Abstract
Despite gathering evidence that ubiquitylation can direct non-degradative outcomes, most investigations of ubiquitylation in T cells have focused on degradation. Here, we integrated proteomic and transcriptomic datasets from primary mouse CD4+ T cells to establish a framework for predicting degradative or non-degradative outcomes of ubiquitylation. Di-glycine remnant profiling was used to reveal ubiquitylated proteins, which in combination with whole-cell proteomic and transcriptomic data allowed prediction of protein degradation. Analysis of ubiquitylated proteins identified by di-glycine remnant profiling indicated that activation of CD4+ T cells led to an increase in non-degradative ubiquitylation. This correlated with an increase in non-proteasome-targeted K29, K33 and K63 polyubiquitin chains. This study revealed over 1,200 proteins that were ubiquitylated in primary mouse CD4+ T cells and highlighted the relevance of non-proteasomally targeted ubiquitin chains in T cell signaling.
Engagement of the T cell antigen receptor (TCR) and the costimulatory receptor CD28 initiates proliferation and effector T cell differentiation. Altering the abundance—via transcriptional, translational and post-translational events—of proteins involved in signal transduction, proliferation and differentiation is critical for activated T cell fate specification.
Quantitative proteomics indicates that over 1,000 proteins change in abundance following TCR stimulation, but correlation between RNA and protein abundance is limited1. This lack of correlation, also observed in other systems2,3, may reflect post-translational control of protein abundance. In T cells, post-translational modification with ubiquitin can regulate protein abundance or activity following TCR-CD28 engagement4–10. Ubiquitin is covalently attached to a lysine residue on a protein substrate in a reaction involving ubiquitin-activating enzymes, ubiquitin-conjugating enzymes and E3 ubiquitin ligases. The seven lysine residues of ubiquitin and its C-terminal methionine can then be ubiquitylated to form polyubiquitin chains with distinct downstream effects. Ubiquitylation is often associated with K48-linked chains, which direct the substrate toward proteasomal degradation. K11 chains also direct proteasomal degradation, while K63-linked chains can result in lysosomal degradation11. Ubiquitylation also promotes non-degradative fates: monoubiquitylation, multimonoubiquitylation and non-K48 ubiquitin chains can drive non-degradative outcomes11–13.
Many reports have detailed how ubiquitylation regulates the activation of T cells via ubiquitin-mediated protein degradation4–10. Examples of non-degradative ubiquitylation also exist, including ubiquitylation of the p85 subunit of PI3K, which impacts its recruitment to CD28 and TCRζ14, and K33 polyubiquitylation of TCRζ, which alters activation of the tyrosine kinase ZAP-70 (ref. 15). In innate immune cells, K63-linked ubiquitylation plays a critical role in activation of the transcription factor NF-κB16. Roles for free, mixed and linear ubiquitin chains have also been described17–19, further illustrating ubiquitin’s importance as a proteasome-independent regulator of T cells.
Here, we have used di-glycine remnant profiling20,21 to quantify changes in the ubiquitylation of over 1,200 proteins in primary mouse CD4+ T cells. We compared ubiquitin modification abundance with whole-cell proteomic and transcriptomic data, generating a predictive framework to analyze the relationship between ubiquitylation and protein abundance. Our results supported that proteins showing increased ubiquitylation following TCR stimulation were more likely to exhibit non-degradative ubiquitylation, which was associated with a global increase in K29, K33 and K63 chains.
Results
Proteasome inhibition by MG132 limits T cell activation
To interrogate ubiquitylation events in activated T cells in a global fashion, we designed an approach centered on di-glycine remnant proteomics. Di-glycine remnants are a result of tryptic cleavage within ubiquitin attached to a substrate lysine— C-terminal ‘GG’ ubiquitin residues remain covalently bound to the substrate lysine, generating the ubiquitin remnant (also known as K-ε-GG peptides, with ε representing the linkage between ubiquitin and the substrate). Antibody enrichment of di-glycine peptides20, coupled with mass spectrometry, identifies peptides modified by ubiquitin. To expand sufficient cells for this approach, which requires large amounts of protein, we used positive selection to isolate primary mouse CD4+ T cells from spleen and lymph nodes of C57BL6/J mice, activated them 72 h in vitro with CD3 + CD28 antibodies, then expanded the cells in IL-2 for 72 h (‘rested’) or restimulated the cells for 4 h following the IL-2 expansion with bead-bound CD3 + CD28 antibodies (‘restimulated’). Our goal was to couple analysis of protein ubiquitylation with an assessment of protein abundance to predict degradation of ubiquitylated proteins in activated CD4+ T cells.
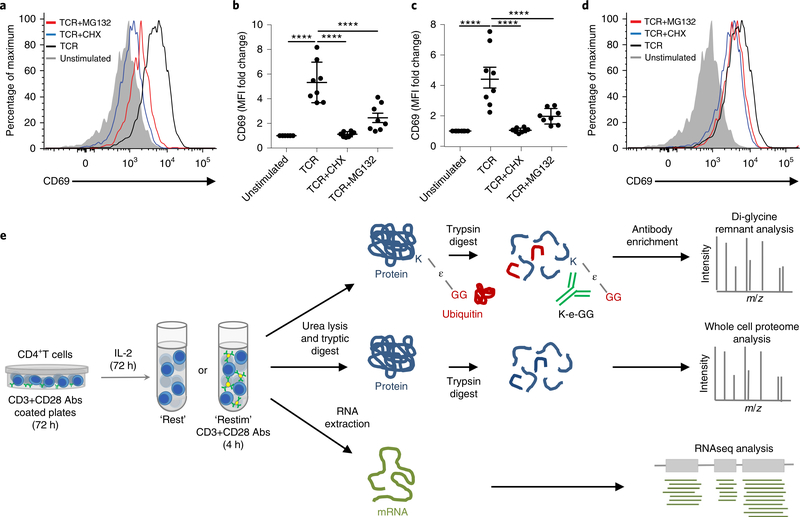
Before undertaking a proteomic approach, we tested whether proteasomal inhibition would aid detection of ubiquitylated proteins in activated CD4+ T cells. In previous di-glycine remnant proteomics analyses20,21, MG132 was found to increase the abundance of modified peptides22 and promote identification of proteins that are constitutively degraded or ubiquitylated at very low abundance23. However, studies using MG132 have reported aberrant cellular responses24. To test how proteasome inhibition impacted in vitro T cell activation, we expanded mouse CD4+ T cells, as described above, restimulated cells with CD3 + CD28 antibodies in the presence or absence of MG132 and assessed T cell activation by quantifying expression of the activation marker CD69 (ref. 25). CD69 upregulation was significantly reduced in CD4+ T cells stimulated in the presence of MG132 compared with CD3 + CD28 alone, exhibiting levels similar to those observed in CD4+ T cells stimulated in the presence of cycloheximide (CHX), an inhibitor of protein translation (Fig. 1a,b). Both extracellular (Fig. 1b) and intracellular CD69 were reduced (Fig. 1c). The reduction in CD69 was negligible when MG132 or CHX were added for the final 2 h of the 4-h restimulation (Fig. 1d), suggesting that its translation was largely completed after 2 h. Similarly, expression of activation-induced CD44 was reduced in CD4+ T cells stimulated in the presence of MG132 or CHX compared with CD3 + CD28 alone (Supplementary Fig. 1a). In contrast, the cell surface expression of CD3ε-γ, which is internalized during TCR engagement26, was reduced equally in the presence of MG132, CHX and CD3 + CD28 alone (Supplementary Fig. 1b). These data indicated that MG132 can block new protein production in CD4+ T cells, as observed in other cells27,28. Because normal protein expression is critical in T cell activation, we did not use proteasomal inhibitors for our subsequent analyses. To assess protein ubiquitylation and to predict ubiquitylation-mediated degradation, we opted to assess ubiquitylation in rested and restimulated CD4+ T cells using diglycine remnant profiling in combination with whole cell proteome (WCP) and RNA sequencing (RNAseq) transcriptome analyses (Fig. 1e).
