Abstract
Background
Human sapovirus (SaV) is an etiologic agent of acute gastroenteritis (AGE) in all age groups worldwide. Genetic recombination of SaV has been reported from many countries. So far, none of SaV recombinant strain has been reported from Thailand. This study examined the genetic recombination and genotype diversity of SaV in children hospitalized with AGE in Chiang Mai, Thailand.
Methods
Stool samples were collected from children suffering from diarrhea who admitted to the hospitals in Chiang Mai, Thailand between 2010 and 2018. SaV was detected by RT-PCR and the polymerase and capsid gene sequences were analysed.
Results
From a total of 3,057 samples tested, 50 (1.6%) were positive for SaV. Among positive samples, SaV genotype GI.1 was the most predominant genotype (40%; 20/50), followed by GII.1 and GII.5 (each of 16%; 8/50), GI.2 (14%; 7/50), GIV.1 (4%; 2/50), and GI.5 (2%; 1/50). In addition, 4 SaV recombinant strains of GII.1/GII.4 were identified in this study (8%; 4/50).
Conclusions
The data revealed the genetic diversity of SaV circulating in children with AGE in Chiang Mai, Thailand during 2010 to 2018 and the intragenogroup SaV recombinant strains were reported for the first time in Thailand.
Keywords: Gastroenteritis, Pediatric, Recombinant, Sapovirus, Thailand
Introduction
Sapovirus (SaV) is one of the important pathogens that cause outbreaks and sporadic cases of acute gastroenteritis (AGE) in people of all ages worldwide (Torner et al., 2016). Prevalences of SaV infection have been reported between 0.2% and 39% in children with AGE (Magwalivha et al., 2018) and between 2.2% and 15.6% in all age groups (Oka et al., 2015; Varela et al., 2019). SaV belongs to genus Sapovirus of the Caliciviridae family. SaV particle is a small, non-enveloped with 30–35 nm in diameter. SaV has a positive-sense, single stranded RNA genome of about 7.1–7.7 kb in length which contains two open reading frames (ORFs) (Green, 2013; Oka et al., 2016). ORF1 encodes for nonstructural proteins (NS1, NS2, NS3, NS4, NS5, and NS6-NS7) and the major capsid protein (VP1). ORF2 encodes for a minor capsid protein (VP2) (Oka et al., 2015). Based on the entire nucleotide sequence of VP1 region, SaV can be classified into 19 genogroups (GI-GXIX) and only four genogroups (GI, GII, GIV, and GV) known to infect human (Yinda et al., 2017). Human SaV can be further divided into 18 genotypes (GI.1 to GI.7, GII.1 to GII.8, GIV.1, GV.1, and GV.2) (Kagning Tsinda et al., 2017; Liu et al., 2016; Oka et al., 2012; Oka et al., 2015; Xue et al., 2019) with one additional genotype of GII.NA that has been reported recently (Diez-Valcarce et al., 2019). Among these, SaV GI and GII are the most prevalent genogroups detected worldwide while other genogroups have been rarely detected (Magwalivha et al., 2018). Like noroviruses, several SaV recombinant strains of both intragenogroup and intergenogroup recombinations have been reported (Chanit et al., 2009; Lasure & Gopalkrishna, 2017; Liu et al., 2015). Within SaV genome, recombination generally occurs in the ORF1, particularly at the junction between the polymerase (NS7) and capsid (VP1) genes (Hansman et al., 2005; Katayama et al., 2004; Phan et al., 2006). Some of SaV recombinant strains have been documented to associate with the outbreaks (Hansman et al., 2007; Lee et al., 2012). In Thailand, SaV infection in children with diarrhea has been reported previously, however, none of SaV recombinant strain has been reported previously. Therefore, this study aimed to investigate the genetic recombination and genotype diversity of SaV circulating in children hospitalized with AGE in Chiang Mai, Thailand from 2010 to 2018.
Materials & Methods
Sample collection
Stool samples were collected from children who admitted to hospitals with AGE during the period 2010 to 2018 from five major hospitals in Chiang Mai province, northern Thailand, including Maharaj Nakorn Chiang Mai Hospital, Sriphat Medical Center, Nakornping Hospital, Sanpatong Hospital, and Rajavej Chiang Mai Hospital. The age of patients enrolled in this study ranged from neonate to 15 years old. Acute gastroenteritis was defined by watery diarrhea with three or more stool episodes per day for less than 14 days (Green, 2013). All specimens were stored at −20 °C until investigation. This work was conducted under the approval of the Research Ethics Committee of the Faculty of Medicine, Chiang Mai University (MIC-2557-02710). The written informed consent form was obtained from parents before samples were collected from their children
Detection of SaV by RT-PCR
Viral nucleic acid was extracted from 200 µl of the supernatant of a 10% stool suspension prepared in phosphate-buffered saline (pH 7.4) using the Geneaid Viral Nucleic Acid Extraction Kit II (Geneaid, Taiwan) according to the manufacturer’s protocol. The viral RNA was reverse transcribed to cDNA using random hexamer primers and RevertAid™ reverse transcriptase (https://www.thermofisher.com/, USA) according to the manufacturer’s instruction. SaV was screened by conventional PCR method using GoTaq DNA polymerase (Promega, USA) with primers SLV5731 and SLV5749 (Table 1) targeting capsid region (VP1 gene) as described previously (Yan et al., 2003). The PCR cycling condition was as follows: initial denature at 94 °C for 3 min, 35 cycles step of denature at 94 °C for 1 min, anneal at 58 °C for 1 min, and extend at 72 °C for 1 min, followed by final extension at 72 °C for 10 min. Amplicon size of 434 bp was separated on 1.5% agarose gel electrophoresis and stained with nucleic acid staining solution (RedSafe, INtRON Biotechonology, South Korea) before subjecting to visualize under ultraviolet transilluminator. In addition to SaV screening, the same set of stool specimens were also tested for several other diarrhea-causing viruses, including rotavirus, norovirus, astrovirus, adenovirus, enterovirus, parechovirus, and Aichivirus using the protocol described previously (Khamrin et al., 2011).
