Abstract
Traditionally viewed as an autodigestive pathway, autophagy also facilitates cellular secretion; however, the mechanisms underlying these processes remain unclear. Here, we demonstrate that components of the autophagy machinery specify secretion within extracellular vesicles (EVs). Using a proximity-dependent biotinylation proteomics strategy, we identify 200 putative targets of LC3-dependent secretion. This secretome consists of a highly interconnected network enriched in RNA-binding proteins (RBPs) and EV cargoes. Proteomic and RNA-profiling of EVs identifies diverse RBPs and small non-coding RNAs requiring the LC3-conjugation machinery for packaging and secretion. Focusing on two RBPs, heterogeneous nuclear ribonucleoprotein K (HNRNPK) and scaffold-attachment factor B (SAFB), we demonstrate these proteins interact with LC3 and are secreted within EVs enriched with lipidated LC3. Furthermore, their secretion requires the LC3-conjugation machinery, neutral sphingomyelinase 2 (nSMase2), and LC3-dependent recruitment of Factor-associated with nSMase2 activity (FAN). Hence, the LC3-conjugation pathway controls EV cargo loading and secretion.
Introduction
Although autophagy is classically viewed as a lysosomal degradation process1, genetic evidence implicates autophagy pathway components (ATGs) in secretion, including the conventional secretion of inflammatory cytokines2, extracellular release of lysozyme3, efficient egress of secretory lysosomes4, extracellular vesicle (EV) production5, 6 and unconventional secretion of proteins lacking N-terminal leader peptides or signal sequences7–10. These processes, collectively termed secretory autophagy, implicate the autophagy pathway in non-cell autonomous control of cell fate decisions and tissue microenvironments, both normally and during disease11–13. Nevertheless, our understanding of secretory autophagy remains rudimentary. First, apart from a limited number of protein targets, the autophagy-dependent secretome remains uncharacterized. Furthermore, studies to date largely rely on phenotypic analysis following ATG genetic loss-of-function, which fail to discern whether secretory defects represent a direct versus indirect consequence of impaired autophagy. Here, we describe a secretory autophagy pathway in which LC3/ATG8 mediates the loading of protein and RNA cargoes into extracellular vesicles (EVs) for secretion outside of cells.
Results
LC3 proximity-dependent biotinylation identifies proteins secreted via autophagy-dependent pathways
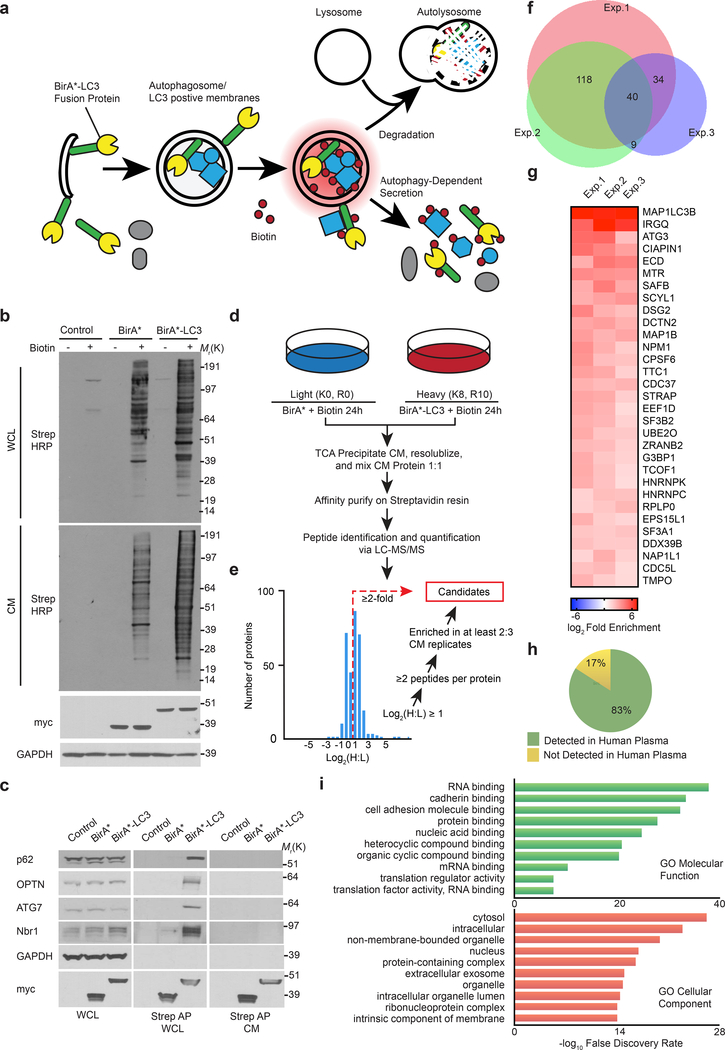
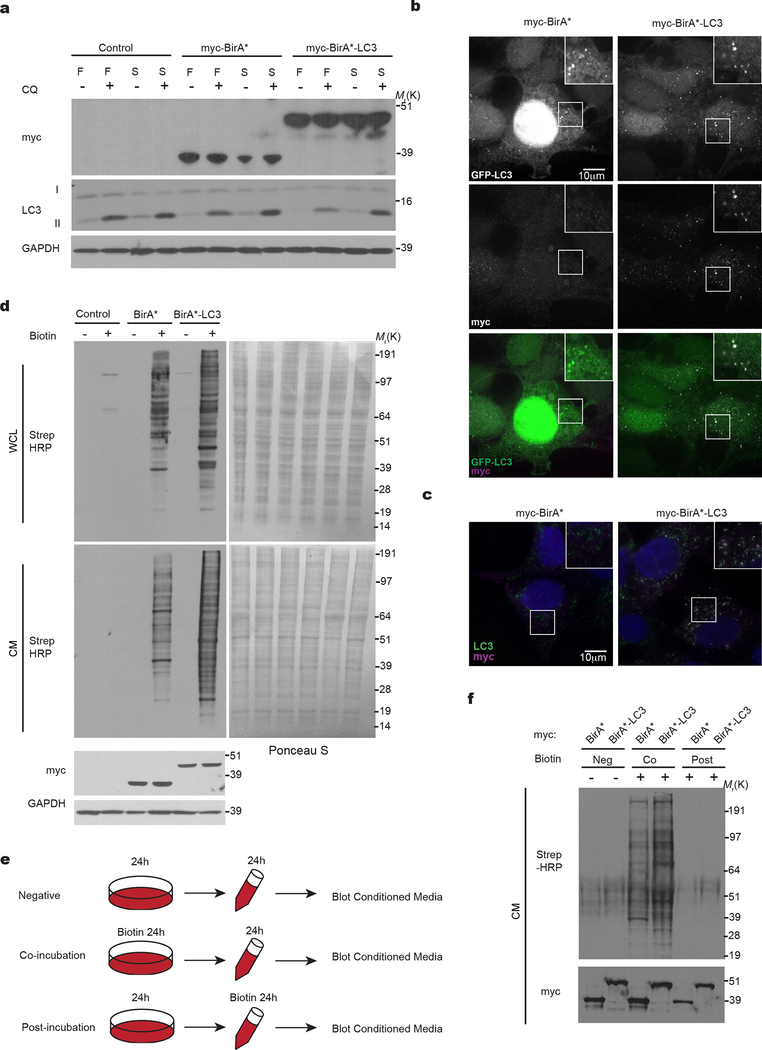
We developed a proximity-dependent biotinylation (BioID)14 strategy to label proteins within autophagic intermediates that are subsequently secreted outside of cells (Fig. 1a). Hypothesizing such secreted proteins interact with or reside near MAP1LC3B (LC3), an ATG8 orthologue that captures substrates for autophagy, we fused the mutant E. coli biotin ligase (BirA*) to the LC3 N-terminus. BirA*-LC3 (myc epitope-tagged) was lipidated with phosphatidylethanolamine (PE), localized at autophagosomes, and degraded within lysosomes (Extended Data Fig. 1a,b,c). Biotin incubation triggered robust labelling of intracellular targets in BirA*-LC3 cells (Fig. 1b, Extended Data Fig. 1d) including multiple well-known LC3-interacting intracellular proteins (Fig. 1c). However, these molecules were not detectably secreted into conditioned media (CM). Instead, numerous unique biotin-labelled proteins were detected in CM of BirA*-LC3 cells compared to BirA* controls (Fig. 1b). Importantly, the BirA*-LC3-labeled secretome represented secretion of proteins that were biotin-labelled inside cells, not promiscuous biotinylation following extracellular release (Extended Data Fig. 1e,f).
Figure 1. Identification of proteins secreted via autophagy-dependent pathways using LC3 proximity-dependent biotinylation and quantitative secretomics.
a, Proximity-dependent biotinylation strategy to label secretory autophagy targets. b, Protein biotinylation in whole cell lysate (WCL, intracellular) and conditioned media (CM, secreted) harvested from HEK293T cells stably expressing myc-BirA*-LC3, myc-BirA* or empty vector (Control) following 24h incubation with (+) or without (−) 50 μM biotin. Equal amounts of protein from trichloroacetic acid precipitated CM or WCL were probed with Streptavidin-HRP (Strep-HRP) to detect biotinylated proteins, myc or GAPDH (n=3 biologically independent experiments). c, Streptavidin affinity purification (Strep AP) and immunoblotting to detect known LC3-interacting proteins within WCL and CM of cells expressing myc-BirA*-LC3 (n=2 biologically independent experiments). d, Autophagy-dependent secretion substrate enrichment and quantitative secretomics workflow. e, Log2(H:L) histogram for CM proteins identified in bioreplicate #2 and scheme for identification of autophagy-dependent secretion candidates. f, Putative secretory autophagy candidates identified in n=3 independent experiments (Exp.). Among the 40 hits enriched in all three experiments, 31 were statistically significant overall (see Extended Data Fig. 2) and classified as Class I candidates. The remaining proteins along with hits enriched in 2 out of n=3 experiments (170 proteins total) were designated Class II candidates. Full list of candidates provided in Supplementary Table 1. g, Log2(BirA*-LC3:BirA*) heat map of Class I candidates. h, Proportion of secreted candidates (Class I, II) detected in human plasma. i, Gene Ontology (GO) enrichment analysis of secreted candidates (Class I, II) with the top terms for molecular function and cellular component plotted according to -log10 False Discovery Rate. Statistical significance was calculated by one-way Fisher’s exact test. Sample size, n=3 independent biological replicates, yielding 200 enriched proteins in the Class I + Class II datasets. Data and unprocessed blots available in Source Data Fig. 1.
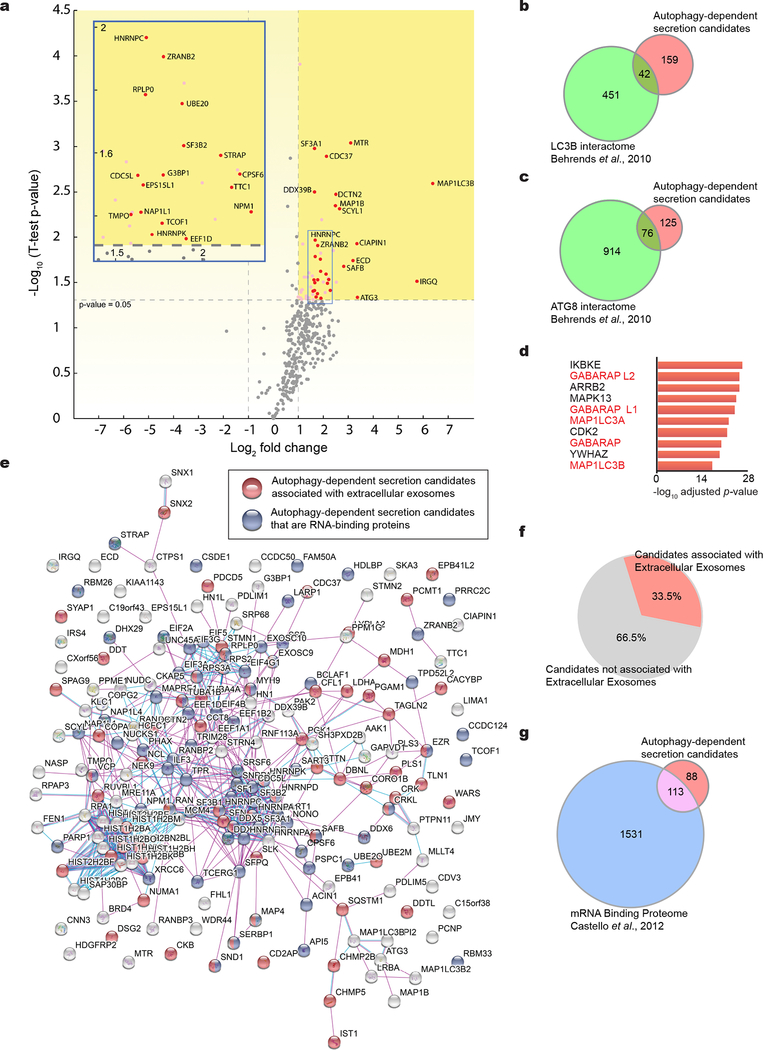
Combining this strategy with stable isotope labelling in cell culture (SILAC) as a quantitative proteomics approach to detect differences in secreted protein abundance between BirA*-LC3 and BirA* via mass spectrometry (Fig. 1d), we identified >350 secreted proteins in three independent biological replicates (Supplementary Table 1). Candidates were selected based on significantly increased log2(BirA*-LC3:BirA*) ratios and the presence of two or more peptides. A total of 31 proteins exhibited statistically significant enrichment (log2(BirA*-LC3:BirA*)>1; p<0.05) in all three replicates and an additional 170 proteins were enriched in two of three replicates (Fig. 1e,f,g, Extended Data Fig. 2a). We refer to hits enriched in all replicates as class I proteins and those in two of three as class II proteins (Supplementary Table 1).
In the class I dataset, we identified two autophagy proteins, MAP1LC3B and ATG3, along with multiple RNA-binding proteins (RBPs) previously shown to interact with LC3 or other ATG8 family proteins15 (Fig. 1g, Extended Data Fig. 2a,b,c). Together, the class I + II datasets contained 42 and 76 proteins in common with the LC3 interactome and entire ATG8 interactome, respectively15. Notably, five LC3/ATG8 family members ranked amongst the top proteins connected to the identified secreted targets (Extended Data Fig. 2d). 83% of the proteins enriched in the BirA*-LC3 labelled secretome were previously identified in proteomics of human plasma, consistent with a role for autophagy in controlling secretion in vivo16 (Fig. 1h). Importantly, an interaction map of the class I + II datasets revealed a highly interconnected network enriched in protein-protein interactions (PPIs)17 (Extended Data Fig. 2e). Gene Ontology (GO) analyses demonstrated the BirA*-LC3 labelled secretome was highly enriched in RBPs and proteins released within EVs (Fig. 1i; Extended Data Fig. 2e); 33% of the candidates identified in our proteomic screen were previously detected in EVs and 113 out of 200 proteins have functions in mRNA binding18 (Extended Data Fig. 2e,f,g; Supplementary Table 1). Collectively, these results broached that LC3 and the autophagy machinery control the loading and secretion of specific proteins such as RBPs within EVs.