Fig. 1 |. Proteasome inhibitor Mg132 prevents robust T cell activation.
a, Histogram of extracellular CD69 expression determined by flow cytometry on CD4+ T cells rested or restimulated for 4 h with CD3 + CD28 antibody-coated beads (1:1 cell/bead ratio, “TCR”). Stimulated cells were untreated or treated with MG132 (red) or CHX (blue) for the entire 4-h stimulation. A representative plot of distributions observed in three independent experiments is shown. b, Extracellular CD69 MFI fold change, as determined by flow cytometry, during CD3 + CD28 activation relative to rested CD4+ T cells, treated as described in a. c, Flow cytometry measurement of extracellular + intracellular CD69 MFI fold change during TCR activation relative to unstimulated CD4+ T cells, treated as described in a. b,c, Data are compiled from three independent experiments comprising eight mice; mean ± s.e.m. is shown. P values calculated by one-way analysis of variance with Holm–Sidak test for multiple comparisons; ****P < 0.0001. d, Flow cytometry measurement of CD69 surface expression on CD4+ T cells, treated as described in a. Stimulated cells were untreated or treated with MG132 (red) or CHX (blue) for the final 2 h of the 4-h stimulation. A representative plot of distributions observed in three independent experiments is shown. e, Schematic of integrated ‘omics’ workflow: primary mouse CD4+ T cells were isolated and stimulated with plate-bound CD3 + CD28 antibody for 72 h, expanded in IL-2 without stimulation for 72 h, then restimulated for 4 h with CD3 + CD28 antibody (3:1 cell/bead ratio). The ‘Rested’ and ‘Restimulated’ cells were then prepared for proteomic di-glycine remnant profiling, WCP or RNA-seq analysis.
WCP analysis identifies over 5,500 proteins in primary CD4+ T cells
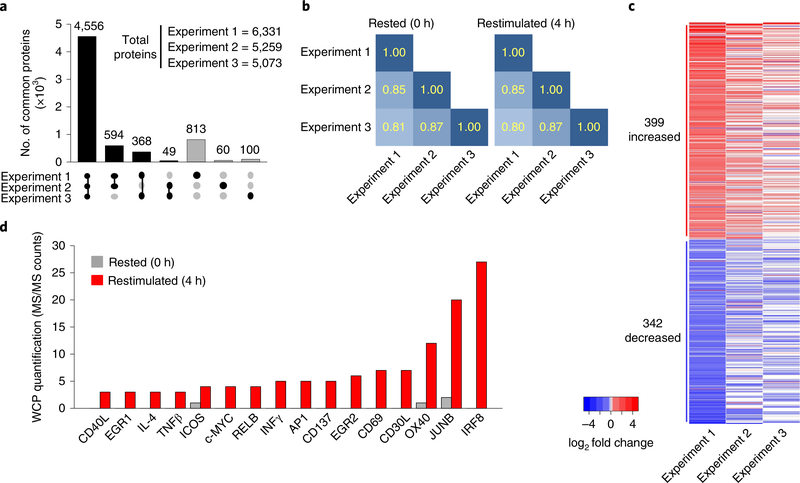
We performed WCP analysis to quantify and compare protein abundance in CD4+ T cells, expanded in culture as described above, ‘rested’ in IL-2 or ‘restimulated’ for 4 h with CD3 + CD28 antibodies. This experimental workflow (cell isolation through WCP analysis of rested and restimulated cells) was done three times (experiment 1, experiment 2, experiment 3), from three independent groups of C57BL6/J mice with two forms of protein quantification. CD69 upregulation, as a measure of T cell activation25, was observed in >65% of restimulated CD4+ T cells in all 3 experiments (Supplementary Fig. 1c). In rested and restimulated CD4+ T cells, we identified 5,567 proteins in at least two of the three experiments; 4,556 proteins (71% of total proteins) were identified in all of the three experiments (Fig. 2a and Supplementary Table 1). Correlation coefficients ranged between 0.80 and 0.87 for pair-wise comparisons of the log2-normalized protein abundances in rested and restimulated CD4+ T cells (Fig. 2b and Supplementary Fig. 2a). Thus, WCP analysis reproducibly identified proteins and quantified their relative abundances across independent experiments.
Fig. 2 |. Quantitative mass spectrometry reproducibly identifies over 5,500 proteins in the CD4+ T cell proteome during TCR stimulation.
a, Number of proteins that are quantified in each of 3, 2 or 1 WCP mass spectrometry experiments in CD4+ T cells rested or restimulated for 4 h with CD3 + CD28 antibody-coated beads (3:1 cell/bead ratio). Black bars indicate protein sets that are identified in two or three experiments and, therefore, are not filtered from the dataset. Gray bars indicate the sets that are identified in only one experiment and are filtered from further analysis. Figure was made in the style of the UpSetR package48. b, Correlation coefficients, based on protein abundance measured for three WCP mass spectrometry experiments (abundance quantified by log2 transformed, normalized intensities) in CD4+ T cells, as described in a. Correlation coefficients were calculated for all-by-all pair-wise comparisons of three experiments, using the Pearson’s method. c, Proteins differentially increased (red) or decreased (blue) in WCP abundance measured by mass spectrometry in CD4+ T cells unstimulated or stimulated, as described in a. Differential expression was defined for proteins that exhibit mean log2 fold change greater than ±1 s.d. from the mean of the distribution of average log2 fold changes. d, MS/MS counts from the label-free quantified WCP mass spectrometry experiment in CD4 + T cells unstimulated or stimulated, as described in a. MS/MS counts were used as a measure of protein abundance for proteins known to be induced on CD4+ T cell activation.
To quantify changes in protein expression during CD4+ T cell activation, we compared protein abundance in restimulated and rested CD4+ T cells using label-free (intensity-based absolute quantification (iBAQ)29) or isotope labeling quantification (stable isotope labeling with amino acids in cell culture (SILAC)). Three hundred ninety-nine proteins were increased, and 342 proteins were decreased, in restimulated compared with rested CD4+ T cells (Fig. 2c). Pathway analysis indicated enrichment of TCR and CD28 signaling pathways among activation-induced proteins in restimulated CD4+ T cells compared with rested cells (Supplementary Fig. 2b). Proteins induced on T cell activation—including CD44, JUNB and CD69—were measurably increased or exclusively identified in restimulated compared with rested CD4+ T cells (Fig. 2d). We observed a similar distribution of global protein abundance in CD4+ T cells restimulated with CD3 + CD28 for either 1 h or 4 h relative to rested cells (Supplementary Fig. 2c). We observed a less robust increase in proteins associated with T cell activation in CD4+ T cells restimulated for 1 h compared with 4-h restimulation (Supplementary Fig. 2d). Together, this indicated that WCP analysis in CD4+ T cells restimulated for 4 h reproducibly captures stimulation-induced changes in protein abundance.
Di-glycine profiling identifies ~1,200 proteins in primary CD4+ T cells
We next used di-glycine remnant profiling20–22,30 to identify ubiquitylation events in activated CD4+ T cells. We used SILAC labeling to uniquely label rested versus restimulated CD4+ T cells, prepared as described above. This allowed stringent quantification and simultaneous enrichment of di-glycine remnant peptides from equivalent protein amounts of rested and restimulated CD4+ T cell lysates. We performed two independent enrichment experiments (experiment 1 and experiment 2) from cell isolation through di-glycine remnant analysis, using independent groups of C57BL6/J mice.
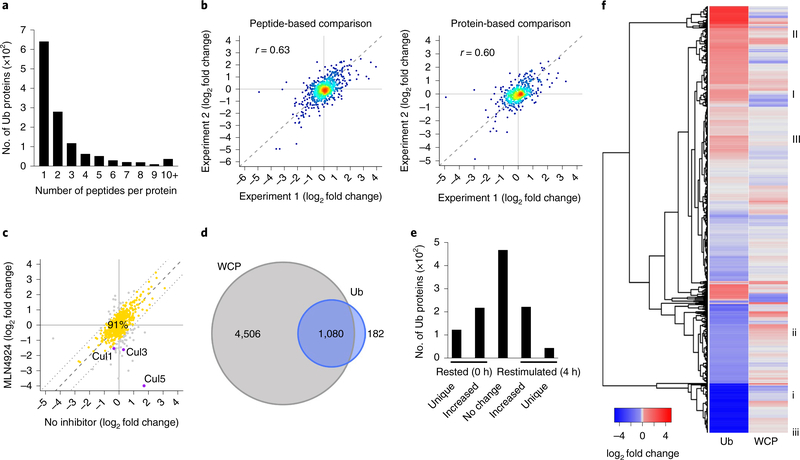
Di-glycine remnant profiling of rested and restimulated CD4+ T cells identified 3,075 unique di-glycine peptides, mapping to 1,262 unique proteins across the two independent experiments (Supplementary Fig. 3a and Supplementary Tables 1 and 2). We observed a single ubiquitylation site (unique di-glycine remnant peptide) for 53% of proteins; multiple unique di-glycine remnant peptides were identified for 47% of proteins (Fig. 3a). SILAC labeling enabled comparison of ubiquitylation site abundance in rested compared with restimulated CD4+ T cells with no additional computation. If only a single unique di-glycine remnant peptide was quantified, this single ratio represented the modified protein in rested compared with restimulated CD4+ T cells. In the case of multiple, unique di-glycine remnant peptides per protein, we calculated a ‘protein-level’ di-glycine remnant ratio, defined as the abundance-weighted average of the SILAC ratio for each unique peptide in stimulated relative to rested CD4+ T cells (see Methods). We evaluated the reproducibility of di-glycine quantification by comparing the SILAC ratios obtained in the two experiments at both the peptide and protein level (Fig. 3b). The Pearson correlation coefficient for comparison of the peptide-based fold changes (r = 0.63) and protein-based fold changes (r = 0.60) in restimulated relative to rested CD4+ T cells indicated a significant correlation between the two experiments (P < 2.2 × 10−16). Thus, numerous unique ubiquitylation sites for many proteins can be reproducibly identified and quantified in rested or restimulated CD4+ T cells.