Table 1. Oligonucleotide primers used for amplification of SaV capsid and polymerase genes.
| Primer | Sequence (5′–3′) | Direction | Target gene | Position in SaV genome (Accession no: X86560) | Reference |
|---|---|---|---|---|---|
| SLV5317 | CTCGCCACCTACRAWGCBTGGTT | Forward | VP1 | 5083–5105 | Yan et al. (2003) |
| SLV5749 | CGGRCYTCAAAVSTACCBCCCCA | Reverse | VP1 | 5494–5516 | Yan et al. (2003) |
| Sapp36 | GTTGCTGTTGGCATTAACA | Forward | RdRp | 4273–4291 | Berke et al. (1997) |
| Sapp128 | GATTACACCAAATGGGATTCCAC | Forward | RdRp | 4354–4376 | Martinez et al. (2002) |
| SaV1245R | CCCTCCATYTCAAACACTA | Reverse | RdRp | 5159–5177 | Harada et al. (2013) |
| SV-r-c | GCATTGTAGGTGGCGAGAGCC | Reverse | RdRp | 5079–5099 | Honma et al. (2001) |
Sequence analysis and genotype identification
A Gel/PCR DNA Fragment Extraction Kit (Geneaid, Taiwan) was used to purify amplicons of SaV-capsid gene according to the manufacturer’s protocol. All purified PCR products were direct sequenced using Applied Biosystems BigDye® Terminator Cycle Sequencing Kit v3.1 (Life Technologies, USA) with forward and reverse primers SLV5731 and SLV5749, and analyzed by using Applied Biosystems 3100 Genetic Analyzer (Life Technologies, USA). Nucleotide sequences of partial capsid gene were analyzed manually using BioEdit and Clustal X softwares. Identification of virus genotype was initially determined by using the BLAST Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Human Calicivirus Typing Tool (https://norovirus.ng.philab.cdc.gov/) and confirmed by phylogenetic analysis using MEGA7 software package (Kumar, Stecher & Tamura, 2016). The tree was statistically supported by bootstrapping with 1,000 replicates.
Recombination analysis
To examine genetic recombination of SaV strains detected in this study, the polymerase (RdRp) region was amplified by using primers shown in Table 1. The PCR amplification was carried out under the same condition as used for the VP1 amplification except for the annealing step which was performed at 45 °C for 1 min. Amplicon sizes of the amplified RdRp region were varied depending on the used primers (746 bp, 824 bp, 827 bp, or 905 bp) shown in Table 1 (Berke et al., 1997; Harada et al., 2013; Honma et al., 2001; Martinez et al., 2002). Phylogenetic tree of the partial capsid gene sequences was constructed by using the Maximum Likelihood method based on the Kimura 2-parameter model. Phylogenetic tree of the partial RdRp region was inferred by using the Maximum Likelihood method based on the General Time Reversible model. To predict the putative recombination point for SaV recombinant strains, nucleotide sequences of the polymerase and capsid genes, spanning the RdRp-VP1 junction region (positions 5078-5265 according to SaV genome of accession no. X86560), were analyzed. The possible recombination point of SaV strain was determined by using SimPlot software v.3.5.1 (Lole et al., 1999). In addition, the confidence interval for recombination between the query strain and parent strains was examined by the Recombination Detection Program v.4.71 (RDP4) complemented with the Max-Chi test to confirm the significant events (p < 0.01) (Martin et al., 2015).
Nucleotide accession number
Nucleotide sequences of the polymerase and capsid regions of SaV strains detected in this study were deposited in the GenBank database under the accession numbers MN245671 to MN245579 and MN253492 to MN253541 for the polymerase and capsid gene sequences, respectively.
Results
Prevalence of SaV
A total of 3,057 stool samples collected between 2010 and 2018 were screened for the presence of SaV. The average of SaV detection rate over the study period of nine years was 1.6% (50 out of 3057) (Table 1). From 2010 to 2015, SaV was detected at low prevalence (0-1.8%). After that, the prevalence of SaV infection increased year by year to 2.2%, 2.5%, and 2.8% in 2016, 2017, and 2018, respectively. There was no specific seasonal pattern of SaV detection observed in this study. The age of SaV-infected patients varied from 8 months up to 11 years (Table 2). Among 50 SaV positive cases, 26 (52%) were male and 24 (48%) were female. All SaV positive samples were also tested for the presence of other enteric viral pathogens. The results showed that majority of cases were single infection with SaV (74%, 37 out of 50) whereas the rest of cases (26%, 13 out of 50) were co-infected with other enteric viruses including rotavirus, norovirus, adenovirus, parechovirus, enterovirus, or astrovirus (Table 3).
Table 2. Prevalence and genotype distribution of sapovirus detected in children with acute gastroenteritis in Chiang Mai, Thailand from 2010 to 2018.
| Years | Total samples | Positive samples (%) | SaV genotypes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GI.1 | GI.2 | GI.5 | GII.1 | GII.5 | GIV.1 | GII.1/GII.4 | |||
| 2010 | 109 | 2 (1.8) | 2 | – | – | – | – | – | – |
| 2011 | 302 | 0 | – | – | – | – | – | – | – |
| 2012 | 341 | 2 (0.6) | – | – | – | – | – | 1 | 1 |
| 2013 | 280 | 4 (1.4) | 2 | – | – | 1 | – | – | 1 |
| 2014 | 268 | 0 | – | – | – | – | – | – | – |
| 2015 | 335 | 6 (1.8) | 3 | 3 | – | – | – | – | – |
| 2016 | 508 | 11 (2.2) | 6 | 3 | – | – | 2 | – | – |
| 2017 | 278 | 7 (2.5) | 3 | – | – | 2 | – | – | 2 |
| 2018 | 636 | 18 (2.8) | 4 | 1 | 1 | 5 | 6 | 1 | – |
| 9 years | 3057 | 50 (1.6) | 20 | 7 | 1 | 8 | 8 | 2 | 4 |
Table 3. General information of patients and genotypes of sapovirus based on polymerase and capsid nucleotide sequences.
