Abstract
Recent discovery of ultra-high binding affinities in the cucurbit[7]uril (CB7) based host-guest pairs in aqueous environment has rendered the CB7 a rather attractive material in analytical and biomedical applications. Due to the lack of a molecular platform that can follow the same host-guest complex during repetitive mechanical measurements, however, mechanical stabilities of the CB7 system have not been revealed, hindering its potential to rival widely used conjugation pairs such as streptavidin–biotin complex. Here, we assembled a DNA template in which a flexible DNA linker was exploited to keep the host (CB7) and guest (adamantane) in proximity. This platform not only increased the efficiency of the single-molecule characterization in optical tweezers, but also clearly revealed mechanical features of the same host-guest complex. We found that positively charged adamantane guest demonstrated higher mechanical stability (49 pN) than neutral adamantane (44 pN), a trend consistent with the chemical affinity between guest molecules and the CB7 host. Surprisingly, we found that a hexyl group adjacent to the adamantane served as a chaperone to assist the formation of the adamantane-CB7 pairs. The discovery of unprecedented chaperone assisted interaction mechanism provides new approaches to efficiently assemble host-guest based supramolecules with increased mechanical stabilities, which can be exploited in various biomedical, biosensing, and materials fields.
Graphical Abstract
Synthetic host-guest binding pairs in aqueous environments represent an exciting area of scientific research since such “ligation” pairs have potential utility in biomedical (e.g., imaging, drug delivery and release) and materials (e.g., self-assembly of nano/microparticles, biomaterials, and hydrogels) applications.1–3 However, many host-guest interactions are limited by relatively low binding affinities in water. Notable exceptions to the low-affinity host-guest pairs are cucurbit[7]uril (CB7) 4–6 based systems where ultra-tight binding are achieved via the hydrophobic effect in addition to the electrostatic interactions. 7–9
Many approaches, such as NMR and Isothermal Titration Calorimetry (ITC), have been used to investigate the affinity between host-guest pairs.5 These ensemble methods report average properties of many host-guest pairs. As a result, the signal-to-noise is often compromised, giving difficulties to reveal subtle and transient actions related to subdomains of a molecule, for example. Due to their superior sensitivities, single-molecule techniques offer more precise description for these subtle interactions. Among different single-molecule techniques, force-based tools furnish a unique mechanical perspective to individual host-guest pairs. 10–12 Given the attractive features of CB7-guest pairs for materials applications, investigation of the mechanical strength of the CB7-guest interactions is essential. Only scattered studies have characterized mechanical properties of host-guest interactions, none of which is related to the highly stable CB7 system.10–12 The difficulty of mechanical investigations lies in the noncovalent nature of the host-guest complex. Upon applying mechanical force, the host-guest complex is forced apart. To continuously probe the strength of host-guest complexes, new host-guest pairs need to be identified after each binding complex is disassembled by force. The process is rather tedious while it is not possible to follow the same host-guest complex during each association and dissociation cycle. This uncertainty leads to measurement error since it is impossible to distinguish each rupture event originated from the host-guest complex or from the pulling handles, the latter representing the noise.
Here, we fully addressed this uncertainty by connecting host and guest molecules using a single-stranded (ss) DNA linker inside a single molecular DNA platform. To build such a platform, we took advantage of DNA self-assembly to form a construct in which host and guest molecules were separately linked to the termini of single-stranded DNA fragments (Figure 1). The linkages were accomplished by click chemistry or activated ester-amine coupling reactions (see Figure 1B and SI)13,14. Each linked DNA fragment was then self-assembled with a complementary DNA overhang at the end of a duplex DNA (dsDNA) handle. Two such dsDNA handles were further attached to two optically trapped beads via streptavidin/biotin and digoxigenin/antibody interactions, respectively. These two dsDNA handles were also directly connected by a polythymine (T50) linker (see Figure S4 for agarose gels of the final DNA construct) so that the separated host and the guest molecules can remain in proximity after the host-guest complex was disrupted mechanically.
Figure 1.
