Summary
Regulation of transcription is the main mechanism responsible for precise control of gene expression. While the majority of transcriptional regulation is mediated by DNA-binding transcription factors that bind to regulatory gene regions, an elegant alternative strategy employs small RNA guides, piwi-interacting RNAs (piRNAs) to identify targets of transcriptional repression. Here we show that in Drosophila the small ubiquitin-like protein SUMO and the SUMO E3 ligase Su(var)2–10 are required for piRNA-guided deposition of repressive chromatin marks and transcriptional silencing of piRNA targets. Su(var)2–10 links the piRNA-guided target recognition complex to the silencing effector by binding the piRNA/Piwi complex and inducing SUMO-dependent recruitment of the SetDB1/Wde histone methyltransferase effector. We propose that in Drosophila, the nuclear piRNA pathway has co-opted a conserved mechanism of SUMO-dependent recruitment of the SetDB1/Wde chromatin modifier to confer repression of genomic parasites.
Keywords: piRNA, Su(var)2–10, SUMO, chromatin, heterochromatin, silencing, transposon
Graphical Abstract.
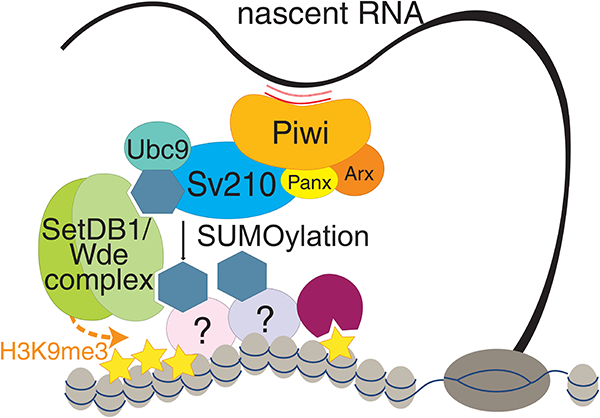
Model of piRNA-guided H3K9me3 deposition. piRNA-loaded Piwi complexes recognize nascent transcripts. Su(var)2–10 interacts with the piRITS complex and promotes SUMOylation of itself and yet-to-be identified chromatin factor(s). The SUMO moiety of Su(var)2–10 and chromatin factors recruits the SetDB1/Wde complex through its SIMs, which installs H3K9me3 at target loci.
eTOC Blurb
The piRNA pathway induces transcriptional silencing of transposons by recruiting the histone methyltransferase SetDB1/Wde, which puts the repressive mark on genomic piRNA targets. Ninova et al. found that in Drosophila, SUMO and the SUMO ligase Su(var)2–10 are required for transposon repression by linking the piRNA-guided target recognition complex to SetDB1/Wde.
Introduction
The majority of transcriptional control is achieved by transcription factors that bind short sequence motifs on DNA. In many eukaryotic organisms, transcriptional repression can also be guided by small RNAs, which - in complex with Argonaute proteins - recognize their genomic targets using complementary interactions with nascent RNA (Holoch and Moazed, 2015). Small RNA-based regulation provides flexibility in target selection without the need for new transcription factors and as such is well suited for genome surveillance systems to identify and repress the activity of harmful genetic elements such as transposons.
Transcriptional repression guided by small RNAs correlates with the deposition of repressive chromatin marks, particularly histone 3 lysine 9 methylation (H3K9me) in S. pombe, plants and animals (Bernatavichute et al., 2008; Enke et al., 2011; Gu et al., 2012; LeThomas et al., 2013; Pezic et al., 2014; Volpe et al., 2002). In addition, plants and mammals also employ CpG DNA methylation for target silencing (Aravin et al., 2008; Mette et al., 2000). Small RNA/Ago induced transcriptional gene silencing is best understood in S. pombe, where the RNA-induced transcriptional silencing complex (RITS) was studied biochemically and genetically (Holoch and Moazed, 2015; Verdel et al., 2004). In contrast to yeast, the molecular mechanism of RNA-induced transcriptional silencing in Metazoans remains poorly understood. Small RNA-induced transcriptional repression mechanisms might have independently evolved several times during evolution and thus might mechanistically differ from that of S. pombe.
In Metazoans, small RNA guided transcriptional repression is mediated by Piwi proteins, a distinct clade of the Argonaute family, and their associated Piwi-interacting RNAs (piRNAs). Both in Drosophila and in mouse, the two best-studied metazoan systems, nuclear Piwis are responsible for transcriptional silencing of transposons (Aravin et al., 2008; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008; LeThomas et al., 2013; Manakov et al., 2015; Pezic et al., 2014; Rozhkov et al., 2013; Sienski et al., 2012). Based on the current model targets are recognized through binding of the Piwi/piRNA complex to nascent transcripts of target genes. In both Drosophila and mouse, piRNA-dependent silencing of transposons correlates with accumulation of repressive chromatin marks (H3K9me3 and, in mouse, CpG methylation of DNA) on target sequences (Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008; LeThomas et al., 2013; Pezic et al., 2014; Rozhkov et al., 2013; Sienski et al., 2012). These marks can recruit repressor proteins, such as HP1 (Maison and Almouzni, 2004), providing a mechanism for transcriptional silencing. However, how recognition of nascent RNA by the Piwi/piRNA complex leads to deposition of repressive marks at the target locus is not well understood. Several proteins, Asterix (Arx)/Gtsf1, Panoramix (Panx)/Silencio and Nxf2, were shown to associate with Piwi and are required for transcriptional silencing (Batki et al., 2019; Donertas et al., 2013; Fabry et al., 2019; Muerdter et al., 2013; Murano et al., 2019; Ohtani et al., 2013; Sienski et al., 2015; Yu et al., 2015; Zhao et al., 2019). Accumulation of H3K9me3 on Piwi/Panx targets requires the activity of the histone methyltransferase SetDB1 (also known as Egg) (Rangan et al., 2011; Sienski et al., 2015; Yu et al., 2015), however, a mechanistic link between the Piwi/Arx/Panx/Nxf2 complex, which recognizes targets, and the effector chromatin-modifier has not been established.
We identified Su(var)2–10/dPIAS to provide the link between the Piwi/piRNA and the SetDB1 complex in piRNA-induced transcriptional silencing. In Drosophila, Su(var)2–10 mutation causes suppression of position effect variegation, a phenotype indicative of its involvement in chromatin repression (Elgin and Reuter, 2013; Reuter and Wolff, 1981). Su(var)2–10 associates with chromatin and regulates chromosome structure (Hari et al., 2001). It also emerged in screens as a putative interactor of the central heterochromatin component HP1, a repressor of enhancer function, and a SUMO pathway component (Alekseyenko et al., 2014; Stampfel et al., 2015; Stielow et al., 2008a). However, its molecular functions in chromatin silencing were not investigated. Su(var)2–10 belongs to the conserved PIAS/Siz protein family (Betz et al., 2001; Hari et al., 2001; Mohr and Boswell, 1999), of which the yeast, plant, and mammalian homologs act as E3 ligases for SUMOylation of several substrates (Garcia-Dominguez et al., 2008; Johnson and Gupta, 2001; Kahyo et al., 2001; Kotaja et al., 2002; Sachdev et al., 2001; Schmidt and Muller, 2002; Takahashi et al., 2001). We studied the role of Su(var)2–10 in germ cells of the ovary, where chromatin maintenance and transposon repression are essential to grant genomic stability across generations. Germ cell depletion of Su(var)2–10 phenocopies loss of Piwi: both lead to strong transcriptional activation of transposons, and loss of repressive chromatin marks over transposon sequences. Su(var)2–10 genetically and physically interacts with Piwi and its auxiliary factors, Arx and Panx. We demonstrated that the repressive function of Su(var)2–10 is dependent on its SUMO E3 ligase activity and the SUMO pathway. Our data points to a model in which Su(var)2–10 acts downstream the piRNA/Piwi complex to induce local SUMOylation, which in turn leads to the recruitment of the SetDB1/Wde complex. SUMO modification was shown to play a role in the formation of silencing chromatin in various systems from yeast to mammals, including the recruitment of the silencing effector SETDB1 and its co-factor MCAF1 by repressive transcription factors (Ivanov et al., 2007; Maison et al., 2011, 2016; Shin et al., 2005; Stielow et al., 2008b; Thompson et al., 2015; Uchimura et al., 2006). Together, these findings indicate that the piRNA pathway utilizes a conserved mechanism of silencing complex recruitment through SUMOylation to confer transcriptional repression.
Results
Germline-specific knockdown of Su(var)2–10 induces embryonic lethality
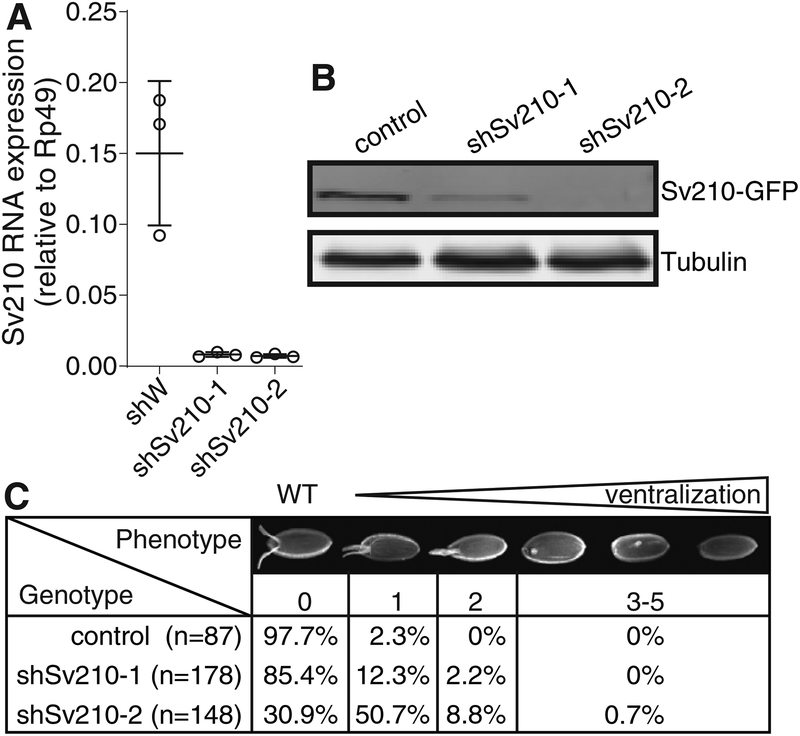
The essential Su(var)2–10 gene encodes a conserved protein that is highly expressed in the Drosophila ovary (FlyAtlas (Chintapalli et al., 2007)). To investigate the role of Su(var)2–10 in the germline, we employed germline-specific knockdown (GLKD) in the Drosophila ovary using two different short hairpin RNAs (shRNAs) that target the Su(var)2–10 mRNA, shSv210–1 and shSv210–2, driven by the maternal tubulin-GAL4. Expression of each shRNA resulted in ~95% reduction of the Su(var)2–10 mRNA (Fig. 1A). The level of ectopically expressed GFP-tagged Su(var)2–10 protein was also reduced by both shRNAs, with shSv210–2 showing stronger depletion than shSv210–1 (Fig. 1B).
Figure 1. Su(var)2–10 depletion in the Drosophila female germline leads to embryo ventralization.
(A) Su(var)2–10 GLKD using two different shRNAs (shSv210–1 and shSv210–2) leads to reduced transcript level. Plot shows the relative expression of Su(var)2–10 in control and Su(var)2–10 depleted ovaries (RT-qPCR). Dots correspond to three independent biological replicates; bars indicate the mean and SD.
(B) Su(var)2–10 protein level is reduced upon GLKD. Western blot shows the levels of MT-Gal4 driven GFP-Su(var)2–10 in ovaries of control (shW) and Su(var)2–10 GLKD flies. Tubulin (Tub) is used as loading control.
(C) Su(var)2–10 GLKD causes egg shell ventralization. Table shows the proportion of eggs from control (shW) and Su(var)2–10 GLKD ovaries displaying each class of ventralization phenotype ordered by severity (Images adopted from (Meignin and Davis, 2008)).
Next, we examined the effect of Su(var)2–10 knockdown on germline development. Ovaries from females with GLKD of Su(var)2–10 did not show gross phenotypic difference in ovarian morphology. Such females produce eggs, but no viable progeny, indicating that Su(var)2–10 plays an important role during gametogenesis. Embryos produced by females with GLKD of Su(var)2–10 showed varying degree of ventralization, a sign of axis specification defect (Fig. 1C). Consistent with the stronger protein depletion, shSv210–2 had a stronger penetrance, with 60% of the embryos showing mild to severe ventralization.