| Sample ID | Collection date | Age | Gender | RdRp genotype | VP1 genotype | Mix-infection |
|---|---|---|---|---|---|---|
| CMH-N018-10 | 12-Dec-2010 | – | Female | GI.1 | GI.1 | – |
| CMH-S050-10 | 14-Dec-2010 | – | Female | GI.1 | GI.1 | – |
| CMH-N061-12 | 11-Feb-2012 | – | Female | GIV.1 | GIV.1 | – |
| CMH-N145-12 | 10-Aug-2012 | – | Male | GII.1 | GII.4 | – |
| CMH-S004-13 | Jan-2013 | – | Male | GI.1 | GI.1 | – |
| CMH-S034-13 | Apr-2013 | – | Female | GII.1 | GII.1 | Rotavirus |
| CMH-N021-13 | 02-May-2013 | – | Male | GII.1 | GII.4 | Rotavirus |
| CMH-N131-13 | 03-Oct-2013 | – | Female | GI.1 | GI.1 | – |
| CMH-S152-15 | 16-Jun-2015 | 28 months | Male | GI.2 | GI.2 | – |
| CMH-S166-15 | 13-Aug-2015 | 35 months | Female | GI.1 | GI.1 | – |
| CMH-S252-15 | 20-Dec-2015 | 54 months | Female | GI.2 | GI.2 | – |
| CMH-S254-15 | 31-Dec-2015 | 32 months | Female | GI.2 | GI.2 | Norovirus |
| CMH-N006-15 | 09 -Jul-2015 | 7 months | Male | GI.1 | GI.1 | Adenovirus |
| CMH-N007-15 | 09 -Jul-2015 | 9 months | Male | GI.1 | GI.1 | Parechovirus |
| CMH-S108-16 | 29-Mar-2016 | 20 months | Female | GI.2 | GI.2 | – |
| CMH-S198-16 | 11-Aug-2016 | 32 months | Male | GI.1 | GI.1 | Enterovirus |
| CMH-S229-16 | 12-Sep-2016 | 20 months | Male | GI.1 | GI.1 | – |
| CMH-ST004-16 | 11-Jan-2016 | 27 months | Female | GI.2 | GI.2 | – |
| CMH-ST016-16 | 02-Feb-2016 | 22 months | Male | GI.1 | GI.1 | – |
| CMH-ST029-16 | 07-Feb-2016 | 143 months | Female | GI.2 | GI.2 | – |
| CMH-ST090-16 | 21-Mar-2016 | 26 months | Female | GI.1 | GI.1 | Norovirus |
| CMH-ST091-16 | 21-Mar-2016 | 8 months | Female | GII.5 | GII.5 | – |
| CMH-ST095-16 | 23-Mar-2016 | 8 months | Female | GII.5 | GII.5 | – |
| CMH-ST163-16 | 13-Aug-2016 | 24 months | Female | GI.1 | GI.1 | Parechovirus |
| CMH-ST199-16 | 12-Dec-2016 | 27 months | Male | GI.1 | GI.1 | Parechovirus |
| CMH-S003-17 | 22-Feb-2017 | 43 months | Male | GI.1 | GI.1 | Astrovirus |
| CMH-S023-17 | 04-Apr-2017 | 34 months | Male | GI.1 | GI.1 | – |
| CMH-S050-17 | 16-Jun-2017 | 16 months | Female | GII.1 | GII.4 | – |
| CMH-S057-17 | 21-Jun-2017 | 22 months | Female | GII.1 | GII.4 | – |
| CMH-S089-17 | 21-Jun-2017 | 12 months | Female | GI.1 | GI.1 | Enterovirus |
| CMH-S120-17 | 21-Nov-2017 | 16 months | Male | GII.1 | GII.1 | – |
| CMH-ST028-17 | 09-May-2017 | 24 months | Male | GII.1 | GII.1 | – |
| CMH-N028-18 | 02-Feb-2018 | 16 months | Female | GIV.1 | GIV.1 | Rotavirus, Parechovirus |
| CMH-N061-18 | 18-Feb-2018 | 26 months | Female | GI.5 | GI.5 | Rotavirus |
| CMH-N091-18 | 24-Jun-2018 | 18 months | Male | GII.1 | GII.1 | – |
| CMH-N104-18 | 10-Sep-2018 | 11 months | Male | GII.1 | GII.1 | – |
| CMH-R031-18 | 21-Oct-2018 | 24 months | Female | GI.1 | GI.1 | – |
| CMH-R076-18 | 21-Oct-2018 | 24 months | Male | GI.2 | GI.2 | – |
| CMH-R089-18 | 21-Oct-2018 | 43 months | Male | GII.5 | GII.5 | – |
| CMH-R140-18 | 21-Oct-2018 | 12 months | Female | GII.1 | GII.1 | – |
| CMH-S174-18 | 21-Nov-2018 | 48 months | Male | GI.1 | GI.1 | – |
| CMH-S175-18 | 21-Nov-2018 | 31 months | Male | GII.1 | GII.1 | – |
| CMH-ST097-18 | 16-Apr-2018 | 25 months | Male | GI.1 | GI.1 | – |
| CMH-ST169-18 | 27-Jun-2018 | 12 months | Male | GI.1 | GI.1 | – |
| CMH-ST189-18 | 27-Jul-2018 | 12 months | Male | GII.1 | GII.1 | – |
| CMH-ST202-18 | 15-Aug-2018 | 20 months | Male | GII.5 | GII.5 | – |
| CMH-ST207-18 | 17-Aug-2018 | 22 months | Male | GII.5 | GII.5 | – |
| CMH-ST247-18 | 10-Nov-2018 | 44 months | Male | GII.5 | GII.5 | – |
| CMH-ST266-18 | 15-Dec-2018 | 24 months | Female | – | GII.5 | – |
| CMH-ST270-18 | 29-Dec-2018 | 32 months | Female | GII.5 | GII.5 | – |
Notes.
The highlighted samples are the ones in which recombinant sapoviruses were found.