LC3-II and BirA*-LC3 biotinylated targets are secreted within small EVs
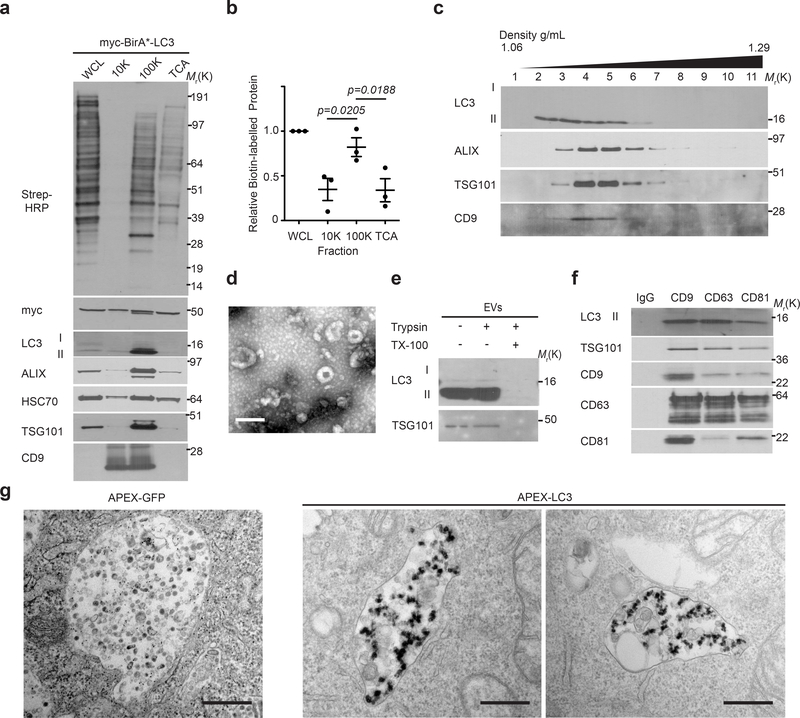
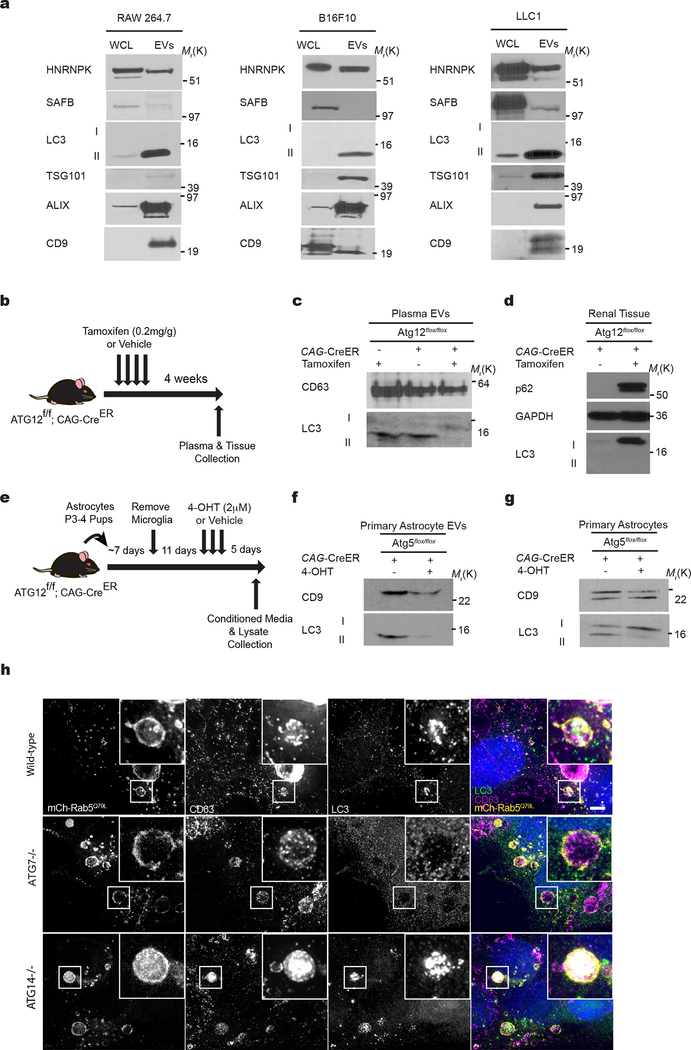
CM from biotin-pulsed BirA*-LC3 cells was subject to serial differential ultracentrifugation to recover large EVs (at 10,000g), small EVs (at 100,000g)19 and soluble proteins precipitated from the remaining sample. BirA*-LC3-labelled secreted proteins were enriched in the 100,000g pellet along with multiple EV markers (Fig. 2a,b). Remarkably, these 100,000g fractions were also enriched in the lipidated, membrane-bound form of endogenous LC3 (LC3-II), suggesting LC3-II itself was secreted via EVs. Upon further purifying EVs via linear sucrose density gradient, we found endogenous LC3-II co-fractionated with well-defined EV markers at characteristic buoyant densities, with a slightly broader distribution into less dense fractions19 (Fig. 2c). Transmission electron microscopy (TEM) further corroborated that samples isolated by ultracentrifugation were enriched in EVs. Importantly, endogenous LC3-II resided inside the lumen of EVs, evidenced by its protease protection in the absence of detergent, and co-purified with EVs immuno-isolated from concentrated preparations using antibodies against EV-associated tetraspanins (Fig. 2d,e,f). Finally, LC3-II was secreted within EVs from multiple cell types comprising diverse lineages, including primary astrocytes, and detected in vivo within EVs isolated from mouse plasma (Extended Data Fig. 3 a–g).
Figure 2. LC3-II and BirA*-LC3 biotinylated targets are secreted within EVs.
a, Protein biotinylation in whole cell lysates (WCL, intracellular) and fractionated conditioned media (CM) harvested from BirA*-LC3 HEK293T cells incubated with 50 μM biotin for 24h. CM subject to differential ultracentrifugation to recover large extracellular vesicles (10,000g; 10K), small extracellular vesicles (100,000g; 100K), and precipitated free soluble protein (TCA). Equal amounts of protein from WCL and fractionated CM probed with Streptavidin-HRP (Strep-HRP) or antibodies for the indicated extracellular vesicle marker proteins, LC3 and myc-tagged BirA*-LC3 (n=3 biologically independent replicates). b, Quantification of global protein biotinylation in the indicated fractions of CM relative to WCL (mean ± s.e.m.; n=3 biologically independent experiments). Statistical significance between CM fractions calculated by one-way analysis of variance (ANOVA) coupled with Tukey’s post hoc test. c, EVs from CM separated via linear sucrose density gradient ultracentrifugation, fractionated and immunoblotted to detect endogenous levels of the indicated markers and LC3 (n=3 biologically independent experiments). d, Representative transmission electron micrograph of EVs isolated via differential ultracentrifugation (n=3 biologically independent samples). Scale bar= 200nm. e, Representative immunoblots of indicated proteins from untreated EVs or EVs incubated with 100 μg/ml trypsin and/or 1% Triton X-100 (TX-100) for 30 min at 4°C (n=3 biologically independent experiments). f, Representative immunoblots of EVs immuno-purified from concentrated CM fractions using antibodies targeting the tetraspanins CD9, CD63, CD81 or a normal mouse IgG isotype control and immunoblotted to detect endogenous levels of the indicated markers and LC3 (n=3 biologically independent experiments). g, Representative transmission electron micrographs of normal rat kidney epithelial cells expressing an APEX2-LC3 recombinant fusion protein of APEX2-GFP control and stained with 3,3-diaminobenzidine (DAB) and hydrogen peroxide (H2O2) (n=3 biologically independent samples). Scale bar=500nm. Data and unprocessed blots available in Source Data Fig. 2.
A subset of EVs are produced by intraluminal budding from the limiting membrane of multivesicular bodies (MVBs)20–23. To determine if intracellular LC3 localized to MVBs, we employed an APEX-LC3 recombinant probe24 to visualize LC3 via TEM, which revealed numerous MVBs containing subpopulations of LC3-positive intraluminal vesicles (ILVs) (Figure 2g). To determine whether endogenous LC3 is delivered into the lumen to MVBs, we immunostained for LC3 in cells expressing constitutively active mutant mCherry-Rab5Q79L, which impairs endosomal trafficking and promotes the formation of enlarged terminal early endosomes exhibiting intraluminal budding25, 26. We detected endogenous LC3 at the limiting membrane and in ILVs within mCherry-Rab5Q79L endosomes of wild-type cells, but not in cells lacking ATG7, an autophagy regulator essential for LC3/ATG8 lipidation27 (Fig. 3a). Furthermore, LC3 co-localized with CD63 in these enlarged vesicular intermediates (Extended data 3h). In contrast, ATG14, an ATG needed for classical degradative autophagy28, 29, did not reduce formation of LC3-positive ILVs in mCherry-Rab5Q79L endosomes (Fig 3a), broaching a specific requirement for the LC3-conjugation pathway in incorporating LC3 into ILVs. Finally, in the absence of Rab5Q79L expression, we observed significant co-localization of endogenous LC3 and CD63, further supporting that LC3 is packaged and released within EVs by an ATG7, but not ATG14 dependent pathway (Fig 3b,c,d).
Figure 3. Endogenous LC3 localizes with endosomes and EV-associated tetraspanins.
a, Representative fluorescence micrographs from WT, ATG7−/−, and ATG14−/− HEK293T cells transfected with mCherry-Rab5Q79L (magenta). Cells were immunostained for endogenous LC3 (green) (n=3 biologically independent experiments). Scale bar=10μm. b, Representative fluorescence micrographs from wild-type, ATG7−/− and ATG14−/− cells immunostained for endogenous LC3 (green) and CD63 (magenta) (n=3 biologically independent experiments). Scale bar=10μm. c, Scatter plot of p-values obtained from Costes significance tests to assess whether the overlap of LC3 and CD63 staining observed in Panel b exceeds thresholds of random co-occurrence. Statistical significance calculated by one-way ANOVA coupled with Tukey’s post hoc (mean ± s.e.m.; WT, n=28; ATG7−/−, n=33; ATG14−/−, n=27 biologically independent samples). d, Scatter plot of Mander’s coefficients for co-occurrence of LC3 with CD63 in immuno-stained cells in Panel b. Statistical significance calculated by unpaired two-tailed t-test (mean ± s.e.m.; WT, n=27; ATG14−/−, n=27 biologically independent samples). Numerical data available in Source Data Fig. 3.
LC3-conjugation pathway is required for EV loading and secretion of RNA-binding proteins
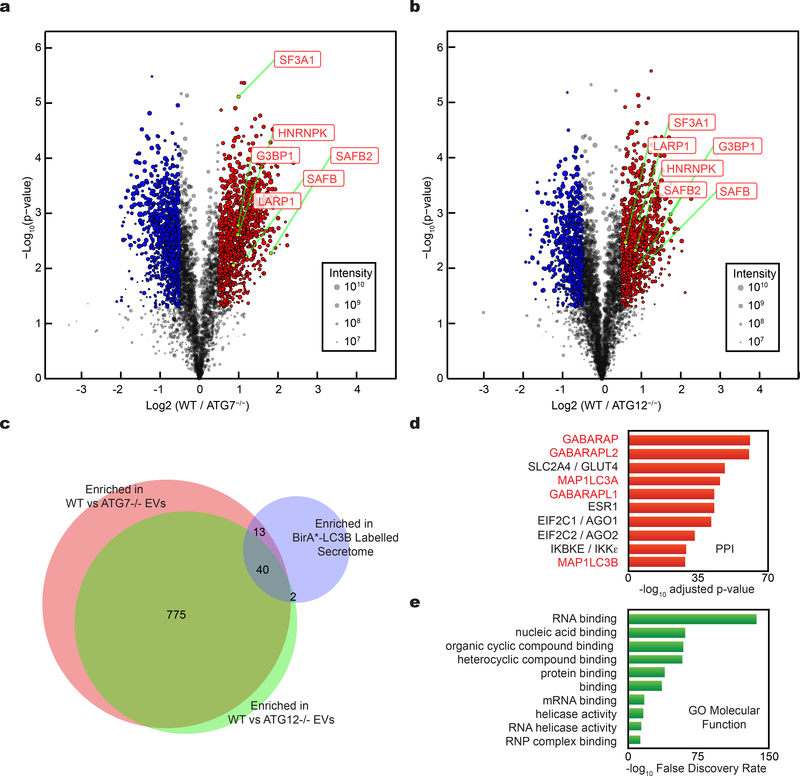
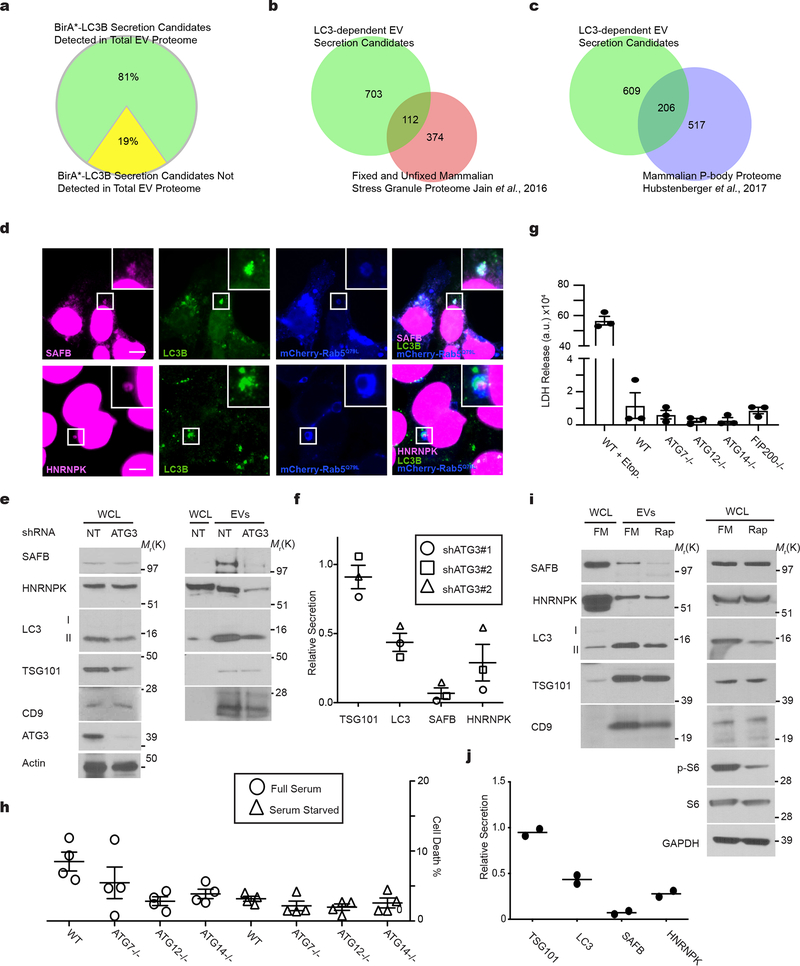
These phenotypic differences between ATG7 versus ATG14 deletion suggested that LC3-conjugation pathways specifically packaged proteins into EVs for secretion outside the cell1. To further elucidate the LC3-dependent EV proteome, we performed tandem-mass tag (TMT)-based quantitative proteomics comparing EVs purified from wild-type versus cells lacking ATG7 or ATG12, two essential components of the LC3-conjugation machinery27 (Fig. 4a,b; Supplementary Table 2). Overall, 81% of targets enriched in the BirA*-LC3B-labelled secretome were detected in the global EV proteome, further supporting EVs as a principal route for autophagy-dependent unconventional secretion (Fig 2a,b; Extended Data Fig. 4a). 815 proteins were enriched in EVs from wild-type relative to ATG7 and ATG12 deficient cells (Fig. 4c; Supplementary Table 2), including 55 proteins overlapping with the BirA*-LC3B-labeled secretome, such as the RNA-binding proteins SAFB, HNRNPK, LARP1, SF3A1 and G3BP1. Although we were unable to detect LC3B, likely due its small size and lipid modification, five LC3/ATG8 family members ranked amongst the top proteins connected to the 815 proteins enriched in the LC3-dependent EV proteome (Fig. 4c). Moreover, similar to the BirA*-LC3 secretome, GO analyses highlighted a profound enrichment in RBPs and proteins that function in RNA metabolism (Fig. 1i, Fig. 4e). Among the 815 EV proteins requiring the LC3-conjugation machinery for secretion, 112 have been identified in stress granules and 206 in processing bodies (P-bodies), two ribonucleoprotein assemblies that functionally sequester RNAs to control gene expression in response to diverse cellular stresses30 (Extended Data Fig. 4b,c; Supplementary Table 2). Finally, among the proteins enriched within the BirA*-LC3 secretome, but not the LC3-dependent EV proteome, histones made up a significant proportion, consistent with reports that autophagy may facilitate the secretion of histones and DNA via EV-independent mechanisms31 (Supplementary Table 2).
Figure 4. Tandem mass tag (TMT) quantitative secretomics identifies EV proteins secreted via the LC3-conjugation machinery.
a, Volcano plot of proteins identified within EVs from wild-type (WT) and ATG7−/− HEK293T cells quantified by TMT mass spectrometry. TMT labelled proteins plotted according to their -log10 p-values as determined by two-tailed t-test and log2 fold enrichment (WT/ATG7−/−; n=4 biologically independent samples). Grey dots: Proteins not relatively enriched in EVs from WT or ATG7−/− cells identified with p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5). Red dots: Proteins significantly enriched in EVs from WT cells relative to ATG7−/− cells. Blue dots: Proteins significantly enriched in EVs from ATG7−/− cells relative to WT cells. Dot size proportional to sum of the signal intensity for identified proteins. b, Volcano plot of proteins identified within EVs from WT and ATG12−/− cells. TMT labelled proteins according to their -log10 p-values as determined by two-tailed t-test and log2 fold enrichment (WT/ATG12−/−; n=4 biologically independent samples). Grey dots: Proteins not relatively enriched in EVs from WT or ATG7−/− cells identified with p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5). Red dots: Proteins significantly enriched in EVs from WT cells relative to ATG12−/− cells. Blue dots: Proteins significantly enriched in EVs from ATG12−/− cells relative to WT cells. Dot size proportional to sum of the signal intensity for identified proteins. c, Venn diagram showing overlap of proteins enriched in EVs from WT cells relative to ATG7−/− cells, EVs from WT cells relative to ATG12−/− cells, and proteins enriched within the BirA*-LC3B labelled secretome. d, Ranked list of proteins with greatest connectivity to the 815 proteins enriched in EVs from WT cells relative to ATG7−/− and ATG12−/− cells. Statistical significance calculated in Enrichr by one-way Fisher’s exact test and adjusted using the Benjamini–Hochberg method. LC3/ATG8 family members highlighted in red. e, Gene Ontology (GO) enrichment analysis of 815 proteins enriched in EVs from WT cells relative to ATG7−/− and ATG12−/− cells with the top terms for molecular function plotted according to -log10 False Discovery Rate. Statistical significance calculated by one-way Fisher’s exact test. Data available in Source Data Fig. 4.