We also addressed Su(var)2–10 function in the somatic follicular cells of the ovary, using shRNA under the control of traffic jam (Tj) GAL4. Depletion of Su(var)2–10 in follicular cells caused severe morphological defects and collapse of oogenesis phenocopying effects Piwi depletion in follicular cells. Together, these data indicate that Su(var)2–10 plays an important role in both germ cells and the ovarian somatic cells that support germline development.
Depletion of Su(var)2–10 induces transposon derepression
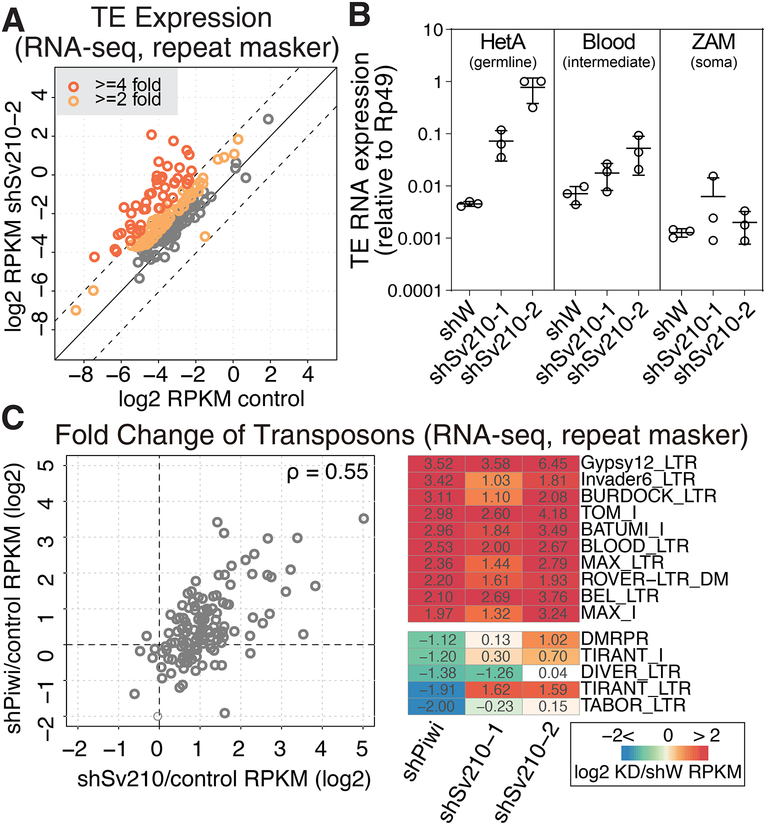
Embryonic ventralization is a known consequence of DNA damage in the ovary (Abdu et al., 2002; Ghabrial et al., 1998) that can be induced by several mechanisms including activation of endogenous transposable elements (Klattenhoff et al., 2007). We analyzed global changes in transposon and gene expression upon GLKD of Su(var)2–10 by RNA sequencing (RNA-seq). We observed strong upregulation of many transposon families upon Su(var)2–10 knockdown with both hairpins (Fig. 2A, S1A). Consistent with its stronger knockdown efficiency and phenotypic effect, the Sv210–2 hairpin induced broader and stronger TE upregulation (Fig. 2A, S1A). We validated these results by performing a further RNA-seq experiment of shSv210–2 and control ovaries in two independent biological replicates. Results showed a highly reproducible upregulation of TEs (Fig. S1B). Differential expression analysis showed that ~60% (118 of 195) of all TE families were more than 2-fold upregulated, with 30% more than 4-fold upregulated (FDR<0.05; Fig. S1C). To independently confirm changes in TE expression we measured expression of several transposon families by RT-qPCR. We found that the germline-specific transposon HetA is strongly upregulated in ovaries of both Su(var)2–10 GLKD lines (~15 and ~150 fold) (Fig. 2B). In contrast, Blood, a transposon expressed in both ovarian germline and soma, and ZAM, a transposon restricted to the somatic cells in the ovary, were not significantly affected. Together, these data suggest that Su(var)2–10 plays a major role in suppressing the activity of many transposon families in the Drosophila germline. As the piRNA pathway plays a central role in TE silencing in the germline (LeThomas et al., 2013; Rozhkov et al., 2013), we compared transposon expression upon Su(var)2–10 and Piwi GLKD. This analysis revealed that Su(var)2–10 and Piwi have similar target repertoires (Fig. 2C), suggesting that Su(var)2–10 may play a role in piRNA-mediated transposon repression. In addition, Su(var)2–10 GLKD affected the expression of approximately 10% of the host genes (Fig S1C). Detailed investigation of the role of Su(var)2–10 in host transcriptome regulation revealed complex TE-dependent and TE-independent effects that we describe in detail in the accompanying manuscript (Ninova et al., accompanying manuscript).
Figure 2. Germline depletion of Su(var)2–10 leads to transcriptional upregulation of transposons similar to those in Piwi depletion.
(A) Su(var)2–10 GLKD leads to global transposon derepression. Scatter plot shows log2-tansformed RPKM values for TEs (RepeatMasker) in RNA-seq data from Su(var)2–10 GLKD vs control (shW) ovaries. Dashed lines indicate 4-fold change. See also Figure S1.
(B) Su(var)2–10 GLKD de-represses germline-specific transposons. Dots correspond to three independent biological replicates (qRT-PCR); bars indicate the mean and SD.
(C) Piwi and Su(var)2–10 GLKD lead to de-repression of similar TEs. (Left) Fold changes in TE expression upon Piwi and Su(var)2–10 GLKD vs. control ovaries (shW) estimated by RNA-seq. Piwi GLKD data is average of two biological replicates, and Su(var)2–10 GLKD is average of shSv210–1 and shSv210–2. The Spearman’s correlation coefficient (rho) is shown. (Right) Fold changes of RNA expression in knockdown (Piwi or Su(var)2–10) versus control ovaries for the 10 most upregulated and 5 most downregulated TEs upon Piwi GLKD.
Su(var)2–10 depletion correlates with loss of repressive and gain of active histone marks over transposons
In the nucleus, piRNA-guided PIWI proteins induce co-transcriptional repression associated with trimethylation of histone H3 lysine 9 (H3K9me3), a repressive histone modification that serves as a binding site for heterochromatin protein 1 (HP1) (Bannister et al., 2001; Jacobs et al., 2001; Lachner et al., 2001). To test whether Su(var)2–10 is involved in transcriptional silencing of transposons, we analyzed the effect of Su(var)2–10 depletion on H3K9me3 and HP1 levels by ChIP-seq employing the stronger shSv210–2 hairpin.
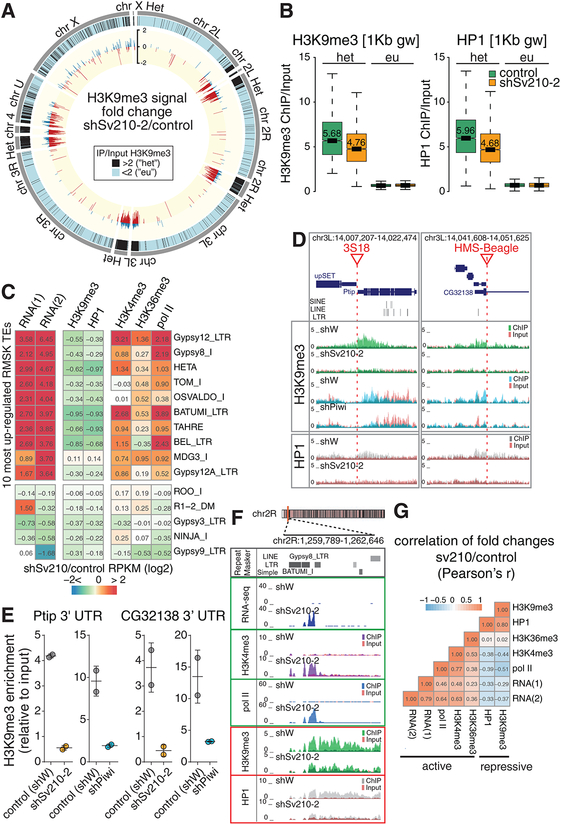
In wild-type flies we found high H3K9me3 levels in heterochromatic genomic regions, including pericentromeric and telomeric regions of the chromosomal arms (Fig. 3A, black bars). In addition, we identified H3K9me3 peaks scattered across euchromatic regions. Regions with elevated H3K9me3 signal are highly enriched in transposon sequences: a median of 70% of their sequence is annotated by RepeatMasker (Fig. S2), while transposons occupy less than 20 % of the whole reference genome (Kaminker et al., 2002). A subset of the detected H3K9me3 peaks in euchromatin lacks transposon sequences in the reference genome within 10 kb proximity. However, transposition of mobile elements might lead to new transposon insertions that are absent in the reference genome sequence but might be present in the genome of the strain used in our experiments. Analysis of de novo TE insertions in the genome of strains used in this study using the TIDAL pipeline(Rahman et al., 2015) identified 119 new transposon integrations, which were absent in the reference genome, residing within 5 kb of euchromatic H3K9me3 peaks (n=479). The association of H3K9me3 islands with non-reference TE integration is more frequent than expected by chance (p<1×10−6, permutation test). Thus, the H3K9me3 mark correlates with TE sequences in both eu- and heterochromatin.
Figure 3. Su(var)2–10 depletion induces ubiquitous H3K9me3 and HP1 loss associated with transposon de-repression.
(A) Genome-wide distribution and fold change of H3K9me3 upon Su(var)2–10 depletion. Outer grey tiles represent chromosome arms (dm3). Black tiles indicate 1Kb genomic windows where H3K9me3 ChIP/Input signal in two biological replicates >2, defined as “heterochromatic” (“het”). Inner circle shows the log2-transpormed H3K9me3 signal change (positive: blue; negative: red) in “het” regions in Su(var)2–10 GLKD vs. control ovaries. Data is average of two biological replicates.
(B) Su(var)2–10 depletion induces loss of H3K9me3 and HP1 from heterochromatic regions. Boxplots show the distributions of H3K9me3 (left) and HP1 (right) ChIP signal in H3K9me3-enriched (het, >2fold H3K9me3 enrichment in control ovaries), and euchromatic 1Kb genomic windows for Su(var)2–10 GLKD and control (shW) ovaries. Data is average of two biological replicates. Median values are shown.
(C) TE upregulation in Su(var)2–10 GLKD correlates with loss of repressive and gain of active transcription marks. Heatmap shows fold changes in RNA and chromatin marks (ChIP-seq) upon Su(var)2–10 GLKD for the 10 most upregulated and 5 most downregulated TEs upon Su(var)2–10 depletion. For RNA-seq, two independent samples using different Su(var)2–10 hairpins are shown. ChIP-seq data is average of two biological replicates using the shSv210–2 hairpin.
(D) H3K9me3 and HP1 loss upon Su(var)2–10 and Piwi KD at genomic regions adjacent to non-reference TE insertions. UCSC browser snapshots of two euchromatic loci with non-reference TE insertions (red lines). Tracks show RPM-normalized ChIP and Input signal (uniquely mapping reads) with tracks overlaid.
(E) qRT-PCR validation of the H3K9me3 loss near the euchromatic TE insertions shown in (D). Dots correspond to two independent biological replicates; bars indicate the mean and SD.
(F) Su(var)2–10 GLKD leads to increase of active marks and loss of repressive marks proximal to TEs. UCSC browser snapshot shows RPM-normalized RNA-seq and ChIP-seq signals (uniquely mapping reads) for indicated marks in Su(var)2–10 depleted (shSv210–2) and control (shW) ovaries. ChIP and input signal tracks are overlaid.
(G) Correlation between changes in active and repressive marks upon Su(var)2–10 GLKD. Heatmap shows all-versus-all correlation coefficients for fold changes of steady state RNA levels, active and repressive chromatin marks (ChIP-seq) upon Su(var)2–10 GLKD. Correlations are calculated based on values for TE families annotated by RepeatMasker with at least 10 RNA-seq reads in at least one condition (n=186 elements).
See also Figure S2.