Molecular characterization of SaV and phylogenetic analysis
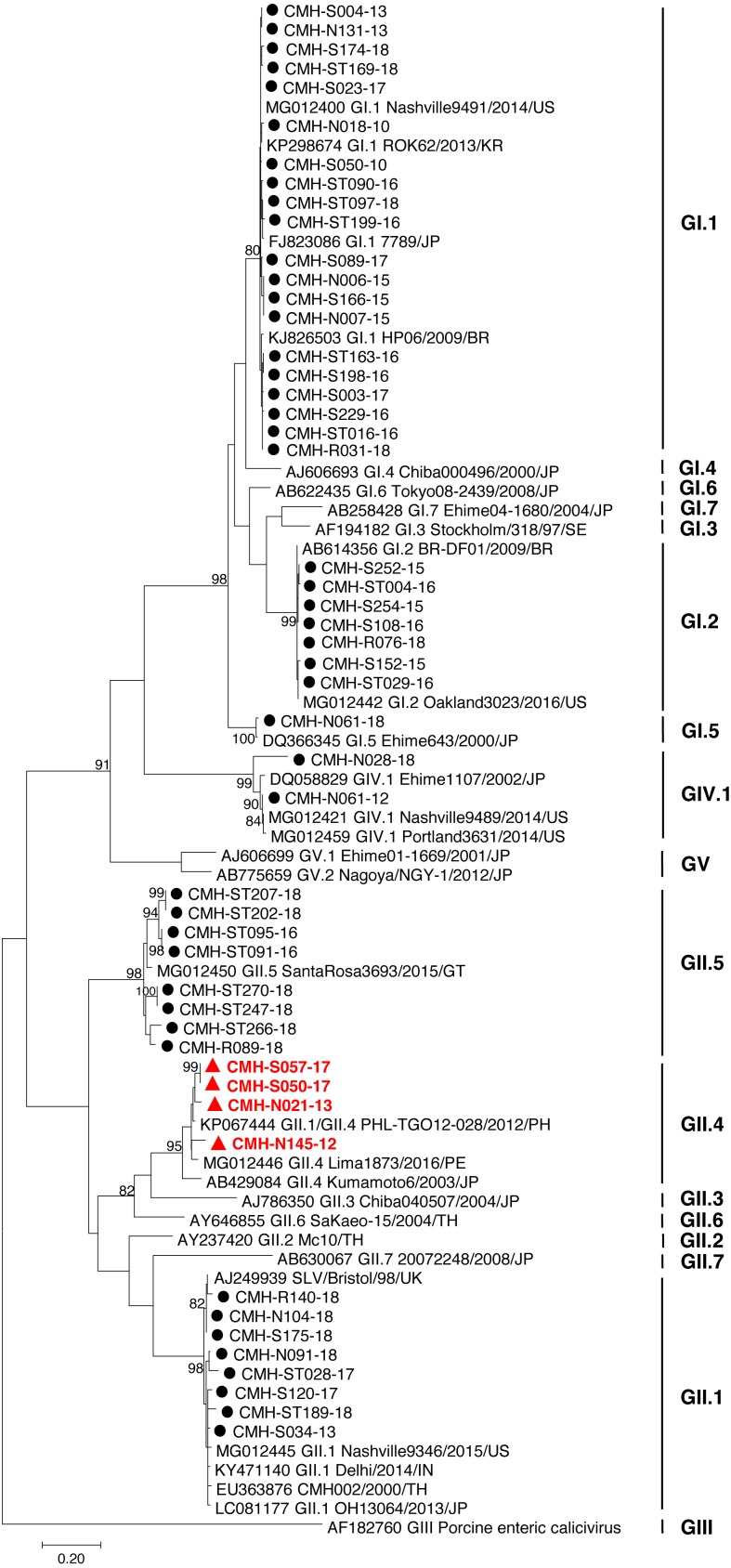
The partial capsid gene sequence (308 nucleotides) of 50 SaV strains were analyzed to identify genotype of the virus. Seven different genotypes were identified in the present study: GI.1 (20), GI.2 (7), GI.5 (1), GII.1 (8), GII.4 (4), GII.5 (8), and GIV.1 (2). Phylogenetic analysis revealed that 20 SaV GI.1 strains were clustered with the GI.1 strains reported previously from USA, Korea, Japan, and Brazil (Fig. 1) and showed 97.4–99.6% nucleotide homology to theses strains. Seven SaV GI.2 strains were grouped together with the GI.2 strains reported previously from Brazil and USA with highest nucleotide sequence similarity (98.7–100%). GI.5 strain was closely related to the GI.5 Ehime643 strain detected previously in Japan (99.0% nucleotide sequence identity). Eight GII.1 strains shared high similarity (90.3–95.0% nucleotide sequence identities) with GII.1 strains detected previously in United Kingdom, USA, India, Japan, and Thailand. Four GII.4 strains were closely related to GII.4 strains reported from Philippines and Peru (94.5–97.1% nucleotide identities). All GII.5 strains shared highest similarity (90.3–95.0%) with the GII.5 SataRosa3693/2015/GT strain detected in Guatemala. The CMH-N061-12 SaV GIV.1 strain displayed 97.9–99.6% nucleotide identities to GIV.1 strains reported previously from USA and Japan while CMH-N028-18 SaV GIV.1 strain shared 87.1–87.8% sequence homology with those of the SaV GIV.1 reference strains.
Figure 1. Phylogenetic tree of the partial capsid gene sequences (296 nucleotides).
Fifty SaV strains detected in this study are indicated by black circle (non-recombinant strains) and red triangle (recombinant strains). The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model. Scale bar indicates nucleotide substitutions per site and bootstrap values (>80) are indicated for the corresponding nodes.
Recombination analysis
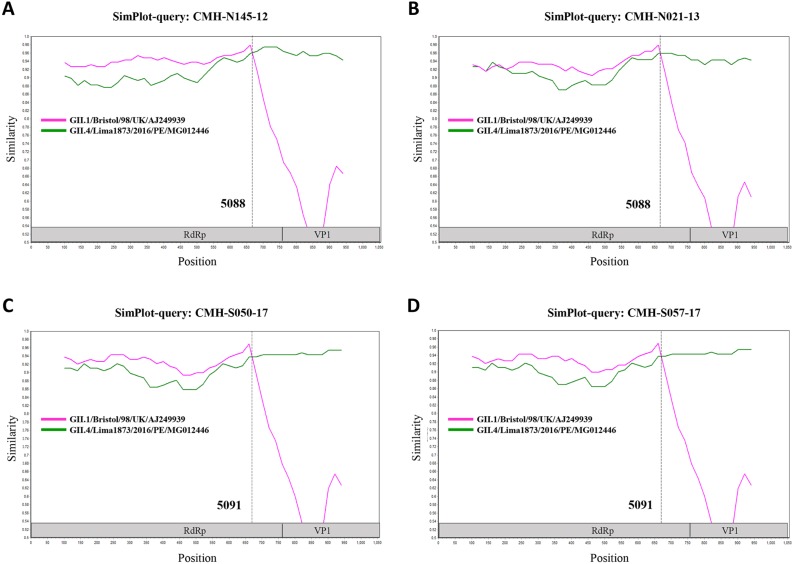
In the SaV genome, recombination event typically occurs between the polymerase (RdRp) and capsid (VP1) genes. To investigate the genetic recombination of SaV strains detected in this study, we further amplified the polymerase gene of the viruses. The partial RdRp gene sequence (745 nucleotides) was successfully obtained from 49 of 50 SaV-positive samples. Phylogenetic tree of the partial RpRp region of 49 SaV strains was constructed (Fig. 2) and the results showed that all strains, except for four GII.4 strains (CMH-S057-17, CMH-S050-17, CMH-N021-13, and CMH-N145-12), were clustered into the same genotypes assigned by the capsid gene sequence as shown previously in Fig. 1. Based in the capsid gene sequence (Fig. 1), CMH-S057-17, CMH-S050-17, CMH-N021-13, and CMH-N145-12 were assigned as the GII.4 genotype whereas based on the RdRp sequence they clustured together with SaV GII.1 reference strains and shared 91.8–95.0% nucleotide sequence identities. The data suggested that these four SaV strains were the SaV GII.1/GII.4 recombinant strains. To predict the putative recombination point for SaV recombinant strains, SimPlot software was used. The similarity plot of SaV recombinant strains detected in this study are shown in Fig. 3. Two recombination break points were identified at positions 5088 and 5091 within the RdRp-VP1 junction region (nucleotide positions 5078-5265). The CMH-145-12 and CMH-N021-13 strains showed the same recombination point at position 5088 (p = 1.572 × 10−14 and p = 1.496 × 10−12, respectively) while other 2 strains (CMH-S050-17 and CMH-S057-17) displayed the recombination point at position 5091 (p = 2.334 × 10−14 and p = 2.322 × 10−14, respectively).