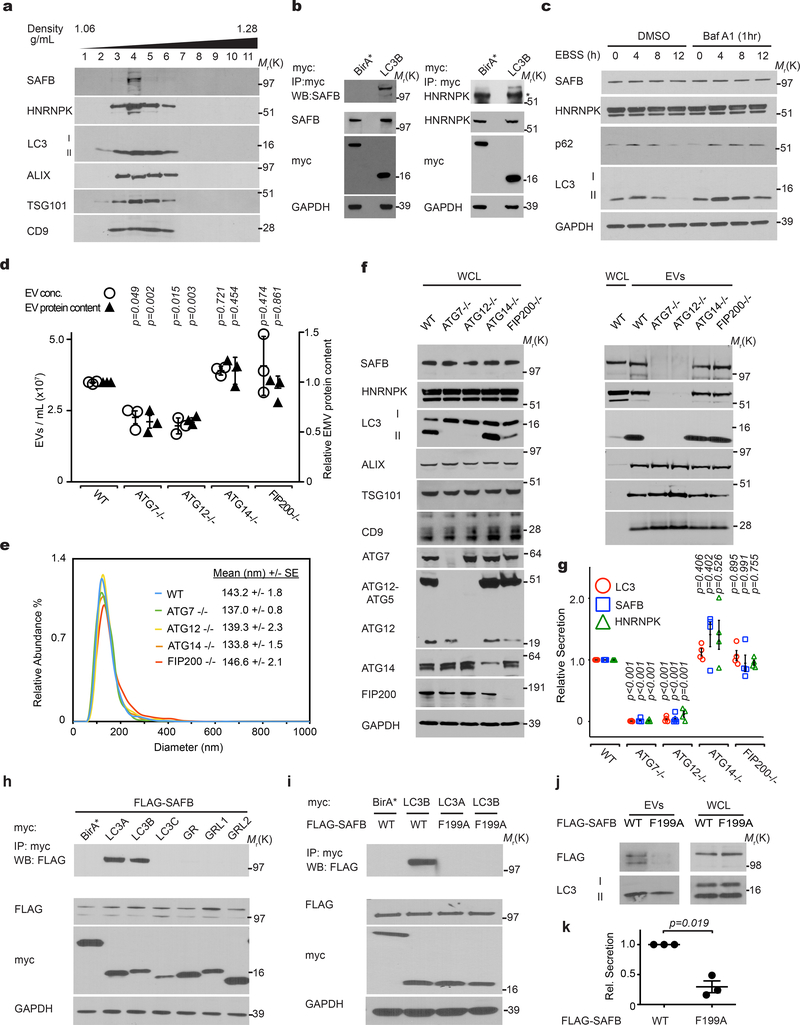
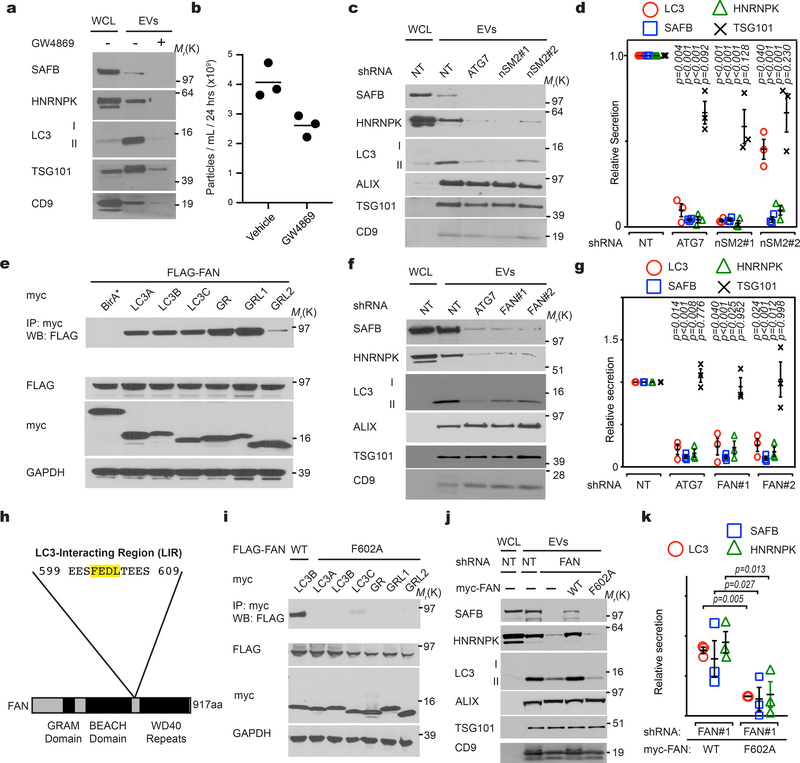
Little is known regarding the pathways specifying cargo loading into EVs20–23. Analogous to its role in sequestering cargo during degradative autophagy32, we hypothesized that lipidated LC3-II captures proteins at the MVB limiting membrane, incorporating them into intraluminal vesicles (ILVs) for subsequent release as EVs. To test this prediction, we focused on SAFB and HNRNPK, two RBPs enriched in both the BirA*-LC3B secretome and LC3-dependent EV proteome (Fig. 1g; Fig. 4a,b,c; Extended Data Fig. 4a). Endogenous SAFB and HNRNPK co-fractionated with membrane-bound LC3-II in purified EVs (Fig. 5a), interacted with LC3 (Fig. 5b), co-localized with LC3 in the lumen of a subset of mCherry-Rab5Q79L endosomes (Extended Data Fig. 4d) and were detected in EVs purified from multiple cell types (Extended Data Fig. 3a). In contrast to p62/SQSTM1, these LC3-binding RBPs were not degraded during starvation-induced autophagy (Fig. 5c).
Figure 5. LC3-conjugation machinery is required for EV loading and secretion of SAFB and HNRNPK.
a, EVs from HEK293T CM separated via linear sucrose density gradient fractionation were immunoblotted to detect endogenous levels of indicated markers (n=2 biologically independent experiments). b, HEK293T cells transfected with myc-tagged LC3B or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody, and immunoblotted with indicated antibodies (n=3 biologically independent experiments). c, Cells were EBSS starved for the indicated times, lysed and immunoblotted for the indicated proteins. Baf A1=50nM Bafilomycin A1 for 1h prior to lysis (n=2 biologically independent experiments). d, CM from equal numbers of indicated cell types subject to nanoparticle tracking analysis (open circles, left axis) to determine EV number or relative protein content measured using BCA from 100,000g EV fractions (black triangles, right axis). (mean ± s.e.m.; n=3 biologically independent experiments). Statistical significance calculated by one-way ANOVA coupled with Dunnett’s test. e, EV size distribution from a representative experiment for the indicated cell types in Panel d (SE=standard error; n=3 biologically independent experiments). f, WCL and 100,000g EV fractions harvested from indicated cell types were collected, normalized for protein concentration, and immunoblotted to detect indicated proteins at endogenous levels (n=4 biologically independent experiments). g, Quantification of LC3 (red circles), SAFB (blue squares), and HNRNPK (green triangles) levels in EVs from the indicated ATG−/− cell lines relative to WT (mean ± s.e.m.; n=4 biologically independent experiments). Statistical significance calculated by one-way ANOVA coupled with Tukey’s post hoc test. h, HEK293T cells co-transfected with FLAG-tagged SAFB and myc-tagged LC3A, LC3B, LC3C, GABARAP (GR), GABARAPL1 (GRL1), GABARAPL2 (GRL2) or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody and immunoblotted with indicated antibodies (n=2 biologically independent experiments). i, Cells co-transfected with FLAG-tagged wild-type SAFB (WT) or mutant SAFB (F199A) and myc-tagged LC3A, LC3B or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody and immunoblotted with indicated antibodies (n=2 biologically independent experiments). j, Whole cell lysate (WCL) and EVs harvested from cells expressing WT or LIR mutant SAFB (F199A) were collected and immunoblotted for FLAG-SAFB and LC3 (n=3 biologically independent experiments). k, Quantification WT and LIR mutant SAFB (F199A) secretion in EVs from cells (mean ± s.e.m.; n=3 biologically independent experiments). Statistical significance calculated by paired two-tailed t-test. Data and unprocessed blots available in Source Data Fig. 5.
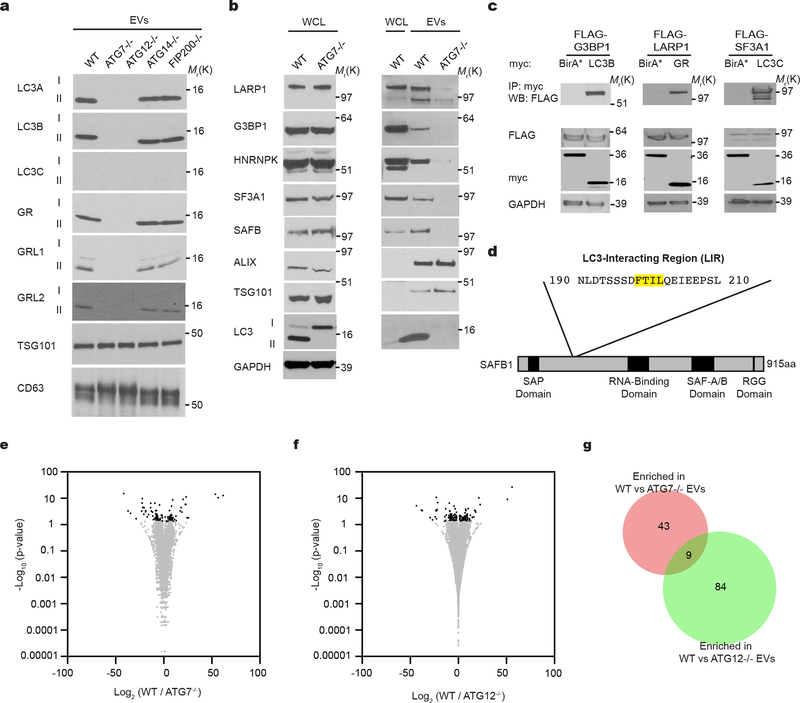
We next assessed whether various autophagy pathway components were required for SAFB and HNRNPK secretion. Deletion of the LC3-conjugation pathway components, ATG7 and ATG12, reduced overall EV production and protein content relative to controls (Fig. 5d), but did not impact the size of EVs (Fig. 5e). To control for these differences in EV production among ATG null cells, we normalized EV lysates based on protein concentration and assayed for LC3-binding RBPs. EVs from ATG7 and ATG12 deleted cells were devoid of LC3-II, HNRNPK, and SAFB, but still contained EV marker proteins (Fig. 5f,g). Similarly, reduced EV secretion of these targets was observed in cells deficient for ATG3, another essential component of the LC3-conjugation machinery27 (Extended Data Fig. 4e,f). Impaired secretion was not due to changes in intracellular HNRNPK and SAFB protein levels or cell death (Fig. 5f,g; Extended Data Fig. 4e,f,g,h). In contrast, loss of ATG14 and FIP200, two ATGs required for the early initiation steps of classical autophagy, but dispensable for LC3-conjugation28, 29, 33, did not attenuate EV production (Fig. 5d) or EV release of LC3-II and LC3-binding RBPs (Fig. 5f,g). Taken together with the results above that ATG14 deletion does not impair LC3-positive ILV formation or LC3 co-localization with tetraspanins (Fig. 3a,b,c,d; Extended Data Fig. 3a), our results substantiate a specific requirement for the LC3-conjugation machinery in loading LC3-II and LC3-binding RBPs into EVs. In further support, LC3-II is not secreted within plasma EVs in vivo in mice systemically deleted for Atg12 (Extended Data Fig. 3b–d). Because proteins that bind LC3/ATG8 frequently contain a motif called an LC3 interaction region (LIR), we asked whether the loading of LC3-binding RBPs into EVs involved LIR-dependent interactions. Primary sequence analyses revealed a putative LIR consensus motif within SAFB (Extended Data Fig. 5d), whereas HNRNPK only contained regions with minimal overlap. Mutation of the core hydrophobic amino acid within this motif to alanine (F199A) was sufficient to disrupt LC3 binding (Fig. 4h,i), which potently suppressed EV secretion of SAFB (Fig. 4j,k).
Overall, this secretory autophagy pathway, which we term LC3-Dependent EV Loading and Secretion (LDELS), is distinct from classical autophagy. In support, treating cells with rapamycin stimulates classical autophagy, but conversely reduces EV secretion of LC3-II and LC3-binding RBPs (Extended Data Fig. 4i,j). In addition, other LC3/ATG8 family members are released in EVs via mechanisms requiring the LC3-conjugation machinery (Extended Data Fig. 5a) and recent unbiased proteomic analyses have detected multiple LC3/ATG8 family members in EVs from diverse cell lines34. Notably, we corroborated several additional RBPs identified from our proteomic screens, including G3BP1, LARP1 and SF3A1, to be secreted in EVs in an ATG7-dependent manner; similar to SAFB and HNRNPK, these targets interacted with diverse LC3/ATG8 family members (Extended Data Fig. 5b,c). Together, these data support that the LC3-conjugation machinery and LC3/ATG8 family proteins mediate the cargo loading and secretion of RBPs via EVs.
LDELS Regulates Extracellular RNA Secretion via EVs
We further reasoned that LDELS influences extracellular RNA secretion via EVs. EVs contain diverse nucleic acids including mRNA, non-coding RNAs, and DNA, but the mechanisms incorporating genetic material into EVs remain poorly defined23, 35. To scrutinize how LDELS impacted extracellular RNA secretion, we performed quantitative RNA sequencing (RNA-seq) from EVs isolated from wild-type, ATG7 and ATG12 deficient cells; in parallel, RNA-seq of corresponding cell samples was performed to assess intracellular gene expression differences. We observed profound differences in the small RNA bio-types isolated from EVs from wild-type versus LDELS deficient cells, with minimal differences in the intracellular small RNA species (Fig. 6a,b; Supplementary Table 3). EVs derived from ATG7 and ATG12 deficient cells had relatively fewer RNA-seq reads mapping to small nucleolar RNAs (snoRNAs) and microRNAs (miRNAs), particularly in ATG7 knockout conditions. Whereas 23% and 15% of the small RNA sequences in EVs from wild-type cells were snoRNAs and miRNAs, respectively, snoRNAs and miRNAs constituted 6% and 5% of EV small RNA sequences from ATG7 deficient cells and 18% and 5% from ATG12 deficient cells. Reduced levels of snoRNA and miRNA sequences in EVs from LDELS deficient cells correlated with a relatively increased proportion of reads mapping to transfer RNA (tRNA) species. In contrast, ATG7/12 deletion minimally impacted extracellular release of large RNAs within EVs (Extended Data Fig. 5e,f,g; Supplementary Table 3). More detailed analysis revealed 252 and 105 small non-coding RNAs to be statistically enriched in EVs from relative to ATG7 and ATG12 deficient cells, respectively (Fig. 6c,d); overlap between these two datasets highlighted 88 distinct RNAs requiring the LC3-conjugation machinery for efficient EV secretion (Fig. 6e; Supplementary Table 3). Most significantly, 76% of EV small non-coding RNAs regulated by LDELS were snoRNAs (or fragments thereof), a class of small RNAs canonically involved the ribosomal RNA modification36 (Fig. 6f). Furthermore, 30% of snoRNAs secreted in an ATG7/12-dependent manner were enriched within EVs relative to total cellular RNA, consistent with their incorporation into EVs through an active packaging mechanism (Fig. 6g). Hence, LDELS influences the spectrum of extracellular small non-coding RNA species secreted within EVs.
Figure 6. LC3-dependent EV loading and secretion (LDELS) regulates the small non-coding RNA composition of EVs.
a, Proportion of RNA sequencing (RNA-seq) reads in total cellular RNA from WT, ATG7−/− and ATG12−/− HEK293T cells from the different classes of small non-coding RNA (n=3 biologically independent samples). b, Proportion of RNA-seq reads in total EV RNA from WT, ATG7−/− and ATG12−/− HEK293T cells from the different classes of small non-coding RNA (n=3 biologically independent samples). c, Volcano plot of small non-coding RNAs detected in EVs from WT and ATG7−/− cells quantified by RNA-seq. Results plotted according to -log10 p-values as determined by DESeq2 and log2 fold enrichment (n=3 biologically independent samples; WT/ATG7−/−). Grey dots: RNAs identified with p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5), and thus, not relatively enriched in EVs from WT or ATG7−/− cells. Log2 fold change reflects WT EV RNA/WT Cell RNA to ATG7−/− EV RNA/ATG7−/− Cell RNA ratio. Black dots: Small non-coding RNAs significantly enriched in EVs from WT or ATG7−/− cells. Red dots: snoRNAs significantly enriched in EVs from WT or ATG7−/− cells. d, Volcano plot of small non-coding RNAs detected in EVs from WT and ATG12−/− cells quantified by RNA-seq. Results were plotted according to -log10 p-values as determined by DESeq2 and log2 fold enrichment (n=3 biologically independent samples; WT/ATG12−/−). Grey dots: RNAs with a p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5), and thus, not relatively enriched in EVs from WT or ATG12−/− cells. Log2 fold change reflects WT EV RNA/WT Cell RNA to ATG12−/− EV RNA/ATG12−/− Cell RNA ratio. Black dots: Small non-coding RNAs significantly enriched in EVs from WT or ATG12−/− cells. Red dots: snoRNAs significantly enriched in EVs from WT or ATG12−/− cells. e, Venn diagram showing overlap of small non-coding RNAs enriched in EVs from WT cells relative to ATG7−/− or ATG12−/− cells. f, Proportion of different classes of small RNAs enriched in EVs from WT vs. ATG-deficient cells. g, Heatmap of the 67 snoRNAs enriched in EVs from WT cells across all genetic conditions and sample types. Scale indicates intensity, defined as Δ(read counts -mean read count)/SD. Data available in Source Data Fig. 6.
