Abstract
Prediction of HLA epitopes is important for the development of cancer immunotherapies and vaccines. However, current prediction algorithms have limited predictive power, in part because they were not trained on high-quality epitope datasets covering a broad range of HLA alleles. To enable prediction of endogenous HLA class I–associated peptides across a large fraction of the human population, we used mass spectrometry to profile >185,000 peptides eluted from 95 HLA-A, B, C and G mono-allelic cell lines. We identified canonical peptide motifs per HLA allele, unique and shared binding submotifs across alleles, and distinct motifs associated with different peptide lengths. By integrating these data with transcript abundance and peptide processing, we developed HLAthena, providing allele-and-length-specific and pan-allele-pan-length prediction models for endogenous peptide presentation. These models predicted endogenous HLA class I–associated ligands with 1.5-fold improvement in positive predictive value compared with existing tools and correctly identified >75% of HLA-bound peptides that were observed experimentally in 11 patient-derived tumor cell lines.
The HLA genes are the most polymorphic across the human population, with more than 16,200 distinct class I alleles as of May 20191,2. Short peptides (8–11mers) bound to the diverse array of HLA class I molecules (HLA-A, -B, -C and -G) arise from intracellular proteins that are cleaved by the proteasome and peptidases prior to loading and display by surface HLA class I proteins to cytotoxic T cell lymphocytes. Given the diversity in HLA binding preferences, an important question is whether one can accurately predict if a peptide is presented by a specific HLA allele. The accuracy of computational models that predict binding between peptides and HLA alleles, especially HLA-A and -B alleles, has been improving3–7. In the field of cancer, these tools are now increasingly used in conjunction with next-generation DNA sequencing of tumors to identify immunogenic cancer neoantigens, which arise from tumor-specific somatic mutations. They have accelerated epitope discovery, as they enable experimental efforts to focus on a narrower list of epitopes with good predicted binding. However, even with widely used algorithms such as NetMHCpan3,8, the numbers of falsely discovered binders increase once the predicted binding affinity decreases (i.e. IC50>100 nM)9. Furthermore, while these algorithms are designed to predict the binding affinity of peptides to individual HLA molecules – the final step of antigen presentation, they do not account for intracellular availability of the peptide precursors or their processing by proteases. Finally, because previous research has focused on the few alleles highly expressed by Caucasian populations, existing algorithms have uneven accuracy in the prediction of epitopes binding to less common alleles in Caucasians, or those highly prevalent in other populations.
Detection and sequencing of HLA-bound peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS) has the unique advantage that information on endogenously processed and presented peptides from a cell can be directly learned. Our previous proof-of-concept study demonstrated that characterization of HLA-bound peptides eluted from a limited set of cell lines engineered to express single HLA alleles could reveal allele-specific peptide motifs and be used to train predictive algorithms for endogenous allele-specific peptide presentation4. Here, we expand our initial dataset of >24,000 peptides from 16 cell lines and identify and characterize 186,464 eluted peptides from 95 HLA-A, -B, -C and -G alleles. We included HLA-G peptidomes because this HLA is implicated in maternal-fetal tolerance and is also upregulated in many cancers10,11. These data allow us to compare peptide length preferences and the spectrum of distinct and shared submotifs across HLA class I alleles, revealing the diversity and complexity of endogenous HLA ligands. Using this information, we trained allele-and-length-specific and pan-allele-pan-length predictors, which identify 1.5-fold more peptides than conventional prediction tools when evaluating ligands directly detected by LC-MS/MS from 11 patient-derived tumor cell lines. The datasets of HLA-binding peptides from mono-allelic cells and patient-derived tumors, as well as the prediction models (HLAthena) and interactive web tools are all made publicly available.
Results
Systematic LC-MS/MS profiling of HLA class I ligands from mono-allelic cell lines
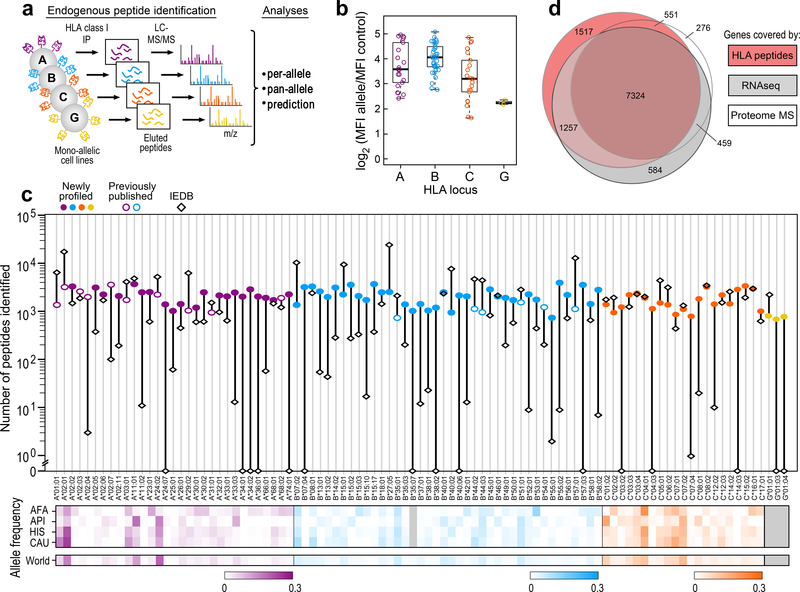
We engineered 79 cell lines expressing a single HLA class I allele by stably transfecting individual HLA-A, -B, -C, or -G alleles into the HLA-null B721.221 cell line (Fig. 1a), adding to the 16 lines we previously reported4. Surface expression of the alleles was confirmed by flow cytometry (Fig. 1b; Supplementary Fig. 1a; Supplementary Table 1a). Altogether, the collection of 95 cell lines (31 HLA-A, 40 HLA-B, 21 HLA-C and 3 HLA-G) covered at least one allele in 95% of individuals worldwide for each HLA-A, -B and -C alleles12–14 (Supplementary Table 1b; Supplementary Note 1).