Figure 2. Phylogenetic tree of the partial RdRp gene sequences (745 nucleotides).
Forty-nine SaV strains detected in this study are indicated by black circle (non-recombinant strains) and blue triangle (recombinant strains). The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model. Scale bar indicates nucleotide substitutions per site and bootstrap values (>80) are indicated for the corresponding nodes.
Figure 3. The similarity plot of four SaV recombinant strains, CMH-N145-12 (A), CMH-N021-13 (B), CMH-S050-17 (C), CMH-S057-17 (D), was constructed using SimPlot software.
The similarity score of 200 nucleotides sliding window and 20 site step were used. The junction of RdRp and capsid sequences of SaV recombinant strains (1,055 nucleotides) were plotted against the nucleotide sequences of the reference strains, GII.1/Bistol/98/UK and GII.4/Lima1873/2016/PE. The x-axis represents the nucleotide position and the y-axis indicates the similarity between the query strain and the reference strains.
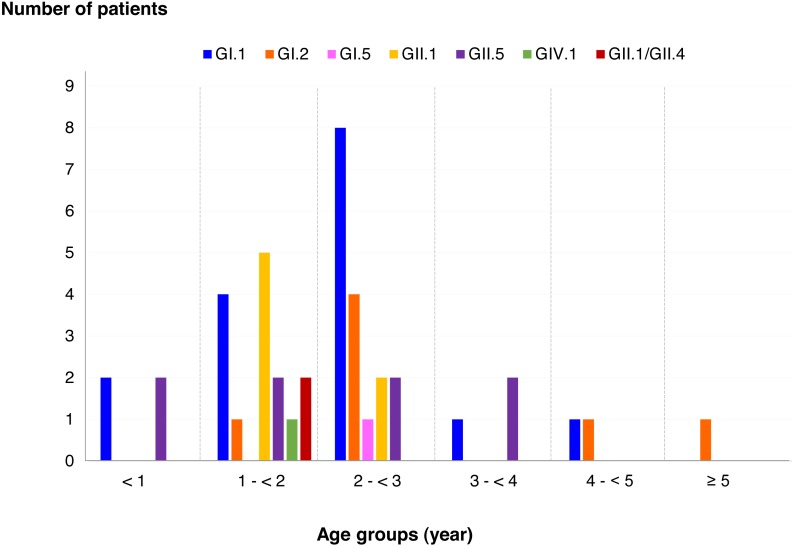
SaV infection in different age groups of patients
Among SaV positive cases, the highest detection rate was seen in patients with the age of 2 to <3 years (40.5%), followed by 1 to <2 years (35.7%), less than 1 year (9.5%), 3 to <4 years (7.1%), 4 to <5 years (4.8%), and more than 5 years (2.4%). In addition, this study identified 6 different genotypes and one recombinant pattern of SaV. Distribution of SaV genotypes detected in different age groups of patients is shown in Fig. 4. It was found that GI.1 genotype was detected in all age groups except for patients with more than 5 years of age. The GII.5 was also identified in patients with age groups of less than 4 years. Interestingly, recombinant SaV GII.1/GII.4 was detected in patients between 1 and 2 years of age.
Figure 4. Distribution of SaV genotypes detected in different age groups of patients.
Discussion
Enteric caliciviruses including noroviruses and sapoviruses are the major causes of AGE of human in all age group worldwide (Khamrin et al., 2017; Lu et al., 2014; Neo et al., 2017; Sala et al., 2014). The detection rate of SaV infection in many countries around the world have been reported with a range from 0.2 to 39% in children and 2.2 to 15.6% in all age groups (Magwalivha et al., 2018; Oka et al., 2015; Varela et al., 2019). In Thailand, the prevalence of SaV was detected between 0% and 15.0% (Kumthip & Khamrin, 2018). The overall SaV-positive rate reported in the present study during the period 2010 to 2018 (1.6%, ranged from 0 to 2.8%) is more or less the same as other similar studies including Bangladesh (2.7%) (Dey et al., 2007), China (1.9%) (Ren et al., 2013), Vietnam (1.4%) (Trang et al., 2012), and Thailand (1.2 and 1.9%) (Khamrin et al., 2017; Supadej et al., 2019). However, when compared to the studies conducted in different geographical regions, the prevalence of SaV detected in this study was lower than those reported from Japan (4.8%), Philippines (7%), USA (5.4%), Italy (6%), Denmark (8.8%), Finland (9.3%), Spain (15.6%), and Nicaragua (17%) (Biscaro et al., 2018; Bucardo et al., 2014; Chhabra et al., 2013; Oka et al., 2015; Thongprachum et al., 2015; Varela et al., 2019). The variation of SaV prevalence in different studies could be explained, at least in part, by the difference in study locations, the detection methods, and the emergence of new epidemic strains. It was noticed that our study as well as other studies that had similar SaV prevalence used single round PCR as a screening method while other studies that reported higher SaV-positive rate performed real-time PCR for the screening process.
