Darpp in the selected cells resulted in a reversal of the glycolytic phenotype, indicating that the phenotype in selected cells (SK.HerR) and transfected cells (SK.tDrp) is likely due to a direct effect of t-Darpp on glycolysis.
Supplementary Material
Translational relevance.
Most patients with HER2+ breast cancer treated with trastuzumab acquire resistance within the first year of therapy. Resistance is attributed to activation of alternative signaling pathways and persistent HER2 signaling despite the presence of trastuzumab. One of the signaling pathways involved in trastuzumab resistance is the IGF-1R pathway involving unique heterodimerization between IGF-1R and EGFR or HER2 in resistant cells. Here we describe a mechanism in which t-Darpp directly interacts with IGF-1R and stimulates heterodimer formation. This activates IGF-1R signaling, which in turn results in increased glycolytic capacity and confers sensitivity to glycolytic inhibitors. Our findings demonstrate t-Darpp as a potential therapeutic target to inhibit IGF-1R signaling in trastuzumab-resistant cancers and thereby reverse metabolic reprogramming required for the resistance phenotype.
ACKNOWLEDGMENTS
The authors would like to acknowledge M. Kane, A. Celis, and Y. Feng for their technical assistance with t-Darpp cloning and bacterial expression and purification. We are grateful for assistance from Brian Armstrong in the Light Microscopy Digital Imaging core. This work was supported by grants from NIGMS (R01GM105898 to S.E. Kane and J. Momand) and NCI (P30CA33572 to S.T. Rosen). G. Lenz is the recipient of the City of Hope – Israel Fellowship in Biomedical Research.
REFERENCES
- 1.Nahta R, Esteva FJ. Trastuzumab: triumphs and tribulations. Oncogene 2007;26(25):3637–43 doi 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 2.Frampton JE. Lapatinib: a review of its use in the treatment of HER2-overexpressing, trastuzumab-refractory, advanced or metastatic breast cancer. Drugs 2009;69(15):2125–48 doi 10.2165/11203240. [DOI] [PubMed] [Google Scholar]
- 3.Chan CT, Metz MZ, Kane SE. Differential sensitivities of trastuzumab (Herceptin)-resistant human breast cancer cells to phosphoinositide-3 kinase (PI-3K) and epidermal growth factor receptor (EGFR) kinase inhibitors. Breast cancer research and treatment 2005;91(2):187–201 doi 10.1007/s10549-004-7715-1. [DOI] [PubMed] [Google Scholar]
- 4.Gu L, Waliany S, Kane SE. Darpp-32 and its truncated variant t-Darpp have antagonistic effects on breast cancer cell growth and herceptin resistance. PloS one 2009;4(7):e6220 doi 10.1371/journal.pone.0006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamel S, Bouchard A, Ferrario C, Hassan S, Aguilar-Mahecha A, Buchanan M, et al. Both t-Darpp and DARPP-32 can cause resistance to trastuzumab in breast cancer cells and are frequently expressed in primary breast cancers. Breast cancer research and treatment 2010;120(1):47–57 doi 10.1007/s10549-009-0364-7. [DOI] [PubMed] [Google Scholar]
- 6.Belkhiri A, Dar AA, Peng DF, Razvi MH, Rinehart C, Arteaga CL, et al. Expression of t-DARPP mediates trastuzumab resistance in breast cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research 2008;14(14):4564–71 doi 10.1158/1078-0432.CCR-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vangamudi B, Peng DF, Cai Q, El-Rifai W, Zheng W, Belkhiri A. t-DARPP regulates phosphatidylinositol-3-kinase-dependent cell growth in breast cancer. Molecular cancer 2010;9:240 doi 10.1186/1476-4598-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Rifai W, Smith MF Jr., Li G, Beckler A, Carl VS, Montgomery E, et al. Gastric cancers overexpress DARPP-32 and a novel isoform, t-DARPP. Cancer research 2002;62(14):4061–4. [PubMed] [Google Scholar]