Figure 1: Mass spectrometric characterization of peptides eluted from HLA proteins in mono-allelic cell lines.
a) Schematic of the experimental design: HLA-null B721.221 cells transfected to express a single HLA allele (31 HLA-A, 40 HLA-B, 21 HLA-C and 3 HLA-G) were subjected to HLA class I-immunoprecipitation with W6/32 antibody from 50–300 million cells per allele followed by identification of eluted peptides by LC-MS/MS, in order to generate endogenous peptide binding data used to characterize allele-specific or pan-allele binding preferences and train neural network predictors of antigen processing and presentation. b) Surface expression of each transfected HLA-alleles was confirmed by flow cytometric detection against parental cells transfected with an empty vector (MFI: Mean fluorescence intensity; n=21 HLA-A, 34 HLA-B, 21 HLA-C, 3 HLA-G biologically independent samples; (boxplots depict median intensity, the box contains 25%−75% of the data, whiskers extend to lowest and highest values no further than 1.5*IQR; profiles of all lines in Supplementary Fig. 1a. c) Overlap of human genes represented by at least two HLA-associated peptides (pink), detected in RNA sequencing (TPM>2, light grey) or identified in deep proteome analysis (≥2 unique peptides, dark grey) of the B721.221 mono-allelic cells lines. d) Top: Numbers of HLA-bound peptides identified per allele by MS-based profiling (circles; filled: newly generated data; open: previously reported4 or recorded in IEDB (diamonds). Bottom: Heatmaps of relative median population frequencies per allele across racial groups (AFA: African American, API: Asian or Pacific Islander, HIS: Hispanic, CAU: Caucasian) in the US population13 and worldwide. See also Supplementary Figure 1.
HLA-bound peptides for each engineered cell line were isolated by HLA immunopurification, analyzed by high-resolution LC-MS/MS and sequences identified by a ‘no-enzyme’ specificity database search at 1% FDR (Supplementary Data 1). We identified a median of 1,860 peptides per allele (range 692–4,033), for a total of 186,464 peptides after excluding non-specifically bound peptides (Supplementary Table 1c–e; Supplementary Fig. 1b; Methods). Most observed modifications, found in 12% of identified peptides, could be explained by sample processing artifacts like methionine oxidation; adding carbamidomethylation of cysteine into the sample processing workflow recovered more cysteine-containing peptides (Supplementary Fig. 1c,d). HLA-bound peptides mapped to 10,649 human genes (≥2 peptides per gene), representing 91% of human gene products detected by LC-MS/MS in an extensively fractionated B721.221 proteome (≥2 peptides per gene), and 89% of transcribed genes (>2 transcripts per million (TPM) from RNA-seq) (Fig. 1c; Supplementary Fig. 1e; Supplementary Note 3). The top 50 proteins with high HLA peptide coverage were large, highly abundant, and consistently observed across HLA-A, -B and -C alleles (Supplementary Fig. 1f). Notably, 1,517 genes represented by HLA-bound peptides were not detected in either of the two expression data sets, suggesting they had very low transcript and protein levels. These peptides had identification metrics comparable to the rest of the dataset, and are thus reliable identifications (Supplementary Fig. 1g). Peptides identified from sets of 6 alleles matched to patient genotypes15 amounted to 4,000–5,000 presented genes (Supplementary Fig. 1h). We conclude that all expressed proteins can undergo processing and presentation by HLA class I, a far higher proportion than previously appreciated16.
Our newly generated data nearly doubles the HLA ligands recorded in the Immune Epitope Database (IEDB)17 which holds 208,885 ligands from 157 human class I alleles (Supplementary Table 1f; Methods). Peptides for 80 of 95 alleles are available in IEDB; however, 33 of 95 alleles have fewer than 100 known binders, which hinders reliable motif deduction and accurate prediction (Fig. 1d). For the 15 previously uncharacterized alleles, we identified 1,845 peptides on average (range 693–4,022). We systematically assessed the length distribution, positional entropy, residue frequencies, binding motif, and submotif clusters of HLA-bound peptides per allele (Supplementary Fig. 1i–l; Methods; Supplementary Note 4) and created an interactive website for data exploration (http://mhc.tools). Altogether, these data and tools greatly expand the current knowledge of HLA class I-bound peptides.