Among different genogroups of SaV, the SaV GI (GI.1 and GI.2) and GII (GII.1 and GII.2) are the most predominant genogroups circulating worldwide while other genotypes are rarely detected in some particular countries (Diez-Valcarce et al., 2018; Magwalivha et al., 2018). Similar to other reports, SaV GI.1, GI.2, and GII.1 were the most common genotypes detected in our study. Nonetheless, there was no GII.2 strain observed in the present study. The SaV GII.5 is not often detected. The occurrence of GII.5 in human stool samples was reported in some particular areas such as Guatemala, Peru, South Africa, and USA (Diez-Valcarce et al., 2018; Liu et al., 2016; Murray et al., 2016). In addition to these countries, it should be noted that a high proportion of GII.5 strain (16%, 8 out of 50) was observed in our study. Interestingly, 7 out of 8 SaV GII.5-infected patients were from the same hospital (Sanpatong hospital) and 5 of them were detected in the same year of 2018, suggesting that this particular genotype is circulating in a particular location.
RNA recombination plays a key role in virus evolution and leads to its pathogenicity and virus diversity (Worobey & Holmes, 1999). At least, two types of recombination events of SaV including intergenogroup and intragenogroup have been reported previously (Chanit et al., 2009; Hansman et al., 2007; Hansman et al., 2005; Katayama et al., 2004; Oka et al., 2015; Phan et al., 2006). In the present study, intragenogroup recombinant GII.1/GII.4 SaV was detected in four samples, accounting for 8% (4/50) of all SaV infected cases. The recombinant GII.1/GII.4 SaV has been reported previously in Philippines, Vietnam, and USA (Diez-Valcarce et al., 2018; Liu et al., 2015; Nguyen et al., 2008). However, it has not been described elsewhere in Thailand. To our knowledge, this is the first report demonstrating the presence of recombinant SaV in Thailand. Generally, characterization of SaV is based on the nucleotide sequence of capsid gene (Oka et al., 2012) and many other previous studies have identified SaV genotypes based only on this gene. To date, both capsid and polymerase genes are used to classify genotype of noroviruses (Chhabra et al., 2019). Since SaV is a very similar pathogen in many aspects, therefore, future classification and characterization of SaV should rely on both polymerase and capsid sequences to identify the virus diversity. In addition, continued surveillance on SaV is important to monitor the emergence of new virus strains.
Conclusions
In summary, the results of this study highlight the impact of SaV in diarrheal diseases among children in Chiang Mai, Thailand over the period of nine years and is the first report to describe recombinant SaV infection in Thai children suffering with AGE. The data of nucleotide analysis of both polymerase and capsid genes from this study provide useful information for a better understanding on the caliciviruses other than noroviruses.
Supplemental Information
Acknowledgments
We are grateful to the physicians and nursing staff who helped to collect the specimens used in this study.
Funding Statement
This work was supported by the Center of Excellence (Emerging and Re-emerging Diarrheal Viruses Research), Chiang Mai University, Chiang Mai, Thailand. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Kattareeya Kumthip conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Pattara Khamrin conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Hiroshi Ushijima conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Limin Chen and Shilin Li analyzed the data, prepared figures and/or tables, and approved the final draft.
Niwat Maneekarn conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Human Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
This work was conducted under the approval of the Research Ethics Committee of the Faculty of Medicine, Chiang Mai University (MIC-2557-02710)
Data Availability
References
- Berke et al. (1997).Berke T, Golding B, Jiang X, Cubitt DW, Wolfaardt M, Smith AW, Matson DO. Phylogenetic analysis of the Caliciviruses. Journal of Medical Virology. 1997;52:419–424. doi: 10.1002/(SICI)1096-9071(199708)52:4<419::AID-JMV13>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Biscaro et al. (2018).Biscaro V, Piccinelli G, Gargiulo F, Ianiro G, Caruso A, Caccuri F, De Francesco MA. Detection and molecular characterization of enteric viruses in children with acute gastroenteritis in Northern Italy. Infection, Genetics and Evolution. 2018;60:35–41. doi: 10.1016/j.meegid.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Bucardo et al. (2014).Bucardo F, Reyes Y, Svensson L, Nordgren J. Predominance of norovirus and sapovirus in Nicaragua after implementation of universal rotavirus vaccination. PLOS ONE. 2014;9(5):e98201. doi: 10.1371/journal.pone.0098201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanit et al. (2009).Chanit W, Thongprachum A, Khamrin P, Okitsu S, Mizuguchi M, Ushijima H. Intergenogroup recombinant sapovirus in Japan, 2007–2008. Emerging Infectious Diseases. 2009;15:1084–1087. doi: 10.3201/eid1507.090153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra et al. (2019).Chhabra P, De Graaf M, Parra GI, Chan MC, Green K, Martella V, Wang Q, White PA, Katayama K, Vennema H, Koopmans MPG, Vinjé J. Updated classification of norovirus genogroups and genotypes. Journal of General Virology. 2019;100(10):1393–1406. doi: 10.1099/jgv.0.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra et al. (2013).Chhabra P, Payne DC, Szilagyi PG, Edwards KM, Staat MA, Shirley SH, Wikswo M, Nix WA, Lu X, Parashar UD, Vinjé J. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. Journal of Infectious Diseases. 2013;208(5):790–800. doi: 10.1093/infdis/jit254. [DOI] [PubMed] [Google Scholar]
- Dey et al. (2007).Dey SK, Phan TG, Nguyen TA, Nishio O, Salim AF, Yagyu F, Okitsu S, Ushijima H. Prevalence of sapovirus infection among infants and children with acute gastroenteritis in Dhaka City, Bangladesh during 2004–2005. Journal of Medical Virology. 2007;79:633–638. doi: 10.1002/jmv.20859. [DOI] [PubMed] [Google Scholar]