Fig. 3 |. Di-glycine remnant enrichment reveals that ubiquitylation is associated with increases or decreases in total protein abundance during TCR stimulation.
a, Distribution of the number of di-glycine remnant peptides identified per protein in di-glycine remnant mass spectrometry experiments in CD4+ T cells rested or restimulated for 4 h with CD3 + CD28 antibody-coated beads (3:1 cell/bead ratio). Peptides identified from two di-glycine mass spectrometry experiments were combined to generate the distribution. b, Di-glycine abundance log2 fold changes for di-glycine peptides (left) and proteins (right) measured from two di-glycine mass spectrometry experiments in CD4+ T cells unstimulated or stimulated, as described in a. Correlation coefficients were calculated using the Pearson’s method. c, Comparison of log2 fold changes in di-glycine abundance measured from di-glycine mass spectrometry experiments in CD4+ T cells stimulated, as described in a, with or without MLN4924 neddylation inhibitor added for the final 2 h of stimulation. Proteins that exhibited <1 log2 fold change difference in untreated and MLN4924-treated cells were considered unaffected by the neddylation inhibitor (yellow). d, Intersection of proteins quantified in the WCP proteome and the di-glycine enrichment ubiquitin proteome. e, Numbers of proteins that exhibited changes di-glycine abundance in di-glycine mass spectrometry experiments in CD4+ T cells unstimulated or stimulated, as described in a. Unique proteins at rest or restimulation were identified by only a heavy or light SILAC intensity, respectively. Differentially ubiquitylated proteins were identified by a ±25% normalized log2 fold change in abundance. f, Normalized di-glycine and WCP abundance log2 fold changes for proteins identified in both the WCP proteome and di-glycine enrichment ubiquitin proteome. Global patterns of increased or decreased ubiquitylation associated with decreased, increased or constant WCP expression are indicated (I-III and i-iii, respectively). Ub, ubiquitylated (a,e) or di-glycine remnant proteome (d,f).
Di-glycine remnant peptides also result from post-translational modification with the ubiquitin-like proteins NEDD8 and ISG15 (ref. 20). To assess the contribution of neddylated or ISGylated peptides in the di-glycine remnant proteomics, we analyzed the relative abundance of these ubiquitin-like proteins in the WCP. The relative abundances of ISG15, NEDD8 and ubiquitin, quantified by their statistical z-scores, were −0.645, 0.681 and 2.659, respectively, in rested CD4+ T cells. We also observed several di-glycine remnant peptides of NEDD8 (but not ISG15), indicative of polyneddylation, NEDD8 ubiquitylation or heterotypic ubiquitin-NEDD8 chains18,31. Because cullin E3 ligases are activated by NEDD832, we assessed the contribution of protein neddylation to the di-glycine remnant proteome by adding the neddylation inhibitor MLN492433 during the final 2 h of the CD4+ T cell restimulation. Rested CD4+ T cells were not treated with MLN4924. Immunoblotting indicated that MLN4924 treatment reduced neddylation of the NEDD8 target cullin1 (Cul1) compared with no inhibitor control (Supplementary Fig. 3b), indicating that MLN4924 impaired neddylation in restimulated CD4+ T cells. Addition of MLN4924 for the final 2 h of the 4-h restimulation did not impact CD69 expression on restimulated CD4+ T cells (Supplementary Fig. 3c). In the di-glycine remnant proteome, addition of MLN4924 to restimulated CD4+ T cells had no effect on 91% of di-glycine remnant peptide SILAC ratios comparing restimulated with rested CD4+ T cells (Fig. 3c). Several cullin ligases (Cul1, Cul3, Cul5) showed decreased di-glycine remnant peptide abundance in restimulated CD4+ T cells treated with MLN4924 (Fig. 3c), indicating that the assay was sufficient to detect loss of NEDD8 di-glycine remnant peptides for known NEDD8 targets. Together, these data support that di-glycine remnant profiling in CD4+ T cells reproducibly and quantitatively identified ubquitylation events following T cell activation.
We next examined the relationship between di-glycine remnant and protein abundances. One thousand eighty proteins were quantified in both the WCP and di-glycine remnant proteomics datasets (Fig. 3d). We normalized the fold change in protein-level di-glycine remnant abundance (SILAC ratio, or weighted average SILAC ratio) in restimulated versus resting CD4+ T cells to the fold change in protein abundance determined by WCP analysis. Utilizing a normalized di-glycine remnant fold change threshold of ±25%, we found that ubiquitylation was increased in 221 proteins following CD3 + CD28 antibody restimulation, while 43 proteins were ubiquitylated only in restimulated CD4+ T cells (Fig. 3e). Ubiquitylation was decreased in 217 proteins, and 121 proteins were ubiquitylated only in resting CD4+ T cells (Fig. 3e). Inversely correlated abundances in WCP (decreased) and di-glycine remnant (increased), as observed for CD3ε, ZAP-70 and LAT, might indicate degradative ubiquitylation. In contrast, in cases in which protein expression drives ubiquitylation changes, both protein abundance and ubiquitylation would change in the same direction (for example CBL-B, LCK). Overall, increased or decreased ubiquitylation did not show strong inverse correlation with total protein abundance (Fig. 3f), leading us to conclude that changes in TCR-induced ubiquitylation were often not associated with degradation. Thus, di-glycine remnant analysis revealed numerous ubiquitylation sites in mouse CD4+ T cell proteins, of which almost half were altered in abundance in restimulated compared with rested CD4+ T cells.
TCR-induced transcriptional changes correlate poorly with protein abundance
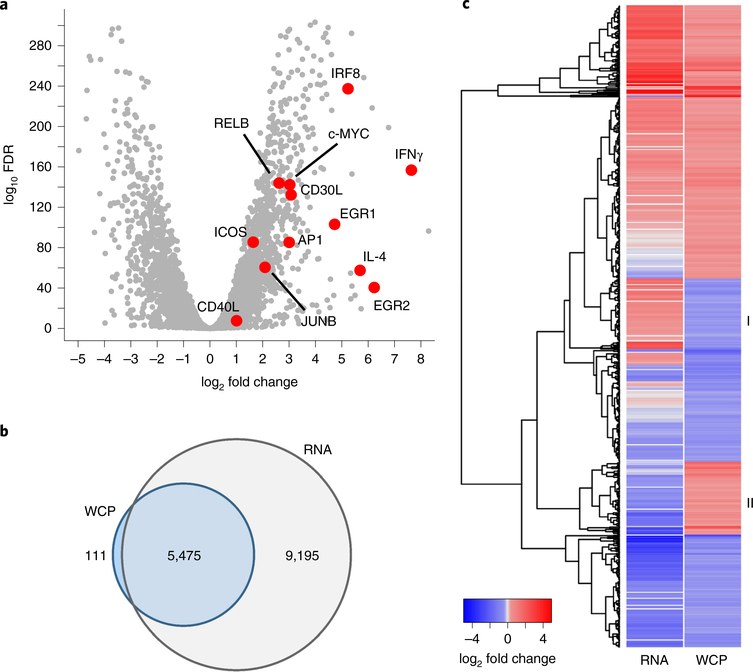
Protein abundance reflects the outcome of protein production and protein degradation. To predict whether decreased protein abundance resulted from degradation or decreased transcription, we performed transcriptomic analysis in rested and restimulated CD4+ T cells, cultured as in our proteomic analyses. RNA transcripts of proteins associated with T cell activation, including Icos and Junb, were increased in restimulated compared with rested CD4+ T cells (Fig. 4a). The RNA-seq data covered 5,475 (98%) of the proteins quantified in the WCP dataset (Fig. 4b and Supplementary Table 1). As observed previously1, we found limited correlation between transcript and associated protein abundance in restimulated and rested CD4+ T cells (Pearson correlation coefficient = 0.25; Supplementary Fig. 4a). The correlation was higher when transcript and protein were compared only for proteins that were significantly changed in the WCP analysis of restimulated and rested CD4+ T cells (Pearson correlation coefficient = 0.45; Supplementary Fig. 4a). The correlation improved further if transcript and protein relative abundances, rather than fold changes, were compared for all proteins (Pearson correlation coefficient = 0.50) or ubiquitylated proteins only (Pearson correlation coefficient = 0.49; Supplementary Fig. 4b). However, approximately one-third of proteins changed in the opposite direction of their transcript (for example, SOS1, RHOA) (Fig. 4c). Thus, comparing the transcriptome and WCP of restimulated versus rested CD4+ T cells suggested post-transcriptional mechanisms drive discordance between transcript and protein abundance.