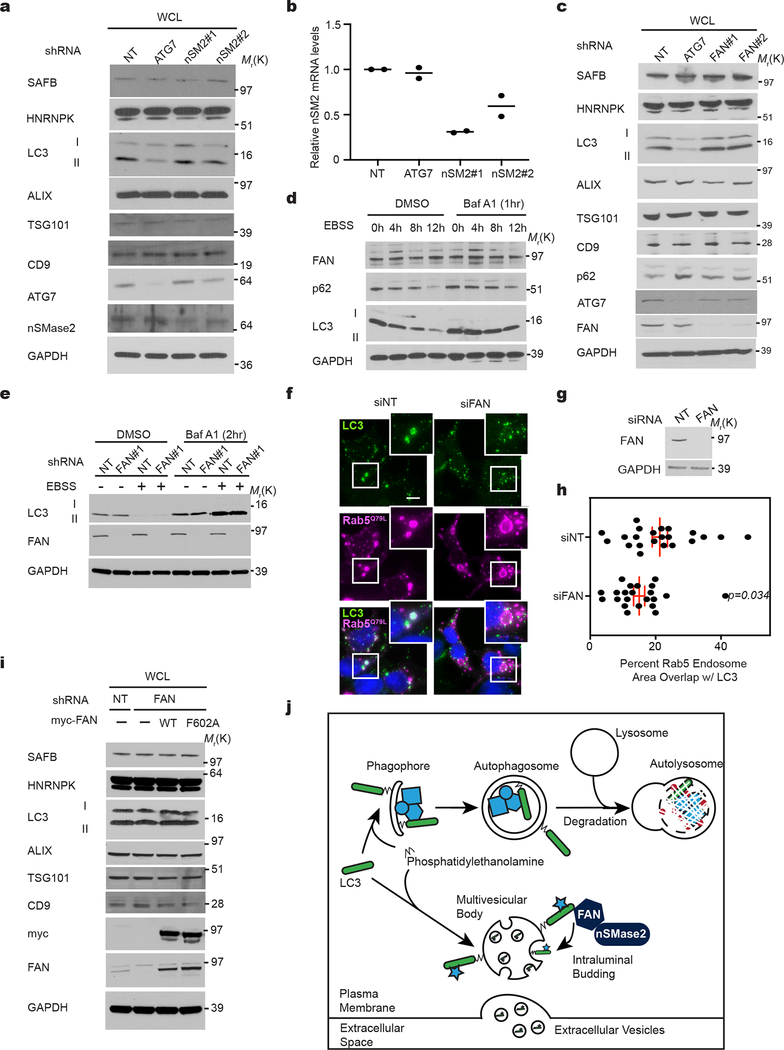
LDELS requires neutral sphingomyelinase 2
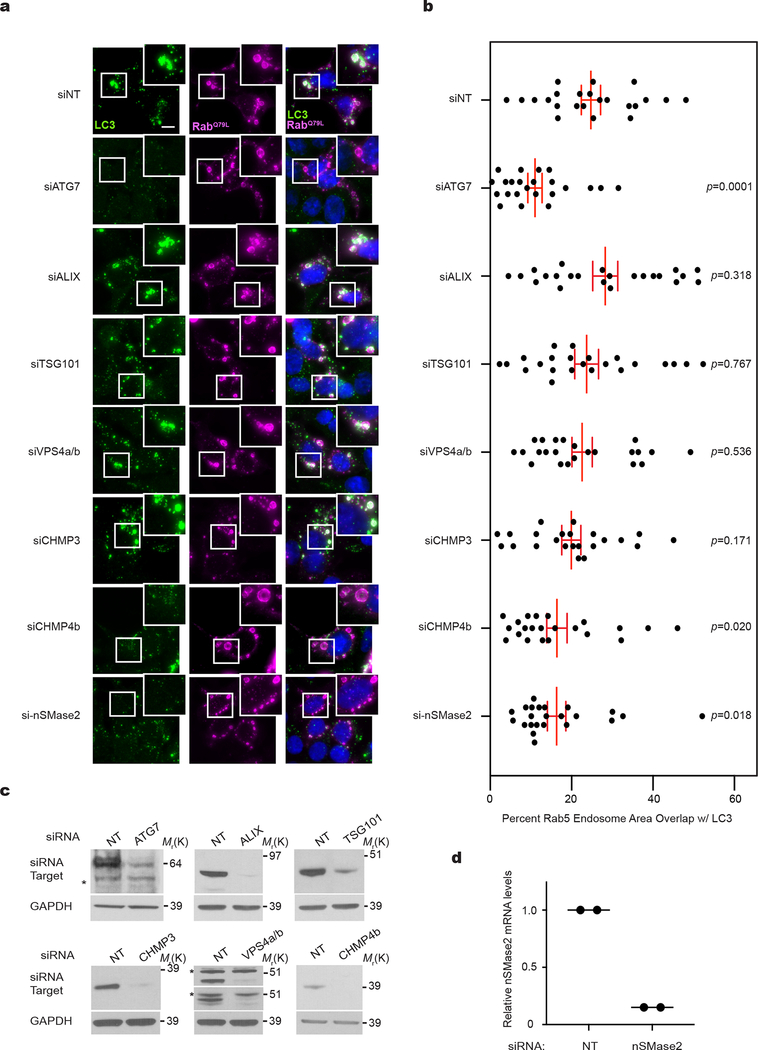
We next scrutinized whether pathways promoting inward budding and ILV formation from the MVB limiting membrane contribute to LDELS. Although the best-characterized process involves the endosomal sorting complexes required for transport (ESCRT) machinery20–23, siRNA against most ESCRT components, except CHMP4b, failed to abrogate incorporation of endogenous LC3 into mCherry-Rab5Q79L endosomes (Extended Data Fig. 6a,b,c). In parallel, we evaluated an alternative pathway in which ceramide produced by neutral sphingomyelinase 2 (nSMase2) induces vesicle curvature and inward budding from the MVB26. Indeed, siRNA-mediated depletion of nSMase2 functionally impaired the incorporation of endogenous LC3 into mCherry-Rab5Q79L endosomes (Extended Data Fig. 6a,d). Furthermore, treatment with the nSMase2 catalytic inhibitor GW4869 or shRNA depletion targeting nSMase2 potently suppressed secretion of LC3-II and LC3-binding RBPs (Fig. 7a,b,c,d; Extended Data Fig. 7a,b). Although nSMase2 ceramide production has been implicated in ILV formation, the mechanisms regulating nSMase2 activity and cargo selection remain obscure32. Bioinformatic analyses of the intracellular LC3/ATG8 interactome15 identified Factor-associated with nSMase2 activity (FAN/NSMAF)37 as a potential LC3/ATG8-associated regulator of nSMase2. Indeed, FAN robustly interacted with multiple LC3/ATG8 isoforms, including LC3B, but itself was not degraded via autophagy (Fig. 7e; Extended Data Fig. 7d). Moreover, FAN knockdown suppressed EV secretion of LC3-II, SAFB and HNRNPK, whereas classical autophagy was unaltered (Fig. 7f,g; Extended Data Fig. 7e); siRNA-mediated depletion of FAN also impeded the incorporation of endogenous LC3 into mCherry-Rab5Q79L endosomes (Extended Data Fig. 7f,g,h). Finally, we identified a putative LIR motif within FAN, whose mutation (F602A) impaired binding to LC3/ATG8 isoforms (Fig. 7h–i). Re-expression of FANF602A in FAN-deficient cells failed to rescue LDELS (Fig. 7j,k; Extended Data Fig. 7i), whereas wild-type FAN fully restored EV secretion of LC3 and LC3-binding RBPs (Fig. 7j,k; Extended Data Fig. 7i). Thus, FAN interaction with LC3 is crucial for an LC3-dependent circuit that coordinates cargo loading and biogenesis for nSMase pathway-derived EVs (Extended Data Fig. 7j).
Figure 7. LC3-dependent EV loading and secretion (LDELS) requires neutral sphingomyelinase 2 (nSMase2) and FAN.
a, Whole cell (WCL) and EV lysates from cells treated in the absence or presence of 5μM GW4869 for 24h and immunoblotted for the indicated marker proteins (n=2 biologically independent experiments). b, Nanoparticle counting for a representative experiment in Panel a (line=mean; n=1, 3 technical replicates). c, Whole cell (WCL) and EV lysates harvested from equal numbers of HEK293T cells stably expressing non-targeting (NT), ATG7 or nSMase2 (nSM2) shRNAs were immunoblotted for indicated proteins (n=3 biologically independent experiments). d, Quantification of indicated protein levels in EVs from equal numbers of stable knockdown cells in Panel c relative to non-targeting (NT) shRNA (mean ±s.e.m.; n=3 biologically independent experiments). Statistical significance calculated by one-way analysis of variance (ANOVA) coupled with Tukey’s post hoc. e, HEK293T cells co-transfected with FLAG-tagged FAN and myc-tagged LC3A, LC3B, LC3C, GABARAP (GR), GABARAPL1 (GRL1), GABARAPL2 (GRL2) or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody, and immunoblotted (WB) with indicated antibodies (n=2 biologically independent experiments). f, Whole cell (WCL) and extracellular vesicle (EV) lysates harvested from equal numbers of HEK293T cells stably expressing non-targeting (NT), ATG7 or FAN shRNAs were immunoblotted for indicated proteins (n=3 biologically independent experiments). g, Quantification of indicated protein levels in EVs from equal numbers of stable knockdown cells in Panel c relative to non-targeting (NT) shRNA (mean ± s.e.m.; n=3 biologically independent experiments). Statistical significance calculated by one-way ANOVA coupled with Tukey’s post hoc test. h, Domain map and primary LC3-interaction region (LIR) in FAN. i,, Cells co-transfected with FLAG-tagged FAN and myc-tagged LC3A, LC3B, LC3C, GABARAP (GR), GABARAPL1 (GRL1), GABARAPL2 (GRL2) or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody, and immunoblotted (WB) with indicated antibodies (n=2 biologically independent experiments). j, Whole cell lysate (WCL) and EV fractions from cells stably co-expressing non-targeting (NT) or FAN shRNA along with FLAG-tagged wild-type FAN (WT) or mutant FAN (F602A) were immunoblotted for indicated markers (n=2 biologically independent experiments). k, Quantification of indicated proteins in EVs from equal numbers of FAN knockdown HEK293T cells expressing FLAG-tagged wild-type FAN (WT) versus mutant FAN (F602A) (mean ± s.e.m.; n=3 biologically independent experiments). Statistical significance calculated by paired two-tailed t-test. Data and unprocessed blots available in Source Data Fig. 7.
Discussion
Here, we demonstrate that LC3/ATG8 and the LC3-conjugation machinery specifies cargo loading into extracellular vesicles (EVs). Previous work has intimated interconnections between the autophagy and EV production. For example, we previously demonstrated a complex between ATG12 and ATG3 that interacts with the ESCRT accessory protein ALIX (PDCDIP6), and regulates small EV release6. In addition, the autophagy proteins ATG5 and LC3 were found to co-ordinately regulate MVB acidification and exosome production via control of ATP6V1E1 interactions with the V1V0-ATPase complex5. Moreover, because late endosomes can fuse to immature autophagosomes, EV biogenesis and autophagy are proposed to be functionally connected 38, 39. In this study, we delineate a process called LDELS, highlighting a unique and previously unappreciated role for the LC3-conjugation machinery in specifying the cargo packaged into EVs.
Importantly, these functions for LC3/ATG8 and the conjugation machinery in LDELS are distinct from their established roles in classical autophagy. We propose that instead of using LC3/ATG8 located at early autophagic intermediates, LDELS employs a pool of LC3-II located at the MVB limiting membrane to directly capture RBPs and package them into ILVs; these are subsequently released as EVs via MVB fusion with the plasma membrane (Extended Data Fig. 7j). Hence, LDELS bears similarity to a growing list of autophagy-related pathways, in which LC3-II is delivered to single membranes in the endolysosomal system, including LC3-associated endocytosis (LANDO), LC3-associated phagocytosis (LAP), entosis, and macroendocytic processing40–43. LDELS also requires nSMase2 and LC3-dependent recruitment of FAN. Indeed, ceramides produced by nSMase2 can drive formation of membrane microdomains that impose negative membrane curvature and facilitate ESCRT-independent intraluminal budding at the MVB limiting membrane26. Unlike many ESCRT components, we have not detected FAN within EVs, suggesting FAN can only transiently interact with LC3 at the limiting membrane of MVBs or may act remotely to facilitate LDELS from other compartments such as the ER or Golgi apparatus, consistent with emerging roles for inter-organelle communication in MVB cargo sorting44, 45.
Our studies also suggest that LDELS is largely independent of the ESCRT machinery; only genetic depletion of CHMP4b leads to reduced LC3 incorporation into Rab5Q79L endosomes. CHMP4b has recently been implicated in endosomal microautophagy42, suggesting that these two pathways may be interconnected. However, in contrast to endosomal microautophagy, LDELS does not appear to require VPS4a/b. Further scrutinizing whether LDELS requires additional components of the ESCRT machinery and its relationship to degradative pathways such as endosomal microautophagy and LC3-associated endocytosis (LANDO) remains an important topic for future study41, 42.
Amongst the array of identified LDELS cargoes, RBPs represent the most prominent functional class of proteins. Though EVs are known to contain diverse RBPs, the mechanisms specifying their loading and secretion remain largely unclear23, 35. We demonstrate here that LC3 and ATG8 family members interact with multiple secreted RBPs, many of which contain LIR consensus motifs, and show that mutation of the LIR within SAFB (SAFBF199A) is sufficient to impair its secretion via EVs. Interestingly, many LDELS targets are detected in RNA granules, including stress granules and P-bodies46, 47. Autophagy is proposed to regulate stress granule clearance48, but its role in RNA granule homeostasis appears more complex and nuanced beyond simple turnover of stress granule components49–52. We are actively investigating how LDELS interacts with classical autophagy to regulate the dynamic remodelling of RNA granules.
Consistent with its role in RBP secretion, the loss of LDELS via genetic ablation of ATG7 and ATG12 leads to profound changes in the extracellular small non-coding RNA profiles present in EVs. EVs contain diverse small RNA bio-types and the mechanisms that contribute to the packaging and secretion of small RNAs are only beginning to be unraveled35, 53–56. Although LDELS affects secretion of multiple small RNA bio-types, its greatest impact is on snoRNAs, a class of nuclear small non-coding RNAs involved in ribosomal RNA modification36. Indeed, snoRNAs (or fragments thereof) are readily detected in EVs purified from human plasma and cell culture conditioned media57–63. However, the mechanisms specifying snoRNA secretion and the functions of extracellular snoRNAs remain largely unclear. Interestingly, a number of RBPs secreted via LDELS can preferentially bind snoRNAs, including SAFB64. Furthermore, processed forms of snoRNAs interact with hnRNPs and cellular stresses such as starvation and oxidative damage promote the accumulation of snoRNAs in the cytoplasm65–67. Deciphering whether LDELS directs the EV secretion of snoRNAs as a means of regulating protein homeostasis during cell stress remains an intriguing question for future investigation.
Originally described as a mechanism to selectively eliminate proteins, lipids and nucleic acids from cells, EVs are emerging as an important mode of intercellular communication in diverse physiological and pathological processes20–23. Elaborating how LDELS impacts intercellular communication and non-cell autonomous functions in vivo remains an important topic for future study. Overall, our studies expand the repertoire of putative targets of secretory autophagy and define a previously unrecognized mechanism by which the LC3-conjugation machinery specifies cargo loading into a subpopulation of EVs.
Materials and Methods
Cell culture
HEK-293T cells (ATCC, CRL-3216), including all cell derivatives generated in this study, were cultured in DMEM (ThermoFisher, 11995065) supplemented with 10% FBS, 25 mM HEPES, 100U/ml penicillin, and 100μg/ml streptomycin (ThermoFisher, 15140163). Murine RAW 264.7 macrophages and murine B16F10 were gifts from Matthew Krummel (UCSF) and were cultured in DMEM (ThermoFisher, 11995065) supplemented with 10% FBS, 25 mM HEPES, penicillin, and streptomycin. Murine LLC1 cells were purchased from ATCC (CRL-1642) and were cultured in DMEM (ThermoFisher, 11995065) supplemented with 10% FBS, 25 mM HEPES, penicillin, and streptomycin. All cell lines were authenticated using STR profiling (IDEXX BioResearch) and routinely tested for mycoplasma contamination (Sigma, MP0025).
To induce biotin-labelling, HEK293T cells expressing myc-BirA* or myc-BirA*-LC3B were incubated with 50 μM biotin in DMEM with all supplements except FBS for 24h. Unless indicated, conditioned media and EV preparations were collected following 24h incubation in DMEM containing all supplements except FBS. For autophagy flux assays, cells were incubated with 50 μM chloroquine (Sigma, C6628) or 50 μM Bafilomycin A1 (Sigma, B1793) as indicated for 1 h prior to lysis. Treatment with 5μM GW4869 (Cayman, 13127) or vehicle (DMSO, Sigma) in serum free DMEM for 24h was used to inhibit nSMase activity.
Plasmid constructs
The following plasmids were obtained or are available on Addgene: pcDNA3.1-myc-BirA* (mycBioID; Addgene #35700), pBABE-GFP-LC3B (Addgene #22405), pBABE-HRasV12 (Addgene #9051), mCherry-Rab5Q79L (Addgene #35138). Human LC3B (NM_022818.4), SAFB (NM_001201338.1), HNRNPK (NM_002140.4), LC3A (NM_032514.3), LC3C (NM_001004343.2), GABARAP (NM_007278.1), GABARAPL1 (NM_031412.2), GABARAPL2 (NM_007285.6), SF3A1 (NM_005877.5), LARP1 (NM_015315.4), G3BP1 (NM_005754), FAN (NM_003580.3) were subcloned from mRNA isolated from human cell lines that was reverse transcribed using AccuScript High Fidelity Reverse Transcriptase (Agilent) and cDNA amplified using PfuUltra II Hot Start DNA polymerase and gene specific primers (LC3B Fwd: agtcggatccatgccgtcggagaagacct; LC3B Rev: gactctcgagttacactgacaatt tcatcccg; SAFB Fwd: agctggatccatggcggagactctgtcag; SAFB Rev: agctgtcgac tcagtagcggcgagtgaag; HNRNPK Fwd: agtcggaattcgcatggaaactgaacagccagaag; HNRNPK Rev: agtcctcgagttagaatccttcaacatctgcata; LC3A Fwd: agtcggatccatgccctcagaccggcct; LC3A Rev: gact ctcgagtcagaagccgaaggtttcct; LC3C Fwd: agtcggatccatgccgcctccacagaaaat LC3C Rev: gact ctcgagctagagaggattgcagggtc GABARAP Fwd: agtcggatccatgaagttcgtgtacaaagaaga GABARAP Rev: gactctcgagttaaagaccgtagacactttc; GABARAPL1 Fwd: agtcggatccatgaagttccagtacaaggac; GABARAPL1 Rev: gactctcgagtcatttcccatagacactctc; GABARAPL2 Fwd: agtcagatctatgaagtggatgttcaaggag; GABARAPL2 Rev: gactctcgagtcagaagccaaaagtgttctc; SF3A1 Fwd: agtctgatcaatgccggccggacccgtg; SF3A1 Rev: gatcctcgagctacttcttcctcccgcctc; LARP1 Fwd: agctagatctatgctttggagggtgcttttg; LARP1 Rev: agctctcgagtcactttcccaaagtctgtgt; G3BP1 Fwd: agct ggatccatggtgatggagaagcctagt; G3BP1 Rev: agctgtcgactcactgccgtggcgcaagc; FAN Fwd: agcttgatcaatggcgtttatccggaagaag; FAN Rev: agctctcgagttaatactgcaatttccagaatata). Subsequently, the cDNAs were subcloned into pcDNA3 between the BamHI and XhoI or EcoRI and XhoI restriction sites downstream of an N-terminal myc-tag or 3xFlag-tag. All constructs were verified by sequencing.