- Diez-Valcarce et al. (2018).Diez-Valcarce M, Castro CJ, Marine RL, Halasa N, Mayta H, Saito M, Tsaknaridis L, Pan CY, Bucardo F, Becker-Dreps S, Lopez MR, Magana LC, Ng TFF, Vinje J. Genetic diversity of human sapovirus across the Americas. Journal of Clinical Virology. 2018;104:65–72. doi: 10.1016/j.jcv.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Valcarce et al. (2019).Diez-Valcarce M, Montmayeur A, Tatusov R, Vinjé J. Near-complete human sapovirus genome sequences from Kenya. Microbiology Resource Announcements. 2019;8(7):e01602–e01618. doi: 10.1128/MRA.01602-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green (2013).Green KY. Caliciviridae: the noroviruses. In: Knipe DM, Howley P, editors. Field’ s virology. 6th edition Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 584–587. [Google Scholar]
- Hansman et al. (2007).Hansman GS, Ishida S, Yoshizumi S, Miyoshi M, Ikeda T, Oka T, Takeda N. Recombinant sapovirus gastroenteritis, Japan. Emerging Infectious Diseases. 2007;13:786–788. doi: 10.3201/eid1305.070049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman et al. (2005).Hansman GS, Takeda N, Oka T, Oseto M, Hedlund KO, Katayama K. Intergenogroup recombination in sapoviruses. Emerging Infectious Diseases. 2005;11:1916–1920. doi: 10.3201/eid1112.050722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada et al. (2013).Harada S, Tokuoka E, Kiyota N, Katayama K, Oka T. Phylogenetic analysis of the nonstructural and structural protein encoding region sequences, indicating successive appearance of genomically diverse sapovirus strains from gastroenteritis patients. Japanese Journal of Infectious Diseases. 2013;66:454–457. doi: 10.7883/yoken.66.454. [DOI] [PubMed] [Google Scholar]
- Honma et al. (2001).Honma S, Nakata S, Sakai Y, Tatsumi M, Numata-Kinoshita K, Chiba S. Sensitive detection and differentiation of Sapporo virus, a member of the family Caliciviridae, by standard and booster nested polymerase chain reaction. Journal of Medical Virology. 2001;65:413–417. doi: 10.1002/jmv.2050. [DOI] [PubMed] [Google Scholar]
- Kagning Tsinda et al. (2017).Kagning Tsinda E, Malasao R, Furuse Y, Gilman RH, Liu X, Apaza S, Espetia S, Cama V, Oshitani H, Saito M. Complete coding genome sequences of uncommon GII.8 Sapovirus strains identified in diarrhea samples collected from peruvian children. Genome Announcements. 2017;5(43):e01137-17. doi: 10.1128/genomeA.01137-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama et al. (2004).Katayama K, Miyoshi T, Uchino K, Oka T, Tanaka T, Takeda N, Hansman GS. Novel recombinant sapovirus. Emerging Infectious Diseases. 2004;10:1874–1876. doi: 10.3201/eid1010.040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamrin et al. (2017).Khamrin P, Kumthip K, Supadej K, Thongprachum A, Okitsu S, Hayakawa S, Ushijima H, Maneekarn N. Noroviruses and sapoviruses associated with acute gastroenteritis in pediatric patients in Thailand: increased detection of recombinant norovirus GII.P16/GII.13 strains. Archives of Virology. 2017;162:3371–3380. doi: 10.1007/s00705-017-3501-3. [DOI] [PubMed] [Google Scholar]
- Khamrin et al. (2011).Khamrin P, Okame M, Thongprachum A, Nantachit N, Nishimura S, Okitsu S, Maneekarn N, Ushijima H. A single-tube multiplex PCR for rapid detection in feces of 10 viruses causing diarrhea. Journal of Virological Methods. 2011;173(2):390–393. doi: 10.1016/j.jviromet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumthip & Khamrin (2018).Kumthip K, Khamrin P. Molecular epidemiology and genotype distributions of noroviruses and sapoviruses in Thailand 2000–2016: a review. Journal of Medical Virology. 2018;90:617–624. doi: 10.1002/jmv.25019. [DOI] [PubMed] [Google Scholar]
- Lasure & Gopalkrishna (2017).Lasure N, Gopalkrishna V. Epidemiological profile and genetic diversity of sapoviruses (SaVs) identified in children suffering from acute gastroenteritis in Pune, Maharashtra, Western India, 2007–2011. Epidemiology and Infection. 2017;145:106–114. doi: 10.1017/s0950268816001953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2012).Lee LE, Cebelinski EA, Fuller C, Keene WE, Smith K, Vinje J, Besser JM. Sapovirus outbreaks in long-term care facilities, Oregon and Minnesota, USA, 2002–2009. Emerging Infectious Diseases. 2012;18:873–876. doi: 10.3201/eid1805.111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2016).Liu X, Jahuira H, Gilman RH, Alva A, Cabrera L, Okamoto M, Xu H, Windle HJ, Kelleher D, Varela M, Verastegui M, Calderon M, Sanchez G, Sarabia V, Ballard SB, Bern C, Mayta H, Crabtree JE, Cama V, Saito M, Oshitani H. Etiological role and repeated infections of sapovirus among children aged less than 2 years in a cohort study in a peri-urban community of Peru. Journal of Clinical Microbiology. 2016;54:1598–1604. doi: 10.1128/jcm.03133-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2015).Liu X, Yamamoto D, Saito M, Imagawa T, Ablola A, Tandoc 3rd AO, Segubre-Mercado E, Lupisan SP, Okamoto M, Furuse Y, Saito M, Oshitani H. Molecular detection and characterization of sapovirus in hospitalized children with acute gastroenteritis in the Philippines. Journal of Clinical Virology. 2015;68:83–88. doi: 10.1016/j.jcv.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Lole et al. (1999).Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology. 1999;73:152–160. doi: 10.1128/JVI.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2014).Lu L, Zhong H, Xu M, Su L, Cao L, Dong N, Xu J. Molecular epidemiology of human calicivirus infections in children with acute diarrhea in Shanghai: a retrospective comparison between inpatients and outpatients treated between 2006 and 2011. Archives of Virology. 2014;159:1613–1621. doi: 10.1007/s00705-013-1881-6. [DOI] [PubMed] [Google Scholar]