Fig. 4 |. RNA-seq data reveal disparate changes in RNA and corresponding protein abundances during TCR stimulation.
a, Log2 fold change in RNA transcript abundance and corresponding statistical significance identified in RNA-seq experiments in CD4+ T cells unstimulated or stimulated for 4 h with CD3 + CD28 antibody-coated beads (3:1 cell/bead ratio). Differential expression and adjusted P value were calculated by the DESeq2 algorithm49. Proteins known to be induced on CD4+ T cell activation (red) exhibit increased transcription during restimulation (included in Fig. 2d). Transcript expression for CD69, LTA (TNFβ), TNFRSF9 (CD137) and TNFRSF4 (OX40) (included in Fig. 2d) is identified only during restimulation. b, Intersection of proteins quantified in the WCP proteome and which contain associated RNA-seq transcript data. c, RNA transcript abundance log2 fold change and WCP abundance log2 fold change for proteins that are differentially increased or decreased in the WCP. WCP differential expression was defined for proteins that exhibit mean log2 fold change greater than ±1 s.d. from the mean of the distribution of average log2 fold changes. Disparate changes in RNA transcript and corresponding WCP protein expression during CD4+ T cell stimulation are indicated (I, II). FDR, false discovery rate (DEseq2 adjusted P value).
T cell activation drives degradative and non-degradative ubiquitylation
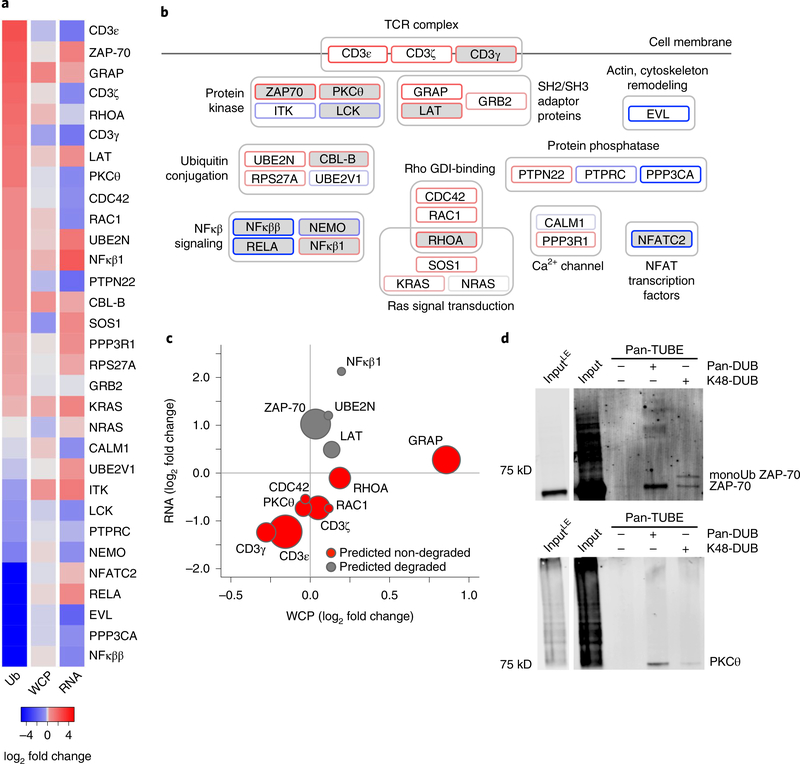
We next integrated the RNA-seq, WCP and ubiquitin remnant datasets to specifically interrogate patterns for proteins in the TCR signaling pathway. A set of 155 TCR signaling proteins was defined from the union of pathway annotations from the Kyoto Encyclopedia of Genes and Genomes34 (103 proteins), Reactome35 (107 proteins), MetaCore (portal.genego.com; 78 proteins) and Ingenuity Pathway Analysis (https://www.qiagenbioinformatics.com; 31 proteins) databases. Thirty-one proteins within the TCR pathway were ubiquitylated in rested and/or restimulated CD4+ T cells (Fig. 5a,b). Among these, 12 proteins (CD3ε, GRAP, ZAP-70, the CD3ζ precursor CD247, RHOA, CD3γ, PKCθ, LAT, CDC42, RAC1, UBE2N, NFKB1) showed increased ubiquitylation, defined as greater than 25% normalized log2 fold increase, in restimulated compared with rested CD4+ T cells (Fig. 5a,b). Eight proteins (CBL-B, PTPN22, PPP3R1, RPS27A, KRAS, GRB2, SOS1, NRAS) showed minor increases in ubiquitylation in restimulated compared with rested CD4+ T cells (normalized log2 fold change between 0% and 25%; Fig. 5a,b). Several of these proteins are known ubiquitylation targets in T cells10,15,36–42 (Fig. 5b).
Fig. 5 |. Ubiquitin targets multiple proteins within the TCR pathway and elicits both degradative and non-degradative outcomes.
a, Ubiquitin-normalized log2 fold changes, WCP protein log2 fold changes and RNA transcript log2 fold changes for TCR pathway proteins identified in di-glycine, WCP and RNA-seq experiments for CD4+ T cells rested or restimulated for 4 h with CD3 + CD28 antibody-coated beads (3:1 cell/bead ratio). b, Ubiquitylated TCR pathway proteins grouped by function within the TCR pathway. Outlines are colored by ubiquitin-normalized log2 fold change, as in a. Gray fill indicates that ubiquitylation of the protein has been previously described in T cells. c, Ubiquitylation, WCP and RNA-seq expression changes for TCR proteins exhibiting TCR-induced ubiquitylation (>25% increase) in CD4+ T cells rested or restimulated, as described in a. Circle size corresponds to increase in ubiquitylation-normalized log2 fold change. Proteins exhibiting consistent protein abundance and increased RNA are predicted to be degraded by ubiquitin (gray) while the remaining proteins are predicted to undergo non-degradative outcomes of ubiquitylation (red). d, pan-TUBE enrichment experiments with pan-DUB or K48-specific DUB added postenrichment shows ubiquitylation of ZAP-70 (top) and PKCθ (bottom) and effects of DUB activity on these substrates in CD4+ T cells restimulated for 4 h with CD3 + CD28 plate-bound antibody. The pan-DUB lane shows the endogenous ZAP-70 and PKCθ. The K48-specific DUB lane shows a mono-ubiquitylated band for ZAP-70 and almost no signal for PKCθ, suggesting that PKCθ is ubiquitylated by polyubiquitin chains other than K48-linkages. Input is ~1% of enrichment. The ‘input LE’ indicates a low exposure image of the input lane. Immunoblot images are representative examples from three independent experiments.
Among the 12 proteins in the TCR pathway that exhibited increased ubiquitylation in restimulated compared with rested CD4+ T cells, comparison of di-glycine profiling, WCP and transcript abundances predicted degradative ubiquitylation of NFKB1, UBE2N, ZAP-70 and LAT (Fig. 5c). In contrast, CD3ε, CD3γ, CD3ζ, PKCθ, CDC42, RAC1, RHOA and GRAP exhibited similar changes in the expression of RNA and the corresponding protein (Fig. 5c), suggesting that their ubiquitylation did not result in protein degradation. To test these predictions, we analyzed the half-life for proteins that we predicted to be ubiquitylated and degraded (SIN3B, MYCBP2, SNC18, ZAP-70 and LAT) or not degraded (PKCθ and GRAP). Expanded CD4+ T cells were restimulated for 4 h, with or without addition of CHX after 1 h, to determine protein loss in the absence of new translation. Immunoblot analysis confirmed the predictions for LAT, SIN3B and MYCBP2 (degraded), as well as GRAP and PKCθ (not degraded) (Supplementary Figs. 5 and 6). Both ZAP-70 and SNX18 were predicted to be degraded; however, there was no evidence of protein loss on addition of CHX (Supplementary Figs. 5 and 6).