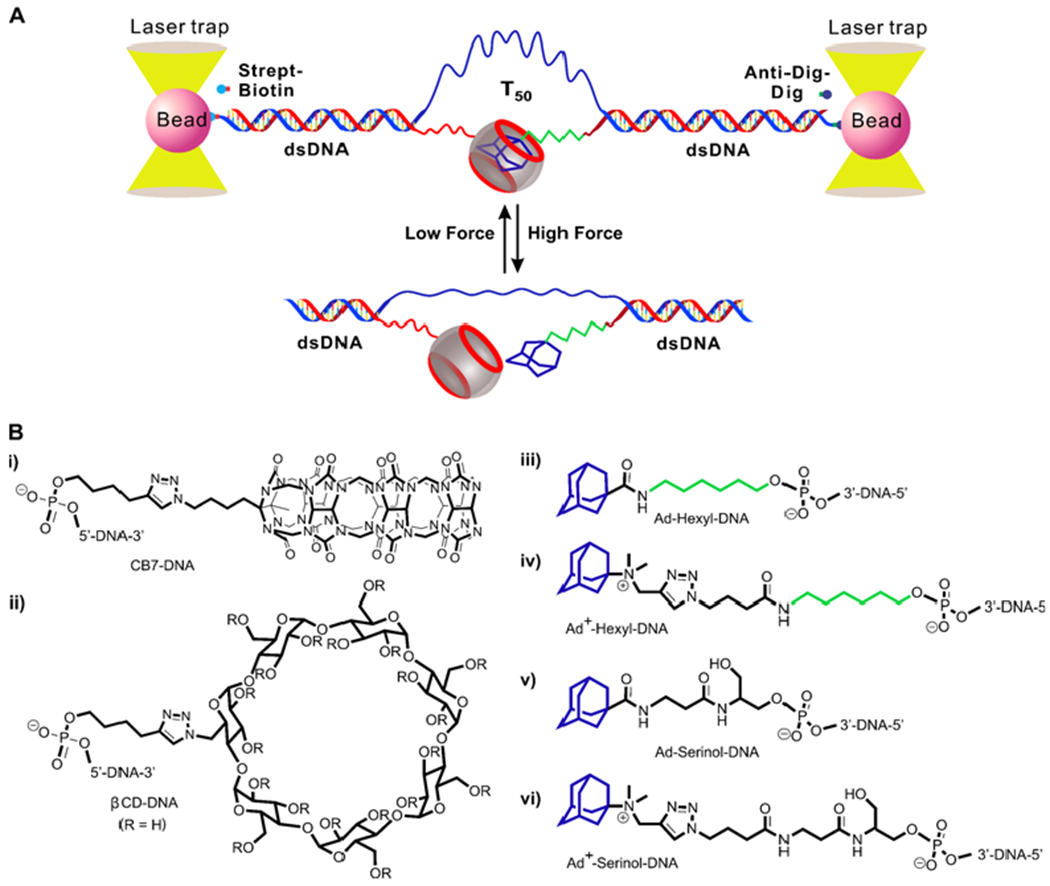
High-throughput single-molecule assay for host-guest chemistry. (A) Experimental setup. Barrels depict host molecules. (B) Structures of host (i & ii) and guest (iii-vi) molecules used in the mechanical unfolding experiments. CB7 stands for cucurbit[7]uril, βCD stands for β-cyclodextrin, and Ad stands for adamantane (blue). Hexyl group is shown in green.
We started by testing the interaction between adamantane and CB7 (Figure 1B). During each experiment, we first let the host-guest complex form in the DNA template prepared above. We then moved one bead away from the other using a steerable mirror that controls one of the traps. During this movement the tensile force in the DNA template increased until a rupture event was observed, which indicated the break of the host-guest complex (Figure 2A). Since the two dsDNA handles were still connected by the T50 linker, the tension inside DNA template did not relax to zero, which was a strong indication that the rupture occurred at the host-guest complex instead of the dsDNA handles or DNA-bead connections. Indeed, when we compared change-in-contour-length (ΔL) for the rupture events, we found ΔL values fell within the range expected to fully stretch T50 linkage after host-guest complex was disassembled (see Figure S2 for ΔL histograms and calculations).
Figure 2.