Su(var)2–10 depletion caused a genome-wide reduction of the H3K9me3 mark: the majority (~80%) of H3K9me3-enriched genomic intervals displayed a decreased H3K9me3 signal and a concomitant decrease in HP1 level upon Su(var)2–10 GLKD (Fig. 3A, B). In line with the global loss of H3K9me3, analysis revealed a widespread decrease of H3K9me3 and HP1 signal on individual TE families, especially at transposons that show a strong derepression upon Su(var)2–10 knockdown (Fig. 3C). Regions flanking non-reference TE insertions in euchromatin also show prominent loss of H3K9me3 and associated HP1 upon Su(var)2–10 GLKD (Fig. 3D). Notably, the same regions exhibited a reduction of H3K9me3 upon Piwi depletion, as demonstrated by both ChIP-seq and -qPCR (Fig. 3D, E), indicating that they are controlled by both Piwi and Su(var)2–10.
We also assessed the effect of Su(var)2–10 GLKD (shSv210–2) on chromatin marks associated with active transcription, including the H3K4me3 mark that is present at active promoters, the elongation mark H3K36me3, as well as RNA polymerase II occupancy. Genome-wide ChIP-seq analysis of two biological replicas revealed that depletion of Su(var)2–10 results in an increase of active marks over transposon sequences and flanking regions (Fig. 3C, F). These changes correlate with the increase of transposon RNA levels, and the decrease of H3K9me3 and HP1 (Fig. 3G). Together, the loss of repressive histone marks and the gain of active marks upon Su(var)2–10 GLKD imply that Su(var)2–10 controls TE expression in the germline through transcriptional silencing.
Su(var)2–10 interacts with the piRNA/Piwi/Panx silencing complex and is required for its ability to induce transcriptional repression
The molecular mechanism of Piwi-induced transcriptional silencing remains poorly understood. Recent studies identified two proteins, Arx and Panx, that form a complex with Piwi and are required for transcriptional silencing of Piwi targets (Donertas et al., 2013; Muerdter et al., 2013; Ohtani et al., 2013; Sienski et al., 2015; Yu et al., 2015). Recruitment of Panx to a reporter locus in a piRNA-independent manner results in transcriptional repression and H3K9me3 deposition at the reporter (Sienski et al., 2015; Yu et al., 2015), providing a model system to study Piwi-induced silencing. We took advantage of this system to test whether Su(var)2–10 is required for Piwi repression downstream of Panx. As in previous reports, Panx tethering resulted in local H3K9me3 deposition at the reporter locus (Fig 4A). Su(var)2–10 GLKD did not alter Panx protein level (Fig. S3A) but reduced the ability of Panx to induce H3K9 trimethylation (Fig 4A). Thus, Su(var)2–10 acts downstream of Panx and is required for Panx-induced H3K9me3 deposition.
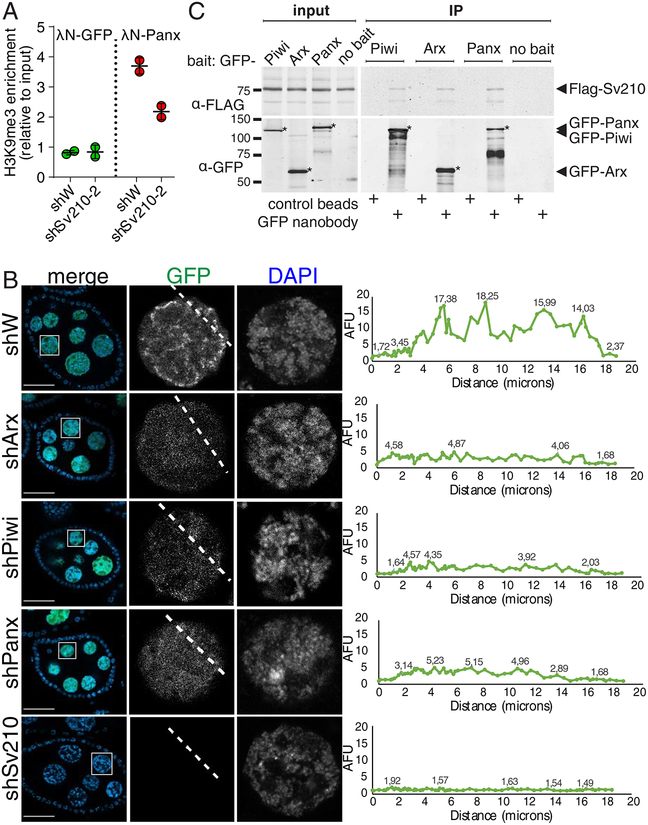
Figure 4. Su(var)2–10 genetically and biochemically interacts with Piwi, Arx and Panx.
(A) Su(var)2–10 GLKD abolishes H3K9me3 deposition induced by Panx tethering to luciferase reporter (Yu et al., 2015) locus. Plot shows the H3K9me3 enrichment (ChIP-qPCR) at the reporter upon tethering of Panx or GFP control in control (shW) or Su(var)2–10 GLKD ovaries. Dots correspond to two independent biological replicates; bars indicate the mean and SD.
(B) Nuclear localization of Su(var)2–10 depends on Arx, Panx and Piwi. Images show nurse cell nuclei (DAPI; red) of ovaries from flies expressing MT-Gal4-driven GFP-Su(var)2–10 (green) and short hairpins against Asterix (shArx), Piwi (shPiwi), Panoramix (shPanx), Su(var)2–10 (shSv210–2) or white (shW, control). Scale bar 30 μm.
(C) Su(var)2–10 interacts with Piwi, Arx and Panx. Protein lysates from ovaries expressing MT-Gal4 driven FLAG-Su(var)2–10 and GFP-fusion Piwi (endogenous promoter), Arx or Panx, were used for co-immunoprecipitation using GFP nanotrap or control beads. Ovaries expressing FLAG-Su(var)2–10 but no GFP partner (no bait) were used as an additional negative control.
To explore the genetic interactions between Su(var)2–10 and components of the Piwi-induced transcriptional silencing complex, we analyzed the subcellular localization of Su(var)2–10 in wild-type flies and upon disruption of Piwi-induced transcriptional silencing. Consistent with the reported chromatin association of Su(var)2–10 (Hari et al., 2001), we found that MT-Gal4 driven GFP-tagged Su(var)2–10 localizes to nurse cell nuclei, where it concentrates at discrete foci, possibly indicating binding at specific genomic sites. Depletion of Piwi, Panx or Arx in germ cells altered the localization of GFP-Su(var)2–10 to a uniform nuclear distribution (Fig 4B). While Su(var)2–10 GLKD dramatically reduced GFP-Su(var)2–10 level, KD of Piwi, Arx or Panx did not affect Su(var)2–10 protein level (Fig. S3B). The requirement of Piwi and its auxiliary factors Arx and Panx for proper localization of Su(var)2–10 further suggests that Su(var)2–10 acts downstream of these factors.
To test if Su(var)2–10 physically interacts with components of the Piwi/Arx/Panx complex, we employed a co-immunoprecipitation assay using ovaries from transgenic flies expressing tagged proteins. Su(var)2–10 co-purified with all three factors, Piwi, Panx and Arx (Fig. 4C), indicating that Su(var)2–10 associates with the Piwi/piRNA transcriptional silencing complex, although we cannot completely exclude that the interaction is caused by driving expression with a heterologous driver. The interaction of Su(var)2–10 with Piwi, Panx and Arx was further validated by co-expression and co-immunoprecipitation of tagged proteins from Drosophila S2 cells (Fig. S4A–C). Overall, our results indicate that Su(var)2–10 interacts with the Piwi-induced silencing complex both genetically and physically, and that it is required for Piwi-induced transcriptional repression.
Su(var)2–10 recruitment to a genomic locus induces transcriptional repression and H3K9me3 accumulation
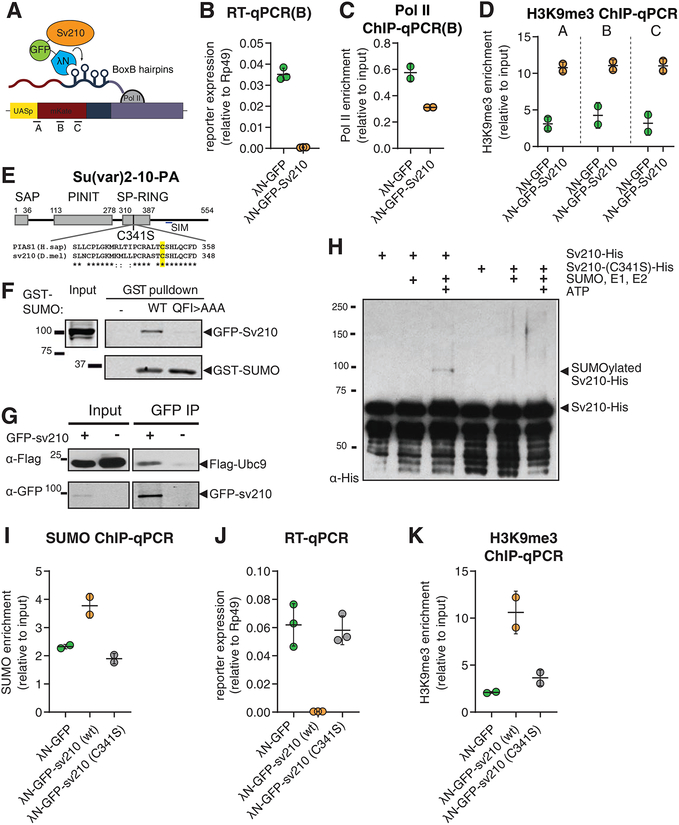
The requirement of Su(var)2–10 for transposon repression and Panx-induced reporter silencing suggests that Su(var)2–10 plays a role in transcriptional repression downstream of target recognition by the piRNA/Piwi/Panx complex. To test if Su(var)2–10 is able to induce local transcriptional repression when recruited to chromatin in germ cells, we used transgenic flies expressing Su(var)2–10 fused to the λN RNA-binding domain, and a reporter encoding BoxB hairpins in its 3’UTR region, under the control of UASp promoter. The λN domain has a high affinity for BoxB hairpins, allowing artificial tethering of Su(var)2–10 to the reporter (Fig. 5A). Tethering of λN–GFP-Su(var)2–10 caused severe (~130 fold) decrease in reporter mRNA expression compared to λN–GFP control, indicating that recruitment of Su(var)2–10 is sufficient to induce strong repression (Fig. 5B). Recruitment of Su(var)2–10 also resulted in an increase in the repressive H3K9me3 mark and a decrease in Pol II occupancy on the reporter as measured by ChIP-qPCR (Fig. 5C, D). Similar results were obtained when Su(var)2–10 was recruited to an alternate reporter containing a different sequence and integrated at another locus, indicating that the results are independent of reporter sequence and local genomic environment (Fig. S5A). These results indicate that recruitment of Su(var)2–10 to a genomic locus induces strong transcriptional repression associated with accumulation of the repressive H3K9me3 mark.
Figure 5. Su(var)2–10 recruitment induces transcriptional repression that depends on the SUMO pathway.
(A) Schematic diagram of the reporter used to study the effect of Su(var)2–10 recruitment to RNA. GFP-Su(var)2–10 fused to the RNA-biding λN domain, and the mKate reporter encoding 4 BoxB hairpins in the 3’UTR, are co-expressed in germ cells of the ovary (using the MT-Gal4 driver), resulting in Su(var)2–10 recruitment to the reporter’s nascent transcript. A-C denote different amplicons used for qPCR analysis.
(B-D) Tethering of Su(var)2–10 leads to reduced reporter mRNA level and Pol II occupancy, and increased H3K9me3 signal. Plots show reporter expression (B), Pol II occupancy (C), and H3K9me3 enrichment (D) upon tethering of λN-GFP-Su(var)2–10 or λN-GFP control. Dots correspond to independent biological replicates; bars indicate the mean and SD.
(E) Diagram of Drosophila Su(var)2–10 protein structure (PA isoform). Grey boxes mark conserved domains. Alignment of the SP-RING domain between human PIAS1 and Su(var)2–10 is shown, highlighting the catalytic cysteine residue identified in human PIAS1 (Kahyo et al., 2001).
(F) Su(var)2–10 interacts with SUMO. S2 cell lysates expressing GFP-Su(var)2–10 were incubated with bacterially expressed GST-SUMO (wild type), interaction-deficient mutant GST-SUMO (QFI>AAA), or no GST bait control. GST-SUMO was affinity purified using glutathione sepharose beads.
(G) Su(var)2–10 interacts with Ubc9. S2 cell lysates expressing FLAG-Ubc9 and GFP-Su(var)2–10 were immunoprecipitated using GFP nanotrap beads.
(H) Su(var)2–10 is SUMOylated in vitro. Bacterially purified His-Su(var)2–10 or His-Su(var)2–10 C341 mutant were incubated with SUMO, SUMO E1 and E2 ligases in the presence or absence of ATP.