To further test the effect of ubiquitylation on ZAP-70, we used ubiquitin chain enrichment and treatment with deubiquitylating enzyme (DUB) to reveal types of chain modifications. We used a pan-TUBE (tandem-ubiquitin binding entity, binds all ubiquitin chain linkages) to enrich for ubiquitylated proteins from restimulated CD4+ T cells and then treated with no DUB, a pan-DUB (cleaves all polyubiquitin chains) or a K48 chain-specific DUB (K48-DUB). Degraded proteins should be deubiquitylated by both the pan-DUB and the K48-DUB, while non-degraded substrates should be deubiquitylated by only the pan-DUB. Analysis of ZAP-70 by immunoblotting revealed that pan-TUBE enrichment with pan-DUB treatment generated a band corresponding to the molecular weight of the unmodified ZAP-70 (Fig. 5d and Supplementary Fig. 6). K48-DUB treatment revealed a band consistent with the molecular weight of mono-ubiquitylated ZAP-70 (Fig. 5d and Supplementary Fig. 6), consistent with K48 chain degradation up to the substrate proximal ubiquitin, which is a lysine-methionine linkage. Thus, ZAP-70 was, to some extent, K48-polyubiquitylated in restimulated CD4+ T cells.
The proteomic and transcriptomic data-driven prediction indicated that PCKθ was ubiquitylated but not degraded, and the CHX pulse chase showed that PKCθ expression is stable during TCR stimulation. Therefore, we implemented the TUBE-DUB method to examined ubiquitin chain modifications on PKCθ in restimulated CD4+ T cells. We detected a prominent band corresponding to the molecular weight of unmodified PKCθ in restimulated CD4+ T cells subjected to pan-TUBE enrichment and pan-DUB treatment (Fig. 5d and Supplementary Fig. 6). We did not observe mono-ubiquitylated PKCθ after K48-DUB treatment (Fig. 5d and Supplementary Fig. 6), confirming our previous prediction. These observations validated the TUBE-DUB method to examine substrate-specific ubiquitylation and ubiquitin chain linkages. Thus, our data-driven predictions were confirmed by protein-specific half-life analysis in combination with the more sensitive analysis of ubiquitin chain specificity using DUBs.
Non-proteasomally targeted ubiquitin chains increase in T cell activation
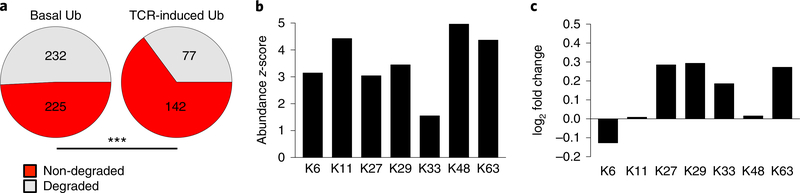
We next asked whether there was a difference in total ubiquitylation events, across all proteins quantified, in restimulated CD4+ T cells relative to rested CD4+ T cells. Protein ubiquitylation that was unchanged following CD4+ T restimulation, or ‘basal ubiquitylation,’ was defined as normalized di-glycine remnant abundance that changed less than 25% in restimulated CD4+ T cells relative to rested CD4+ T cells (457 proteins) (Fig. 3e). ‘TCR-induced ubiquitylation’ was defined as ubiquitylation that showed an increase of at least 25% in restimulated CD4+ T cells relative to rested CD4+ T cells (219 proteins) (Fig. 3e). Following the predictive framework used to analyze the TCR signaling pathway, we found that basal ubiquitylation events had a predicted degradative outcome for approximately 51% of the ubiquitylated proteins (Fig. 6a), similar to ratios reported previously22,30. TCR-induced ubiquitylation showed predicted non-degradative outcomes for approximately 65% of the ubiquitylated proteins (Fig. 6a), a significantly higher proportion than that predicted for basal ubiquitylation (P < 0.006). The relative abundance of ubiquitin peptides in the diglycine remnant dataset, quantified by their statistical z-score as a proxy for the abundance of different ubiquitin chains, indicated that all of the ubiquitin lysince were relatively abundant in both rested and restimulated CD4+ T cells (Fig. 6b). When assessing the average change in relative abundance of the ubiquitin lysine di-glycine peptides in restimulated CD4+ T cells, we found that K29, K33 and K63 peptides were reproducibly increased relative to rested CD4+ T cells (Fig. 6c). The abundances of K48 and K11 ubiquitin remnant peptides, associated with proteasomal degradation, were unchanged in restimulated CD4+ T cells compared with rested CD4+ T cells (Fig. 6c). Similar changes—including increased K29 and K33 peptides and limited changes in the abundances of K11 or K48 peptides—were observed in 1-h restimulated CD4+ T cells compared with rested CD4+ T cells (Supplementary Fig. 7), suggesting that earlier TCR signaling events also generated increased non-degradative events. Together, this analysis suggested an increase of non-degradative ubiqiquitylation in activated CD4+ T cells associated with an increase in non-proteasomally targeted ubiquitin chains.
Fig. 6 |. Integrating ubiquitin proteomics, WCP and RNA-seq data reveals a prevalence of non-degradative ubiquitylation.
a, Numbers of predicted degradative (gray) and non-degradative (red) ubiquitylation events for all proteins exhibiting basal (left) or TCR-induced (right) ubiquitylation in CD4+ T cells rested or restimulated for 4 h with CD3 + CD28 antibody-coated beads (3:1 cell/bead ratio). TCR-induced ubiquitylation produces a significantly higher proportion of non-degradative ubiquitylation events compared with basal ubiquitylation (P < 0.006) according to a two-tailed two-proportion test. b, Relative abundance of ubiquitin lysine peptides identified in the ubiquitin proteome of resting CD4+ T cells. Relative abundance is represented by the z-score of the peptide abundance quantified by mass spectrometry intensity values. c, The log2 fold change in abundance of ubiquitin lysine peptides identified in the ubiquitin proteome during TCR stimulation. b,c, Relative abundance and log2 fold changes are averaged for two di-glycine immunoprecipitation mass spectrometry experiments, as described in a.
Discussion
We integrated di-glycine remnant profiling, WCP and transcriptome analysis to identify TCR stimulation-dependent ubiquitylation events in rested and/or restimulated primary mouse CD4+ T cells. This revealed over 3,000 ubiquitylated peptides, representing approximately 1,200 substrates of ubiquitylation in CD4+ T cells. Integration of the proteomics and transcriptomics datasets provided a framework for predicting the fate of ubiquitylated substrates. We validated our predictions with a combination of pulse-chase experiments and a TUBE-DUB approach which allowed substrate-specific detection of ubiquitylation patterns, and revealed a complexity that cannot be discerned using di-glycine remnant profiling alone. Finally, we found an increase in non-degradative ubiquitylation in restimulated CD4+ T cells, coincident with an increase in K29, K33 and K63 ubiquitin remnant peptides.
The most well characterized outcome of ubiquitylation is proteasomal degradation. Studies characterizing global ubiquitylation have estimated that about 50% of ubiquitylated peptides increase abundance in response to the proteasomal inhibitor MG132 (refs. 22,30), suggesting that these proteins undergo proteasomal degradation. However, MG132 treatment affects proteins in pathways related to cell cycle, transcription and cell stress, often with negative impacts on protein translation43,44. Similarly, we found that proteasomal inhibition adversely affected T cell activation. Therefore, we used di-glycine remnant proteomics combined with WCP analysis and RNA-seq to assess the impact of ubiquitylation on protein abundance and predict degradative outcomes. Because we did not use proteasomal inhibition, our dataset represents physiologically relevant ubiquitylation events. However, proteasome inhibition increases the number and diversity of ubiquitylated peptides that can be identified22,23,30, and, thus, our di-glycine remnant dataset probably represents the most abundantly modified peptides. Furthermore, use of SILAC quantification in the di-glycine remnant proteome for stringent quantification restricted our analysis to ubiquitin remnant peptides (and proteins) that were identified in both rested and restimulated CD4+ T cells. Thus, label-free di-glycine analysis may yield additional, dynamically regulated ubiquitylation events. Ubiquitylation events are likely to occur on subpopulations of the protein substrate. The approach we present here is unlikely to detect changes in ubiquitylation if a small subset of the protein is modified, though these modifications may be of great functional impact. Alternative approaches would be needed to capture changes in subpopulation ubiquitylation. These data could be further expanded through use of reagents to identify linear polyubiquitin chains, which are known to have roles in T cell development and homeostasis45–47 but cannot be detected by di-glycine remnant proteomics.
The prevalence of predicted non-degradative ubiquitylation in restimulated CD4+ T cells is higher than that reported in other di-glycine studies of ubiquitylation, which were performed without analogous physiological stimulation22,30. We analyzed cells after 4 h of TCR stimulation—after proximal phosphorylation events have occurred, but preceding cytokine signaling and cell cycle entry. Whether the observed shift away from proteasomally targeted ubiquitin chains is unique to early T cell activation remains to be seen. A time course of CD4+ T cell activation would be beneficial. This approach could also be applied to other T cell signaling cascades, such as cytokine receptor signaling.