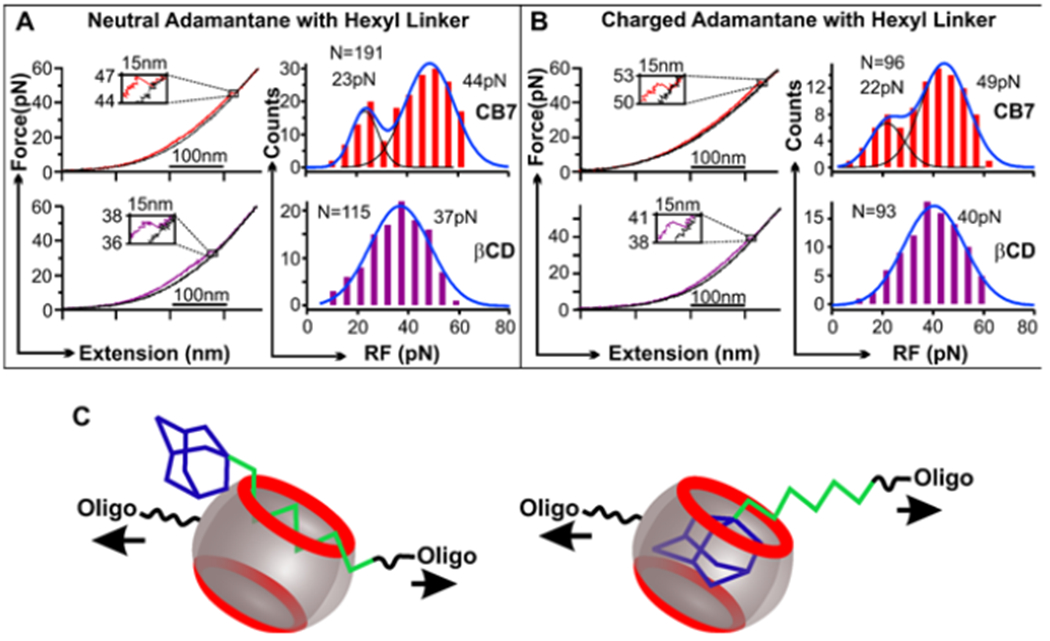
Mechanical unfolding of host-guest complexes. (A) Typical extending (colored) and relaxing (black) force-extension curves (left) and rupture force (RF) histograms (right) for cucurbit[7]uril (CB7)-neutral adamantane (Ad, compound iii in Figure 1B) complex (upper panel) and β-cyclodextrin (βCD)–Ad complex (lower panel). (B) Typical extending (colored) and relaxing (black) force-extension curves (left) and rupture force (RF) histograms (right) for the CB7 – positively charged Ad+ (Figure 1B, iv) complex (upper panel) and βCD–Ad+ complex (lower panel). (C) Schematics of two distinct models of host-guest interactions. Hexyl group is shown in green. N depicts number of data points.
When we plotted the unfolding force histograms for CB7 and neutral adamantane complexes, we observed two populations of rupture forces (Figure 2A). While a minor population was centered at 23 pN, a major population had an average force of 44 pN. This was surprising since we expected only one rupture event should occur between this simple host-guest complex. To confirm this finding, we repeated the same mechanical unfolding experiments (Figure 2A) on the host-guest interactions between a positively charged adamantane and CB7 (Figure 1B). As shown in Figure 2B, we again observed two rupture force populations. This time, the minor population remained the same rupture force (22 pN) whereas the major population increased the force to 49 pN. Due to the charge-dipole interaction, it is known that the binding between the CB7-adamantane pair is significantly increased when adamantane is positively charged 15. Therefore, these experiments suggest that the higher force populations are likely due to the rupture between the adamantane and CB7.
To reveal the nature of the lower force (22/23 pN) population, we inspected the guest molecules attached to oligonucleotide fragments. For both neutral and positively charged guests, we found there is a hexyl linker between the adamantane and the DNA fragment. It has been reported that hydrocarbon chains can be accommodated in the CB7 cavity, although with a weaker interaction compared to the better fitting adamantane guest.5,16,17 Therefore, it is possible that populations with the weaker rupture force (22/23 pN) can be due to the binding of the hexyl group to the CB7 (Figure 2C). When we used a control guest molecule that only contains the hexyl group (Figure S5), we found the hexyl-CB7 binding required ~20 pN to disassemble. This confirmed the weaker rupture force populations in Figure 2 A&B are due to the hexyl-CB7 binding.
Compared to cucurbituril, cyclodextrin binds with rather weak association to hexyl groups.5,17 Hence, we may observe only one rupture population in the force histogram when cyclodextrin is used as a host. To test this, we repeated the unfolding experiments using β-cyclodextrin as a host molecule (Figure 1B). As expected, only one rupture force population was observed (Figure 2 A&B, lower panels). Both neutral and charged adamantanes in the β-cyclodextrin showed reduced forces (37 and 40 pN) compared to those in the CB7 host (44 and 49 pN respectively). These results confirmed our assignment that the higher rupture force populations are due to the accommodation of the adamantane inside the CB7 whereas the smaller-force species are likely due to the hexyl linker binding in the CB7 host. It is rather significant that mechanical stabilities of the CB7-adamantane complexes (44/49 pN) fall in the range of many biomolecules including DNA G-quadruplexes18,19 and proteins 20 as well as peptides 21.