To generate pBABE-myc-BirA*, myc-BirA* was amplified with flanking primers (Fwd: agctaagcttaccggtgccaccatggaacaaaaactc; Rev: gactctcgagttacttctctgcgcttctcagg) from pcDNA3.1-myc-BirA* and sub-cloned into pBABE-GFP-LC3 between AgeI and SalI, entirely replacing the GFP-LC3 open reading frame. To generate pBABE-myc-BirA*-LC3, myc-BirA* was amplified with flanking primers (Fwd: agctaagcttaccggtgccaccatggaacaaaaactc; Rev: gactggatcccttctctgcgcttctcagg) from pcDNA3.1-myc-BirA* and sub-cloned into the HindIII and BamHI sites in front of pcDNA3-FLAG-LC3, replacing the FLAG sequence. Subsequently, the myc-BirA*-LC3 fragment was sub-cloned from pcDNA3-myc-BirA*-LC3 using AgeI (introduced 3’ to the HindIII site) and XhoI into pBABE-GFP-LC3 between AgeI and SalI, replacing the GFP-LC3 open reading frame. All constructs were verified by sequencing.
Site-directed mutagenesis of FLAG-tagged SAFB and FAN was performed via QuikChange PCR. Overlapping primers carrying the desired mutation (SAFB Fwd: ttagatacttca tcatctgacgccactatattacaggaaattgaa; SAFB Rev: ttagatacttcatcatctgacgccactatattacaggaaattgaa; FAN Fwd: gattccccaggtgaagagtctgccgaagacctgaccgaagaaagc; FAN Rev: gctttcttcggtcaggtcttcggcagactcttcacctggggaatc) were used to amplify pcDNA3 FLAG-SAFB and FLAG-FAN, respectively, and template plasmid was eliminated via DpnI digestion. Subsequently, individual clones were sequenced to verify mutagenesis of the desired sites. Finally, for FAN rescue experiments, wild-type FAN and FAN F602A were amplified with primers incorporating an N-terminal myc-tag (myc-FAN/F602A Fwd: agcttgatcaaccatggaacaaaaacttatttctgaagaggatctgggctccatggcgttt atccggaagaag; Rev: agctctcgagttaatactgcaatttccagaatata) and cloned between the BamHI and XhoI of pLenti-CMV-blast (Addgene: #17486).
Retroviral and lentiviral packaging, infection and selection
Retroviral pBABE expression vectors were packaged and target cells transduced according to established protocols68. Briefly, Phoenix-AMPHO cells (gift from Craig McCormick, Dalhousie University) were seeded and transfected with retroviral vectors using polyethylenimine (PEI). Virus-containing conditioned media was collected 2 days after transfection and clarified using a 0.45 μM filter. Prior to infection, virus-containing media was diluted 1:4 in DMEM growth media and the mix was supplemented with polybrene to a final concentration of 8 μg/ml. Subsequently, the viral transduction mix (5 ml total volume/10 cm culture dish) was incubated with HEK293T cells for 24h. Cells were selected 24 h post-transduction with 1μg/ml puromycin for 2 days. To package lentivirus, HEK293T cells were seeded and co-transfected with packaging vectors psPAX2 and pMD2.G, and individual pLKO.1 transfer vectors. Virus collection, infection, and puromycin selection of stable cell pools was carried as above.
Stable isotope labelling with amino acids in cell culture (SILAC), proximity-dependent biotinylation, and affinity purification of biotin-labelled secretome
For experimental replicates #1 and #2, HEK293T stably expressing myc-BirA* were grown in Lys- and Arg-free DMEM supplemented with 10% dialysed FBS (ThermoFisher), 25 mM HEPES, penicillin-streptomycin, and light (K0) Lys (50 μg/ml) and light (R0) Arg (50 μg/ml), whereas HEK293T stably expressing myc-BirA*-LC3 were grown in Lys- and Arg-free DMEM media with supplements and the heavy isotopes K8 Lys (Cambridge Isotopes) and R10 Arg (Cambridge Isotopes). For experimental replicate #3, a label swap was performed to rule out potential labelling artefacts; the heavy and light labelling was reversed between myc-BirA* and myc-BirA*-LC3 cells. Cells were labelled in SILAC media and expanded into 18 × 15 cm culture dishes of equal cell density (80% confluent).
Cells were gently washed twice in 20 ml EBSS and incubated with 15 ml serum free SILAC media supplemented with 50 μM biotin to trigger biotin labelling according to standard protocols69. Conditioned media (CM) was collected 24 h later and centrifuged serially at 200g for 10 min to pellet cells, followed by 2000g for 20 min to pellet cellular debris and apoptotic bodies. CM was routinely stored at −80oC prior to analysis. CM was precipitated by adding trichloroacetic acid (TCA) to a final concentration of 15% and incubating on ice for 1 h. Subsequently, precipitated protein was pelleted by ultracentrifugation at 200,000g for 18 h. Pellets were washed with 10 ml of ice-cold acetone, re-centrifuged at 200,000g for 1 h, and air dried. To solubilize protein, pellets were resuspended in 8M guanidine hydrochloride (Sigma G3272), 100 mM Tris hydrochloride pH 8.0 for 1 h with manual trituration every 15 min. Re-solubilized protein from myc-BirA* or myc-BirA*-LC3 was pooled separately and protein concentrations were quantified via BCA protein assays (ThermoFisher, 23225), diluting samples below BCA threshold limits for chaotropic agents. Subsequently, samples from light myc-BirA* and heavy myc-BirA*-LC3 were mixed in equal protein proportions and the combined sample was diluted to 2.5 mM guanidine hydrochloride with 50 mM Tris pH 8.0. Biotin-labelled proteins were captured on 120 μl of packed high capacity neutravidin sepharose (ThermoFisher, 29204) overnight at 4°C, followed by 3 washes with 5 ml 2.5 mM guanidine hydrochloride, 50 mM Tris HCl pH 8.0 prior to analysis by immunoblotting or mass spectrometry.
Mass spectrometry of affinity purified proteins—Sample preparation, LC-MS/MS, peptide identification, and protein quantification
Neutravidin sepharose and affinity purified proteins were resuspended and mildly denatured in 1 M guanidine hydrochloride, 1 mM calcium chloride and 100 mM Tris buffer pH 8.0. Disulfide bonds were reduced with 4 mM tris(2-carboxyethyl)phosphine (Sigma, C4706), and free thiols were alkylated with 10 mM iodoacetamide (Sigma, I1149). Subsequently, 10 μg of mass spectrometry (MS) grade trypsin (ThermoFisher) dissolved in 10 μL 50 mM acetic acid was added to proteins on beads, and incubated at 37° C for at least 18 h. The eluate was transferred to a new tube, acidified to a final concentration of 0.3% trifluoroacetic acid (pH < 3) and desalted by reversed phase C18 solid phase extraction (SPE) cartridge, using either a Sep-pak (Waters Corp.) or SOLA SPE (ThermoFisher), and then dried down in a speed-vac. Dried peptides were stored at −20°C, and resuspended in 2% acetonitrile, 0.1% formic acid in a bath sonicator for 5 min to a concentration of 2 μg/μL before MS analysis.
Peptides (0.5 – 1 μg) were submitted for nano-LC-ESI-MS/MS, using a 195 min reversed phase (2.4 – 32% acetonitrile, 0.1 – 0.08% formic acid) buffer gradient with a 15 cm Acclaim™ PepMap™ 100 C18 analytical column (2 μm beads, 75 μm i.d.), running at 200 nL/min on a Dionex Ultimate 3000 RSLCnano pump, in-line with a hybrid quadrupole-Orbitrap Q-Exactive Plus mass spectrometer (ThermoFisher). A data dependent method with a parent ion scan at a resolving power of 70,000 was run on each sample and a top 12 method for replicates 1 and 2 and top 15 method for replicate 3 selected for the 12 and 15 most intense peaks for MS/MS using HCD fragmentation (normalized collision energy 27), respectively. Dynamic exclusion was activated such that parent ions are excluded from MS/MS fragmentation for 20s after initial selection.
For protein identification and quantification, Thermo.RAW files were analyzed by Maxquant70. The recorded spectra from three independent biological replicates were searched against the human reference proteome from UniProt (2014–11-16 release, with 20,193 unique proteins, including isoforms, on SwissProt and 47,673 entries in TrEMBL) using MaxQuant, version 1.5.1.2. Maxquant uses the Andromeda search engine to identify peptides. Search parameters allowed for 4.5 ppm peptide ion search tolerance, 7.5 ppm centroided fragment ion tolerance, and 2 missed tryptic cleavages. Oxidation of methionine, phosphorylation of serine/threonine/tyrosines, and N-terminal acetylation were allowed as variable modifications, while carbamidomethylation of cysteines was selected as a constant modification and a threshold peptide spectrum match (PSM) false discovery rate (FDR) and protein FDR of 1% was allowed. Quantification of SILAC ratios was performed by Maxquant on the MS1 level and the resulting ratios for all replicates were compared using statistical tools found in the Perseus bioinformatics analysis package71. Proteins with ratio quantification in only one replicate were filtered out. Statistical significance was determined by applying a one-sample student T-test to the replicates with a p-value cut off of p = 0.05. Protein group ratios and p-values are included in Supplementary Table 1.
Extracellular Vesicle Preparation and Characterization
Extracellular vesicles were purified according to standard differential centrifugation protocols72. Briefly, cells seeded in 15 cm culture dishes at approximately 70% confluence were incubated with serum free DMEM for 24 h. Conditioned media was collected and centrifuged serially at 200g for 10 min to pellet cells, 2000g for 20 min to pellet cellular debris and apoptotic bodies, 10,000g for 30 min to pellet large extracellular vesicles, and 100,000g in an ultracentrifuge for 2 h to pellet extracellular vesicles (EVs). Crude EVs pellets were then gently triturated in PBS using a P1000 pipette, diluted further in PBS (12 ml), and ultracentrifuged for an additional 70 min at 100,000g to generate EV preparations for further analysis as described below. Importantly, for all comparisons of EVs between experimental conditions, results from individual cohorts were corrected as indicated based on total cell number or whole cell lysate protein concentration to ensure that EV or EV protein quantification was not confounded by seeding differences.
Sucrose density gradient separation was utilized to generate highly purified EV preparations and to analyse the co-fractionation of LC3-II and LC3 binding proteins with EV marker proteins on linear sucrose gradients. Briefly, the 100,000g EV pellets generated via differential centrifugation as described above were thoroughly resuspended in 100 μl 10% sucrose solution and gently layered on to a continuous 10–60% sucrose gradient formed on a gradient station (BioComp Instruments) and then ultracentrifuged at 210,000g for 18h. Subsequently, 1 ml fractions from the gradient were top unloaded, weighed, and diluted in 10 ml of PBS. The diluted fractions were spun at 100,000 for 70 min and pellets resuspended in urea lysis buffer for analysis by immunoblotting.
For protease protection assays, equal amounts of EVs were resuspended in PBS or PBS containing 1% TritonX-100 in the absence or presence of 100 μg/ml trypsin for 30 min at 4°C. Subsequently, the reactions were stopped by the addition of 2x protein sample buffer and the lysates subjected to immunoblotting. Established protocols were used to perform transmission electron microscopy of HEK293T EVs prepared by differential centrifugation5.
Nanoparticle tracking analysis (NTA) for EV preparations was performed with a LM10 nanoparticle analyser (NanoSight, Worchestershire, UK). Camera level was set at 9 for all recordings. Camera focus was adjusted to make the particles appear as individual dots with surrounding refractory rings. Five 60 s videos were recorded for each sample with a delay of 10 seconds between each recording. All post-acquisition functions were set at automatic, with the exception of detection threshold, which was set to 2. Protein content in EV pellets was quantified via BCA assay (ThermoFisher, 23225), diluting samples below the threshold limits for interfering detergents and chaotropic agents.
Mass spectrometry of Extracellular Vesicle Proteins—Sample preparation, Tandem Mass Tag (TMT) Labelling, LC-MS/MS, peptide identification, and protein quantification
Serum free media conditioned for 24 h from 4 bioreplicates of approximately 5 × 109 wild-type, ATG7−/− and ATG12−/− HEK293T cells was collected and extracellular vesicles (EVs) were isolated using the standard differential centrifugation protocols described above. Upon purification, EVs from each condition were lysed in 800 μL of radioimmunoprecipitation (RIPA) buffer (25mM Tris•HCl pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), supplemented with 2% SDS and sonicated with a probe sonicator set to an amplitude of 8 for 10 bursts of 10s. Each 800 μL sample was then diluted in 3.5 mL ice-cold 100% acetone and incubated at −20° for 12 h. Subsequently, samples were spun in an ultracentrifuge at 200,000g for 18 h, the acetone decanted, and pelleted protein was left to air dry. Dried pellets were stored at −80° prior to solubilization and tandem mass tag (TMT) labelling.
Biological replicates of HEK293T WT, ATG7−/−, and ATG12−/− of precipitated EV proteins were resuspended in 30 μL 6 M guanidine-chloride (Gdn), 100 mM Tris pH 8.0, 10 mM TCEP, 40 mM 2-Chloroacetamide (CAA). Proteins were denatured for 1h at 37°C and quantified with 660nm Protein Assay Reagent (Thermo). The 4 bioreplicates from each condition were pooled into 2 separate samples and subsequently diluted six-fold with 150 μL 100 mM Tris pH 8.0. 200 μg of protein for each sample was trypsinized with 4 μg trypsin (Thermo) in an orbital shaker at 250 rpm, 37°C for ~ 20 h. After 20 h, trypsin digestion was stopped by acidifying samples with 10% trifluoroacetic acid (TFA) to a final concentration of 0.5% TFA, then desalted with SOLA C18 reverse phase SPE columns (Thermo), where the samples were washed with 1.5 mL 0.1% TFA, then 0.5 mL 0.1% formic acid (FA), 2% acetonitrile (ACN), and eluted with 450 μL 0.1% FA, 50% ACN. These samples were dried by speed-vac, then resuspended in 10 μL 50 mM HEPES (pH 8.5) and quantified by peptide quantification kit (Thermo).
For Tandem Mass Tag (TMT) labelling, 800 μg of each TMT six-plex (Thermo #90061) isobaric compound was resuspended with 41 μL 100% anhydrous acetonitrile (Sigma). 15 μg of peptides for each one replicate were individually combined with 7.69 μL (or 150 μg) of the TMT isobaric tags. Samples were incubated at 25°C for 1 h, and then the reaction was quenched with 8 μL 5% hydroxylamine for 15 min. After labelling, the six individually labelled samples are pooled and 1/3 of the pooled sample (30 μg is fractionated by high-pH reverse phase fractionation into 8 fractions and dried down by speed-vac. The fractions are then resuspended in 0.1% FA, 2% ACN before LC-MS/MS analysis.
For LC-MS/MS analysis, 1 μg TMT-labelled peptides were analysed on a 15 cm C18 analytical column, in-line with a Q-Exactive Plus mass spectrometer. The peptides were separated on a multi-slope, 100 min gradient (6.4% - 27.2% ACN with 0.1% FA for 80 min at 0.2 uL/min, then 27.2% - 40% ACN with 0.09% FA for 15 min at 0.3 uL/min, then 40% - 56% ACN with 0.09% FA for 5 min at 0.3 μL/min, and then washed for 3 min). Data dependent acquisition with MS1 resolution of 70,000, top15 method, and HCD normalized collision energy of 32 was used, with MS2 resolution of 35,000 and an isolation window of 0.7 m/z. Dynamic exclusion was activated for 30s after initial parent ion selection.
Eight injections of the different fractions of TMT-labelled exosome peptides, were analysed together via Maxquant (v1.6.0.16). Search parameters for peptide search tolerance was 4.5 ppm, for centroided MS2 tolerance was 7.5 ppm, and allowed for 2 missed tryptic cleavages. Constant modification of carbamidomethylation of cysteines and variable modifications of N-terminal acetylation, methionine oxidation, and Ser/Thr/Tyr phosphorylation were allowed. Peptide spectrum match FDR and protein FDR was set at 1%. Match between runs was enabled to increase peptide identifications. Type is set to “Reporter ion MS2,” and TMT6plex-Nter126 to 131, TMT6plex-Lys126 to 131 labels are selected. The resulting quantifications are then median normalized for each protein group and statistical analysis (two-sample two-sided T-test) was applied in Perseus, with a statistical significance threshold of p < 0.05.
Isolation and sequencing of small and large RNA species in EVs
Serum free media conditioned for 24h from 4.5 × 108 wild-type (WT), ATG7−/− and ATG12−/− HEK293T cells was collected and extracellular vesicles (EVs) isolated according to the standard differential centrifugation protocols described above. Small and large RNA was isolated from EVs and cells using the Macherey-Nagel NucleoSpin kit (# 740971) according to the manufacturer’s protocol.