Identification of HLA binding motifs and submotifs that are shared across alleles
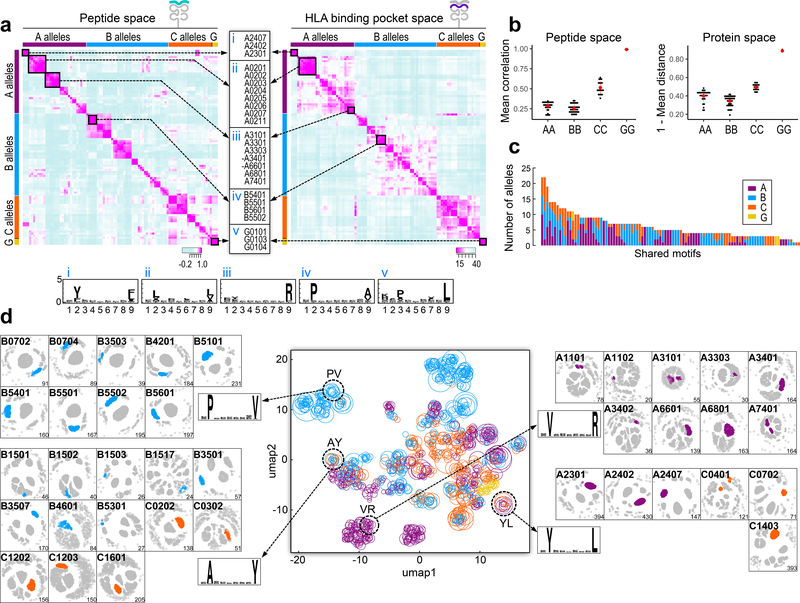
Since the numbers of peptide identifications per allele were only weakly correlated with surface HLA levels, differential binding potential likely contributes to the variation in peptide numbers (Supplementary Fig. 1m,n). To better understand the basis for differential binding, we compared HLA alleles based on the motifs of their observed ligands and the physicochemical properties of binding pocket residues in the HLA protein. By computing pairwise correlations between the peptide binding motifs of each allele, we found groups of alleles sharing well defined HLA-A and -B motifs (Fig. 2a–left; Supplementary Table 2a). As expected, HLA alleles belonging to supertypes such as HLA-A*02 clustered together (Fig. 2a-inset ii) as did split antigen serotypes such as HLA-B*54,55,56 and HLA-A*23,24 (Fig. 2a-inset i, iv)2,18,19. There was minimal motif sharing outside of the dominant groups (mean motif correlation of each HLA-A allele to all other -A alleles; and each HLA-B allele to all other -B alleles was 0.28 and 0.25, respectively, Fig. 2b–left). In contrast, HLA-C motifs were more similar to each other (mean correlation 0.51), thus sharing more overlapping motifs, consistent with previous studies indicating that HLA-C (and HLA-G) alleles are more evolutionarily recent, with less divergence amongst alleles20. The patterns of similarity revealed by binding motifs were mirrored by similarities in the HLA binding clefts, quantified by physicochemical properties of HLA residues in contact with the ligand (Fig. 2a–right, 2b–right; Supplementary Table 2b; Methods). To assess the agreement of the two approaches, for each allele, we counted the number of neighboring alleles in motif space analysis that were also proximal in pocket space (Supplementary Fig. 2a; Methods). This correspondence maps the rules of ligand preference onto HLA protein sequence and serves as the basis for creating pan-allele predictors that rely on transfer learning from characterized to uncharacterized alleles21.
Figure 2: Identification of shared motifs and submotifs amongst HLA-A, B, C and G alleles.
a) Pairwise correlations between 95 HLA-A, -B, -C and -G binding motifs, each represented as a vector of frequencies of the 20 amino acids at every peptide position) (left), and pairwise distances between the 95 HLA binding pockets, each represented by the properties of amino acids that are in contact with the peptide (right). Examples of groups of alleles with high similarity (middle; negative sign indicates that the allele was part of the peptide space group but not the HLA pocket group) and the corresponding binding motif of each group (bottom). b) Average correlations of A to A, B to B, C to C and G to G alleles show that C and G alleles are more similar to each other than A and B alleles in both peptide motif (left) and protein (right) space. Each dot represents an HLA allele and the y-axis is the mean of the correlations between that allele and all other alleles in that group. c) Number of alleles sharing a submotif colored according to HLA locus (A: purple, B: blue, C: orange, G: yellow). d) 2D-visualization of submotifs identified across the 95 alleles (middle), colored according to HLA locus (A:purple; B:blue; C:orange; G:yellow) and scaled in size according to the number of underlying peptides making up the sub-cluster. The collection of all allele-specific submotifs was clustered to identify groups of alleles that share a submotif (Supplementary Fig. 2b). Four examples of clusters of submotifs are highlighted with dashed circles, along with the respective motifs they represent and the allele-specific clusters that contribute to each shared motif. Motif xVxxxxxxR was found to be shared across subclusters amongst the −11:01, −11:02, −31:01, −33:03, −34:01, −34:02, −66:01, −68:01 and −74:01 alleles of HLA-A; likewise, motif xPxxxxxxV was shared by subclusters amongst the −07:02, −07:04, −35:03, −42:01, −51:01, −54:01, −55:01, −55:02 and −56:01 alleles of HLA-B. Motif xYxxxxxxL is shared amongst A*23:01, A*24:02, A*24:07, C*04:01, C*07:02, C*14:03; xAxxxxxxY is shared amongst B*15:01, B*15:02, B*15:03, B*15:17, B*35:03, B*35:07, B*46:01, B*53:01, C*02:02, C*03:02, C*12:01, C*12:03, C*16:01). See also Supplementary Figure 2.
To delineate allele similarity at finer granularity, we decomposed each aggregate motif per allele into submotifs by computing inter-peptide distances, projecting them onto 2-dimensional space and clustering the peptides, obtaining 1,133 submotifs (≥20 peptides per submotif) across the 95 alleles. Distinct motifs were then identified by clustering the allele-specific submotifs, revealing 101 clusters (Supplementary Fig. 2b; Methods). While half of the clusters containing HLA-A/B submotifs were contributed solely by HLA-A/B alleles, respectively (Fig. 2c), most of the HLA-C submotifs overlapped with submotifs from HLA-A and/or B alleles (Fig. 2d; Supplementary Fig. 2c), consistent with HLA-A and -B alleles having more divergent structure than the evolutionarily ‘younger’ HLA-C alleles. Submotif overlap across alleles enables selection of a minimal set of epitopes covering an optimal set of alleles and thus individuals.