In another control, we replaced the hexyl linker with a serinol linker between the adamantane molecule and the DNA oligo (see Figure 1B and Figure 3 A&B). When we evaluated the mechanical stabilities of the CB7-adamantane pairs, we observed the rupture force histograms reduced to one population (Figure 3 A&B) with an average force close to the higher-force population of the hexyl-adamantane-host complexes discussed above (Figure 2). In comparison, the guest molecule that only contains the serinol group did not show any CB7 binding (Figure S5). These results are consistent with the assignment of the smaller force population as the binding of the hexyl linker to the CB7 cavity. Comparison of the rupture force histograms revealed that unfolding force is slightly smaller for the adamantane without the hexyl linker. This suggests that in the presence of the hexyl group, the mechanical stability of the host-guest complex increases. One possibility is that the hexyl linker may serve as a chaperone for better orientation of the guest molecule when it approaches to the host, resulting in sturdier host-guest interaction (Figure 4).
Figure 3.
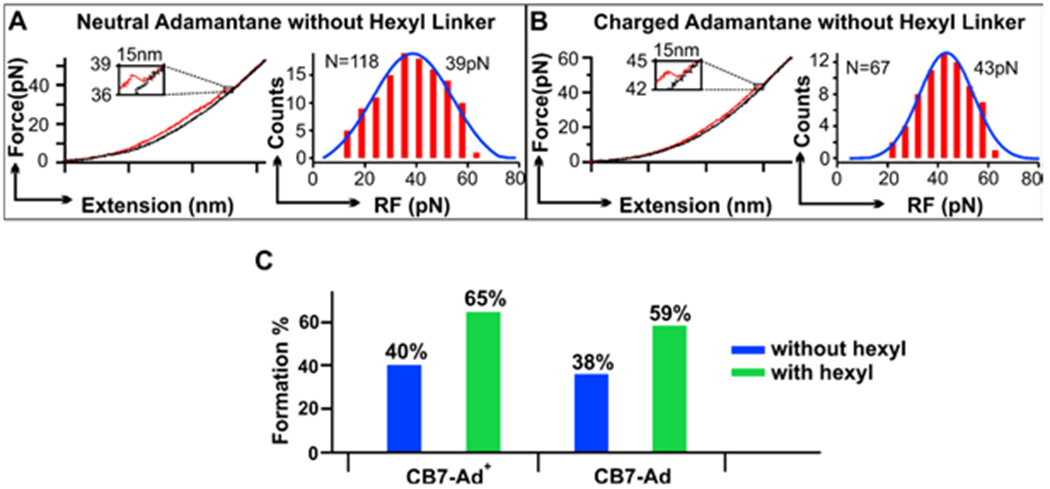
Effect of hexyl group on the host-guest interaction. (A) Typical extending (red) and relaxing (black) force-extension curves (left) and rupture force (RF) histograms (right) for the interaction between cucurbit[7]uril (CB7) and adamantane without ((A), Ad) and with ((B), Ad+) a positive charge. Adamantane is attached to the oligo without hexyl linker. (C) Percentage formation of the CB7 – Ad+ and CB7 – Ad complexes with (purple)
Figure 4.