- 9.Hong J, Katsha A, Lu P, Shyr Y, Belkhiri A, El-Rifai W. Regulation of ERBB2 receptor by t-DARPP mediates trastuzumab resistance in human esophageal adenocarcinoma. Cancer research 2012;72(17):4504–14 doi 10.1158/0008-5472.CAN-12-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckler A, Moskaluk CA, Zaika A, Hampton GM, Powell SM, Frierson HF Jr., et al. Overexpression of the 32-kilodalton dopamine and cyclic adenosine 3’,5’-monophosphate-regulated phosphoprotein in common adenocarcinomas. Cancer 2003;98(7):1547–51 doi 10.1002/cncr.11654. [DOI] [PubMed] [Google Scholar]
- 11.Belkhiri A, Zaika A, Pidkovka N, Knuutila S, Moskaluk C, El-Rifai W. Darpp-32: a novel antiapoptotic gene in upper gastrointestinal carcinomas. Cancer research 2005;65(15):6583–92 doi 10.1158/0008-5472.CAN-05-1433. [DOI] [PubMed] [Google Scholar]
- 12.Denny EC, Kane SE. t-Darpp Promotes Enhanced EGFR Activation and New Drug Synergies in Her2-Positive Breast Cancer Cells. PloS one 2015;10(6):e0132267 doi 10.1371/journal.pone.0132267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkhiri A, Dar AA, Zaika A, Kelley M, El-Rifai W. t-Darpp promotes cancer cell survival by up-regulation of Bcl2 through Akt-dependent mechanism. Cancer research 2008;68(2):395–403 doi 10.1158/0008-5472.CAN-07-1580. [DOI] [PubMed] [Google Scholar]
- 14.Gu L, Lau SK, Loera S, Somlo G, Kane SE. Protein kinase A activation confers resistance to trastuzumab in human breast cancer cell lines. Clinical cancer research : an official journal of the American Association for Cancer Research 2009;15(23):7196–206 doi 10.1158/1078-0432.CCR-09-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). Journal of the National Cancer Institute 2001;93(24):1852–7. [DOI] [PubMed] [Google Scholar]
- 16.Nahta R. Molecular Mechanisms of Trastuzumab-Based Treatment in HER2-Overexpressing Breast Cancer. ISRN oncology 2012;2012:428062 doi 10.5402/2012/428062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer research 2005;65(23):11118–28 doi 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 18.Sachdev D, Yee D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. Molecular cancer therapeutics 2007;6(1):1–12 doi 10.1158/1535-7163.MCT-06-0080. [DOI] [PubMed] [Google Scholar]
- 19.Insulin Pollak M., insulin-like growth factors and neoplasia. Best practice & research Clinical endocrinology & metabolism 2008;22(4):625–38 doi 10.1016/j.beem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Pollack MN. Insulin, insulin-like growth factors, insulin resistance, and neoplasia. The American journal of clinical nutrition 2007;86(3):s820–2. [DOI] [PubMed] [Google Scholar]
- 21.Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, et al. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes & Diseases 2015;2(1):13–25 doi 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warburg O. On the origin of cancer cells. Science 1956;123(3191):309–14. [DOI] [PubMed] [Google Scholar]
- 23.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2008;49 Suppl 2:24S–42S doi 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 24.Locasale JW, Cantley LC, Vander Heiden MG. Cancer’s insatiable appetite. Nature biotechnology 2009;27(10):916–7 doi 10.1038/nbt1009-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Current opinion in genetics & development 2008;18(1):54–61 doi 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism 2008;7(1):11–20 doi 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Pan C, Guo L, Wu M, Guo J, Peng S, et al. A new mechanism of trastuzumab resistance in gastric cancer: MACC1 promotes the Warburg effect via activation of the PI3K/AKT signaling pathway. Journal of hematology & oncology 2016;9(1):76 doi 10.1186/s13045-016-0302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer research 2011;71(13):4585–97 doi 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment 2010;123(3):725–31 doi 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.Riedemann J, Takiguchi M, Sohail M, Macaulay VM. The EGF receptor interacts with the type 1 IGF receptor and regulates its stability. Biochemical and biophysical research communications 2007;355(3):707–14 doi 10.1016/j.bbrc.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Belfiore A, Frasca F. IGF and insulin receptor signaling in breast cancer. Journal of mammary gland biology and neoplasia 2008;13(4):381–406 doi 10.1007/s10911-008-9099-z. [DOI] [PubMed] [Google Scholar]