Short RNA libraries were prepared using the SMARTer smRNA-Seq Kit for Illumina (Takara #635030). Briefly, input RNA from 3 bio-replicates of wild-type, ATG7−/− and ATG12−/− cell and EV RNA samples were first polyadenylated and then reverse transcribed using Primescript Reverse Transcriptase and oligo(dT) and SMART smRNA oligos. Subsequently, primers with unique Illumina i5 and i7 adapter sequences were used to PCR amplify the cDNA from the different samples for 12 cycles. Libraries were then subjected to size selection to enrich for small RNA species using SPRI AMPure Beads (Beckman Coulter, #A63880) and the yield and concentration were assessed using a Qubit 2.0 Fluorometer (Invitrogen) and DNA 1000 chips on an Agilent 2100 Bioanalyzer (Applied Biosystems), respectively. Libraries were then diluted to 8 nM and pooled together.
Large RNA libraries were prepared using the SMARTer Stranded Total RNA Sample Prep Kit - HI (Takara # 634875). Input RNA from 3 bio-replicates of wild-type, ATG7−/− and ATG12−/− cell and EV RNA samples were depleted of ribosomal RNA, chemically fragmented and then reverse transcribed using Primescript Reverse Transcriptase, random primers and SMART stranded oligos. The cDNA was then purified using SPRI AMPure Beads (Beckman Coulter, #A63880). Subsequently, primers with unique Illumina i5 and i7 adapter sequences were used to PCR amplify the cDNA from the different samples for 22 cycles. Libraries were then purified using SPRI AMPure Beads (Beckman Coulter, #A63880) and the yield and concentration were assessed using a Qubit 2.0 Fluorometer (Invitrogen) and DNA 1000 chips on an Agilent 2100 Bioanalyzer (Applied Biosystems), respectively.
Each library was diluted a final concentration of 4.2 nM and pooled prior to single-end, 50-bp sequencing on two separate lanes of a HiSeq 4000 (UCSF Center for Advanced Technology, San Francisco, CA). For small RNAs, raw reads were first trimmed using cutadapt (v2.3) by removing the polyA tail and the tri-nucleotide template switch. Trimmed reads were then mapped to the human reference genome (GRCh38/hg38) using bowtie2 (v2.3). Bedtoold (intersectBed) was used to count the number of reads that overlap annotated small RNA species (DASHRv2.073). Differential expression analysis was performed using DESeq2. For RNA-seq, salmon (v0.14) was used to map reads to the human transcriptome (Gencode v28). Tximport was used to import the data into R and DESeq2 were then used to perform differential gene expression analysis.
Bioinformatic Analyses
Candidate proteins identified by mass spectrometry of the BirA*-LC3 labelled secretome were subdivided into Class I proteins, which were statistically enriched (log2 BirA*-LC3/BirA* > 1) in the three quantitative mass spectrometry bioreplicates, and Class II proteins, which were enriched in 2 out of 3 biological replicates. Unless indicated, higher stringency Class I and lower stringency Class II data sets were combined to generate the BirA*-LC3 labelled secretome for all bioinformatic analyses. The proportion of candidates detected in human plasma was established by searching the BirA*-LC3 labelled secretome against the Plasma Proteome Database74. Gene Ontology (GO) analysis was performed using protein analysis through evolutionary relationships (PANTHER) and the top 10 terms for molecular function and cellular component plotted according to their -log10 false discovery rate75. The network map of the BirA*-LC3 labelled secretome was plotted with Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) using experiments and databases for interaction sources with a minimum interaction confidence score of 0.40076. Extracellular exosome and RNA-binding proteins within the network were identified using PANTHER GO analyses for individual candidates. The number of candidate proteins associated with the GO term extracellular exosome was obtained using PANTHER. The top ten proteins with the greatest connectivity to the combined class I and class II data were identified using the protein-protein interaction tool (PPI) in the Enrichr gene set enrichment analysis suite and plotted according to their adjusted p-values77. Overlap between data sets, including individual Q-MS bioreplicates, LC3 and broader ATG8 interactome15 and mRNA binding proteome78 were performed using Biovenn79.
For tandem mass tag (TMT) Q-MS, the overlap between candidates statistically enriched in EVs from wild-type versus EVs from ATG7−/− and ATG12−/− (p-value <0.05; log2 WT/ATG7−/− or WT/ATG12−/− >0.5; n=2) and the BirA*-LC3 labelled secretome (class I and II), as well the ATG7 and ATG12-dependent EV proteome versus the stress granule47 and processing body46 proteomes were performed using Biovenn13. The top ten proteins with the greatest connectivity to the ATG7 and ATG12-dependent EV proteome were identified using the protein-protein interaction tool (PPI) in the Enrichr gene set enrichment analysis suite and plotted according to their adjusted p-values10. GO analysis of the ATG7 and ATG12-dependent EV proteome was performed with PANTHER and the top 10 non-redundant terms for molecular function plotted according to their -log10 false discovery rate 8.
For RNA sequencing data, the overlap between RNA candidates statistically enriched in EVs from wild-type (WT) versus EVs from ATG7−/− and ATG12−/− (p-value <0.05; log2 WT EV RNA/ WT cell RNA vs ATG7−/− EV RNA/ATG7−/− cell RNA or WT EV RNA/WT cell RNA vs ATG12−/− EV RNA/ ATG12−/− cell RNA >0.5) using were performed using Biovenn13. The classes of different RNA species identified were annotated using metascape80 and the heatmap of snoRNA species enriched in WT EVs relative to ATG7−/− and ATG12−/− EVs was plotted using Prism statistical analysis software.
Antibodies
Immunoblotting: Streptavidin-HRP (Thermo Fisher, Cat#21130, Lot# TA263511, SB241752A; 1:40000)), rabbit anti-MAP1LC3 is commercially available (Millipore, Cat#ABC232; 1:1,000), rabbit anti-myc (Cell Signaling Technology, Cat#2278S. Clone#71D10; Lot#5 1:1000), mouse anti-myc (Sigma, Cat#M5546, Clone#9E10, Lot#128M4898V; 1:1000), mouse anti-GAPDH (Millipore, Cat#MAB374, Lot#3075597; 1:1000), guinea pig anti-p62/SQSTM1 (Progen/Cedarlane, Cat#GP62-C, Lot# 703241–1; 1:1000), rabbit anti-OPTN (Abcam, Cat#ab23666, Lot# GR3270251; 1:1000), goat anti-ATG7 (Santa Cruz Biotechnology, Cat#sc-8668, Clone#N-20, Lot#B0316; 1:1000), mouse anti-Nbr1 (Santa Cruz Biotechnology, Cat#sc-130380, Clone#4BR, Lot#B2019; 1:1000), mouse anti-ALIX (Cell Signaling Technology, Cat#2171S, Clone#3A9, Lot#5; 1:1000), rabbit anti-Hsc70 (Cell Signaling Technology, Cat#8444, Clone#D12F2, Lot#1; 1:1000), rabbit anti-TSG101 (BD Biosciences, Cat#612696, Lot#8172853; 1:1000), mouse anti-CD9 (Millipore, Cat#CBL162, Clone#MM2–57, Lot#2691299; 1:1000),mouse anti-FLAG-M2 (Sigma-Aldrich, Cat#F3165, Clone#M2, Lot#SLBN8915V, SLCC4005, SLBF1225; 1: 5000), rabbit anti-FLAG/DYKDDDDK (Cell Signaling Technology, Cat#2368S, Lot#12; 1:1000), rabbit anti-SAFB (Abcam, Cat#ab187650, Clone#EPR13588, Lot#GR167184–1, GR167184–4, GR167184–5; 1:500), mouse anti-HNRNPK (Abcam, Cat#ab39975, Clone#3C2, Lot#GR275360–20, GR275360–6, GR275360–8, GR148352–24; 1:1000), rabbit anti-ATG12 (Cell Signaling Technology, Cat#2010S, Clone#human specific, Lot#2, 5; 1:1000), rabbit anti-ATG14 (MBL, Cat#PD026, Lot#5; 1:1000), rabbit anti-VPS24/CHMP3 (Abcam, Cat#ab175930, Clone#EPR12821, Lot# YK032912CS; 1:1000), rabbit anti-VPS4a (Sigma, Cat#SAB4200022, Lot#119K4789; 1:1000), rabbit anti-VPS4b (Abcam, Cat#ab102687, Lot# GR60499–4; 1:1000), rabbit anti-CHMP4b (Abcam, Cat#ab76334, Lot# GR26679–7; 1:1000), rabbit anti-FIP200/RB1CC1 (Proteintech, Cat#17250–1-AP, Lot#/Barcode#00012978; 1:1000), rabbit anti-phospho-S6 (Cell Signaling Technology, Cat#4858S, D57.2.2E, Lot#3; 1:1000), rabbit anti-S6 (Cell Signaling Technology, Cat#2217L, Clone#5G10, Lot#3; 1:1000), rabbit anti-nSMase2 (Santa Cruz Biotechnology, Cat#sc-67305, Clone#H-195, Lot#E1116; 1:1000), rabbit anti-CD9 (Abcam, Cat#ab92726, Clone#EPR2949, Lot#GR260186–21, 1:1000), rabbit anti-CD63 (Abcam, Cat#ab134045, Clone#EPR5702, Lot#GR3212162–2; 1:1000), rabbit anti-CD81/TAPA (Abcam, Cat#ab109201, Clone# EPR4244, Lot#GR181359; 1:1000), rabbit anti-FAN/NSMAF (Abcam, Cat#ab81260, Lot#GR32198–5; 1:1000), rabbit anti-ATG3 (Sigma-Aldrich, Cat#A3231, Lot#NA;1:500), rabbit anti-LC3A (Cell Signaling Technology, Cat#4599T, Clone#D50G8, Lot#5; 1:1000), rabbit anti-LC3B (Cell Signaling Technology, Cat#3868T, Clone#D11, Lot#11; 1:1000), rabbit anti-LC3C (Cell Signaling Technology, Cat#14736T, Lot#1; 1:1000), rabbit anti-GABARAP (Cell Signaling Technology, Cat#13733T, Clone#E1J4E, Lot#3; 1:1000), rabbit anti-GABARAPL1 (Cell Signaling Technology, Cat#26632T, Clone#D5R9Y, Lot#1; 1:1000), rabbit anti-GABARAPL2 (Cell Signaling Technology, 14256T, Clone#D1W9T, Lot#3; 1:1000), rabbit anti-LARP1 (Cell Signaling Technology, Cat#14763S, Lot#1; 1:500), mouse anti-G3BP1 (BD Biosciences, Cat#611126, Clone#23/G3BP, Lot#39834, 1:500), mouse anti-SF3A1 (Abcam, Cat#ab139271, Clone#OTI2C4, Lot#GR128640–10, 1:500), Peroxidase-AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson, Cat#711–035-152, Lot#143451; 1:5000), Peroxidase-AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (Jackson, Cat#706–035-148, Lot#108535; 1:5000), Peroxidase-AffiniPure Donkey Anti-Goat IgG (H+L) (Jackson, Cat#705–035-147, Lot#125729; 1:5000), and Peroxidase-AffiniPure Donkey Anti-Mouse IgG (H+L) (Jackson, Cat#715–035-150, Lot#142341;1:5000).
EV immuno-isolation: mouse anti-CD9 (Millipore, Cat#CBL162, Clone#MM2–57, Lot#2691299), mouse anti-CD63 (Abcam, Cat#ab8219, Clone#MEM-259, Lot#GR3196070–12), mouse anti-CD81 (BD Biosciences, Cat#555675, Lot#7096566).
Immunofluorescence
rabbit anti-LC3B (1:500, MBL, Cat#PM036, Lot#031) and mouse anti-CD63 (1:200, Abcam, Cat#ab8219, Clone#MEM-259, Lot#GR3196070–12), mouse anti-LC3(1:200, Nanotools, Cat# 0231–100/LC3–5F10, Clone#5F10, Lot#0231S), rabbit anti-CD9 (1:200, Abcam, Cat#ab92726, Clone#EPR2949, Lot#GR260186–21), rabbit anti-LC3B (1:200, Cell Signaling Technology, Cat#3868T, Clone#D11, Lot#11), AlexaFluor goat anti-rabbit 488 (1:500, ThermoFisher, Cat#A-11034, Lot#52700A), goat anti-mouse 647 (1:500, ThermoFisher, Cat#A21235, Lot#490187).
Immunoblotting
To generate whole cell lysate, cells were lysed in NP40 buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 1% NP40, 5% glycerol) plus protease inhibitor cocktail (Sigma-Aldrich), 2 mM EDTA, 2 mM EGTA, 10 mM β-glycerophosphate, 2.5 mM sodium pyrophosphate, 10 mM NaF, 1 mM Na3VO4. For EV lysate and re-solubilization of precipitated material for immunoblotting, samples were resuspended in Urea buffer (50mM Tris-HCl, pH 8.0, 8M urea, 2% SDS, 10mM Sodium Fluoride, 5mM EDTA) plus protease inhibitor cocktail. Lysates were cleared by centrifugation, quantified by BCA assay (ThermoFisher, 23225), mixed with sample buffer, resolved by SDS-PAGE, and transferred to polyvinylidene fluoride membrane (PVDF). Membranes were blocked for 1 h in 5% milk in PBS with 0.1% Tween 20, incubated in primary antibody overnight at 4°C, washed, incubated for 1h at RT with HRP-conjugated secondary antibodies (1:5,000; Jackson), washed, and visualized via enhanced chemiluminescence (Thermo Fisher) on film. Immunoblots were quantified by densitometry using Fiji.
Immunoprecipitation
For immunoprecipitation of endogenous SAFB and HNRNPK, cells transiently transfected with myc-tagged BirA*, LC3B were lysed 24 h post-transfection in NP40 buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 1% NP40, 5% glycerol) plus protease inhibitor cocktail (Sigma-Aldrich), 2 mM EDTA, 2 mM EGTA, 10 mM β-glycerophosphate, 2.5 mM sodium pyrophosphate, 10 mM NaF, 1 mM Na3VO4. Lysates were pre-cleared with protein A/G beads (Santa Cruz, sc-2003) and normal mouse IgG (Santa Cruz, sc-2025) at 4°C and incubated overnight with mouse anti-myc antibody (Sigma, M5546; 1 μg/200 μg lysate) at 4°C. Immune complexes were captured by incubation with protein A/G beads for 4h at 4°C and then washed 5 times with NP40 buffer plus inhibitors, eluted with sample buffer, and analysed by immunoblotting. For immunoprecipitation epitope-tagged prey, cells transiently transfected with myc-tagged BirA*, LC3B (or individual LC3/ATG8 family members) and FLAG-SAFB, FLAG-G3BP1, FLAG-LARP1, FLAG-SF3A1, FLAG-FAN or mutants thereof were lysed 24 h post-transfection and captured using methods identical to those employed for endogenous protein immunoprecipitations.
Immuno-isolation of EVs
The following antibodies were employed for immune-isolation of EVs: mouse anti-CD9 (Millipore, CBL162), mouse anti-CD63 (Abcam, ab8219), mouse anti-CD81 (BD, 555675). Briefly, EVs purified from approximately 4.5 × 108 cells by differential centrifugation were resuspended in 100 μL of PBS, split equally between 4 eppendorf tubes (25 μL each), and resuspended in 500 μL PBS. Each sample was mixed with 2 μg of normal mouse IgG and 50 μL of MACs microbeads (Miltenyi Biotec) for 1 h and then pre-cleared through a MACs magnetic μ Column (Miltenyi Biotec). Flow through from each of the samples was collected, mixed with 2 μg of antibody against 1 of 3 different EV-associated tetraspanins or normal mouse IgG, 50 μL of MACs microbeads (Miltenyi Biotec) and left to capture overnight at 4oC. The following day, bound EVs were captured separately on MACs magnetic μ Columns, washed with 1500 μL PBS, and then eluted with 100 μL of hot 2x sample buffer. Samples were then resolved via SDS-PAGE and immunoblotted for LC3 and EV marker proteins.
Immunofluorescence and Fluorescence Microscopy
For immunofluorescence of myc-BirA* and myc-BirA*-LC3, cells stably expressing GFP-LC3 and myc-BirA* probes were seeded on coverslips coated with 10 μg/ml fibronectin (BD Biosciences, 356008) diluted in PBS. The next day cells were briefly washed with PBS, fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature (RT), quenched with PBS-glycine and permeabilized with 0.2% Triton X-100 in PBS. Cells were blocked for 1 h in blocking buffer (5% goat serum in PBS), incubated with mouse anti-myc antibody (1:1000) overnight at 4oC, washed, incubated with Alexa Fluor 594 goat anti mouse-secondary antibodies (1:500; Life Technologies) for 1h at RT, washed, and mounted using Prolong Gold Anti-Fade mounting medium (ThermoFisher, P36934). For immunofluorescence of endogenous LC3, cells stably expressing myc-BirA* probes were processed similar to above with the exception that rabbit anti-LC3 antibody (1:500, MBL PM036) and Alexa Fluor 488 goat anti rabbit-secondary antibody (1:500; Life Technologies) were incorporated into primary and secondary staining steps, respectively. Confocal imaging was performed using a TI-inverted microscope stand (Nikon) equipped with a Borealis-modified Yokogawa CSU-X1 confocal head (Spectral Applied Research), solid-state 488-nm (for GFP) and 561-nm (for Alexa Fluor 594) lasers, and a Clara cooled scientific-grade interline CCD camera (Andor) or a CoolSNAP MYO cooled scientific-grade CCD camera (Photometrics). Intracellular fluorescent-tagged protein and immunofluorescence images were acquired at 37°C using a 100× (NA, 1.49; oil) objective (CFI Aprochromat; Nikon). Microscope hardware was controlled with Nikon Elements Advanced (version 4.5.1) and the images were prepared in Fiji and Adobe Photoshop.