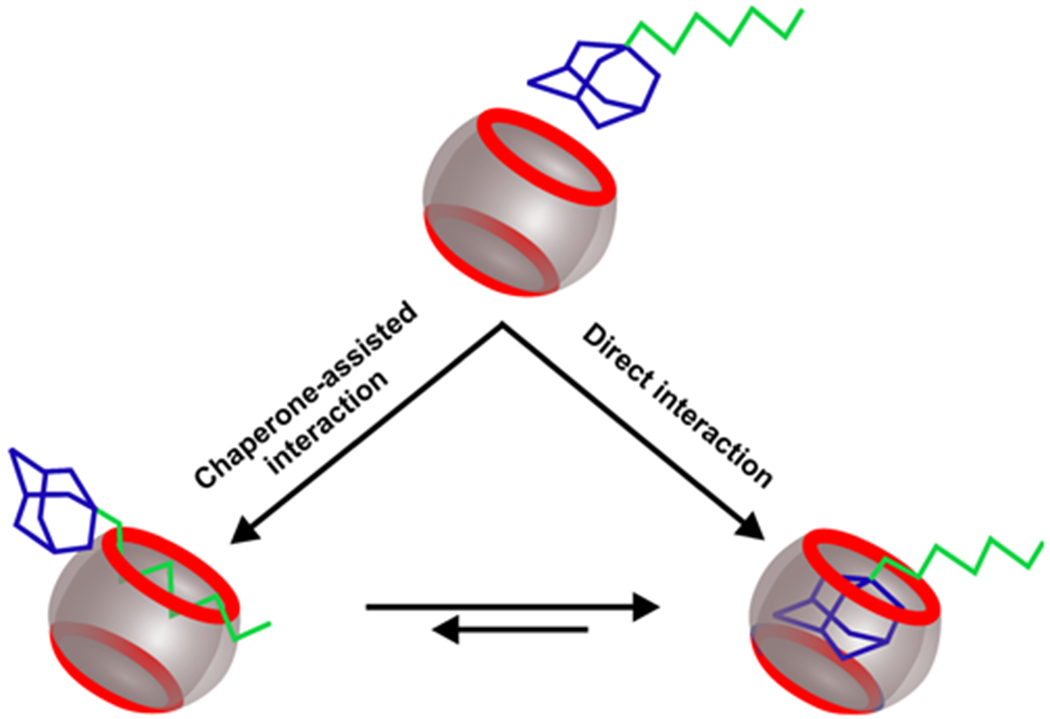
Three-state chaperone assisted host-guest interaction model. Blue and green depict adamantane and alkyl groups respectively. Barrels depict cucurbituril host molecules.
To test whether the hexyl moiety can function as a molecular chaperon for binding events inside the CB7 cavity, we evaluated the percentage formation of the host-guest complex with and without the hexyl group tethered to the adamantane guest. As shown in Figure 3C, we found that when the hexyl linker is present, the higher-force population increases significantly compared to the guest without the hexyl group. Again, this observation corroborated with the chaperone role of the hexyl linker in the host-guest formation (Figure 4). We propose that the weak interaction between the amide NH close to the hexyl group and the C=O of the cucurbituril portal brings the host-guest pair into proximity, which is followed by the binding of the hexyl to the CB7.22 Since the hexyl group has 6 flexible methylene units, there is more chance of the initial interaction between the hexyl group and the CB7 with respect to the spherical adamantane molecule. In addition, the binding of the hexyl chain to the CB7 barrel has an easier contact mode than the penetration of the admantyl group into the CB7 barrel (Figure 4). This interaction brings the adamantane closer to the CB7. Since the binding strength between the adamantane and the CB7 is much higher than that between the hexyl group and the CB7, 4–6 the more stable adamantane-CB7 complex eventually predominates, likely through egression and ingression7 of the hexyl and the adamantane groups, respectively. The much increased high-force populations (Figure 2 A&B) reflect this equilibrium process. Similar mechanism has been proposed for the pH-dependent binding between cyclohexylmethylamine and cucurbit[6]uril.23
Analyses of the change-in-contour-length (∆L) of each rupture event of the host-guest interaction also support this 3-state chaperon model (Figure 4). As shown in Figures S2 and S3, when hexyl group interacts with the portal of the CB7, the initial end-to-end distance (x) of the DNA template becomes smaller. Given that the total contour length (L) is determined by the 50 T linker, which is a fixed value, ∆L is expected to be bigger (∆L = L − x). This was exactly observed in the single-molecule experiment (compare 1st and 3rd rows in Figure S2), confirming different interactions between the adamantane and the hexyl group to the CB7.
Previously, chaperone-like behaviors with different mechanisms have been proposed in the crystallization of cucurbit[8]uril24 in the presence of a single guest molecule. Our unprecedented finding on the chaperone assisted binding has salient implications on the specificity issue in many host-guest systems. Due to the generic nature of hydrophobic forces and electrostatic interactions,7 different non-polar guests can compete for binding pockets in cyclodextrin or cucurbituril systems.25,26 The discovery of chaperone-like behavior here suggests specificity issue may become advantageous in the rapid assembly of materials with strong stabilities.