(I) Tethering of Su(var)2–10 promotes SUMO accumulation at reporter locus. Plots show SUMO enrichment at the reporter locus upon tethering of λN-GFP control, wild type λN-GFP-Su(var)2–10 or λN-GFP-Su(var)2–10 C341 mutant estimated by ChIP-qPCR. Dots correspond to two independent biological replicates; bars indicate the mean and SD.
(J, K) Su(var)2–10 mutant (C341S) is unable to repress reporter transcription or induce H3K9me3 deposition. Plots show reporter expression (J) and H3K9me3 enrichment at the reporter locus (K) upon tethering of λN-GFP control, λN-GFP-Su(var)2–10 (wild type) or λN-GFP-Su(var)2–10 C341S mutant. Dots correspond to independent biological replicates; bars indicate the mean and SD.
See also Figure S5.
Su(var)2–10 is involved in the SUMO pathway
Su(var)2–10 is a member of the Siz/PIAS (Protein Inhibitor of Activated STAT) protein family (Betz et al., 2001; Hari et al., 2001; Mohr and Boswell, 1999). Genetic and biochemical studies showed that members of the PIAS protein family in yeast and mammals function in the SUMO pathway, which covalently attaches SUMO (Small Ubiquitin-like Modifier) to proteins to modify their activity (Johnson and Gupta, 2001; Kahyo et al., 2001; Kotaja et al., 2002; Sachdev et al., 2001; Schmidt and Muller, 2002; Takahashi et al., 2001). Yeast and mammalian PIAS proteins interact with SUMO and the E2 SUMO-conjugating enzyme Ubc9, and facilitate the transfer of SUMO from Ubc9 to substrates, thereby acting as SUMO E3 ligases (Johnson and Gupta, 2001; Kahyo et al., 2001; Kotaja et al., 2002; Sachdev et al., 2001; Schmidt and Muller, 2002; Takahashi et al., 2001). Siz/PIAS proteins have a highly conserved domain structure, which involves a Siz1/PIAS (SP)-RING domain that is responsible for their interaction with Ubc9 (Kahyo et al., 2001) (Fig. S5B). Su(var)2–10 has all the conserved domains present in yeast and mammalian members of the PIAS family, including the Siz/PIAS RING (SP-RING) domain, as well as a SUMO interaction motif (SIM) at its C-terminus, (Fig. S5B, Fig. 5E). In line with this, a yeast two hybrid screen showed that Su(var)2–10 directly interacts with Ubc9, SUMO, as well as the SUMO E1 ligase complex component Uba2 (Table S1). To validate Su(var)2–10 interaction with SUMO, we showed that Su(var)2–10 interacts with purified wild-type SUMO in vitro (Fig. 5F, S5C), however, it does not bind a SUMO mutant generated by changing three conserved residues (Q26A, F27A, I29A) essential for binding to the SUMO interaction motif (Zhu et al., 2008), confirming specificity of the interaction (Fig. 5F, S5C). We also validated the interaction of Su(var)2–10 and the E2 SUMO ligase Ubc9 by co-immunoprecipitation of tagged proteins expressed in S2 cells (Fig. 5G). The interactions of Su(Var)2–10 with SUMO and the E2 SUMO ligase Ubc9 indicate that it acts in the SUMO pathway and likely has a conserved function as a SUMO E3 ligase.
Several SUMO E3 ligases including members of the PIAS family were found to possess activity towards themselves (i.e., to promote self-SUMOylation) (Garcia-Dominguez et al., 2008; Ivanov et al., 2007; Kotaja et al., 2002; Schmidt and Muller, 2002; Takahashi and Kikuchi, 2005). In mammals, auto-SUMOylation of the KAP1 SUMO E3 ligase was shown to promote recruitment of silencing complex (Ivanov et al., 2007). Computational analysis identified three SUMOylation motifs located in the C-terminal domain of Su(var)2–10 adjacent to its SP-RING domain (Fig. S5D). To test whether Su(var)2–10 is SUMOylated, we co-expressed GFP-tagged Su(var)2–10 and 3XFlag-tagged SUMO in S2 cells, and immunopurified GFP-Su(var)2–10 using stringent washing conditions to eliminate non-covalently bound proteins. Western blotting analysis showed additional higher molecular weight bands corresponding to SUMOylated Su(var)2–10 that were dependent on the presence of the SUMO peptidase inhibitor N-ethylmaleimide (NEM) (Fig. S5E). In an in vitro SUMOylation assay using purified Su(var)2–10, SUMO, E1 and E2 enzymes, we observed a higher molecular weight Su(var)2–10 band in the presence of ATP, confirming that Su(var)2–10 is SUMOylated (Fig. 5H). The interaction with Ubc9 and therefore the SUMO E3 ligase activity of PIAS proteins was shown to be abolished by mutating a single conserved cysteine residue in the SP-RING domain in yeast, plant and human (Fig 5E, Fig S5B) (Crozet et al., 2016; Kahyo et al., 2001; Munarriz et al., 2004; Takahashi et al., 2001). This cysteine residue is conserved in the SP-RING domain of Drosophila Su(var)2–10. We generated a mutant carrying a Cys to Ser substitution (C341S) and purified mutant protein after expression in E. coli. In contrast to wild type Su(var)2–10, the mutant did not become SUMOylated in vitro, indicating that the function of the SP-RING domain of Su(var)2–10 is similar to its role in other PIAS proteins (Fig. 5H). Together our results suggest that Su(var)2–10 acts as an intra-molecular SUMO E3 ligase, analogous to KAP1 in mammals.
The repressive function of Su(var)2–10 depends on the SUMO pathway
To explore whether Su(var)2–10 can promote SUMOylation of chromatin in vivo, we assessed the level of SUMO on chromatin upon recruitment of Su(var)2–10 to the reporter locus. ChIP-qPCR using an antibody against the endogenous Drosophila SUMO protein (smt3) (Gonzalez et al., 2014) showed that tethering of Su(var)2–10 leads to increased SUMO signal at the reporter, supporting its function as a SUMO E3 ligase that modifies chromatin targets (Fig. 5I). Next, we asked if the SUMO E3 ligase activity of Su(var)2–10 is important for its function in transcriptional repression. Tethering of Su(var)2–10 C341S mutant failed to promote SUMOylation at the reporter locus, confirming that this mutation abolishes SUMO E3 ligase activity of Su(var)2–10 (Fig. 5I). Tethering of wild-type Su(var)2–10 reproducibly induced over 100-fold reporter repression. In contrast, tethering of the Su(var)2–10 C341S did not affect reporter expression or H3K9me3 level at its genomic locus, indicating that the SUMO-ligase activity of Su(var)2–10 is essential for its function in transcriptional repression (Fig. 5J, K). Next, we addressed the role of different Su(var)2–10 domains in its repressive activity (Fig. S5F). As expected, complete deletion of the SP-RING domain impaired reporter silencing. Deletion of the PINIT domain also abolished the silencing activity. Conversely, deletion of the SAP domain, which was proposed to bring PIAS proteins to some of their targets through binding to DNA (Reindle et al., 2006), caused reporter repression at levels comparable to those induced by the wild-type protein. These results indicate that transcriptional repression by Su(var)2–10 depends on its SP-RING domain and SUMO E3 ligase activity. To directly test if SUMO is required for Su(var)2–10-induced transcriptional repression, we studied the ability of Su(var)2–10 to induce silencing when SUMO is depleted. Knockdown of the single Drosophila SUMO gene (smt3) in germ cells released repression of the reporter caused by Su(var)2–10 recruitment ~10 fold (Fig. 6A). Thus, transcriptional silencing caused by Su(var)2–10 recruitment to chromatin depends on the SUMO pathway and correlates with local accumulation of SUMO.
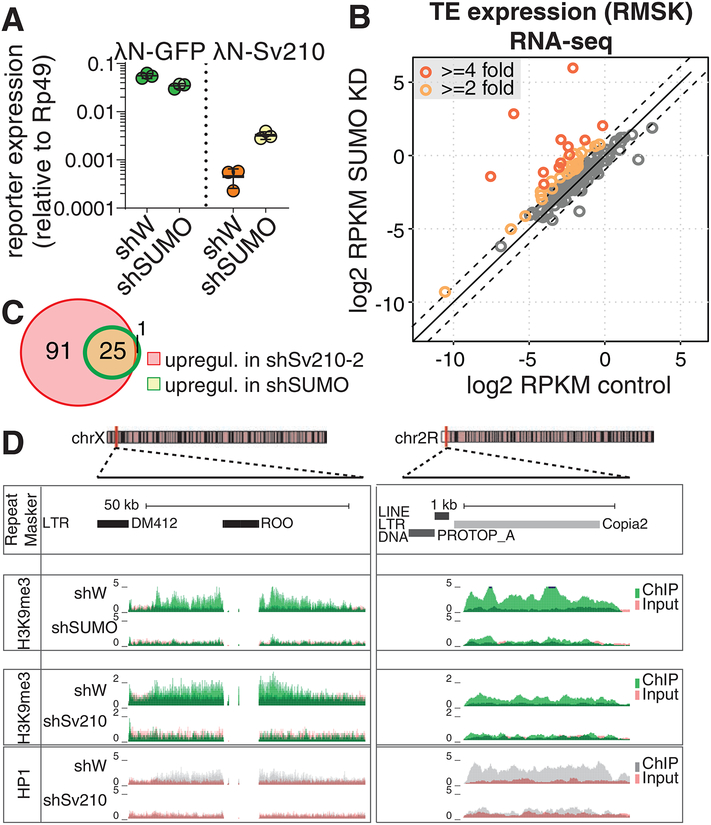
Figure 6. SUMO is required for Su(var)2–10 mediated reporter silencing and global regulation of TEs.
(A) SUMO is required for Su(var)2–10 mediated reporter silencing. Plots show reporter expression upon tethering of λN-GFP or λN-GFP-Su(var)2–10 in SUMO GLKD or control (shW) ovaries measured by RT-qPCR (region B). Dots correspond to three independent biological replicates; bars indicate the mean and SD.
(B) SUMO depletion leads to global transposon de-repression. Scatter plots show log2-tansformed RPKM values for TEs (RepeatMasker annotations) in RNA-seq data from SUMO GLKD versus shW control ovaries. Dashed lines indicate 2-fold change. Data is average from two biological replicates.
(C) Venn diagram shows the numbers and overlap of significantly upregulated TE families upon Su(var)2–10 and SUMO GLKD as determined by DESeq2 (FDR<0.05, log2 fold change ≥2).
(D) SUMO depletion leads to loss of H3K9me3 signal at transposon loci. UCSC browser snapshots showing RPM-normalized H3K9me3 and HP1 ChIP-seq signal (uniquely mapping reads) at TE-rich genomic regions in control (shW), Su(var)2–10 and SUMO GLKD ovaries. ChIP and input signal tracks are overlaid.
To determine whether the SUMO pathway is required for piRNA/Piwi mediated transcriptional silencing, we investigated the effect of SUMO depletion on transposon expression in the germline. Germline depletion of smt3 resulted in sterility, phenocopying the effect of knockdowns of Su(var)2–10 and other piRNA pathway mutants. Global RNA-seq followed by DESeq2 analysis revealed upregulation of 26 transposable element families (>2-fold increase, FDR<0.05) (Fig. 6B). Notably, the set of transposons that are depressed upon SUMO depletion nearly completely overlaps with elements upregulated in Su(var)2–10 knockdown, suggesting that Su(var)2–10 and SUMO act on the same targets (Fig. 6C). Furthermore, ChIP-seq analysis showed loss of H3K9me3 from TE sequences and flanking regions upon smt3 GLKD, as exemplified in Fig. 6D. Thus, SUMO is crucial for transcriptional repression of TEs in the germline.