For immunofluorescence detection of endogenous LC3, CD63, SAFB and HNRNPK in enlarged Rab5-positive endosomes, cells of the indicated genotypes were transiently transfected with mCherry-Rab5CA(Q79L) at 70% confluency with Lipofectamine 2000 (ThermoFisher, 11668027) according to manufacturer’s instructions; in RNA interference experiments, cells were first transfected with siRNA SMARTpools and 24h later mCherry-Rab5Q79L was transfected using Lipofectamine 2000 (ThermoFisher, 11668027). After 6 h, DNA transfected cells were plated onto coverslips coated with fibronectin. Cells were incubated on coverslips for 72 h and then fixed with 4% PFA in PBS for 15 min at RT, permeabilized with ice cold methanol and incubated at −20°C for 5min before quenching with PBS/Glycine. Cells were blocked in blocking buffer (PBS + 0.1% Tween + 10% Goat Serum) for 1h at room temp, incubated with rabbit anti-LC3B (1:200, MBL PM036), mouse anti-CD63 (1:200, Abcam ab8219) or mouse anti-LC3 (1:200, Axxora 5F10) and rabbit anti-SAFB (1:200, Abcam ab187650) or rabbit anti-LC3B (1:200, CST 3868) and mouse anti-HNRNPK (1:200, Abcam ab39975) antibodies diluted in blocking buffer overnight at 4oC, washed with PBS, and then incubated with AlexaFluor goat anti-rabbit 488 (1:500, ThermoFisher, A-11034) and goat anti-mouse 647 (1:500, ThermoFisher, A21235) secondary antibodies in blocking buffer for 1h at RT. Coverslips were washed with PBS and mounted using Prolong Gold (ThermoFisher, P36934). For immunofluorescence of endogenous LC3 and CD63 in the absence of Rab5CA(Q79L) perturbation, cells were processed as above using rabbit anti-LC3B (1:200, CST 3868) and mouse anti-CD63 (1:200, Abcam ab8219) antibodies for primary staining. Cells were visualized using a DeltaVision microscope (Applied Precision Ltd.) fitted with a 60Å~, 1.4-NA objective and CoolSnap HQ camera (Photometrics). Images were acquired using softWoRx software (Applied Precision Ltd.) and prepared in Fiji and Adobe Photoshop. Costes significance tests for co-occurance81 and the Mander’s overlap co-efficient for LC3 with CD63 was performed by drawing a region of interest (ROI) around individual cells and then employing the Coloc 2 analysis function within Fiji (PSF:20, Costes Randomizations:10). Furthermore, the proportion of mCherry-Rab5Q79L endosomal area that overlaps with endogenous LC3 in siRNA treated cells was quantified using Fiji.
APEX Staining and Transmission Electron Microscopy
APEX staining and transmission electron microscopy was performed as previously described82. Briefly, cells transfected with APEX-LC3 or APEX-GFP were fixed using room temperature 2.5% glutaraldehyde (Electron Microscopy Sciences) in buffer (100 mM sodium cacodylate with 2 mM CaCl2, pH 7.4), then transferred to ice. All subsequent steps until resin infiltration were performed at 4°.After 30 min, cells were rinsed 5 × 2 min in cold buffer (100 mM sodium cacodylate with 2 mM CaCl2, pH 7.4), then incubated for 5 min in buffer containing 20 mM glycine, followed by 5 × 2 min washes in cold buffer. Freshly prepared 1 mg/mL 3,3′-diaminobenzidine (DAB) tetrahydrochloride (Sigma, 32750) dissolved in 0.1 M HCl was combined with 0.03% (v/v) (10 mM) H2O2 in chilled buffer, and the solution was added to cells for 2 min. The reaction was terminated by removal of the DAB solution, and cells were rinsed 5 × 2 min with cold buffer. Post-fixation staining was performed with 2% osmium tetroxide (Electron Microscopy Sciences, 19152) for 30 min in chilled buffer. Cells were washed 5 × 2 min in ice-cold distilled water, then placed in 2% aqueous uranyl acetate (Electron Microscopy Sciences, 22400) at 4° overnight. Samples were then incubated in cold graded ethanol series (20%, 50%, 70%, 90%, 100%, 100%) 2 min each, washed once in RT anhydrous ethanol, and infiltrated in Durcupan ACM resin (Electron Microscopy Sciences, 14040) using 1:1 (v/v) anhydrous ethanol and resin for 30 min, then 100% resin 2 × 1 h, and finally into fresh resin and polymerized in a vacuum oven at 60°C for 48 h.
For transmission electron microscopy, DAB-stained areas of resin-embedded cultured cells were identified by bright-field microscopy. Areas of interest were sawed out using a jeweler’s saw and mounted onto dummy blocks using cyanoacrylic glue, with the glass coverslip opposite the dummy block. The coverslip was carefully removed, the block trimmed, and ultrathin (80 nm thick) sections were cut using an ultramicrotome (Leica Ultracut UTC6). Electron micrographs were recorded using a JEOL 1200 TEM operating at 80 keV.
In vivo EV Isolation from Mice
All experimental procedures and treatments were conducted in compliance with UCSF Institutional Animal Care and Use Committee (IACUC) guidelines under an approved animal protocol (#AN170608). Compound transgenic C57bl/6 male and female mice of harbouring floxed alleles of Atg12 (Atg12f/f) or Atg5 (Atg5f/f) and Cre-recombinase under the ubiquitous CAG promoter (Cag-CreER) were generated by cross-breeding of Atg12f/f or Atg5f/f mice15 and CagCreER mice83. Offspring were genotyped with the following polymerase chain reaction (PCR) primers: ATG12 wild-type and f/f: 5’-atgtgaatcagtcctttgccc-3’, 5’-actctgaaggcgttcacggc-3’, 5’-ctctgaaggcgttcacaaca-3’; ATG5 wild-type and f/f: 5’-gaatatgaaggcacacccctgaaatg-3’, 5’-acaacgtcgagcacagctgcgcaagg-3’, 5’-gtactgcataatggtttaactcttgc-3’; Cag-CreER: 5’- gcctgcattaccggtcgatgc-3’, 5’-cagggtgttataagcaatccc-3’.
Primary astrocyte cultures were prepared as previously described84. Briefly, cerebral cortices were harvested from P3–4 Atg5f/f and Cag-CreER;Atg5f/f pups (8 male and 8 female across conditions). The meninges were removed and the cerebral cortices were dissected into smaller pieces with forceps and then triturated in DMEM, 20% FBS, Penicillin/Streptomycin (P/S), and 20 ng/ml of GM-CSF (PeproTech, Rocky Hill, NJ). The cells were grown in poly-L-lysine coated flasks. The media was replenished 3d after the initial harvest. Microglia were removed from the astrocyte layer 6–10d later by shaking flasks at 200 rpm for 1–2 hours at 37° C. The remaining primary astrocytes were trypsinized and expanded in DMEM, 20% FBS, Penicillin/Streptomycin (P/S), and 20 ng/ml of GM-CSF (PeproTech, Rocky Hill, NJ) for 11 days. Finally, astrocyte cultures were treated with media containing 2 μM 4-hydroxytamoxifen (4-OHT) for 3d, washed with PBS, and replenished with Neurobasal media supplemented with N2, L-glutamine, and P/S. After 5d the conditioned media from primary astrocyte cultures was collected for EV purification by differential centrifugation (see section on EV preparation above) and cells were lysed to assess genetic deletion.
For plasma and kidney analysis, 6-week old Cag-CreER;Atg12f/f animals or controls received either Tamoxifen (TAM; 0.2mg/gram mouse) or vehicle (peanut oil) via oral gavage for 5 consecutive days (8 male and 8 female across conditions). At 10 weeks after the first TAM treatment, tissues were collected. Kidney was homogenized and lysed prior to immunoblotting. Whole blood was collected in heparin coated tubes (Sarstedt, CB 300 LH), centrifuged at 2,000xg for 5 minutes at RT and the plasma phase was collected. For purification of plasma EVs, approximately 1 mL of platelet-free murine plasma (pooled from 4 mice per cohort) was diluted in 10 mL of PBS and clarified through a 0.22 μm filter. Subsequently, filtered samples were ultracentrifuged at 100,000g for 2 h at 4° and purified EV pellets resuspended in lysis buffer for analysis.
Cell Viability Assays
To quantify EV-independent release of lactate dehydrogenase (LDH), cells were seeded in 6 well culture plates and serum starved for 24h; cells treated with 100 μM etoposide for 24h served as a positive control for cell death. After 24h, conditioned media was collected and EVs pelleted via differential centrifugation. The supernatant from these samples was then collected and soluble proteins precipitated by adding trichloroacetic acid (TCA) to a final concentration of 20% and incubating on ice for 1h. Subsequently, precipitated protein was pelleted by ultracentrifugation at 200,000g for 1h. Pellets were washed with 5 ml of ice-cold acetone, re-centrifuged at 200,000g for 1h, and air dried. Urea lysis buffer was added to re-solubilize the pellets and the levels of LDH quantified by western blot.
To quantify viability following serum starvation, cells were seeded on to coverslips coated with fibronectin and subject to serum starvation or control media with full serum for 24h. Cells were then stained with for viability using Calcein AM and Ethidium homodimer-1 (ThermoFisher, MP 03224) according to manufacturer’s instructions, visualized on Axiovert 200 epifluorescent microscope, and dead cells (Ethidium positive, Calcein negative) were enumerated.
CRISPR/Cas9 gene-deletion
HEK293T knockout cell lines were generated by transient transfection of pSpCas9(BB)-2A-Puro (Addgene #48139) encoding U6 driven expression of sgRNAs (Scramble Guide: GCACTACCAGAGCTAACTCA; ATG7 Guide: ACACACTCGAGTCTTTCAAG; ATG12 Guide: CCGTCTTCCGCTGCAGTTTC; ATG14 Guide: CTACTTCGACGGCCGCGACC; FIP200 Guide: AGAGTGTGTACCTACAGTGC). Cells were selected 48–72 hours post-transfection with 1μg/ml puromycin for 48 h. Polyclonal populations were collected for Surveyor analysis (IDT, 706020) and were sorted into single-cell populations by limiting dilution at 1.5 cells/well per 96-well plate. For DNA analysis, genomic DNA samples were prepared using QuickExtract (Epicentre). The PCR products were column purified and analysed with Surveyor Mutation Detection Kit (IDT). For genotyping of single-sorted cells, PCR amplified products encompassing the edited region (ATG7 Fw: TGGGGGACAGTAGAACAGCA, ATG7 Rev: CCTGGATGTCCTCTCCCTGA; ATG12 Fw: AGCCGGGAACACCAAGTTT, ATG12 Rev: GTGGCAGCCAAGTATCAGGC; ATG14 Fw: AAAATCCCACGTGACTGGCT, ATG14 Rev: AATGGCAGCAACGGGAAAAC; FIP200 Fw: ATTCTCTGGCTTGACAGGACAG, FIP200 Rev: AAATACTGAGCGTGCACATTGC) were cloned into pCR™4-TOPO® TA vector using the TOPO-TA cloning kit (Thermo Fisher #450030) and sequence verified. Sequencing is available upon request.
RNA interference
For transient siRNA-mediated knockdown, cells were first transfected with siRNA and using DharmaFECT #1 (Dharmacon T-2001–03) according to the manufacturer’s instructions, and after 24h transfected with plasmid encoding mCherry-Rab5Q79L. ON-TARGETplus smart pools against ATG7 (10533; L-020112–00-0005), ALIX (10015; L-004233–00-0005), TSG101 (7251; L-003549–00-0005), CHMP3/VPS24 (51652; L-004696–00-0005), VPS4a (27183; L-013092–00-0005), VPS4b (9525; L-013119–00-0005), CHMP4b (128866; L-018075–01-0005), nSMase2/SMPD3 (55512, L-006678–00-0005), FAN/NSMAF (8439; L-017920–00-0005) and non-targeting siRNA (D-001810–01-20) were purchased from Dharmacon. To generate stable knockdowns, cells were transduced with pLKO.1 lentiviral vectors (Sigma-Aldrich) expressing shRNAs targeting ATG7 (TRCN0000007584), ATG3 (#1 TRCN0000148120; #2 TRCN0000146846; #3 TRCN0000147381), SMPD3/nSMase2 (#1; TRCN0000048944; #2 TRCN0000048945), FAN/NSMAF (#1 TRCN0000145430; #2 TRCN0000143925) and non-targeting shRNA (SHC002).
Quantitative PCR
For qPCR analysis, total RNA from cells isolated using QIAzol lysis reagent (Qiagen). RNA was reverse transcribed using MMLV reverse transcriptase (Roche) and the resulting cDNA was analysed via real-time quantitative PCR using QuantiTect SYBR Green PCR Master Mix and gene specific primers (GAPDH Fwd: TGTCAAGCTCATTTCCTGGTATG, GAPDH Rev: CTCTCTTCCTCTTGTGCTCTTG; SMPD3/nSMase2 Fwd: CAACAAGTGTAACGACGATGCC, nSMase2 Rev: CGATTCTTTGGTCCTGAGGTGT) according to manufacturer’s instructions. Ct values for intracellular transcripts were converted to fold expression changes (2−ΔΔCt values) following normalization to intracellular GAPDH. Quantitative real-time PCR was performed using an Applied Biosystems StepOneplus Real-Time PCR System (ThermoFisher).
Statistics and Reproducibility
Statistical analyses were performed using Prism GraphPad 5 software. Groups were compared using unpaired or paired Student’s t-test where indicated for pairwise comparisons or one-way analysis of variance followed by Tukey’s post-hoc test for multiple comparisons. Sample size was chosen based upon the size of the effect and variance for the different experimental approaches. Details regarding the statistical analysis of proteomic data and bioinformatics analysis of the BirA*-LC3 labelled secretome (both Class I and Class II datasets), ATG7 and ATG12-dependent EV proteome and RNA profiles are provided in the corresponding figure legends and/or methods sections above. P values of less than 0.05 are considered to be significant.
Data Availability
The mass spectrometry proteomics data associated with this study has been deposited into the ProteomeXchange Consortium via the PRIDE85 partner repository with the dataset identifier PXD015479. RNA sequencing data has been deposited in GEO: GSE137618. Furthermore, the data and/or reagents that support the findings of this study are available from the corresponding author, JD, upon reasonable request.
Extended Data
Extended Data Fig. 1: Functional validation of the BirA*-LC3 recombinant probe.
a, Cells stably expressing myc-BirA*-LC3, myc-BirA* or vector control were incubated in either full (F) or serum free media (S) for 4h in the absence or presence of 50 μM chloroquine (CQ) for the last 1h. Cells were lysed and subject to immunoblotting for indicated proteins (n=2 biologically independent experiments). b, Representative images of cells stably expressing GFP-LC3 and myc-BirA*-LC3 or myc-BirA* and immunostained with anti-myc antibody (n=3 biologically independent samples). c, Representative images of cells stably expressing myc-BirA*-LC3 or myc-BirA* and co-immunostained with anti-LC3 (green) and anti-myc (magenta) antibody (n=3 biologically independent samples). d, Biotinylation blots reproduced from Fig. 1b with accompanying Ponceau S stained membranes of whole cell lysate (WCL) and conditioned media (CM) (n=3 biologically independent experiments). e, Schematic of experiment to test for intracellular versus extracellular origin of BirA* and BirA*-LC3-mediated biotinylated targets isolated from CM. f, Representative Strep-HRP blot for biotinylated proteins in the precipitated CM from myc-BirA*-LC3 or myc-BirA* cells co-incubated (Co) or post-incubated (Post) with 50 μM biotin for 24h and negative control (Neg). CM was probed to validate expression and secretion of the myc-tagged recombinant proteins (n=2 biologically independent experiments). Unprocessed blots available in Source Data Extended Data Fig. 1.
Extended Data Fig. 2: BirA*-LC3B-labelled secretome is enriched in RBPs.
a, Volcano plot of BirA*-LC3-labeled secretome quantified by mass spectrometry. SILAC labelled biotin-tagged proteins plotted according to -log10 p-values as determined by two-tailed t-test and log2 fold enrichment (BirA*-LC3/BirA*) (n=3 biologically independent samples). Grey horizontal dotted line: significance cut-off with p-value of 0.05. Log2 fold change reflects LC3-BirA* to BirA* alone ratio. Grey vertical dotted line: 2-fold enriched and de-enriched cut-off. Pink: significantly enriched proteins relative to BirA* alone. Red: Class I enriched proteins represented in heat map in Figure 1. Inset: Expanded view of significantly enriched proteins. b, Venn diagram showing overlap of secretory autophagy candidates (Class I and II hits) with the LC3B intracellular interactome defined in Behrends et al. 2010. c, Venn diagram showing the overlap of secretory autophagy candidates (Class I and II hits) with the entire ATG8 intracellular interactome defined in Behrends et al. 2010. d, Ranked list of proteins with greatest connectivity to secretory autophagy candidates as determined by the Enrichr gene enrichment analysis tool (n=3 biologically independent samples; 200 enriched proteins in Class I + II datasets). Statistical significance calculated by one-way Fisher’s exact test and adjusted using the Benjamini–Hochberg method. LC3/ATG8 family members highlighted in red. e, Network map of autophagy-dependent secretion candidates. Class I and II secretory autophagy candidates mapped to zero-order protein interaction network using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and proteins associated with extracellular exosomes or with RNA-binding functions coloured in red and blue, respectively. f, Pie chart plotting percentage of Class I and II secretory autophagy candidates assigned to Gene Ontology (GO) term extracellular exosome by PANTHER. g, Venn diagram showing overlap of class I and II secretory autophagy candidates with the mRNA binding proteins from Castello et al. 2012. Data available in Source Data Extended Data Fig. 2.