- Magwalivha et al. (2018).Magwalivha M, Kabue JP, Traore AN, Potgieter N. Prevalence of human sapovirus in low and middle income countries. Advances in Virology. 2018;2018:5986549. doi: 10.1155/2018/5986549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin et al. (2015).Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evolution. 2015;1(1):vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez et al. (2002).Martinez N, Espul C, Cuello H, Zhong W, Jiang X, Matson DO, Berke T. Sequence diversity of human caliciviruses recovered from children with diarrhea in Mendoza, Argentina, 1995–1998. Journal of Medical Virology. 2002;67:289–298. doi: 10.1002/jmv.2220. [DOI] [PubMed] [Google Scholar]
- Murray et al. (2016).Murray TY, Nadan S, Page NA, Taylor MB. Diverse sapovirus genotypes identified in children hospitalised with gastroenteritis in selected regions of South Africa. Journal of Clinical Virology. 2016;76:24–29. doi: 10.1016/j.jcv.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Neo et al. (2017).Neo FJX, Loh JJP, Ting P, Yeo WX, Gao CQH, Lee VJM, Tan BH, Ng CG. Outbreak of caliciviruses in the Singapore military, 2015. BMC Infectious Diseases. 2017;17(1):719. doi: 10.1186/s12879-017-2821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen et al. (2008).Nguyen TA, Hoang L, Pham le D, Hoang KT, Okitsu S, Mizuguchi M, Ushijima H. Norovirus and sapovirus infections among children with acute gastroenteritis in Ho Chi Minh City during 2005–2006. Journal of Tropical Pediatrics. 2008;54:102–113. doi: 10.1093/tropej/fmm096. [DOI] [PubMed] [Google Scholar]
- Oka et al. (2016).Oka T, Lu Z, Phan T, Delwart EL, Saif LJ, Wang Q. Genetic characterization and classification of human and animal sapoviruses. PLOS ONE. 2016;11:e0156373. doi: 10.1371/journal.pone.0156373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka et al. (2012).Oka T, Mori K, Iritani N, Harada S, Ueki Y, Iizuka S, Mise K, Murakami K, Wakita T, Katayama K. Human sapovirus classification based on complete capsid nucleotide sequences. Archives of Virology. 2012;157:349–352. doi: 10.1007/s00705-011-1161-2. [DOI] [PubMed] [Google Scholar]
- Oka et al. (2015).Oka T, Wang Q, Katayama K, Saif LJ. Comprehensive review of human sapoviruses. Clinical Microbiology Reviews. 2015;28:32–53. doi: 10.1128/cmr.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan et al. (2006).Phan TG, Yan H, Khamrin P, Quang TD, Dey SK, Yagyu F, Okitsu S, Muller WE, Ushijima H. Novel intragenotype recombination in sapovirus. Clinical Laboratory. 2006;52(7–8):363–366. [PubMed] [Google Scholar]
- Ren et al. (2013).Ren Z, Kong Y, Wang J, Wang Q, Huang A, Xu H. Etiological study of enteric viruses and the genetic diversity of norovirus, sapovirus, adenovirus, and astrovirus in children with diarrhea in Chongqing, China. BMC Infectious Diseases. 2013;13:412. doi: 10.1186/1471-2334-13-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala et al. (2014).Sala MR, Broner S, Moreno A, Arias C, Godoy P, Minguell S, Martinez A, Torner N, Bartolome R, De Simon M, Guix S, Dominguez A. Cases of acute gastroenteritis due to calicivirus in outbreaks: clinical differences by age and aetiological agent. Clinical Microbiology and Infection. 2014;20:793–798. doi: 10.1111/1469-0691.12522. [DOI] [PubMed] [Google Scholar]
- Supadej et al. (2019).Supadej K, Khamrin P, Kumthip K, Malasao R, Chaimongkol N, Saito M, Oshitani H, Ushijima H, Maneekarn N. Distribution of norovirus and sapovirus genotypes with emergence of NoV GII.P16/GII.2 recombinant strains in Chiang Mai, Thailand. Journal of Medical Virology. 2019;91:215–224. doi: 10.1002/jmv.25261. [DOI] [PubMed] [Google Scholar]
- Thongprachum et al. (2015).Thongprachum A, Takanashi S, Kalesaran AF, Okitsu S, Mizuguchi M, Hayakawa S, Ushijima H. Four-year study of viruses that cause diarrhea in Japanese pediatric outpatients. Journal of Medical Virology. 2015;87(7):1141–1148. doi: 10.1002/jmv.24155. [DOI] [PubMed] [Google Scholar]
- Torner et al. (2016).Torner N, Martinez A, Broner S, Moreno A, Camps N, Dominguez A. Epidemiology of acute gastroenteritis outbreaks caused by human calicivirus (Norovirus and Sapovirus) in catalonia: a two year prospective study, 2010–2011. PLOS ONE. 2016;11:e0152503. doi: 10.1371/journal.pone.0152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang et al. (2012).Trang NV, Luan le T, Kim-Anh le T, Hau VT, Nhung le TH, Phasuk P, Setrabutr O, Shirley H, Vinje J, Anh DD, Mason CJ. Detection and molecular characterization of noroviruses and sapoviruses in children admitted to hospital with acute gastroenteritis in Vietnam. Journal of Medical Virology. 2012;84:290–297. doi: 10.1002/jmv.23185. [DOI] [PubMed] [Google Scholar]
- Varela et al. (2019).Varela MF, Rivadulla E, Lema A, Romalde JL. Human sapovirus among outpatients with acute gastroenteritis in Spain: a one-year study. Viruses. 2019;11(2):E144. doi: 10.3390/v11020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey & Holmes (1999).Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. Journal of General Virology. 1999;80(Pt 10):2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- Xue et al. (2019).Xue L, Cai W, Gao J, Jiang Y, Wu H, Zhang L, Zuo Y, Dong R, Pang R, Zeng H, Wu S, Wang J, Zhang J, Wu Q. Genome characteristics and molecular evolution of the human sapovirus variant GII.8. Infection, Genetics and Evolution. 2019;73:362–367. doi: 10.1016/j.meegid.2019.05.017. [DOI] [PubMed] [Google Scholar]
- Yan et al. (2003).Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H. Detection of norovirus (GI, GII), Sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. Journal of Virological Methods. 2003;114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Yinda et al. (2017).Yinda CK, Conceicao-Neto N, Zeller M, Heylen E, Maes P, Ghogomu SM, Van Ranst M, Matthijnssens J. Novel highly divergent sapoviruses detected by metagenomics analysis in straw-colored fruit bats in Cameroon. Emerg Microbes Infect. 2017;6:e38. doi: 10.1038/emi.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The nucleotide sequences of polymerase and capsid genes are available at GenBank: MN245671–MN245579 and MN253492–MN253541.