Length-specific differences in ligand preferences are detectable among HLA alleles and loci
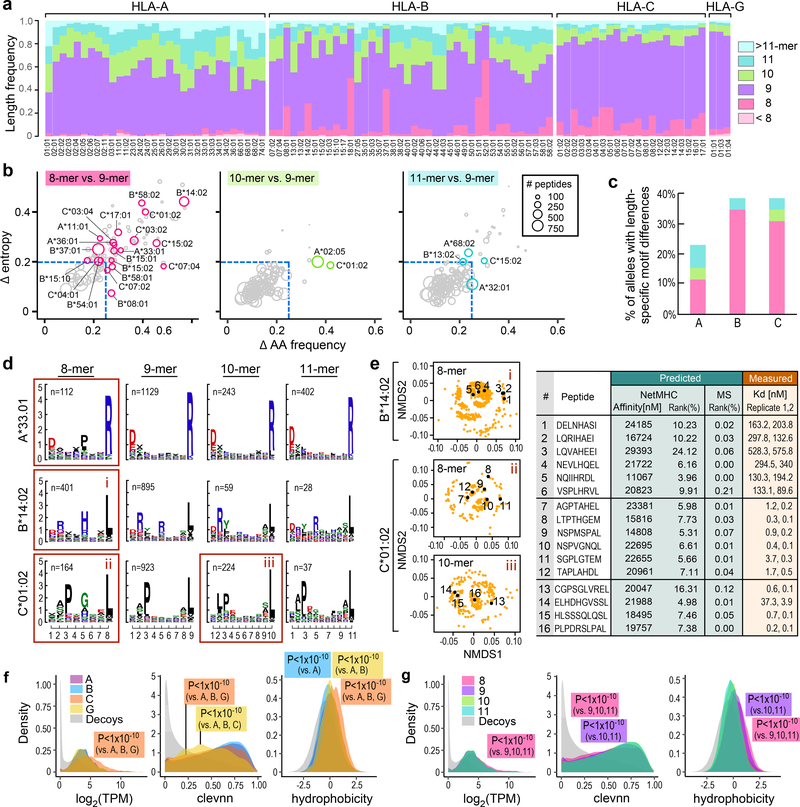
Unbiased evaluation of length distributions revealed that 9-mers were dominant for all but 3 HLA-B alleles (which preferentially bound 8-mers) and that 8-mers were more common for HLA-B/C alleles while 10/11-mers were more common for HLA-A/B (Fig. 3a; Supplementary Table 3a; Table 2a). To systematically identify potential length-specific binding motifs, we compared 8/10/11-mers to 9-mers based on residue frequency and entropy at every peptide position (Fig. 3b; Methods), and found 26 differences across HLA-A, -B and C alleles (20 8-mer, 2 10-mer, and 4 11-mer) out of 178 motifs with at least 100 identified 8/10/11-mer peptides (Fig. 3b–d; Supplementary Table 3b). The most notable changes in entropy were at position 5 for 8-mers compared to 9-mers (Supplementary Fig. 3a). This residue position has been implicated in structural changes of certain HLA-alleles upon binding as it allows embedding of these short peptides in the cleft22,23. Selected peptides were confirmed as strong binders in in vitro binding assays, despite poor predicted affinity by NetMHCpan4.0.BA (Fig. 3e; Supplementary Fig. 3c). Collectively, these observations motivate a more explicit approach of modeling length-specific HLA binding characteristics.
Figure 3: Mono-allelic data uncovers lengths-specific HLA-binding preferences.
a) Frequencies of peptide lengths observed across alleles (8: pink; 9: violet; 10: green; 11: cyan). All but two HLA-B alleles preferentially present 9-mers. HLA-A alleles bind longer peptides more frequently than B and C alleles, while B and C alleles have a higher propensity for short peptides. b) 8-, 10- and 11-mer binding motifs were compared to 9-mer motifs by dropping middle residues (positions 4, 5, 6 or 7 depending on the length) to create pseudo motifs of the same length (8-mers: pseudo 8-mer from 9-mers vs true 8-mer motif; 10- and 11-mers: pseudo 9-mer from 10- and 11-mers vs true 9-mer motif) and selecting the pseudo motif which was most similar to the corresponding true motif. The maximum difference amongst peptide residue positions between the 8-, 10- and 11-mer pseudo motifs and the corresponding true motifs in amino acid frequency (x-axis) and entropy (y-axis) are shown. Circle size reflects number of peptides, dashed lines indicate cutoff values. Circles in color and label denote alleles with >100 peptides with change in amino acid frequency or entropy greater than the selected cutoffs (absolute difference in residue frequency with the true motif of >0.25 or an absolute difference in entropy of >0.2 at any position). c) Percent motif changes within each HLA type colored by length. d) Length dependent logo plots for A*33:01, B*14:02 and C*01:02; red boxes outline the changing motifs. e) Experimental validation of selected peptides (indicated with black dots on the NMDS plots) by in vitro binding assays compared to their predicted scores by NetMHCpan4–0.BA and MS models. f), g) Expression, predicted cleavability (clevnn) and hydrophobicity stratified by HLA loci (n=95 alleles, (31 HLA-A, 40 HLA-B, 21 HLA-C and 3 HLA-G, 1× 1e6 decoys) and peptide length (n=12,970 8-mers, 111,898 9-mers, 29,956 10-mers, 18.202 11-mers; all comparisons Welch’s two sample t-test, two-sided, provided in Supplementary Table 3c). See also Supplementary Figure 3.
Peptide-extrinsic properties vary per HLA and length
Since HLA-bound peptides captured from the cell surface reflect the cell-endogenous processes that shape the ligandome, we assessed whether HLA-A, -B, -C and -G ligands of different peptide lengths are preferentially derived from peptides with variable extrinsic properties. We found that HLA-C bound peptides were biased towards higher expression and hydrophobicity (Fig. 3f; Supplementary Table 3c; Methods). HLA-C also showed a preference toward peptides with poorer proteasome cleavability scores, likely driven by the higher frequency of 8-mers which were observed to have lower cleavage scores (Fig. 3f,g; Supplementary Fig. 3a). These observations agree with prior structural analyses that have reported more shallow HLA-C binding clefts24, as higher abundance and elevated hydrophobicity of cognate peptides could compensate for decreased binding stability. We noted that HLA-G peptides had an even stronger bias towards lower cleavability scores, possibly due to the lack of HLA-G training data for the cleavability predictor and may suggest differential protease activity in shaping the HLA-G ligandome25. Other differences with smaller effect sizes were also observed which altogether prompted us to model extrinsic properties per HLA loci in pan-allele predictors.