Extended Data Fig. 3: Endogenous LC3-II is secreted within EVs isolated from cultured cells and murine plasma.
a, Whole cell lysate (WCL) and extracellular vesicle lysates (EVs) from murine RAW264.7 macrophages treated with 100 ng/ml LPS for 24h and 20 μM nigericin for 1 h, murine B16F10 melanoma cells, and murine LLC1 cells were immunoblotted for LC3, SAFB, HNRNPK and extracellular vesicle marker proteins (n=2 biologically independent experiments). b, Workflow employed to obtain plasma and tissue from CAG-CreER;Atg12flox/flox mice in which Atg12 was systemically deleted via tamoxifen treatment. c, Extracellular vesicles (EVs) isolated from the plasma of naïve wild-type mice (CAG-CreER or Atg12flox/flox) and mice in which Atg12 was systemically deleted by 4-OHT treatment (CAG-CreER; Atg12flox/flox) were lysed and immunoblotted for LC3 and the indicated extracellular vesicle marker proteins (n=2 biologically independent experiments). d, Whole cell lysates (WCL) derived from the renal tissue of mice in Panel c were immunoblotted for LC3 and the indicated marker proteins (n=2 biologically independent experiments). e, Workflow employed to obtain CM from murine astrocytes (CAG-CreER;Atg5flox/flox) in which Atg12 was deleted ex vivo via 4-OHT treatment. f, Extracellular vesicles (EVs) isolated from the conditioned media of naive wild-type (Atg5flox/flox) primary astrocytes and astrocyte cultures in which Atg12 was deleted ex vivo (CAG-CreER; Atg5flox/flox) by 4-OHT treatment were lysed and immunoblotted for LC3 and CD9 (n=2 biologically independent experiments). g, Whole cell lysates (WCL) primary astrocyte cultures in Panel d were immunoblotted for LC3 and CD9 (n=2 biologically independent experiments). h, Representative fluorescence micrographs from wild-type, ATG7−/− and ATG14−/− HEK293T cells transfected with mCherry-Rab5Q79L (yellow). Cells were immunostained for endogenous LC3 (green) and CD63 (magenta) (n=3 biologically independent samples). Scale bar=10μm. Unprocessed blots available in Source Data Extended Data Fig. 3.
Extended Data Fig. 4: Components of stress granules and P-bodies secreted in EVs through mechanisms requiring the LC3-conjugation machinery.
a, Proportion of BirA*-LC3B- labelled secretome (Class I, II candidates) detected in the total extracellular vesicle (EV) proteome defined by TMT quantitative mass spectrometry. b, Venn diagram showing overlap of EV components requiring ATG7 and ATG12 for secretion with the fixed and unfixed stress granule proteome from Jain et al., 2016. c, Venn diagram showing the overlap of EV components requiring ATG7 and ATG12 for secretion with the P-body proteome from Hubstenberger et al., 2017. d, Representative fluorescence micrographs from wild-type HEK293T cells transfected with mCherry-Rab5Q79L (blue) and immunostained for endogenous LC3 (green) and SAFB or HNRNPK (magenta) (n=3 biologically independent samples). Scale bar=10μm. e, Whole cell (WCL) and EV lysates harvested from equal numbers of cells stably expressing non-targeting (NT) or ATG3 shRNA were immunoblotted for indicated proteins (n=3 biologically independent samples). f, Quantification of indicated protein levels in EVs from cells stably expressing shRNAs targeting ATG3 relative to non-targeting shRNA (mean ±s.e.m.; n=3 biologically independent samples). g, Quantification of Lactate Dehydrogenase (LDH) in EV-depleted conditioned media from wild-type (WT) HEK293T cells treated 100 μM Etoposide (Etop) for 24h or WT and ATG knockout cells serum starved for 24h (mean ±s.e.m.; n=3 biologically independent experiments). h, Cell death in wild-type (WT) and ATG knockout cells (KO) after 24h in full serum media (FM) or serum starved media (SS) quantified using Calcein-AM and ethidium bromide staining (mean ±s.e.m.; n=3 biologically independent experiments). i, Whole cell and EV lysates from wild-type cells grown in EV-depleted full serum media (FM) or EV-depleted FM with 100 nM Rapamycin (Rap) for 24h. Immunoblots probed against the indicated proteins (n=2 biologically independent experiments). j, Quantification of the relative levels of indicated proteins in EVs from Rap-treated cells in Panel d (line=mean; n=2 biologically independent experiments ). Data and unprocessed blots available in Source Data Extended Data Fig. 4.
Extended Data Fig. 5: LC3-conjugation machinery controls EV-mediated secretion of diverse RBPs.
a, EVs from WT and ATG deficient cells normalized for protein concentration and immunoblotted to detect endogenous LC3A, LC3B, LC3C, GABARAP (GR), GABARAPL1 (GRL1), GABARAPL2 (GRL2), and indicated marker proteins (n=2 biologically independent experiments). b, Whole cell (WCL) and EV lysates from WT and ATG7−/− cells were normalized for protein concentration and immunoblotted for indicated proteins (n=3 biologically independent experiments). c, HEK293T cells co-transfected with FLAG-tagged G3BP1, LARP1 or SF3A1, and myc-tagged LC3B, GABARAP (GR), LC3C respectively, or myc-BirA* were lysed, immunoprecipitated (IP) with anti-myc antibody and immunoblotted (WB) with indicated antibodies (n=3 biologically independent experiments). d, Diagram mapping the domains and primary LC3-interaction region (LIR) in SAFB. e, Volcano plot of mRNA and long non-coding RNA (large RNA) detected in EVs from WT and ATG7−/− cells. Results plotted according to -log10 p-values as determined by DESeq2 and log2 fold enrichment (n=3 biologically independent samples; WT/ATG7−/−). Grey dots: RNAs not enriched in EVs from WT or ATG7−/− cells identified with a p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5). Black dots: Large RNAs enriched in EVs from WT cells or ATG7−/− cells. f, Volcano plot of mRNA and long non-coding RNA (large RNA) detected in EVs from WT and ATG12−/− cells. Results plotted according to -log10 p-values as determined by DESeq2 and log2 fold enrichment (n=3 biologically independent samples; WT/ATG12−/−). Grey dots: RNAs not enriched in EVs from WT or ATG12−/− cells identified with a p-value >0.05 and/or log2 fold change between −0.5 and 0.5 (−0.5<log2FC<0.5). Black dots: Large RNAs enriched in EVs from WT or ATG12−/− cells. g, Venn diagram showing the overlap of mRNA and long non-coding RNAs (large RNAs) enriched in EVs from WT relative to ATG7−/− cells and EVs from WT relative to ATG12−/− cells. Data and unprocessed blots available in Source Data Extended Data Fig. 5.
Extended Data Fig. 6: LC3 delivery into ILVs of Rab5Q79L endosomes requires CHMP4b and nSMase2, but is independent of other ESCRT machinery components.
a, Representative fluorescence micrographs from wild-type HEK293T cells co-transfected with mCherry-Rab5Q79L (magenta) and non-targeting (NT) control siRNA or siRNAs targeting ATG7, ALIX, TSG101, VPS4a/b, CHMP3, CHMP4b and nSMase2. Cells were immunostained for endogenous LC3 (green) (n=2 biologically independent experiments). Scale bar=10μm. b, Scatter plot of the proportion of mCherry-Rab5Q79L endosomes that overlap with LC3 in immuno-stained cells in Panel a (mean ± s.e.m.; n=23 biologically independent samples). Statistical significance calculated by one-way ANOVA coupled with Fisher’s least significant difference test. c, Lysates from cells in Panel a were immunoblotted with antibodies the various siRNA targets and GAPDH as a loading control. Representative blots are shown (n=2 biologically independent experiments). Non-specific bands are indicated with an asterisk (*). d, Quantitative PCR (QPCR) measurement of nSMase2 mRNA in HEK293T cells transfected with siRNAs targeting nSMase2, nSMase2 (nSM2) relative to non-targeting siRNA (NT) control cells (line=mean; n=1, 2 technical replicates). Data and unprocessed blots available in Source Data Extended Data Fig. 6.
Extended Data Fig. 7: LC3-dependent EV loading and secretion (LDELS) requires FAN and nSMase2.
a, Whole cell lysate harvested from equal numbers of HEK293Ts stably expressing non-targeting (NT), ATG7 or nSMase2(nSM2) shRNAs immunoblotted for indicated proteins (n=2 biologically independent experiments). b, Quantitative PCR (QPCR) for nSMase2 mRNA in HEK293Ts stably expressing shRNAs targeting ATG7, nSMase2 (nSM2) relative to non-targeting shRNA (NT) control cells (line=mean; n=1, 2 technical replicates). c, Whole cell lysates from HEK293Ts stably expressing non-targeting shRNA (NT) or shRNAs targeting ATG7 or FAN were immunoblotted with antibodies for the indicated proteins (n=2 biologically independent experiments). d, HEK293Ts were EBSS starved for the indicated times, treated with DMSO or 50nM Bafilomycin A1 (Baf A1) for 1 h prior to lysis, lysed and immunoblotted for FAN and the indicated proteins (n=2 biologically independent experiments). e, HEK293Ts expressing non-targeting (NT) or FAN shRNA were starved in EBSS for 4h, treated with DMSO or 50nM Bafilomycin A1 (Baf A1) for 1h prior to lysis, lysed and immunoblotted for the indicated proteins (n=2 biologically independent experiments). f, Representative fluorescence micrographs from wild-type cells co-transfected with mCherry-Rab5Q79L (magenta) and non-targeting (NT) control siRNA or siRNAs targeting FAN. Cells were immunostained for endogenous LC3 (green)(n=2 biologically experiments). Scale bar=10μm. g, Lysates from cells in Panel f were immunoblotted with antibodies against FAN and GAPDH as a loading control (n=2 biologically independent experiments). h, Scatter plot of the proportion of mCherry-Rab5Q79L endosomes overlapping with LC3 in immuno-stained cells in Panel f (mean ±s.e.m.; n=22 biologically independent samples). Statistical significance calculated by unpaired two-tailed t-test. i, Whole cell lysate from HEK293Ts analysed in Fig. 7j that were co-expressing non-targeting (NT) or FAN shRNA along with FLAG-tagged wild-type FAN (WT) or mutant FAN (F199A) were immunoblotted for the indicated proteins (n=2 biologically independent experiments). j, Proposed model for LC3-dependent EV loading and secretion (LDELS) in comparison to classical autophagy. Data and unprocessed blots available in Source Data Extended Data Fig. 7.
Supplementary Material
Acknowledgements
We thank Dr. Max Krummel (UCSF) for generously providing reagents, members of the Debnath laboratory for helpful discussions, and Dr. Rushika Perera (UCSF) for critical reading of the manuscript. We also thank Drs. Seda Kilinc and Andrei Goga (UCSF) for assistance with extracellular vesicle concentration and size analysis, and Dr. Albertas Navickas for advice on RNA sequencing library preparation. Grant support includes the NIH (CA201849, CA126792, CA201849, CA213775 to JD, R00CA194077, R01CA240984 to HG, K08CA184116 to APW and AG057462 to JD and EJH), the DOD BCRP (W81XWH-11-1-0130 to JD), Samuel Waxman Cancer Research Foundation (to J.D.), UCSF QB3 Calico Longevity Fellowship (to JD and AML) and Dale Frey Breakthrough Award from the Damon Runyon Cancer Research Foundation (DFS 14-15 to APW). Fellowship support includes a Banting Postdoctoral Fellowship from the Government of Canada (201409BPF-335868) and Cancer Research Society Scholarship for Next Generation of Scientists to AML, NSF Graduate Student Fellowships (1650113 to TS and 1144247 to JG), Canadian Institutes of Health Research Postdoctoral Fellowship to FK, NRSA awards from the NCI (F31CA217015 to TM Jr., F30CA224693 to JYL), a UCSF IRACDA Postdoctoral Fellowship (K12GM081266) to TM, and a NCI T32 training grant (T32CA108462) to AV.
Footnotes
Conflict of interest: JD is a Scientific Advisory Board Member for Vescor Therapeutics, LLC.
References
- 1.Kaur J & Debnath J Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 16, 461–472 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Lock R, Kenific CM, Leidal AM, Salas E & Debnath J Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov 4, 466–479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bel S et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeSelm CJ et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 21, 966–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo H et al. Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 43, 716–730 e717 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Murrow L, Malhotra R & Debnath J ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 17, 300–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupont N et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J 30, 4701–4711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duran JM, Anjard C, Stefan C, Loomis WF & Malhotra V Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188, 527–536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manjithaya R, Anjard C, Loomis WF & Subramani S Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188, 537–546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torisu T et al. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med 19, 1281–1287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadwell K & Debnath J Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol 217, 813–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhotra V Unconventional protein secretion: an evolving mechanism. EMBO J 32, 1660–1664 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponpuak M et al. Secretory autophagy. Curr Opin Cell Biol 35, 106–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roux KJ, Kim DI, Raida M & Burke B A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrends C, Sowa ME, Gygi SP & Harper JW Network organization of the human autophagy system. Nature 466, 68–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanjappa V et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res 42, D959–965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castello A et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Amigorena S, Raposo G & Clayton A Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3 22 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Maas SLN, Breakefield XO & Weaver AM Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 27, 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathieu M, Martin-Jaular L, Lavieu G & Thery C Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Pegtel DM & Gould SJ Exosomes. Annu Rev Biochem 88, 487–514 (2019). [DOI] [PubMed] [Google Scholar]
- 23.van Niel G, D’Angelo G & Raposo G Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Martell JD, Deerinck TJ, Lam SS, Ellisman MH & Ting AY Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat Protoc 12, 1792–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenmark H et al. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J 13, 1287–1296 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trajkovic K et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Ichimura Y et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Itakura E, Kishi C, Inoue K & Mizushima N Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19, 5360–5372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga K et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11, 385–396 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Ivanov P, Kedersha N & Anderson P Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb Perspect Biol 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeppesen DK et al. Reassessment of Exosome Composition. Cell 177, 428–445 e418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stolz A, Ernst A & Dikic I Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16, 495–501 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Kishi-Itakura C, Koyama-Honda I, Itakura E & Mizushima N Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci 127, 4089–4102 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Zhang H et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 20, 332–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F & Mittelbrunn M Sorting it out: regulation of exosome loading. Semin Cancer Biol 28, 3–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis-Sandoval F, Poirier M & Scott MS The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6, 381–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adam-Klages S et al. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell 86, 937–947 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Berg TO, Fengsrud M, Stromhaug PE, Berg T & Seglen PO Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 273, 21883–21892 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Fader CM, Sanchez D, Furlan M & Colombo MI Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Florey O, Kim SE, Sandoval CP, Haynes CM & Overholtzer M Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13, 1335–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heckmann BL et al. LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551 e514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mejlvang J et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol 217, 3640–3655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puri C et al. The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev Cell 45, 114–131 e118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Progida C & Bakke O Bidirectional traffic between the Golgi and the endosomes - machineries and regulation. J Cell Sci 129, 3971–3982 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Raiborg C, Wenzel EM & Stenmark H ER-endosome contact sites: molecular compositions and functions. EMBO J 34, 1848–1858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubstenberger A et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68, 144–157 e145 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Jain S et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchan JR, Kolaitis RM, Taylor JP & Parker R Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alberti S, Mateju D, Mediani L & Carra S Granulostasis: Protein Quality Control of RNP Granules. Front Mol Neurosci 10, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganassi M et al. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol Cell 63, 796–810 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Mateju D et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36, 1669–1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell 74, 742–757 e748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abels ER & Breakefield XO Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol 36, 301–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skog J et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spinelli C, Adnani L, Choi D & Rak J Extracellular Vesicles as Conduits of Non-Coding RNA Emission and Intercellular Transfer in Brain Tumors. Noncoding RNA 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valadi H et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Bellingham SA, Coleman BM & Hill AF Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 40, 10937–10949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freedman JE et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 7, 11106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur S et al. CD63, MHC class 1, and CD47 identify subsets of extracellular vesicles containing distinct populations of noncoding RNAs. Sci Rep 8, 2577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lasser C et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol 14, 58–72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lefebvre FA et al. Comparative transcriptomic analysis of human and Drosophila extracellular vesicles. Sci Rep 6, 27680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimer JM et al. Long-range function of secreted small nucleolar RNAs that direct 2’-O-methylation. J Biol Chem 293, 13284–13296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shurtleff MJ et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci U S A 114, E8987–E8995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong E et al. Unravelling the RNA-Binding Properties of SAFB Proteins in Breast Cancer Cells. Biomed Res Int 2015, 395816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falaleeva M & Stamm S Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays 35, 46–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holley CL et al. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J Biol Chem 290, 11741–11748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li MW et al. Nuclear export factor 3 regulates localization of small nucleolar RNAs. J Biol Chem 292, 20228–20239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Materials and Methods References
- 68.Pear WS, Scott ML & Nolan GP Generation of high-titer, helper-free retroviruses by transient transfection. Methods Mol Med 7, 41–57 (1997). [DOI] [PubMed] [Google Scholar]
- 69.Roux KJ, Kim DI, Raida M & Burke B A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Thery C, Amigorena S, Raposo G & Clayton A Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3 22 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Kuksa PP et al. DASHR 2.0: integrated database of human small non-coding RNA genes and mature products. Bioinformatics 35, 1033–1039 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nanjappa V et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res 42, D959–965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi H, Muruganujan A, Ebert D, Huang X & Thomas PD PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szklarczyk D et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castello A et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Hulsen T, de Vlieg J & Alkema W BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costes SV et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86, 3993–4003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martell JD, Deerinck TJ, Lam SS, Ellisman MH & Ting AY Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat Protoc 12, 1792–1816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hayashi S & McMahon AP Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 244, 305–318 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Albuquerque C, Joseph DJ, Choudhury P & MacDermott AB Dissection, plating, and maintenance of cortical astrocyte cultures. Cold Spring Harb Protoc 2009, pdb prot5273 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Vizcaino JA et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44, 11033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data associated with this study has been deposited into the ProteomeXchange Consortium via the PRIDE85 partner repository with the dataset identifier PXD015479. RNA sequencing data has been deposited in GEO: GSE137618. Furthermore, the data and/or reagents that support the findings of this study are available from the corresponding author, JD, upon reasonable request.