- 33.Dricu A, Kanter L, Wang M, Nilsson G, Hjertman M, Wejde J, et al. Expression of the insulin-like growth factor 1 receptor (IGF-1R) in breast cancer cells: evidence for a regulatory role of dolichyl phosphate in the transition from an intracellular to an extracellular IGF-1 pathway. Glycobiology 1999;9(6):571–9. [DOI] [PubMed] [Google Scholar]
- 34.Gallardo A, Lerma E, Escuin D, Tibau A, Munoz J, Ojeda B, et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. British journal of cancer 2012;106(8):1367–73 doi 10.1038/bjc.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Meyts P, Urso B, Christoffersen CT, Shymko RM. Mechanism of insulin and IGF-I receptor activation and signal transduction specificity. Receptor dimer cross-linking, bell-shaped curves, and sustained versus transient signaling. Annals of the New York Academy of Sciences 1995;766:388–401. [DOI] [PubMed] [Google Scholar]
- 36.Surinya KH, Forbes BE, Occhiodoro F, Booker GW, Francis GL, Siddle K, et al. An investigation of the ligand binding properties and negative cooperativity of soluble insulin-like growth factor receptors. The Journal of biological chemistry 2008;283(9):5355–63 doi 10.1074/jbc.M707054200. [DOI] [PubMed] [Google Scholar]
- 37.Fiszman GL, Jasnis MA. Molecular Mechanisms of Trastuzumab Resistance in HER2 Overexpressing Breast Cancer. International journal of breast cancer 2011;2011:352182 doi 10.4061/2011/352182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 2012;149(5):1098–111 doi 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komurov K, Tseng JT, Muller M, Seviour EG, Moss TJ, Yang L, et al. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Molecular systems biology 2012;8:596 doi 10.1038/msb.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene 2006;25(34):4633–46 doi 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 41.Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast cancer : basic and clinical research 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 42.Mukohara T, Shimada H, Ogasawara N, Wanikawa R, Shimomura M, Nakatsura T, et al. Sensitivity of breast cancer cell lines to the novel insulin-like growth factor-1 receptor (IGF-1R) inhibitor NVP-AEW541 is dependent on the level of IRS-1 expression. Cancer letters 2009;282(1):14–24 doi 10.1016/j.canlet.2009.02.056. [DOI] [PubMed] [Google Scholar]
- 43.Kostler WJ, Hudelist G, Rabitsch W, Czerwenka K, Muller R, Singer CF, et al. Insulin-like growth factor-1 receptor (IGF-1R) expression does not predict for resistance to trastuzumab-based treatment in patients with Her-2/neu overexpressing metastatic breast cancer. Journal of cancer research and clinical oncology 2006;132(1):9–18 doi 10.1007/s00432-005-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aaltonen KE, Rosendahl AH, Olsson H, Malmstrom P, Hartman L, Ferno M. Association between insulin-like growth factor-1 receptor (IGF1R) negativity and poor prognosis in a cohort of women with primary breast cancer. BMC cancer 2014;14:794 doi 10.1186/1471-2407-14-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yerushalmi R, Gelmon KA, Leung S, Gao D, Cheang M, Pollak M, et al. Insulin-like growth factor receptor (IGF-1R) in breast cancer subtypes. Breast cancer research and treatment 2012;132(1):131–42 doi 10.1007/s10549-011-1529-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.