Interferon stimulation has minimal impact on peptide trimming signatures
Exposure to inflammation can impact proteasomal processing, but the extent to which it alters the HLA peptidome in normal and cancerous human tissues has not been completely elucidated. Therefore, we generated a series of patient tumor cell lines, derived from melanoma (MEL; n=4)15, glioblastoma (GBM; n=3)26 and clear cell renal cell carcinoma (ccRCC; n=1) specimens (Fig. 4a; Supplementary Data 2), and assessed the effect of IFNγ stimulation on the processing of HLA-bound ligands. As expected, full proteome analysis of one of the GBM samples revealed IFNγ treatment to result in elevated expression of immunoproteasome-specific subunits (PSMB8, PSMB9, PSMB10) and genes involved in interferon-regulated pathways (STAT1, STAT2, STAT4, IRF1, IRF9), along with reduced expression of constitutive proteasome subunits (PSMB5, PSMB7; Fig. 4b; Supplementary Note 5). Likewise, for all 7 IFNγ-matched samples, we observed an increase of 2.4–5.6% in peptides derived from IFNγ-response genes post stimulation (Supplementary Table 4). We also analyzed external datasets from lung epithelial cell lines exposed to IFNγ and TNFα27.
Figure 4: Proteasomal and peptidase shaping of the HLA-associated peptidome.
a) Three types of primary tumor cell lines (melanoma [MEL], glioblastoma [GBM] and clear cell renal cell carcinoma [ccRCC]) used to identify HLA-associated peptidomes. b) Changes in relative protein abundance of proteasomal subunits and IFNγ inducible genes in patient-derived GBM cells with or without IFNγ-treatment based on MS proteome analysis. c) Schematic of cleavage signature analysis. d) Peptide processing signatures of HLA ligands presented by primary tumor and cancer cell lines at baseline (top, n=4 MEL, 3 GBM, 1 ccRCC, 1 Lung, biologically independent samples) and following IFNγ treatment (bottom, n=3 MEL, 3 GBM, 1 ccRCC, 1 Lung, biologically independent samples), showing overrepresented (red) or underrepresented (blue) amino acid residues upstream and downstream of the HLA peptide. The number in each cell denotes percent change over a background decoy set; color intensity indicates significance (see key, Chi-squared test, df=1). e) Heatmap of correlations between the processing preferences in untreated and IFNγ-treated samples at upstream and downstream positions. Signatures for peptides from the IFNγ treated cells correlate well with peptides eluted from untreated cells suggesting minimal to no difference between the two patterns (sample sizes as in 4d; Spearman’s rank correlation). See also Supplementary Figure 4.
To assess proteolytic cleavage preferences in the untreated and treated datasets, we calculated the enrichment of residues upstream and downstream of observed peptides in the protein sequence versus a set of decoys drawn from the proteome, controlling for HLA motif biases (Fig. 4c; Supplementary Note 5). As before, in the untreated dataset (Fig. 4d–top), we observed an enrichment for A, K, S and R at downstream positions as well as for peptides derived from protein C-termini (indicated by ‘-’)4. Upstream residues R and K were not enriched after removing potential tryptic peptide contaminants. Proline was depleted at both peptide termini in all samples, likely due to steric hindrance. Acidic residues (E, D), and certain hydrophobic residues (I, L, F, W) were underrepresented downstream of HLA-associated peptides. Proteasomal signatures of untreated and IFNγ-treated samples were strongly correlated (Fig. 4d,e; Spearman’s ρ>0.76), suggesting that immunoproteasome activation has minimal impact on the processing of HLA-presented ligands in malignant cells.
Generation and performance of allele-and-length-specific and pan-allele-pan-length predictive models of antigen presentation
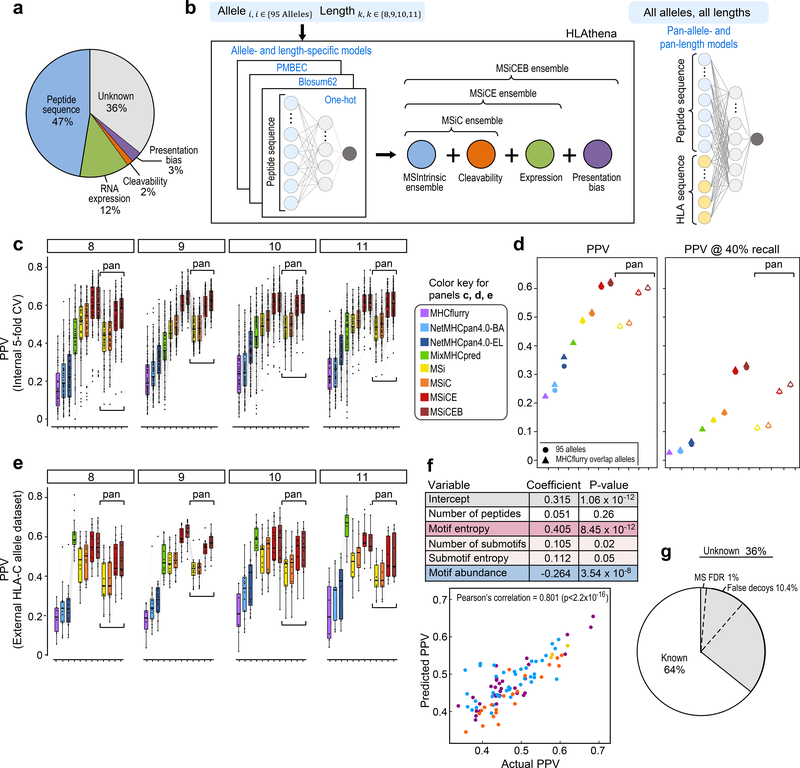
We previously reported that multivariate models incorporating endogenous HLA-presentation descriptors, such as transcript abundance and likelihood of protease cleavage, outperform affinity-trained predictors4. With our extended dataset of 95 alleles, we evaluated the predictive contribution of these variables (Supplementary Fig. 4a,b; Supplementary Note 6,7) along with gene presentation bias and translation quantification via ribosome profiling. Presentation bias quantifies the discrepancy between expected and observed number of MS-identified peptides per gene in a given set of samples and can boost recall of lowly expressed peptides7, while ribosomal profiling captures actively translated mRNA molecules and could provide a more accurate proxy for peptide precursor abundance. To evaluate predictive power, we constructed evaluation datasets with 1:999 ratio of observed binders to random genomic peptides and