In summary, using mechanical unfolding in optical tweezers, we repeatedly measured the mechanical stability of the same host-guest complex between adamantane and CB7. We found that the mechanical stability of this host-guest chemistry is in the range of many biological recognitions. We also revealed that a hexyl group in the guest can serve as a chaperone to promote the formation and to stabilize the CB7-adamantane complex. This chaperone group is specific as it only assists the formation of adamantane-CB7 complex but not adamantane-β-cyclodextrin interaction. Such findings not only elucidate the mechanical property of individual host-guest complexes with high accuracy and much improved throughput, but it also provides a new approach to form mechanically stable host-guest complexes in an efficient manner through chaperone groups. Given the wide-spread applications of cucurbituril based host-guest systems in the biomedical, biosensing, and materials fields,1,2,4 we anticipate that our new findings at the single-molecule level can provide unique insights facilitating the advancement of these fields.
Supplementary Material
ACKNOWLEDGMENT
H.M. thanks NIH (R01 CA236350) and NSF (CBET-1904921) for support. J.J. is grateful for support from the NSF (CHE-1609603). L.I. thanks the NSF (CHE-1807486) for financial support.
Footnotes
Supporting Information
Materials and methods used to prepare adamantane-DNA conjugates with and without hexyl groups, CB- and CD-DNA conjugates, and DNA constructs for single molecule force spectroscopy experiments. Mechanical unfolding procedures. Histograms for the host-guest conjugates. Calculation for change-in-contour-length.
The Supporting Information is available free of charge on the ACS Publications website.
The authors declare no competing financial interests.
REFERENCES
- (1).Ko YH; Hwang I; Lee D-W; Kim K Ultrastable Host–Guest Complexes and Their Applications. Isr. J. Chem 2011, 51, 506. [Google Scholar]
- (2).Liu W; Samanta SK; Smith BD; Isaacs L Synthetic mimics of biotin/(strept)avidin. Chem. Soc. Rev 2017, 46, 2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wenz G; Han B-H; Muller A Cyclodextrin Rotaxanes and Polyrotaxanes. Chem. Rev 2006, 106, 782. [DOI] [PubMed] [Google Scholar]
- (4).Shetty D; Khedkar JK; Park KM; Kim K Can we beat the biotin–avidin pair?: cucurbit[7]uril-based ultrahigh affinity host–guest complexes and their applications. Chem. Soc. Rev 2015, 44, 8747. [DOI] [PubMed] [Google Scholar]
- (5).Barrow SJ; Kasera S; Rowland MJ; del Barrio J; Scherman OA Cucurbituril-Based Molecular Recognition. Chem. Rev 2015, 115, 12320. [DOI] [PubMed] [Google Scholar]
- (6).Liu S; Ruspic C; Mukhopadhyay P; Chakrabarti S; Zavalij PY; Isaacs L The Cucurbit[n]uril Family: Prime Components for Self-Sorting Systems. J. Am. Chem. Soc 2005, 127, 15959. [DOI] [PubMed] [Google Scholar]
- (7).Masson E; Raeisi M; Kotturi K Kinetics Inside, Outside and Through Cucurbiturils. Isr. J. Chem 2018, 58, 413. [Google Scholar]
- (8).Nau WM; Florea M; Assaf KI Deep Inside Cucurbiturils: Physical Properties and Volumes of their Inner Cavity Determine the Hydrophobic Driving Force for Host–Guest Complexation. Isr. J. Chem 2011, 51, 559. [Google Scholar]
- (9).Biedermann F; Uzunova VD; Scherman OA; Nau WM; De Simone A Release of High-Energy Water as an Essential Driving Force for the High-Affinity Binding of Cucurbit[n]urils. J. Am. Chem. Soc 2012, 134, 15318. [DOI] [PubMed] [Google Scholar]
- (10).Naranjo T; Cerrón F; Nieto-Ortega B; Latorre A; Somoza Á; Ibarra B; Pérez EM Mechanical measurement of hydrogen bonded host-guest systems under non-equilibrium, near-physiological conditions. Chem Sci 2017, 8, 6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Walsh-Korb Z; Yu Y; Janeček E-R; Lan Y; del Barrio J; Williams PE; Zhang X; Scherman OA Single-Molecule Force Spectroscopy Quantification of Adhesive Forces in Cucurbit[8]Uril Host–Guest Ternary Complexes. Langmuir 2017, 33, 1343. [DOI] [PubMed] [Google Scholar]