Su(var)2–10-induced transcriptional repression requires the histone methyltransferase complex SetDB1/Wde
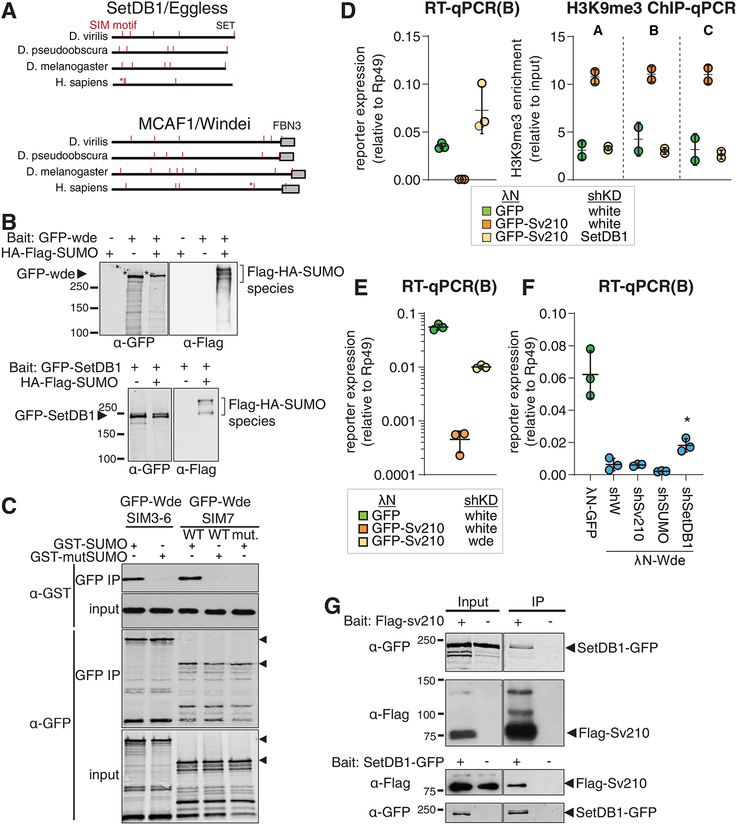
H3K9me3 deposition induced by Piwi and Panx requires the activity of the methyltransferase SetDB1 (Sienski et al., 2015; Yu et al., 2015). We found that repression by Su(var)2–10 depends on the SUMO pathway. Previous reports indicate that the mammalian homolog of SetDB1, as well as its co-factor MCAF1/ATF7IP, have SUMO-interaction motifs (SIM) (Ivanov et al., 2007; Stielow et al., 2008b; Thompson et al., 2015; Uchimura et al., 2006). To explore whether the SUMO pathway plays a role in recruitment of SetDB1 to chromatin in Drosophila, we analyzed the sequences of fly SetDB1 and its conserved co-factor, Windei (Wde) (Koch et al., 2009). Computational analysis (Zhao et al., 2014) identified several canonical SIMs in both proteins (Fig. 7A,S6A), which are conserved between D. melanogaster and other Drosophilid species separated by 30–60 My of evolution (Fig. 7A,S6A). While the fly and human SetDB1 and Wde proteins have little sequence homology outside of conserved domains, the relative position of some SIMs is preserved (Fig. 7A, S6A). To test if Drosophila SetDB1 and Wde interact with SUMOylated proteins, we co-transfected S2 cells with tagged SetDB1 or Wde and SUMO followed by purification of SetDB1 and Wde complexes. Western blotting showed that both SetDB1 and Wde co-purify with several SUMOylated proteins (Fig. 7B). To further explore the interaction between Wde and SUMO, we expressed SIM-containing fragments of Wde in S2 cells and incubated the lysate with purified GST-SUMO. Wde fragments co-purified with wild type, but not mutant SUMO (Q26A, F27A, I29A), which cannot interact with SIMs. Mutations in the SIM of Wde also abolished its interaction with SUMO, indicating that Wde binds SUMO directly via its SIM (Fig. 7C). Thus, the SetDB1/Wde complex directly and specifically binds SUMO.
Figure 7. Drosophila SetDB1 is required for Su(var)2–10 mediated reporter silencing and physically interacts with Su(var)2–10.
(A) Drosophila SetDB1 and Wde contain predicted SIMs. Diagrams show computationally predicted SIMs (red marks) of SetDB1 and Wde homologs in three representative Drosophila species and in human. Asterisks mark the previously described functional SIM of human SetDB1 and Wde (Ivanov et al., 2007; Uchimura et al., 2006). Grey boxes show position of conserved domains.
(B) SetDB1 and Wde interact with SUMOylated proteins. Total protein lysates from S2 cells co-expressing FLAG-HA-SUMO and GFP-SetDB1 or GFP-Wde were immunopurified using anti-GFP nanotrap beads. Cells not expressing FLAG-HA-SUMO were used as negative control.
(C) Wde interacts with SUMO through its SIM motifs. Total protein lysates from S2 cells expressing GFP-Wde fragments including SIM 3–6 and SIM 7 were incubated with recombinant GST-SUMO and immunopurified using anti-GFP nanotrap beads. SIM interaction deficient SUMO mutant and Wde SIM 7 mutant were used to probe specificity of interactions.
(D) SetDB1 is required for Su(var)2–10-induced reporter repression and H3K9me3 deposition at reporter locus. Plots show expression (RT-qPCR) and H3K9me3 enrichment (ChIP-qPCR) at the reporter locus upon tethering of λN-GFP, or λN-GFP-Su(var)2–10 in control (shW) and SetDB1-depleted ovaries. λN-GFP, shW and λN-GFP-Su(var)2–10, shW data is the same as in Fig 5B, D. Dots represent independent biological replicates; bars show mean and SD.
(E) Wde is required for Su(var)2–10 induced reporter repression. Plot shows reporter expression upon tethering of λN-GFP, or λN-GFP-Su(var)2–10 in Wde GLKD or control (shW) ovaries. Dots correspond to three independent biological replicates; bars indicate the mean and SD.
(F) Su(var)2–10 and SUMO are not required for Wde-induced repression. Plot shows reporter expression upon tethering of λN-GFP, or λN-GFP-Wde in ovaries depleted of Su(var)2–10, SUMO or SetDB1 by shRNAs. *λN-GFP-Wde mediated repression is significantly reverted only upon SetDB1 GLKD (p<0.01, single factor ANOVA followed by Tukey post-hoc test). Dots correspond to three independent biological replicates; bars indicate the mean and SD.
(G) Su(var)2–10 interacts with SetDB1. Protein lysates from ovaries expressing FLAG-Su(var)2–10 and GFP-SetDB1 were immunoprecipitated with either anti-FLAG or anti-GFP nanotrap beads. In each experiment, lysate from ovaries not expressing the bait protein was used as negative control.
See also Figure S6.
To define the place of Su(var)2–10 in the Piwi silencing pathway, we tested if Su(var)2–10 induced repression is dependent on SetDB1 and Wde. We tethered Su(var)2–10 to the reporter and depleted SetDB1 or Wde in germ cells using shRNA. SetDB1 GLKD abolished Su(var)2–10-induced reporter repression and H3K9me3 deposition (Fig. 7D). Similarly, Wde GLKD resulted in partial release of reporter silencing (Fig. 7E). Thus, the transcriptional repression caused by Su(var)2–10 depends on H3K9me3 deposition by the SetDB1/Wde histone methyltransferase complex.
As SetDB1 and Wde have SUMO-interaction motifs, SUMOylation of chromatin-associated proteins by Su(var)2–10, including Su(var)2–10 itself, might promote recruitment of the SetDB1/Wde complex to target loci. Alternatively, or in addition, interaction with SUMOylated proteins might enhance the histone methyltransferase activity of SetDB1/Wde. To test the latter possibility, we decided to tether Wde to the reporter locus in a SUMO-independent manner and probe the involvement of Su(var)2–10 in its silencing activity. Tethering of Wde induced strong reporter repression in ovarian germ cells (Fig. 7F). This repression was dependent on SetDB1, but independent of Su(var)2–10 and SUMO, as reporter silencing was unaffected by depletion of the latter two proteins. This result suggests that SUMO and Su(var)2–10 do not impact the enzymatic activity of SetDB1/Wde and are instead involved in its recruitment to chromatin targets. Co-immunoprecipitation from ovarian lysate and from S2 cells showed that Su(var)2–10 and SetDB1 interact in vivo (Fig. 7G, S6B). Taken together, our results suggest that SUMOylation of protein targets by Su(var)2–10 – including SUMOylation of Su(var)2–10 itself – provides a binding platform for the recruitment of SetDB1/Wde to induce H3K9 trimethylation and transcriptional repression.
Discussion
In both insect and mammals, piRNA-guided transcriptional silencing is associated with the deposition of repressive chromatin marks on genomic targets (LeThomas et al., 2013; Pezic et al., 2014; Rozhkov et al., 2013; Sienski et al., 2012). In Drosophila the conserved histone methyltransferase SetDB1 (Egg) is responsible for deposition of the silencing H3K9me3 mark at Piwi targets (Sienski et al., 2015; Yu et al., 2015). However, the molecular mechanism leading to the recruitment of SetDB1 by the Piwi/piRNA complex remained unknown. Here, we showed that in Drosophila SUMO and the SUMO E3 ligase Su(var)2–10 act together downstream of the piRNA-guided complex to recruit the histone methyltransferase complex SetDB1/Wde and cause transcriptional silencing. Our results suggest a model for the molecular mechanism of piRNA-guided transcriptional silencing in which Su(var)2–10 provides the connection between the target recognition complex composed of piRNA/Piwi/Panx/Arx and the chromatin effector complex composed of SetDB1 and Wde.
We identified a new role for the SUMO pathway in piRNA-guided transcriptional silencing. The SUMO pathway plays important roles in heterochromatin formation/maintenance and genome stability in different organisms from yeast to humans. Among different functions, SUMO is required for recruitment and activity of the histone methyltransferase complex composed of SetDB1 and MCAF1 (Wde in Drosophila), which confers transposon silencing in mammals (Ivanov et al., 2007; Stielow et al., 2008b; Thompson et al., 2015; Uchimura et al., 2006). Remarkably, SUMO-dependent recruitment of SetDB1 to TEs in mammalian somatic cells does not require piRNAs but is instead mediated by the large vertebrate-specific family of Krüppel-associated box domain-zinc finger proteins (KRAB-ZFPs) that bind specific DNA motifs (reviewed in (Wolf et al., 2015)). Although distinct members of the KRAB-ZFP family recognize different sequence motifs in target transposons, repression of all targets by various KRAB-ZFPs requires the universal co-repressor KAP1/TIF1b (KRAB-associated protein 1). KAP1 is a SUMO E3 ligase and its auto-SUMOylation leads to SetDB1 recruitment (Ivanov et al., 2007). Our results suggest that Drosophila Su(var)2–10 can be SUMOylated (Fig. 5H, S5E), and SetDB1 and Wde have functional SIMs (Fig 7), suggesting that Su(var)2–10 auto-SUMOylation might induce SetDB1/Wde recruitment (see graphical abstract). Our results suggest that two distinct transposon repression pathways – by DNA-binding proteins and by piRNAs, both rely on SUMO-dependent recruitment of the conserved silencing effector to the target.
Our results in Drosophila and studies in mammals (Ivanov et al., 2007) suggest that in both clades self-SUMOylation of SUMO E3 ligases might be involved in recruitment of SetDB1 to chromatin. However, these results do not exclude the possibility that the recruitment of SetDB1 is facilitated by SUMOylation of additional chromatin proteins by Su(var)2–10. Studies in yeast led to the ‘SUMO spray’ hypothesis that postulates that SUMOylation of multiple different proteins localized in physical proximity promotes the assembly of multi-unit effector complexes (Psakhye and Jentsch, 2012). Local concentration of multiple SUMO moieties leads to efficient recruitment of SUMO-interacting proteins. According to this hypothesis multiple SUMO-SIM interactions within a protein complex act synergistically, thus SUMOylation of any single protein is neither necessary nor sufficient to trigger downstream processes (Jentsch and Psakhye, 2013; Psakhye and Jentsch, 2012). Assembly of such ‘SUMO spray’ on chromatin might be governed by the same principles of multiple weak interactions as was recently recognized for the formation of various phase-separated liquid-droplet compartments in the cell (Shin and Brangwynne, 2017). The presence of Su(var)2–10 on a chromatin locus might lead to SUMOylation of multiple chromatin-associated proteins that are collectively required for the recruitment of effector chromatin modifiers. The SUMOylation consensus (ΨKxE/D) is very simple and therefore quite common in the fly proteome. Consistent with this, several hundred SUMOylated proteins were identified in proteomic studies in Drosophila (Handu et al., 2015; Nie et al., 2009). Thus, it is possible that collective SUMOylation of multiple chromatin-associated proteins contributes to recruitment and stabilization of the SetDB1 complex on chromatin.