considered the fraction of true hits scoring within the top 0.1% (i.e. positive predictive value (PPV)). The MS models trained on peptide sequence features alone achieved an average PPV across the 95 alleles of 47% (Fig. 5a; Supplementary Table 5a). This PPV strongly correlated with the number of decoys with scores higher than 50% of hits, which fit the binding motifs, suggesting that such decoys could be real binders and hence artificially decrease PPV (Supplementary Fig. 5). Integrating RNA-seq, as a proxy for peptide abundance, boosted PPV to 60%, while protein abundance achieved 54%. Combining RNA-seq with Ribo-seq reached a PPV of 61% (data not shown). Protein presentation bias and cleavability were the next most predictive variables adding 2.9% and 1.5% to PPV. Based on these results, we trained prediction models that integrate intrinsic peptide features (MSintrinsic, or MSi) with extrinsic properties: cleavability (C), expression (E) and gene presentation bias (B).
Figure 5: Generation and evaluation of allele-and-length-specific and pan-allele-pan-length MS-based models on mono-allelic data.
a) Incremental contribution of predictor variables (peptide binding, transcript expression, cleavability, and gene presentation bias) to positive predictive value (PPV) as the most informative variables are added one at a time (analysis performed for 9-mer peptides). b) Cartoon schematic of the neural networks used to generate allele-and-length-specific and pan-allele-pan-length predictive models. c) Models are evaluated based on their ability to score MS-observed binders in the top 0.1% amongst a 999-fold excess of random decoys (PPV). Shown are 5-fold cross validation (CV) PPVs across each of the n=95 HLA alleles (grey lines) achieved by MHCflurry (available overlapping alleles = 31), NetMHCpan4.0-BA, NetMHCpan4.0-EL, MixMHCpred (available overlapping alleles = 72), and MS-informed models (boxplots depict median PPV, the box contains 25%−75% of the data, whiskers reach to lowest and highest values no further than 1.5*IQR). d) Average PPVs across alleles and lengths for state-of-the-art and MS-based models resulting in a 2-fold improvement in PPV, or 6–12-fold improvement in PPV at 40% recall in an evaluation dataset with 1:10000 hit:decoy ratio. e) Model evaluation as in d) on an external dataset of HLA-C presented peptides identified by MS28. f) Correlation of actual PPVs achieved by the allele-specific 9-mer MSi models vs PPVs predicted by a multivariate linear regression fit, with variables and their respective effect sizes and significance tabulated (n=95). g) The negative contribution of motif abundance to PPV (i.e. negative regression coefficient) suggests that ~10.4% of ‘Unknown’ PPV (estimated as the average motif abundance scaled by the coefficient) can be attributed to false decoys present in the negative set, which artificially decreases PPV. Similarly, 1% of unexplained PPV could be due to false-positive identifications by MS at the 1% FDR threshold (n=95). See also Supplementary Table 5.
The observed length-specific binding motifs (Fig. 3) in conjunction with the high frequency of non-9-mer presentation for some alleles motivated the generation of length-specific binding predictors, trained exclusively on ligands of specific lengths (8-, 9-,10- or 11-mers), without ‘borrowing’ information from 9-mers (Fig. 5b–left; Methods). Model performance was compared against the most recent version of NetMHCpan, 4.03, which incorporates training data from binding affinity (BA) and MS-sequenced eluted peptide (EL; including our previously published 16 alleles), as well as against MHCflurry5 and MixMHCpred26. We found an average improvement in the MS-based sequence-only models (MSi) across lengths of 2.2-, 1.9-, 1.5-, and 1.2-fold compared to MHCflurry (for overlapping alleles), NetMHCpan4.0-BA, NetMHCpan4.0-EL and MixMHCpred (for overlapping alleles), respectively (Fig. 5c; Supplementary Table 5a). Models which additionally integrated cleavability (MSiC) added +2.6 percentage points to the PPV achieved by MSi, cleavability and expression (MSiCE) an additional +9.3, or cleavability, expression and gene presentation bias (MSiCEB) a further +1.1 PPV. Length-specific models for 8-, 10- and 11-mers outperformed the corresponding non-length-specific models currently used (Fig. 5c) with average increases of +15, +18, +26, and +27 PPV for MSi, MSiC, MSiCE and MSiCEB over NetMHCpan4.0-EL, respectively, and +7, +9, +17, +18 PPV over MixMHCpred. The largest benefits were observed for 8-mer models, which was expected since 8-mer motifs were most different from 9-mer motifs (Fig. 3b–left). Ultimately, MSiCEB achieved 2.7-, 2.4-, 1.8-, and 1.5-fold improvements compared to the four benchmark algorithms.