These data are a valuable resource that can be used to probe for novel TCR-dependent ubiquitylation events or to study the mechanistic effect of ubiquitylation during T cell activation. Together, these datasets shed light on the dynamic ubiquitylation landscape of activated CD4+ T cells and provide a valuable stepping stone for further research into the ubiquitylation processes that occur during T cell activation.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41590-019-0381-6.
Methods
Mice
C57BL6/J mice were maintained in house, in a barrier facility at the Children’s Hospital of Philadelphia. Experiments were approved by the Institutional Care and Use Committee of the Children’s Hospital of Philadelphia. All ethical regulations were followed throughout the course of this study. Mice of both sexes were used at 7–9 weeks of age.
T cell isolation and culture
Spleen and total peripheral lymph nodes were harvested and mashed through 70-μM nylon filters using cold HBSS, subjected to red blood cell lysis using ACK lysis buffer, then washed, filtered again and resuspended in cold HBSS. CD4+ T cells were isolated by positive selection using the Miltenyi Biotech mouse CD4+ T cell isolation kit. For proteomic experiments, total CD4+ T cells were pooled from approximately 20 mice. For RNA-seq, one mouse per sample was used. For immunoblot, one mouse per replicate was used. Cells were resuspended at 1.75 × 106 cells ml–1 before stimulation with 5 μg ml−1 plate-bound αCD3 (145–2C11; BioLegend) and αCD28 (37.51; BioLegend) for 3 d. Cells were expanded in recombinant human IL-2, at 50 U ml−1 (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health) for 3 d. Cells were then collected as ‘resting’ cells or ‘restimulated’ with αCD3/αCD28 beads (Invitrogen) for 4 h. Restimulation for transcriptome, di-glycine proteome and WCP occurred at a 3:1 cell/bead ratio due to large culture sizes required for di-glycine enrichment proteomics. Other experiments utilized the manufacturer-recommended 1:1 cell/bead ratio (MG132 flow cytometry-based experiments) or utilized plate-bound antibody (immunoblots, TUBE-DUB) for restimulation. Cells were then prepared for RNA analysis, whole-proteome analysis or K-ε-GG immunoprecipitation.
Purity was assessed by flow cytometry after isolation, after plate-bound activation and after restimulation as noted below. Cells were greater than 90% CD4+CD3+. For proteomics data, whether or not SILAC quantification was used, all cells were cultured in SILAC media: SILAC RPMI (Hyclone), 12C or 13C l-arginine:HCl and l-lysine:2HCl (Cambridge Isotope Laboratory), 1% MEM NEAA (Invitrogen), 1% sodium pyruvate (Invitrogen), 1% Glutamax (Invitrogen), 1% pen/strep (Invitrogen), 10% Dialyzed FCS (Life Technologies), 2% HEPES (Fisher Scientific), 0.12 mM betamercaptoethanol (Sigma). All cultured cells were cultured at 37 °C, 10% CO2.
TUBE enrichment experiments
CD4+ T cells were isolated and cultured as described above. Cells were restimulated by seeding on 100 × 20 mm2 tissue culture plates precoated for 24 h with αCD3 and αCD28 (5 μg ml−1). Cells were seeded at a concentration of approximately 5 × 106 cells ml–1 in 10 ml media per dish and incubated at 37 °C for 4 h. Cells were collected, washed three times in cold PBS and pelleted. Cells were lysed in a Pierce RIPA (ThermoFisher)-based lysis buffer (RIPA, 50 mM Tris-HCl, 0.15 M NaCl, 1 mM EDTA, 1% NP40, 10% glycerol, Roche complete EDTA-free protease inhibitor and 4× deubiquitylase inhibitors PR619 and OPA), vortexed, incubated for 30 min on ice and centrifuged for 20 min at maximum speed. Bradford assay was used to measure protein concentration of the resulting lysate. Pan-TUBE enrichment was performed using the Lifesensors UbiTest Agarose-TUBE Elution Kit, with modifications to the supplied protocol, as described here. Approximately 3–4 mg total protein was used as input for the pan-TUBE enrichment (approximately 100–120 × 106 cells). Approximately 150 μl agarose slurry was used for enrichment experiment. The input lysate was diluted 1:10 in PBS with additional deubiquitylase and protease inhibitors added at the appropriate volume. The pan-TUBE agarose was added to the diluted lysate and incubated at 4 °C, with rotation, for approximately 12 h. The agarose was washed in 200–400 μl of supplied wash buffer. The captured proteins were eluted in 150 μl of supplied elution buffer. Deubiquitylase inhibitors were added, at appropriate volumes, to the elution. All centrifugation was performed at 1,000g for 5 min with the exception of the postelution centrifugation, which was performed at 4,500g for 5 min. Following elution, the supernatant was collected and divided into 3 parts of equal volume; the first sample received no DUB, the second sample received the supplied non-specific pan-DUB (USP2, supplied with kit) and the third sample received a K48-specific DUB (Otub1, Lifesensors). The samples were incubated at 30 °C for 1.5 h. The reaction was terminated by the addition of SDS sample buffer (Genetex Trident 6X Laemmli SDS sample buffer, 15% 2-mercaptoethanol) and boiled at 100 °C for 5 min. Ubiquitin enrichment and impact of DUB addition was assessed by immunoblot using standard protocol as described below.
CHX and MLN4924 treatments
Cells for CHX time course and MLN4924 pilot experiments were collected in cold PBS, spun at 1,300 r.p.m. for 5 min and resuspended in complete RPMI (cRPMI) at a concentration of 3 × 106 cells ml–1. Cells were seeded on 24-well plates precoated for 24 h with either PBS (for ‘rested’ cells) or αCD3 and αCD28 (for ‘restimulated’ cells) at a concentration of 3 × 106 cells per well in 1 ml media. Plates were centrifuged for at 1,300 r.p.m. for 5 min to promote cell contact with the plate-bound antibody. cRPMI medium (1 ml) supplemented with CHX (Sigma C4859) was added, at times noted, to obtain a total concentration of 1/10,000. cRPMI medium (1 ml) without CHX was added to wells for which the cells were not exposed to translational inhibition. Likewise, cells were treated with MLN4924 (Calbiochem CAS 951950–33-7) at varying concentrations during restimulation (Supplementary Fig. 3), or with 1 μM for 2 h of restimulation for proteomics analysis of neddylation.
Rested or restimulated cells were collected at indicated time points, and washed/resuspended three times in cold PBS. Cell pellets were lysed in 1% NP40 lysis buffer (1% NP40, 150 mM NaCl, 50 mM Tris-HCl, 1 mM sodium orthovanadate, 5 mM sodium fluoride, Roche complete EDTA-free protease inhibitor, MilliQ H20) and 1× sample buffer (NuPAGE LDS Sample Buffer), on ice. Cells were vortexed and incubated on ice for 20 min. Lysed cells were microcentrifuged for 10 min at maximum speed at 4 °C. Supernatant was transferred to a clean microcentrifuge tube. Lysate was boiled for 5 min in tubes at 95 °C before use.
Immunoblotting
Lysates were loaded by cell number equivalents on precast 4–12% Bis-Tris gels (NuPAGE) or 7% Tris-Acetate gel (NuPAGE) (only for Mycbp2 immunoblot). Lysates were subjected to SDS–PAGE using either SDS MOPS running buffer (NuPAGE) for the Bis-Tris gel or Tris-Acetate SDS running buffer (NuPAGE) for the Tris-Acetate gels. Protein was transferred to PVDF (Millipore) using a semi-dry transfer apparatus (100 mA for 1–3 h) or wet cell apparatus (6V-30V, 1 h to overnight). Membrane was blocked in Odyssey Blocking Buffer (LICOR). Protein was visualized on an Odyssey imager (LICOR) using AF680 goat αrabbit (Invitrogen), IRdye 800 donkey αmouse (LICOR) and IRdye 800 donkey αgoat (LICOR) secondary antibodies. Bands were quantified using FIJI software50. Immunoblot quantifications are based on three biological replicates and images are a selected representative replicate.