The cascade of events leading to repression initiated by target recognition by piRNA/Piwi, followed by interaction with Su(var)2–10 and subsequent SUMO-dependent recruitment of SetDB1/Wde suggest that the three complexes tightly cooperate. But do these three complexes (Piwi, Su(var)2–10 and SetDB1) always work together, or does each complex have additional functions independent of the other two? Genome-wide analysis suggests that the vast majority of Piwi targets are repressed through SUMO/Su(var)2–10 and, likely, SetDB1/Wde, suggesting that Piwi always requires these other complexes for its function in transcriptional silencing. On the other hand, we found multiple instances of host genes that are repressed by Su(var)2–10 and SetDB1, but do not require piRNAs (Ninova et al., accompanying manuscript). Su(var)2–10 and SetDB1 are also expressed outside of the gonads and were implicated in chromatin silencing in somatic tissues that lack an active piRNA pathway (Brower-Toland et al., 2009; Hari et al., 2001; Seum et al., 2007; Stampfel et al., 2015; Stielow et al., 2008a; Tzeng et al., 2007). We speculate that Su(var)2–10 might bind to specific targets directly though its SAP domain or get recruited by specific DNA-binding proteins, similarly to the way SetDB1 is recruited to ERVs by KRAB-ZFP in mammals, though specific factors are yet to be uncovered.
Though both Drosophila and mouse have nuclear Piwi proteins involved in transcriptional silencing of transposons, these proteins, PIWI and MIWI2, are not one-to-one orthologs. Unlike Drosophila, other insects including the silkworm Bombyx mori, the flour beetle Tribolium castaneum and the honeybee Apis mellifera encode only two Piwi proteins and, at least in B. mori, these proteins do not localize to the nucleus (Nishida et al., 2015). These observations suggest that the nuclear Piwi pathway in Drosophila has evolved independently in this lineage. In light of this evolutionary interpretation, the interaction of the Piwi complex and the E3 SUMO ligase Su(var)2–10 indicates that in Drosophila the nuclear piRNA pathway co-opted an ancient mechanism of SUMO-dependent recruitment of the histone modifying complex for transcriptional silencing of transposons. The molecular mechanism of piRNA-induced transcriptional repression in other clades such as mammals might have evolved independently of the corresponding pathway in flies. It will be interesting to investigate if mammals also use SUMO-dependent recruitment of silencing complexes for transcriptional repression of piRNA targets.
STAR METHODS
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexei Aravin (aaa@caltech.edu).
Experimental model and subject details
Drosophila stocks
All Drosophila stocks were maintained at 24°C and 80% humidity on standard media. Female flies were put on yeast for 2 to 3 days before ovary dissection and were at age 3–14 days. Females of the same age, genotype and generation were randomly assigned to biological replicates. The stocks for shRNAs of Su(var)2–10 (shSv210–1 and shSv210–2, BDSC #32915 and BDSC #32956, respectively), piwi (shPiwi, BDSC #33724), wde (shWde, BDSC #33339) and white (shWhite, BDSC #33623) were obtained from the Bloomington Drosophila Stock Center. UASp-mKate2-4xBoxB-K10polyA, UASp-λN-GFP-eGFP control, GFP-Piwi, GFP-Arx and shPiwi were described previously (Chen et al., 2016; LeThomas et al., 2013). shSetDB1, shPanx, λN-Panx and Tubulin-BoxB reporter stocks were gifts from Julius Brennecke, the luciferase 10BoxB reporter is a gift from Gregory Hannon. To obtain the shAsterix line, the short hairpin sequence was ligated into the pValium20 vector (Ni et al., 2011) using T4 DNA ligase from NEB (M0202), according to the manual, and then integrated into the attP2 landing site (BDSC #25710). Hairpin sequences are listed in Key Resources/Oligonucleotides. shSmt3 (shSUMO) was reconstructed based on the TRiP line HMS01540 and integrated into the attP2 landing site (BDSC #8622). For all other Drosophila lines generated in this study, respective full length cDNA sequences or mutants were cloned in pENTR™/D-TOPO® (Invitrogen) entry vectors and transferred to Gateway® destination vectors containing attB site, a miniwhite marker followed by UASp promoter sequence, and GFP or λN-GFP upstream the gateway cassette, or GFP downstream the gateway cassette. Transgenic flies carrying these constructs were generated by phiC31 transformation at BestGene Inc. UASp-SetDB1-GFP was integrated into attP-3B landing site (BDSC #9750). UASp-λN-GFP-Su(var)2–10-PA was integrated into attP9A landing site (BDSC #9736). UASp-λN-GFP-Panx, UASp-GFP-Arx, UASp-λN-GFP-Arx, UASp-FLAG-Su(var)2–10, UASp-λN-GFP-Su(var)2–10-C341S, UASp-λN-GFP-Su(var)2–10-ΔSAP, UASp-λN-GFP-Su(var)2–10-ΔPINIT, UASp-λN-GFP-Su(var)2–10-ΔSP-RING were integrated into the attP40 landing site (y1 w67c23; P{CaryP}attP40). The expression of all constructs was driven by maternal alpha-tubulin67C-Gal4 (MT-Gal4) (BDSC #7063 or #7062), except for the experiment of Su(var)2–10 depletion in the ovarian soma where Tj-gal4 (DGRC #104055) driver was used. For eggshell phenotyping, freshly laid eggs were mounted in 1XPBS and manually counted under a dissecting microscope.
S2 cells
S2 cells were cultured at 25°C in Schneider’s Drosophila Medium containing 10% heat-inactivated FBS and 1X Penicillin-Streptomycin.
Method details
Imaging
Ovaries from Drosophila lines expressing UASp-λN-GFP-Su(var)2–10 and UASp-driven shRNAs against white, Arx, Panx and Piwi under the control of the maternal-tubulin-Gal4 (MT-Gal4) driver were fixed in PBS supplemented with 4% formaldehyde for 20 minutes at room temperature with end-to-end rotation. Samples were washed three times 10 minutes with PBS, and mounted in Prolong Gold Antifade Mountant with DAPI. Imaging was performed using a Zeiss LSM 880 confocal microscope and data was processed using Fiji (Schindelin et al., 2012).
Co-immunoprecipitation and western blot from ovaries
For immunoprecipitation (IP), 50–70 pairs of freshly dissected ovaries were lysed with a douncer in 500μl lysis buffer (0.2% NP40, 20 mM Tris pH7.4, 150 mM NaCl, 10% glycerol) supplied with protease inhibitor (Roche, 11836170001) and 20 mM deSUMOylation inhibitor N-Ethylmaleimide (Sigma, E3876). For Piwi/Arx/Panx coIP with Su(var)2–10 the lysis buffer contained 0.4% NP40. For GFP IP, lysates were incubated with GFP-Trap® or control (ChromoTek) magnetic agarose beads for 1–2 hr at 4°C with end-to-end rotation. For FLAG IP, lysates were incubated with anti-FLAG M2 ® magnetic beads (Sigma M8823). After incubation, the beads were washed 5 times with 500μl wash buffer (0.1% NP40, 20 mM Tris pH7.4, 150mM NaCl) containing protease inhibitor and 20 mM N-Ethylmaleimide. For Piwi/Arx/Panx coIP with Su(var)2–10 the wash buffer contained 250mM NaCl and 0.2% NP40. The washed beads were boiled in 75 μl SDS-PAGE sample buffer, and then the supernatant was used for western blot analysis. Western blots were carried out using the following antibodies: anti-FLAG [Sigma, A8592], anti-GFP [ab290], or rabbit polyclonal anti-GFP (Chen et al., 2016), and HRP-conjugated or IRDye® anti-rabbit and anti-mouse secondary antibodies (Li-cor #925–68070 and −68071, 925–32210 and −32211, Cell Signalling 7074, 7076).
Immunoprecipitation from S2 cells
Expression vectors encoding GFP-fusion and FLAG-fusion proteins under the control of the Actin promoter were generated from entry cDNA clones transferred to pAGW or pAFW destination vectors from the Drosophila Gateway™ Vector collection using the Gateway® system. GFP-wde expression vector was a generous gift from Andreas Wodarz (Koch et al., 2009). S2 cells were transfected with TransIT-LT1 (Mirus). 24–48h post transfection cells were harvested and lysed in lysis buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.2% NP-40, 0.2% Triton-X, 5% glycerol), supplemented with protease inhibitor cocktail (Roche, 11836170001) and 20mM N-Ethylmaleimide (NEM). Co-immunoprecipitation experiments and western blots were performed as described for ovarian tissue.
For SUMO-modified Su(var)2–10 detection, cells were lysed in RIPA-like buffer (20mM Tris pH7.4, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail Roche, 11836170001, with or without 20mM NEM), and washed with lysis buffer supplement to 500 mM NaCl, 0.5% SDS, with or without 20 mM NEM.
GST-SUMO interaction assays
pGEX-2TK vectors (GE Healthcare) expressing GST-SMT3 (SUMO) (plasmid was a generous gift from G Suske (Stielow et al., 2008a)) and SIM interaction deficient GST-SMT3 generated in our lab were transformed in E. coli strain BL21 and purified by glutathione affinity chromatography using a standard protocol. In brief, Glutathione Sepharose 4B (GE Healthcare) slurry was equilibrated using five bead volumes of 50 mM Tris-HCl pH 7.4, 150 mM NaCl. Bacteria were lysed in 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM buffer using a French press, and slurry was incubated with bacterial lysate at 4°C with end - over-end rotation. After three washes with the same buffer, the fusion proteins were eluted in a buffer containing 50 mM Tris HCl (pH 8.0), 150 mM NaCl and 20 mM reduced glutathione. Eluates were dialysed with 10 kDa cut-off dialysis tubing against Tris-HCl buffer (50 mM Tris-HCl, pH 7,5, 500 mM NaCl and 1 mM DTT) overnight at 4°C.
S2 cells transfected with plasmid encoding either GFP-Su(var)2–10-PA, GFP-tagged truncated Wde including predicted SIM-s 3 to 6 (aa385–655), GFP-tagged truncated Wde including SIM 7 (aa1058–1310), or GFP-tagged truncated Wde with mutated SIM 7 (1202DL>AA) (see Figure S6A for Wde map and SIM motif annotations) under the control of Actin promoter. Cells were harvested 24–48h post transfection.
For Su(var)2–10-SUMO interaction, cells were lysed in RIPA buffer supplemented with protease inhibitor (Roche 11836170001) and 20 mM NEM. Cleared lysates were diluted 1:10 with binding buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% NP40) and pre-cleared by rotation with Glutathione Sepharose 4B slurry (GE Healthcare) for 1 hour at 4°C with end-to-end r otation. Lysates were divided in equal parts and incubated with 2 μg purified GST-SUMO(wild type), GST-SUMO(mutant) and 10 μl Glutathione Sepharose 4B slurry for 2 hours at 4°C with end-to-end rotati on. Beads were washed 4 times for 10 minutes with binding buffer and boiled in SDS-PAGE protein loading buffer. Bound proteins were analysed by western blot using anti-GFP antibody.
For Wde-SUMO interactions, cells were lysed in lysis buffer (20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.2% NP-40, 0.1% Triton-X, 5% glycerol), supplemented with protease inhibitor (Roche 11836170001) and 20 mM NEM. Cleared lysates were incubated with 2–3 μg GST-SUMO for 2 hr, at 4°C with end-to-end rotatio n. Next, lysates were incubated with GFP Nanotrap beads (Chromotek), pre-blocked with 0.5% BSA and untransfected S2 cell lysates, for 1 hr at 4°C with end-to-end rotation. Beads were washed 5 times for 10 minutes at 4°C with wash buffer (0.1% NP-40, 20 mM Tris-HCl pH 7.4, 150 mM NaCl) and finally boiled in SDS-PAGE protein loading buffer. Bound proteins were analysed by western blot using anti-GFP antibody (ab290, Abcam) and anti-GST antibody (Cell Signalling #2622).
In vitro SUMOylation assay
His-Su(var)2–10 was cloned into the pET24a vector and expressed in E. coli strain BL21. Protein was purified using His-Pur Ni-NTA Resin (Thermo Scientific), equilibrated using 2 bed volumes of PBS pH=7.4. The fusion protein was eluted in a buffer containing PBS pH=7.4 and 250 mM imidazole. The elution fractions were dialysed using 10 kDa cut-off dialysis tubing in PBS supplemented with 1 mM DTT overnight at 4°C. After dialysis protein was concentrated usi ng 10kDa MWCO Amicon filter (Millipore). In vitro SUMOylation assay was performed using SUMO2 Conjugation Kit (BostonBiochem, K-715) according to the manufacturer’s instruction, using 5μl 34μM stock of recombinant Su(var)2–10. Reaction without ATP was set up as a negative control. Results were detected using Western Blotting using anti-His primary antibody (ThermoFisher, His.H8).
Yeast two hybrid screen
Yeast two hybrid screen was performed by Hybrigenics (ULTImate Y2H™ service) using the full-length Su(var)2–10-PA isoform fused at its N-terminus to GAL4 DNA binding domain as a bait and a Drosophila ovary cDNA library as prey.