To enable prediction for any HLA allele (beyond our MS dataset), we built a pan-allele-pan-length model (panMSintrinsic or panMSi, Fig. 5b–right). Although the performance of our pan models was on average highly comparable to our non-pan models (mean and median differences of −2% PPV) and improvements over the non-pan models were observed for over 35% of allele-length combinations (Supplementary Table 5a), we also noted several cases with considerable decrease in predictive power. The subpar performance of pan models largely coincided with alleles for which non-9-mer motifs were different from the 9-mer motif (Fig. 3b). Compared to prior algorithms, the largest gains in PPV were observed for poorly characterized alleles (+20% PPV for HLA-C and +38% for HLA-G for MSi against NetMHCpan4.0-EL as the best scoring pan-allele benchmark algorithm), but gains were also observed for all other alleles (+12% for HLA-A and +14% for HLA-B), even when only the 16 previously profiled alleles were considered (+9% for HLA-A and +5% for HLA-B).
We proposed PPV at 0.1% of the top-scoring peptides as a more suitable metric to evaluate HLA presentation predictors4 because of the importance of assessing true positive rates at the realistic 0.1% prevalence of binders (Methods). A recent study quantified model performance using a different version of PPV, where the fraction of true positive calls out of all predictions necessary to retrieve 40% of the true binders is calculated, and the hits:decoys ratio in the evaluation set was modified from 1:999 to a 1:100007 (i.e. ‘PPV at 40% recall’). Due to these differences, the two metrics are not directly comparable. Using a PPV at 40% recall, we found that MSi outperforms MHCflurry, NetMHCpan-BA, NetMHCpan-EL, and MixMHCpred by 5-, 4-, 2-, and 1.3-fold, while MSiCEB achieved 12-, 10-, 6-, and 3-fold improvements (Fig. 5d; Supplementary Table 5b). Finally, we observed similar gains in PPV compared to MHCflurry, NetMHCpan, and MixMHCpred when evaluating an external dataset of HLA-C and -G binders28, although MixMHCpred performed better on lengths other than 9 (Fig. 5e; Supplementary Table 5c). In summary, we observed 1.5–2.7x improvements in PPV (at the top 0.1% of the dataset) compared to existing predictors, which corresponds to 3–12x gains at 40% recall.
Motif complexity and motif abundance largely explain PPV variability
We observed that some allele preferences were harder to learn than others, with 9-mer-specific models (MSi) PPV values ranging from 37% to 68%. This variation was not readily explained by the amount of training data available as model performance plateaus at several hundred peptides4 and >375 9-mer peptides were identified for all alleles. Since PPVs after the addition of endogenous features (MSiCEB) were strongly correlated with PPVs achieved with the simple model (Pearson’s correlation=0.92, p-value<2.2×10−16), and since we observed differences in the allele-specific motifs and submotifs (Supplementary Fig. 1d; Supplementary Fig. 2b), we posited that PPV variability is driven by differential complexity in the peptide repertoire of each allele. To model complexity, we summed the entropy along each peptide position, to test whether higher information content implies more easily-learned motifs. Similarly, we considered all submotifs identified per allele by summing positional entropies over the submotifs, each weighted by the number of supporting peptides, and the number of submotifs normalized by the total number of peptides. Finally, we approximated motif abundance by the natural frequency of amino acids in the human genome underlying each binding motif. When motifs are more likely to occur by chance, more peptides from the random decoy set are real binders, leading to decreased PPV. To assess if these variables are predictive of PPV, we used a multivariate linear fit, controlling for the size of the training data (Methods), and found a strong correlation between predicted and actual PPV (Fig. 5f; Supplementary Table 5d). The number of peptides per allele was not predictive, while the entropy of the main motif, and the number and entropy of submotifs were positively associated with PPV, whereas motif abundance strongly negatively contributed to PPV. Based on the model coefficients we estimated that motif abundance could be responsible for ~10.4% of the unexplained PPV, although motif abundance likely underestimates the rate of undiscovered binders amongst the decoys. An additional 1% could be due to false positive MS identification at 1% FDR (Fig. 5g). Overall, these findings suggest that limitations in learning motifs can be in large part attributed to motif complexity and abundance.
Peptides proposed to be derived from proteasomal splicing have poor predicted binding scores
Peptides derived from proteasomally-ligated fragments (‘spliced peptides’) have been recently proposed as a major component of the HLA ligandome29,30. Since our collection of mono-allelic data covered the HLA alleles evaluated in those studies, we compared the binding potential of reported linear and proposed spliced peptide sets using our de novo predictors. Consistent with previous analyses31,32, we found that the majority of reported spliced peptides had poor predicted binding: although 81% of canonical linear peptides described in Liepe et al. had an HLA-binding likelihood score >0.75 only 28% of spliced peptides passed the same threshold (Supplementary Fig. 6a–left; Methods). Similar results were obtained for peptides described in Faridi et al.: 84% linear, 36% cis- and 37% trans-spliced (Supplementary Fig. 6a–right). While spliced peptides have been reported to make up 30% of the HLA class I peptidome27, our computational results suggest that no more than 11% (37% of 30%) of presented HLA ligands could be derived from spliced peptides, a number previously shown to be further diminished by factors such as ambiguity in peptide spectral matches and variability in sequence database search strategies31,33.