Flow cytometry and immunoblot antibodies
The following flow cytometry antibodies were purchased from BioLegend: CD4 (GK1.5), CD8α (53–6.7), CD44 (IM7), CD3ε-γ (17A2). Antibody to CD69 (H1.2F3) was purchased from BD Biosciences. For analysis of cultured cells by flow cytometry, cells were collected from culture plates with gentle pipetting, washed in serum-free HBSS or PBS, stained with live/dead fixable blue dead cell stain (L-23105; Invitrogen), Fc blocked (αCD16/32,2.4G2; BD Biosciences) and stained with the appropriate antibody mixtures. After staining for 25 min at 4 °C, samples were fixed using the FoxP3 fix/perm kit (eBioscience) or run fresh. Intracellular staining was done for 1 h at 4 °C. Samples were acquired on an LSRFortessa (BD Biosciences), and analyzed using FlowJo version 9.8 (FlowJo LLC). Events analyzed were live singlets (forward scatter-area by forward scatter-height (FSC-A x FSC-H), viability dye negative). Total live cells (FSC-A x SSC-A (side scatter-area)) were analyzed, with no lymphocyte pregate, for accurate measure of purity. The following antibodies were used for immunoblotting: SIN3B (Novus Biologicals, NBP2–20367, dilution 1:250), MYCBP2 (Abcam, ab86078, dilution 1:800), SNX18 (ThermoFisher, PA5–31460, dilution 1:1,000), GRAP (Abcam, ab178023, dilution 1:500), PKCθ (Abcam, ab185974, dilution 1:1,000), LAT (Origene, TA327925, 1:800), ZAP-70 (Origene TA332508, 1:1250). Antibodies were diluted in Odyssey Blocking Buffer.
Experimental design for proteomic and transcriptomic analysis of primary T cells during TCR stimulation
In our initial efforts, we assessed resting, 1-h restimulation and 4-h restimulation in a single experiment using a ‘super SILAC’ approach. In this experimental set up, a calibrator sample was prepared using CD4+ T cells cultured in heavy carbon SILAC media. After cell collection and lysis (described below), the calibrator sample was mixed at a 1:1:1 ratio of resting, 1-h stimulated and 4-h stimulated CD4+ T cells. This heavy carbon sample was mixed 1:1 with light carbon SILAC-labeled resting 1-h stimulated and 4-h stimulated CD4+ T cells. These mixed lysates were subjected to di-glycine remnant proteomic analysis. Relative di-glycine remnant abundance is represented as a ratio of ratios, with each sample normalized to the calibrator sample. For label-free quantification WCP analysis of these samples, cells were collected separately at rest or after 4-h TCR restimulation and liquid chromatography with tandem mass spectrometry (LC–MS/MS) was performed on each sample. To quantify the changes in WCP protein expression, we utilized iBAQ29 as a proxy of protein abundance in cells stimulated for 0 h, 1 h or 4 h. Comparing protein abundance in rested cells with cells restimulated for either 1 h or 4 h, we determined that minimal effects were observed after only 1 h of stimulation. Therefore, we repeated this experiment comparing rested and 4-h restimulated cells 2 additional times. In these repeat experiments, comparing only two conditions, a simple two-label SILAC approach was sufficient (heavy label, resting cells; light, restimulated). The cells were mixed in a 1:1 ratio and LC–MS/MS was performed on the combined sample before further processing for other analyses. Therefore, we quantified changes in the T cell proteome between rested cells and cells after 4 h of TCR stimulation using SILAC ratios instead of iBAQ. For the di-glycine remnant profiling experiments (n = 2), the SILAC (super SILAC or two label as described above) samples were trypsinized and immunoprecipitation was performed with the α-K-ε-GG antibody to enrich for the di-glycine modified peptides. LC–MS/MS was performed on the di-glycine remnant enriched sample following off-line fractionation and recombination (described below). For proteomics analysis of neddylation, 1 μM MLN4924 was added for the last 2 h of stimulation for light carbon-labeled CD4+ T cells, and compared with heavy carbon-labeled resting T cells.
In-gel digest
Whole-proteome samples lysed in urea buffer, either individually or as SILAC mixed lysates (1:1 protein mix quantified by micro bicinchoninic acid (BCA), were run ~2 cm past the stacking gel in 10% Criterion precast Tris-HCl gels (Biorad). Gels were fixed overnight and stained briefly with Coomassie blue. Each lane of the Coomassie-stained gel was divided into 10 2 × 9 mm2 ‘pixels,’ each cut into 1 mm3 cubes51. They were destained with 50% methanol/1.25% acetic acid, reduced with 5 mM dithiothreitol (Thermo) and alkylated with 20 mM iodoacetamide (Sigma). Gel pieces were then washed with 20 mM ammonium bicarbonate (Sigma) and dehydrated with acetonitrile (Fisher). Trypsin (5 ng μl−1 in 20 mM ammonium bicarbonate, Promega) was added to the gel pieces and proteolysis was allowed to proceed overnight at 37 °C. Peptides were extracted with 0.3% triflouroacetic acid (J.T. Baker), followed by 50% acetonitrile. Extracts were combined and the volume was reduced by vacuum centrifugation.
K-ε-GG immunoprecipitation (di-glycine remnant enrichment)
Cell pellets (and bead-cell pellets in the case of αCD3/αCD28 restimulated cells) were lysed in urea buffer, protein concentration was measured via micro BCA assay (Thermo) and peptides were prepared and immunoprecipitated as described21,52. Approximately 250 × 106 total CD4+ T cells at the start of stimulation yielded ~3 mg total peptide. 12C- and 13C-labeled cells from the rested and stimulated time points were mixed at equal protein ratio after BCA quantification, before peptide preparation, as described above. Peptides were off-line basic reversed-phase fractionated and recombined non-contiguously into 3–4 fractions of ~1 mg peptide per fraction for immunoprecipitation. PTMscan ubiquitin remnant antibody, non-covalently conjugated to beads (Cell Signaling Technologies), was cross-linked as described52, and validated by SDS–PAGE. Cross-linked antibody (31 μg) was used for each 1-mg peptide fraction. The following modifications were made to the published protocol: step 6: alkylation was done with 20 mM iodoacetamide (IAM); step 10: peptides were acidified by 1% formic acid (final concentration); step 37: wash beads 2× with IAP buffer (50 mmol/L MOPS/NaOH, 10 mmol/L Na2HPO4, 50 mmol/L NaCl), followed by 2× IAP buffer plus 0.05% RapiGest SF surfactant (Waters), followed by 3× PBS; step 41: the eluted K-ε-GG peptides were desalted via Oasis HLB uElution plate 30 μM (Waters). All mass spectrometry samples were prepared in 0.1% trifluoroacetic acid (TFA)/water and analyzed as described below. After LC–MS/MS analysis, greater than 50% of peptides were modified.
LC–MS/MS
Tryptic digests were analyzed by LC–MS/MS on a hybrid LTQ Orbitrap Elite mass spectrometer (ThermoFisher) coupled with a nanoLC Ultra (Eksigent). Peptides were separated by reverse-phase HPLC on a nanocapillary column, 75 μM inner diameter × 15 cm Reprosil-pur 3 μM (Dr. Maisch) in a Nanoflex chip system (Eksigent). Mobile phase A consisted of 0.1% formic acid (ThermoFisher) and mobile phase B of 0.1% formic acid/80% acetonitrile. Peptides were eluted into the mass spectrometer at 300 nl min−1 with each reverse-phase HPLC run comprising a 90-min gradient from 10% to 25% B in 65 min, 25% to 40% B in 25 min, followed by column re-equilibration. The mass spectrometer was set to repetitively scan m/z from 300 to 1,800 (R = 240,000 for LTQ Orbitrap Elite) followed by data-dependent tandem MS (MS/MS) scans on the 20 most abundant ions, with a minimum signal of 1,500, dynamic exclusion with a repeat count of 1, repeat duration of 30 s, exclusion size of 500 and duration of 60 s, isolation width of 2.0, normalized collision energy of 33, and waveform injection and dynamic exclusion enabled. Fourier transform mass spectrometry (FTMS) full scan automatic gain control (AGC) target value was 1 × 106, while multi stage mass spectrometry (MSn) AGC was 1 × 104, respectively. FTMS full scan maximum fill time was 500 ms, while ion trap MSn fill time was 50 ms; microscans were set at 1. FT preview mode, charge state screening and monoisotopic precursor selection were all enabled with rejection of unassigned and 1+ charge states.
RNA-seq
Three biological replicates each for rested (0 h) or 4-h TCR restimulation cells were prepared. Total RNA was isolated from each replicate (Qiagen RNeasy kit) and poly-A selection was used to remove ribosomal RNA. Sequencing was performed with paired-end 100-base pair reads at an approximate coverage depth of 30 million reads. Reads were aligned to the Ensembl Mus musculus reference genome (GRCm38) using the spliced transcripts alignment to a reference (STAR) alignment program53.