RNA extraction and RT-qPCR
All RT-qPCR experiments were performed using 3 biological replicates per genotype of hand-dissected 10–20 pairs of ovaries. RNA was isolated with Ribozol (Amresco, N580) and treated with DNaseI (Invitrogen, 18068–015). Reverse transcription was carried out using Superscript III (Invitrogen) with random hexamers. qPCR was performed on a Mastercycler®ep realplex PCR thermal cycler machine (Eppendorf). Primers used in qPCR are listed in Key Resources/Oligonucleotides.
RNA-seq
For RNA-seq, ovarian total RNA (10–15 μg) from shW, shSv210–1, shSv210–2, and shSmt3 GLKD lines was depleted of ribosomal RNA with the Ribo-Zero™ rRNA Removal Kit (Epicentre/Illumina). Initial RNA-seq libraries were made using the TruSeq RNA prep kit by Illumina (shW, shSv210–1, shSv210–2). A second set of libraries from shW, shSv210–2 and shSmt3 in two biological replicates were made using the NEBNext® Ultra™ Directional RNA Library Prep Kit. Libraries were sequenced on the Illumina HiSeq 2000/2500 platform. RNA-seq data from shPiwi and corresponding shW control lines were previously published (LeThomas et al., 2013).
ChIP-seq and ChIP-qPCR
All ChIP experiments involving Su(var)2–10 GLKD were performed using the shSv210–2 hairpin and shW control in two biological replicates. ChIPs were carried out as described previously (LeThomas et al., 2014) with the following antibodies: anti-H3K9me3 [ab8898], anti-RNA Pol II [ab5408], anti-H3K4me2/3 [Ab6000], anti-H3K36me3 [ab9050], HP1 [C1A9, DSHB] and anti-Drosophila SUMO (smt3), a kind gift from G Cavalli(Gonzalez et al., 2014). ChIP-seq library construction was carried out using the NEBNext ChIP-Seq Library Prep Master Mix Set (E6240) with minor modifications. After adaptor ligation and PCR amplification, size selections were done on a 2% agarose gel to select the 200bp-400bp size window. Libraries were sequenced on the Illumina HiSeq 2000/2500 platform (SR 49bp or 50bp). ChIP-qPCR was performed on a Mastercycler®ep realplex PCR thermal cycler machine (Eppendorf). Primers used in ChIP-qPCR are listed in Key Resources/Oligonucleotides. All ChIPs were normalized to respective inputs and to control region rp49. H3K9me3 ChIP-seq data from shPiwi and corresponding shW control lines (biological duplicates) were previously published(LeThomas et al., 2013).
Bioinformatic analyses
The D. melanogaster genome assembly BDGP RP/dm3 (April 2006) was used in all analysis. All alignments were performed using an in-house pipeline employing Bowtie 0.12.17 (Langmead et al., 2009).
RNA-seq datasets were pre-processed to remove adaptor contamination using Cutadapt and reads aligning to rRNA sequences with up to 3 mismatches were removed. The remaining reads were aligned to the D. melanogaster genome allowing 2 mismatches and retaining only uniquely mapping reads. For analysis of transposons, data were mapped allowing up to 10,000 mapping positions and 0 mismatches, and read counts were corrected based on the number of mapped positions as described previously (Manakov et al., 2015). Protein-coding gene annotations and TE annotations were obtained from the RefSeq and RepeatMasker tables, respectively, retrieved from the UCSC genome browser (Karolchik et al., 2004). TE expression values for TE families were calculated as the sum of mappability-corrected reads aligning to individual repeats annotated by RepeatMasker. Similar results were obtained by an alternative approach were RNA-seq reads were directly aligned to RepBase TE consensuses allowing 3 mismatches (data not shown). For scatter plots, read counts per element were normalized as reads per million mapped reads (RPKM). Differential expression analyses were performed using the DESeq2 R package using raw read counts as input (Love et al., 2014).
ChIP-seq data was aligned allowing 2 mismatches and retaining uniquely mapped reads for analysis of unique regions. ChIP signal over 1Kb genomic windows was defined as the ratio of normalized unique ChIP to Input counts (ChIP/Input). For global analysis, 1Kb genomic windows that had less than 1 RPKM in input libraries were excluded. 1Kb genomic windows with ChIP/Input ratio > 2 in all control ChIP-seq datasets (shW) were annotated as “het”. We note that using higher ChIP/Input ratio cutoffs to define heterochromatic windows (stricter H3K9me3 enrichment cutoff) produces similar results (data not shown). For analysis of ChIP-seq signal on TE families, reads were mapped to the genome allowing up to 10,000 mapping positions and 0 mismatches, and read counts were corrected based on the number of mapped positions. Read counts for 1Kb genomic windows (unique mappers) or individual TE families (mappability corrected read counts) were normalized as RPKM.
Heatmaps were generated using the ‘pheatmap’ R package. Normalized genome coverage tracks were generated using BedTools (Quinlan and Hall, 2010) and BigWig tools (Kent et al., 2010), using the total mapped reads as a scaling factor. Circular plot was generated using Circos 0.67.7 (Krzywinski et al., 2009). Non-reference insertions were annotated using the TIDAL pipeline (Rahman et al., 2015) with default parameters, using merged reads that do not map to the reference genome with 2 mismatches from all experiments involving DNA sequencing as input (DNA from Input and ChIPs).
Orthologs of SetDB1 and Wde in Drosophila species were extracted from OrthoDB1(Waterhouse et al., 2011) and UniProt databases. SUMO sites and SIM predictions were performed using the GPS-SUMO online tool (Zhao et al., 2014). Protein sequences were aligned by MUSCLE (Edgar, 2004) or Clustal.
Data and software availability
RNA-seq and H3K9me3 ChIP-seq data from shPiwi and corresponding shW control lines were previously published (LeThomas et al., 2013). All RNA-seq and ChIP-seq data generated in this study have been deposited to the Gene Expression Omnibus database: GSE115277.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Flag (HRP conjugated) | Sigma | Cat#A8592 |
| Rabbit polyclonal anti-GFP | Abcam | Cat#ab290 |
| Rabbit polyclonal anti-GFP | Chen et al., 2016 | |
| Anti-Tubulin | Sigma-Aldrich | Cat#T5168 |
| HRP-conjugated anti-mouse | Cell Signalling | Cat#7074 |
| HRP-conjugated anti-rabbit | Cell Signalling | Cat#7076 |
| IRDye®-conjugated anti-mouse | LiCor | Cat#925–68070 |
| IRDye®-conjugated anti-rabbit | LiCor | Cat#925–68071 |
| Rabbit polyclonal anti-GST | Cell Signalling | Cat#2622 |
| GFP-Trap® | ChromoTek | Cat#gtma-20 |
| Anti-FLAG® M2 Magnetic Beads | Sigma | Cat#M8823 |
| Rabbit polyclonal anti-H3K9me3 | Abcam | Cat#ab8898 |
| Mouse monoclonal anti-RNA Pol II | Abcam | Cat#ab5408 |
| Mouse monoclonal anti-H3K4me2/3 | Abcam | Cat#ab6000 |
| Rabbit polyclonal anti-H3K36me3 | Abcam | Cat#ab9050 |
| Mouse anti-HP1 | DSHB | Cat#C1A9 |
| Anti-Smt3 (D. melanogaster) | G Cavalli | |
| Mouse 6x-His Tag Monoclonal Antibody | ThermoFisher | Cat#His.H8 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| GST-SUMO(wt) and GST-SUMO(mutant) | This paper | |
| His-Su(var)2–10(wt) and C341S mutant | This paper | |
| 16% Formaldehyde solution | ThermoScientific | Cat #28908 |
| N-ethylmaleimide | ThermoScientific | Cat#23030 |
| Critical Commercial Assays | ||
| Ribo-Zero™ rRNA Removal Kit | Epicentre/Illumina | Cat#MRZH11124 |
| NEBNext® Ultra™ Directional RNA Library Prep Kit | NEB | Cat#E7760 |
| NEBNext ChIP-Seq Library Prep Master Mix Set | NEB | Cat#E6240 |
| TruSeq RNA Library Prep Kit | Illumina | FC-122–1001 |
| SUMO2 Conjugation Kit | BostonBiochem | Cat#K-715 |
| Deposited Data | ||
| Raw data and bigwig files | This paper | GSE115277 |
| Raw image data files | This paper | DOI: 10.17632/dbzsn49kmg.1 |
| Experimental Models: Organisms/Strains | ||
| UASp-shSv210–1 | BDSC #32915 | |
| UASp-shSv210–2 | BDSC #32956 | |
| UASp-shSUMO | this study | |
| UASp-shPiwi | BDSC #33724 | |
| UASp-shWde | BDSC #33339 | |
| UASp-shWhite | BDSC #33623 | |
| UASp-shSetDB1 | J Brennecke | |
| UASp-shPanx | J Brennecke | |
| UASp-shAsterix | this study | |
| UASp-mKate2-4xBoxB reporter | Chen et al. 2016 | |
| pTubulin-GFP-5BoxB reporter | J Brennecke | |
| pUbi-Luciferase-10BoxB reporter | G Hannon | |
| GFP-Piwi(BAC) | LeThomas et al., 2013 | |
| UASp-SetDB1-GFP | this study | |
| UASp-3Flag3HA-Su(var)2–10 PA | this study | |
| UASp-λN-GFP-Panx | Rogers et al. 2017 | |
| UASp-λN-G FP-Arx | Rogers et al. 2017 | |
| UASp-λN-G FP-eG FP | Chen et al. 2016 | |
| UASp-λN-G FP-Su(var)2–10-PA | this study | |
| UASp-λN-G FP-Su(var)2–10-C341S | this study | |
| UASp-λN-G FP-Su(var)2–10-ΔSAP | this study | |
| UASp-λN-G FP-Su(var)2–10-ΔPINIT | this study | |
| UASp-λN-GFP-Su(var)2–10-ΔSP-RING | this study | |
| UASp-λN-HA-Panx | J Brennecke | |
| maternal alpha-tubulin67C-Gal4 | BDSC #7063 | |
| maternal alpha-tubulin67C-Gal4 | BDSC #7062 | |
| Traffic jam-Gal4 | DGRC #104055 | |
| Oligonucleotides | ||
| Su(var)2–10-F | IDT | CCAGCACAGGACGAACAGCCC |
| Su(var)2–10-R | IDT | CGTGGAACTGGCGACGGCTT |
| rp49-F | IDT | CCGCTTCAAGGGACAGTATCTG |
| rp49-R | IDT | ATCTCGCCGCAGTAAACGC |
| HetA-F | IDT | CGCGCGGAACCCATCTTCAGA |
| HetA-R | IDT | CGCCGCAGTCGTTTGGTGAGT |
| Blood-F | IDT | TGCCACAGTACCTGATTTCG |
| Blood-R | IDT | GATTCGCCTTTTACGTTTGC |
| ZAM-F | IDT | ACTTGACCTGGATACACTCACAAC |
| ZAM-R | IDT | GAGTATTACGGCGACTAGGGATAC |
| Ptip 3’UTR-F | IDT | CATGTGTGTTTCCGCCACAG |
| Ptip 3’UTR-R | IDT | TTCCCAGCTCGCGAAGAAAT |
| CG32138-F | IDT | CAGGATCTGCGCTACGACAT |
| CG32138-R | IDT | AATCGTCGGTCCAGCTCATC |
| sh-Asterix-F | IDT | CTAGCAGTCCAGTAGTTCGTGTTCATCAATAGTTATATTCAAGCATATTGATGAACACGAACTACTGGGCG |
| sh-Asterix-R | IDT | AATTCGCCCAGTAGTTCGTGTTCATCAATATGCTTGAATATAACTATTGATGAACACGAACTACTGGACTG |
| mKate2-F(A) | IDT | GTGACTGTGCGTTAGGTCCTG |
| mKate2-F(A) | IDT | TGAAGTGGTGGTTGTTCACGG |
| mKate2-F(B) | IDT | TCAGAGGGGTGAACTTCCCA |
| mKate2-R(B) | IDT | CTCCCAGCCGAGTGTTTTCT |
| mKate2-F(C) | IDT | GGCCGACAAAGAGACCTACG |
| mKate2-R(C) | IDT | CCAGTTTGCTAGGGAGGTCG |
| Tub-GFP-BoxB reporter-F | IDT | CTTCCTCCTCATCCACAGCG |
| Tub-GFP-BoxB reporter-R | IDT | ACTTGTGGCCGTTTACGTCG |
| Recombinant DNA | ||
| pValium20 | DRSC/TRiP | |
| Drosophila Gateway Vector collection | DGRC | |
| pENTR™/D-TOPO™ | Invitrogen | |
| pActin-GFP-Sv210 | This study | |
| pActin-Flag-mKate | This study | |
| pActin-3xFlag-Arx | This study | |
| pActin-3xFlag-Piwi | This study | |
| pActin-3xFlag-Panx | This study | |
| pActin-3xFlag-SetDB1 | This study | |
| pActin-3xFlag3xHA-SUMO | This study | |
| pActin-SetDB1-G FP | This study | |
| pActin-GFP-Wde | gift | |
| pActin-GFP-Wde fragments/mutants | This study | |
| pGEX-2TK | ||
| pGEX-2TK-Smt3 | G Suske | |
| pGEX-2TK-Smt3-SIM interface mutant | This study | |
| His-Su(var)2–10 construct1 | This study | |
| His-Su(var)2–10 construct2 | This study | |
| Software and Algorithms | ||
| Bowtie 0.12.17 | Langmead et al., 2009 | https://sourceforge.net/proiects/bowtie-bio/files/bowtie/0.12.7/ |
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| UCSC browser | Karolchik et al., 2004 | https://genome.ucsc.edu/ |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| BEDtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| USCS/BigWig tools | Kent et al., 2010 | http://hgdownload.soe.ucsc.edu/admin/exe/linux.x8664/ |
| Circos 0.67.7 | Krzywinski et al., 2009 | http://circos.ca/software/ |
| TIDAL | Rahman et al., 2015 | https://github.com/laulabbrandeis/TIDAL |
| Jalview | Clamp et al., 2004 | http://www.jalview.org/ |
| MUSCLE | Edgar, 2004 | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| GPS-SUMO | Zhao et al., 2014 | http://sumosp.biocuckoo.orfl/ |
| pheatmap | https://cran.r-project.org/web/packages/pheatmap/index.html | |
| Other | ||
| Schneider’s Drosophila Medium | Gibco (Life Technologies) | 21720–024 |
| Fetal Bovine Serum | GEMINI bio-products | 100–106 |
| Penicillin/Streptomycin | Gibco (Life Technologies) | 15140–122 |
Highlights.