- (12).Zapotoczny S; Auletta T; de Jong MR; Schönherr H; Huskens J; van Veggel FCJM; Reinhoudt DN; Vancso GJ Chain Length and Concentration Dependence of β-Cyclodextrin–Ferrocene Host–Guest Complex Rupture Forces Probed by Dynamic Force Spectroscopy. Langmuir 2002, 18, 6988. [Google Scholar]
- (13).Kolb HC; Finn MG; Sharpless KB Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. Engl 2001, 40, 2004. [DOI] [PubMed] [Google Scholar]
- (14).Zhou X; Su X; Pathak P; Vik R; Vinciguerra B; Isaacs L; Jayawickramarajah J Host-Guest Tethered DNA Transducer: ATP Fueled Release of a Protein Inhibitor from Cucurbit[7]uril. J. Am. Chem. Soc 2017, 139, 13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Moghaddam S; Yang C; Rekharsky M; Ko YH; Kim K; Inoue Y; Gilson MK New Ultrahigh Affinity Host–Guest Complexes of Cucurbit[7]uril with Bicyclo[2.2.2]octane and Adamantane Guests: Thermodynamic Analysis and Evaluation of M2 Affinity Calculations. J. Am. Chem. Soc 2011, 133, 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Carrazana J; Jover A; Meijide F; Soto VH; Vázquez Tato J Complexation of Adamantyl Compounds by β-Cyclodextrin and Monoaminoderivatives. J. Phys. Chem. B 2005, 109, 9719. [DOI] [PubMed] [Google Scholar]
- (17).Tee OS; Gadosy TA; Giorgi JB The binding of alkyl chains to β-cyclodextrin and ‘hydroxypropyl-β-cyclodextrin’. Journal of the Chemical Society, Perkin Transactions 2 1993, 1705. [Google Scholar]
- (18).Yu Z; Schonhoft JD; Dhakal S; Bajracharya R; Hegde R; Basu S; Mao H ILPR G-Quadruplexes Formed in Seconds Demonstrate High Mechanical Stabilities. J. Am. Chem. Soc 2009, 131, 1876. [DOI] [PubMed] [Google Scholar]
- (19).Koirala D; Dhakal S; Ashbridge B; Sannohe Y; Rodriguez R; Sugiyama H; Balasubramanian S; Mao H A Single-Molecule Platform for Investigation of Interactions between G-quadruplexes and Small-Molecule Ligands. Nat. Chem 2011, 3, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cecconi C; Shank EA; Bustamante C; Marqusee S Direct observation of the three-state folding of a single protein molecule. Science 2005, 309, 2057. [DOI] [PubMed] [Google Scholar]
- (21).Xi Z; Gao Y; Sirinakis G; Guo H; Zhang Y Single-molecule observation of helix staggering, sliding, and coiled coil misfolding. Proc. Nat. Acad. Sci. USA 2012, 109, 5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ko YH; Kim Y; Kim H; Kim K U-Shaped Conformation of Alkyl Chains Bound to a Synthetic Receptor Cucurbit[8]uril. Chemistry – An Asian Journal 2011, 6, 652. [DOI] [PubMed] [Google Scholar]
- (23).Marquez C; Nau WM Two Mechanisms of Slow Host–Guest Complexation between Cucurbit[6]uril and Cyclohexylmethylamine: pH-Responsive Supramolecular Kinetics. Angew. Chem. Int. Ed. Engl 2001, 40, 3155. [DOI] [PubMed] [Google Scholar]
- (24).Zhu W; Wang C; Lan Y; Li J; Wang H; Gao N; Ji J; Li G Chaperone-Assisted Formation of Cucurbit[8]uril-Based Molecular Porous Materials with One-Dimensional Channel Structure. Langmuir 2016, 32, 9045. [DOI] [PubMed] [Google Scholar]
- (25).Tootoonchi MH; Yi S; Kaifer AE Detection of Isomeric Microscopic Host–Guest Complexes. A Time-Evolving Cucurbit[7]uril Complex. J. Am. Chem. Soc 2013, 135, 10804. [DOI] [PubMed] [Google Scholar]
- (26).Watfa N; Melgar D; Haouas M; Taulelle F; Hijazi A; Naoufal D; Avalos JB; Floquet S; Bo C; Cadot E Hydrophobic Effect as a Driving Force for Host–Guest Chemistry of a Multi-Receptor Keplerate-Type Capsule. J. Am. Chem. Soc 2015, 137, 5845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