Data analysis
Mass spectrometry data analysis was performed using MaxQuant. The identified peptides were searched against the Uniprot M. musculus proteome containing both canonical and isoform proteins (UP000000589). Label-free quantification was measured by iBAQ intensity values29. SILAC quantification was measured by the ‘normalized light/heavy (L/H) ratio’ calculated for the corresponding heavy and light peptides. The WCP experiments (label-free and SILAC) were combined to produce an n = 3 and the log2 fold changes were averaged to measure the protein expression change for 4-h TCR stimulation compared with baseline. Differential expression was defined for proteins exhibiting an average log2 fold change greater or less than than ±1 s.d., respectively, from the mean of the distribution of average log2 fold changes. Hypothesis testing (P values) was not used to identify differentially expressed proteins since two distinct methods of total protein quantification (label-free quantification and SILAC) were used in the three independent experiments. Our two di-glycine enrichment datasets represent both a super SILAC, comprising a resting, 1-h stimulated and 4-h stimulated sample, and a replicate experiment with SILAC-labeled resting and restimulated cells only. In the case of the super SILAC experiment, fold change in stimulation relative to rest is a ‘ratio of ratios’ as the normalized L/H ratio in each of the three samples represents the light-labeled time point (rest, 1 h, 4 h) against the heavy-labeled calibrator sample (equal parts cell lysate from resting, 1-h and 4-h stimulated cells). For the cases in which a single K-ε-GG peptide is identified for a protein, the change in abundance of that peptide, on restimulation, is used as a measure of the change in K-ε-GG abundance. Since we aim to predict the impact of the ubiquitylation on protein levels, we devised a calculation for protein-level ubiquitylation. For cases for which multiple peptides are identified for a single protein, the changes in abundance of each peptide are weighted by the relative intensity of that peptide and the weighted abundances are averaged to calculate the change in K-ε-GG abundance for the respective protein. Total protein ubiquitylation was quantified from the di-glycine remnant data by calculating the intensity-weighted mean ‘normalized L/H ratio’ of the set of modified peptides identified for the respective protein. The resulting ratios (n = 2) were averaged to measure the log2 fold change of ubiquitylation for 4-h TCR stimulation compared with baseline. The weighted average of the log2 SILAC ratios, where the weighting is based on ubiquitylated peptide intensity, is a robust estimate of protein ubiquitylation based on quantification of individual peptides. This method takes into account that peptides exhibiting higher ubiquitin abundance provide a stronger overall ubiquitin signal, and, therefore, those peptides should have a greater impact on the protein-level estimate. RNA-seq differential transcript expression was quantified using DESeq2 (ref. 49). Data processing and analysis to integrate and analyze the genomics and proteomics data were performed in R and Perl.
Statistics
For flow cytometry experiments, the mean fluorescence intensity (MFI) of each sample (one sample represents cells cultured from one or more mice), including rested cells, was normalized to the average of the rested (no CD3 + CD28 restimulation) samples. Two experiments were pooled for an n = 8. P values were calculated by one-way analysis of variance with Holm–Sidak correction for multiple testing. For immunoblots, quantification of band intensity was averaged across three replicate experiments (one experiment represents cells cultured from one or more mice), and compared with the control sample within the same experiment by use of a paired two-tailed t-test.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus54 and are accessible through GEO Series accession number GSE128154. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE55 partner repository with the dataset identifier PXD012831.
Code availability
The data were analyzed using standard algorithms for data manipulation, quantification and statistical analysis. The implementation of the analysis was performed using the R software. The scripts are available from the corresponding author upon request or can be accessed via GitHub, https://github.com/JosephDybas/TcellReceptorProteomics.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the helpful discussion and methods development insight from H. Fazelinia and D. Taylor at the Children’s Hospital of Philadelphia. This work was supported by NIH grants to P.M.O. (grant nos. R01 AI093566 and R01 AI114515), and an NRSA to C.E.O. (grant no. F31 CA180300).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41590–019-0381–6.
References
- 1.Tan H et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Sousa Abreu R, Penalva LO, Marcotte EM & Vogel C Global signatures of protein and mRNA expression levels. Mol. Biosyst 5, 1512–1526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel C & Marcotte EM Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet 13, 227–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Leary CE et al. Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4+ effector T cells. Nat. Commun 7, 11226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang D et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol 3, 281–287 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Scharschmidt E, Wegener E, Heissmeyer V, Rao A & Krappmann D Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol. Cell. Biol 24, 3860–3873 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B et al. Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat. Immunol 9, 1356–1363 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magnifico A et al. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem 278, 43169–43177 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Chen A et al. The HECT-type E3 ubiquitin ligase AIP2 inhibits activation-induced T-cell death by catalyzing EGR2 ubiquitination. Mol. Cell. Biol 29, 5348–5356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heissmeyer V et al. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol 5, 255–265 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Komander D & Rape M The ubiquitin code. Annu. Rev. Biochem 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Kulathu Y & Komander D Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell. Biol 13, 508–523 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yau R & Rape M The increasing complexity of the ubiquitin code. Nat. Cell Biol 18, 579–586 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Fang D & Liu YC Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat. Immunol 2, 870–875 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Huang H et al. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity 33, 60–70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata Y et al. HTLV-1 tax induces formation of the active macromolecular IKK complex by generating Lys63- and Met1-linked hybrid polyubiquitin chains. PLoS Pathog. 13, e1006162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajsbaum R et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity 40, 880–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swatek KN & Komander D Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park Y et al. SHARPIN controls regulatory T cells by negatively modulating the T cell antigen receptor complex. Nat. Immunol 17, 286–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G, Paige JS & Jaffrey SR Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol 28, 868–873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Udeshi ND, Mertins P, Svinkina T & Carr SA Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc 8, 1950–1960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udeshi ND et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elia AE et al. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell 59, 867–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hjerpe R et al. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J 441, 927–936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara T, Jung L, Bjorndahl J & Fu S Rapid induction of a phosphorylated 28 kD/32 kD disulfide-linked early activation antigen (EA 1) by 12-O-tetradecanoyl phorbol-13-acetate, mitogens, and antigens. J. Exp. Med 164, 1988–1994 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Rhodes M, Wiest DL & Vignali DA On the dynamics of TCR:CD3 complex cell surface expression and downmodulation. Immunity 13, 665–675 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Cowan JL & Morley SJ The proteasome inhibitor, MG132, promotes the reprogramming of translation in C2C12 myoblasts and facilitates the association of hsp25 with the eIF4F complex. Eur. J. Biochem 271, 3596–3611 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Jiang HY & Wek RC Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem 280, 14189–14202 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Schwanhäusser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wagner SA et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh RK et al. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol. Cell. Proteomics 11, 1595–1611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin H.-s, Liao L, Park Y & Liu Y-C Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc. Natl Acad. Sci. USA 110, 624–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soucy TA et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kanehisa M & Goto S KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabregat A et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanova E & Carpino N Negative regulation of TCR signaling by ubiquitination of Zap-70 Lys-217. Mol. Immunol 73, 19–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H et al. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J. Exp. Med 213, 399–414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang HY et al. Cbl promotes ubiquitination of the T cell receptor ζ through an adaptor function of Zap-70. J. Biol. Chem 276, 26004–26011 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Hsu TS, Hsiao HW, Wu PJ, Liu WH & Lai MZ Deltex1 promotes protein kinase Cθ degradation and sustains Casitas B-lineage lymphoma expression. J. Immunol 193, 1672–1680 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Xie JJ, Liang JQ, Diao LH, Altman A & Li Y TNFR-associated factor 6 regulates TCR signaling via interaction with and modification of LAT adapter. J. Immunol 190, 4027–4036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao N et al. Negative regulation of Lck by Cbl ubiquitin ligase. Proc. Natl Acad. Sci. USA 99, 3794–3799 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J et al. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J. Immunol 169, 2236–2240 (2002). [DOI] [PubMed] [Google Scholar]
- 43.van der Wal L et al. Improvement of ubiquitylation site detection by Orbitrap mass spectrometry. J. Proteomics 172, 49–56 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Sap KA, Bezstarosti K, Dekkers DHW, Voets O & Demmers JAA Quantitative proteomics reveals extensive changes in the ubiquitinome after perturbation of the proteasome by targeted dsRNA-mediated subunit knockdown in Drosophila. J. Proteome Res 16, 2848–2862 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Draber P et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teh CE et al. Linear ubiquitin chain assembly complex coordinates late thymic T-cell differentiation and regulatory T-cell homeostasis. Nat. Commun 7, 13353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okamura K et al. Survival of mature T cells depends on signaling through HOIP. Sci. Rep 6, 36135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lex A, Gehlenborg N, Strobelt H, Vuillemot R & Pfister H UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph 20, 1983–1922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevchenko A, Wilm M, Vorm O & Mann M Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Udeshi ND et al. Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–d450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this publication have been deposited in NCBI’s Gene Expression Omnibus54 and are accessible through GEO Series accession number GSE128154. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE55 partner repository with the dataset identifier PXD012831.