Transcriptional silencing by piRNA requires SUMO and the SUMO E3 ligase Su(var)2–10
Su(var)2–10 links the piRNA complex that identifies TEs to the silencing effector
Su(var)2–10 deposits SUMO on chromatin
Su(var)2–10 recruits the SetDB1/Wde histone methyltransferase complex via SUMO
Acknowledgements
We thank members of the Fejes Toth and Aravin labs for discussion. We thank Gary Karpen for suggestions and discussion of some of the experiments. We appreciate the help of Kathy Situ, Zsófia Török, Sivani Vempati, Solomiia Khomandiak, and Angel Galvez Merchan with the experiments. We are grateful to Julius Brennecke, Gregory Hannon and the Bloomington Stock Center for providing fly stocks, Giacomo Cavalli for providing antibodies, Andreas Wodarz for the GFP-wde expression vector, and Guntram Suske for the GST-Smt3 (wild type) plasmid. We thank Igor Antoshechkin (Caltech) for help with sequencing and Sergei Manakov for bioinformatic support. This work was supported by grants from the NIH (R01 GM097363), the Ministry of Education and Science of Russian Federation (14.W03.31.0007) and by the Packard Fellowship Awards to A.A.A., and the NIH (R01GM110217) and the Ellison Medical Foundation Awards to K.F.T. AKR was an NSF GRFP fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Abdu U, Brodsky M, and Schüpbach T (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of gurken through Chk2/Mnk. Curr. Biol 12, 1645–1651. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Zee BM, Fuchs SM, Kharchenko PV, and Kuroda MI (2014). Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28, 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, and Hannon GJ (2008). A piRNA pathway primed by individual transposons is linked to De Novo DNA methylation in mice. Mol. Cell 31, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, and Kouzarides T (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Batki J, Schnabl J, Wang J, Handler D, Andreev VI, Stieger CE, Novatchkova M, Lampersberger L, Kauneckaite K, Xie W, et al. (2019). The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat. Struct. Mol. Biol 26, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, and Jacobsen SE (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, Lampen N, Martinek S, Young MW, and Darnell JE (2001). A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci 98, 9563–9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, and Elgin SCR (2009). Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181, 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJGG, Bourc’his D, Bestor TH, de Rooij DG, and Hannon GJ (2007). MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell 12, 503–514. [DOI] [PubMed] [Google Scholar]
- Chen Y-CA, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. (2016). Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol. Cell 63, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, and Dow JAT (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39, 715–720. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, and Barton GJ (2004). The Jalview Java alignment editor. Bioinformatics 20, 426–427. [DOI] [PubMed] [Google Scholar]
- Crozet P, Margalha L, Butowt R, Fernandes N, Elias CA, Orosa B, Tomanov K, Teige M, Bachmair A, Sadanandom A, et al. (2016). SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 85, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donertas D, Sienski G, and Brennecke J (2013). Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27, 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR, and Reuter G (2013). Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol 5, a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enke RA, Dong Z, and Bender J (2011). Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana. PLoS Genet. 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry MH, Ciabrelli F, Munafò M, Eastwood EL, Kneuss E, Falciatori I, Falconio FA, Hannon GJ, and Czech B (2019). piRNA-guided co-transcriptional silencing coopts nuclear export factors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M, March-Diaz R, and Reyes JC (2008). The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J. Biol. Chem 283, 21469–21477. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, and Schüpbach T (1998). okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12, 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I, Mateos-Langerak J, Thomas A, Cheutin T, and Cavalli G (2014). Identification of regulators of the three-dimensional polycomb organization by a microscopy-based genome-wide RNAi screen. Mol. Cell 54, 485–499. [DOI] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, and Fire A (2012). Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 44, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handu M, Kaduskar B, Ravindranathan R, Soory A, Giri R, Elango VB, Gowda H, and Ratnaparkhi GS (2015). SUMO-Enriched Proteome for Drosophila Innate Immune Response. G3 (Bethesda). 5, 2137–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari KL, Hari KL, Cook KR, Cook KR, Karpen GH, and Karpen GH (2001). The Drosophila Su(var)2–10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15, 1334–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, and Moazed D (2015). RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. (2007). PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing. Mol. Cell 28, 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis Cd., and Khorasanizadeh S (2001). Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, and Psakhye I (2013). Control of Nuclear Activities by Substrate-Selective and Protein-Group SUMOylation. Annu. Rev. Genet 47, 167–186. [DOI] [PubMed] [Google Scholar]
- Johnson ES, and Gupta AA (2001). An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744. [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, and Yasuda H (2001). Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. (2002). The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3, RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, and Theurkauf WE (2007). Drosophila rasiRNA Pathway Mutations Disrupt Embryonic Axis Specification through Activation of an ATR/Chk2 DNA Damage Response. Dev. Cell 12, 45–55. [DOI] [PubMed] [Google Scholar]
- Koch CM, Honemann-Capito M, Egger-Adam D, and Wodarz A (2009). Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/eggless in germ line development. PLoS Genet. 5, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Jänne OA, Jorma J, and Ja OA (2002). PIAS Proteins Modulate Transcription Factors by Functioning as SUMO-1 Ligases. Mol. Cell. Biol 22, 5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, and Marra MA (2009). Circos: An information aesthetic for comparative genomics. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 1, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, and Jenuwein T (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeThomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, and Tóth KF (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeThomas A, Marinov GK, and Aravin AA (2014). A Transgenerational Process Defines piRNA Biogenesis in Drosophila virilis. Cell Rep. 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, and Almouzni G (2004). HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy J-P, et al. (2011). SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet 43, 220–227. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Quivy J-P, and Almouzni G (2016). The methyltransferase Suv39h1 links the SUMO pathway to HP1α marking at pericentric heterochromatin. Nat. Commun 7, 12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, and Aravin AA (2015). MIWI2 and MILI Have Differential Effects on piRNA Biogenesis and DNA Methylation. Cell Rep. 12, 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17.1, 10–12. [Google Scholar]
- Meignin C, and Davis I (2008). UAP56 RNA helicase is required for axis specification and cytoplasmic mRNA localization in Drosophila. Dev. Biol 315, 89–98. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, and Matzke AJ (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, and Boswell RE (1999). Zimp encodes a homologue of mouse Miz1 and PIAS3 and is an essential gene in Drosophila melanogaster. Gene 229, 109–116. [DOI] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, and Hannon GJ (2013). A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in drosophila. Mol. Cell 50, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munarriz E, Barcaroli D, Stephanou A, Townsend P. a, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight R. a, et al. (2004). PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol. Cell. Biol 24, 10593–10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murano K, Iwasaki YW, Ishizu H, Mashiko A, Shibuya A, Kondo S, Adachi S, Suzuki S, Saito K, Natsume T, et al. (2019). Nuclear RNA export factor variant initiates piRNA‐guided co‐transcriptional silencing. EMBO J. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, Yang-Zhou D, Shim H-S, Tao R, Handler D, Karpowicz P, et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, and Courey AJ (2009). Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, and Siomi MC (2015). Respective Functions of Two Distinct Siwi Complexes Assembled during PIWI-Interacting RNA Biogenesis in Bombyx Germ Cells. Cell Rep. 10, 193–203. [DOI] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A, Siomi H, and Siomi MC (2013). DmGTSF1 is necessary for Piwi − piRISC-mediated transcriptional transposon silencing in the Drosophila ovary DmGTSF1 is necessary for Piwi – piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 34283, 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Castro-diaz N, Ecco G, Coluccio A, and Aravin AA (2014). piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 28, 1410–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, and Jentsch S (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R, Chirn G-W, Kanodia A, Sytnikova YA, Brembs B, Bergman CM, and Lau NC (2015). Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. 43, 10655–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, and Lehmann R (2011). piRNA production requires heterochromatin formation in drosophila. Curr. Biol 21, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, and Johnson ES (2006). Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757. [DOI] [PubMed] [Google Scholar]
- Reuter G, and Wolff I (1981). Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. MGG 182, 516–519. [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, and Hannon GJ (2013). Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 27, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, and Grosschedl R (2001). PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, and Muller S (2002). Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ, Spierer P, and Bontron S (2007). Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 3, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- Shin JA, Eun SC, Hyun SK, Ho JCY, Watts FZ, Sang DP, and Jang YK (2005). SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19, 817–828. [DOI] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, and Brennecke J (2012). Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti K-A, Dönertas D, Tirian L, Meixner K, and Brennecke J (2015). Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 29, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfel G, Kazmar T, Frank O, Wienerroither S, Reiter F, and Stark A (2015). Transcriptional regulators form diverse groups with context-dependent regulatory functions. Nature 528, 147–151. [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, Boutros M, and Suske G (2008a). Identification of SUMO-Dependent Chromatin-Associated Transcriptional Repression Components by a Genome-wide RNAi Screen. Mol. Cell 29, 742–754. [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Wink C, Krüger I, and Suske G (2008b). SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9, 899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, and Kikuchi Y (2005). Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J. Biol. Chem 280, 35822–35828. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kahyo T, Toh-E A, Yasuda H, and Kikuchi Y (2001). Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem 276, 48973–48977. [DOI] [PubMed] [Google Scholar]
- Thompson PJ, Dulberg V, Moon K, and Foster LJ (2015). hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng T-Y, Lee C-H, Chan L-W, and Shen C-KJ (2007). Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Natl. Acad. Sci. U. S. A 104, 12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, and Saitoh H (2006). Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem 281, 23180–23190. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, and Moazed D (2004). RNAi-Mediated Targeting of Heterochromatin by the RITS Complex. Science 303, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, and Martienssen RA (2002). Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science 297, 1833 LP–1837. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, and Kriventseva EV (2011). OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 39, D283–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Greenberg D, and Macfarlan TS (2015). Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob. DNA 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, and Hannon GJ (2015). Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Cheng S, Miao N, Xu P, Lu X, Wang M, Zhang Y, Yuan X, Liu W, Lu X, et al. (2019). A Pandas complex adapted for piRNA-guided transposon silencing. BioRxiv 608273. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, and Ren J (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, and Matunis MJ (2008). Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem 283, 29405–29415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and H3K9me3 ChIP-seq data from shPiwi and corresponding shW control lines were previously published (LeThomas et al., 2013). All RNA-seq and ChIP-seq data generated in this study have been deposited to the Gene Expression Omnibus database: GSE115277.