Leading sensitivity performance of MS-trained integrative algorithms validates in HLA peptidomes from patient-derived tumors
To evaluate the utility of our predictive models for clinical samples, we assessed their sensitivity to retrieve HLA-bound peptides observed in patient-derived tumor cell lines. To this end we (i) used LC-MS/MS to identify 51,531 HLA-associated peptides from 11 tumor samples (3 chronic lymphocytic leukemia, 1 ovarian, 3 glioblastoma, 4 melanoma) and utilized external peptide datasets from 4 melanoma34 and 27 ovarian35 tumors; (ii) predicted the likelihood that each observed peptide is presented per HLA allele per sample; (iii) compared the proportions of correctly predicted peptides amongst a large set of random genomic peptides relative to 4 prior tools. Observed ligands which scored better than 99.9% of random peptides for at least one allele (top 0.1 percentile) were considered as correct identifications (Fig. 6a). Our mono-allelic dataset covered 50 of 57 unique HLA alleles found amongst the 42 patient samples used in the evaluation. For covered allele/lengths, predictions were made with our allele-and-length-specific models, while missing alleles were scored by our pan-allele-pan-length predictors. Across malignancies, we consistently observed a higher proportion of observed peptides predicted by the MS-based models compared to existing algorithms (Fig. 6b; Supplementary Fig. 6; Supplementary Table 6a,b). At the 0.1 percentile threshold, 26% of observed peptides were recalled by MHCflurry (for MHCflurry and MixMHCpred we were only able to assess supported alleles), followed by 31%, 46%, and 49% predicted by NetMHCpan4.0-BA, NetMHCpan4.0-EL, and MixMHCpred respectively, compared to 56%, 60%, 77%, and 78% predicted by MSi, MSiC, MSiCE and MSiCEB, on average across all samples. This constitutes a 1.6-fold improvement in recall.
Figure 6: Integrative MS-informed models more accurately predict peptides directly observed on primary tumor cells.
a) Schematic of data generation and model evaluation: peptides displayed on primary tumor specimens are isolated and sequenced by MS, the HLA alleles of the patient sample are clinically typed, matched RNA-seq data is generated; each observed epitope is evaluated for binding against each of the unique HLA alleles in the sample, predictions that are better than 0.1% scores within a large decoy set are considered binders and assigned to the corresponding allele; the performance of 7 algorithms is evaluated as the fraction of observed binders that are successfully predicted as binders. b) MS-based predictor ranks MS detected peptides better than NetMHCpan and MHCflurry. Internal data on 11 patients (n=3 CLL (squares), 1 OV (diamond), 3 GBM (circle), 4 MEL (triangle), all biologically independent), and external data (n=4 MEL and 27 OV biologically independent samples)34,35. c) Peptides were assigned to alleles in each sample based on the best scoring peptide-allele combination. Allele contribution to peptide presentation varies per tumor, IFNγ treatment and per individual. d) Fraction of peptides contributing per allele type +/− IFNγ (n=6, biologically independent samples). Peptide presentation on HLA-B increases with IFNγ stimulation (Wilcoxon signed-rank tests). See also Supplementary Figure 5.
Allele contribution to peptide presentation varies by individual and changes with IFNγ stimulation
Since MS-detected epitopes were assigned to the best-scoring HLA-allele(s), this allowed calculation of allele frequency among the presented peptides (Fig. 6c; Supplementary Table 6c). Notably, 3% of peptides on average were uniquely assigned to HLA-C alleles and an additional 6% of peptides were compatible with a HLA-C allele jointly with other alleles, thus suggesting that HLA-C has the potential to harbor neoantigens. In all 6 samples for which we profiled HLA-associated peptidomes +/− IFNγ treatment, we observed a shift towards HLA-B presentation, consistent with HLA-B having two IFNγ-inducible promoters elements27,36. We further examined the HLA-B allele combinations of each and found elevated presentation for alleles with both tryptic and chymotryptic C-terminal preferences. This suggests that HLA-B upregulation could be in part responsible for a shift in presentation from tryptic-like to chymotryptic-like peptides which was observed in a cell line with a C-terminal chymotryptic-like HLA-B motif37. Finally, the contribution of each allele to the antigen repertoire varied by patient suggesting that MS-profiling of tumor-presented epitopes can reveal allele specific utilization and further guide peptide vaccine selection.
Discussion
We demonstrate the superior performance of HLA class I predictors trained on large-scale data of peptides eluted from cellular HLA proteins, consistent with growing appreciation of MS-derived datasets as a basis for epitope prediction algorithms3,5–7. Using an optimized experimental workflow, we eluted peptides from immunoprecipitated HLA proteins and used high performance mass spectrometry followed by a refined database searching approach to build the largest dataset to date of HLA ligands eluted from single HLA-expressing cell lines. The resulting collection of >185,000 peptides from 95 alleles greatly expands available knowledge of the human HLA-associated peptidome17, such that at least 95% of individuals worldwide have at least one of their A, B, and C alleles covered. The data are publicly available, thus providing a valuable resource for researchers. To facilitate access, we have implemented a web-based tool for data visualization, interactive exploration and prediction.
Our large dataset enables more comprehensive insights into the rules of peptide presentation by HLA-A, -B, -C and -G alleles, each of which impacted our model design. First, we ascertained that peptide presentation does access the entire proteome for potential sources of antigen, in contrast to previous reports16 which relied on a small number of peptides. Second, our analysis revealed 101 binding submotifs, many of which were shared amongst the 95 HLA alleles. We observed strong similarity in physicochemical features of the HLA-C alleles along with their greater promiscuity in binding peptides, compared to the more divergent HLA-A and -B alleles. Moreover, HLA-C alleles only rarely had unique submotif clusters that were not also shared with HLA-A and B alleles, consistent with their recent evolutionary history20. We speculate that this may increase competition with HLA-A and -B alleles for peptides and may explain our observation that HLA-C epitopes originate from more highly expressed genes. Third, we detected not only differences in length distribution, but also that ~10% of alleles displayed length-based epitope preferences. Altogether, the detailed knowledge gained from our extensive dataset enabled us to generate allele-and-length-specific and pan-allele-pan-length prediction models that we demonstrate to outperform state-of-the-art algorithms, especially for understudied alleles or those with length-specific preferences. Fourth, while IFNγ signaling broadly modulates gene expression and thus alters the genes HLA ligands derive from, we did not observe prominent differences in cleavage preferences across primary tumor cells of various lineages when exposed to IFNγ. This is consistent with the expression of both constitutive and immunoproteasome subunits in cancers and supports the application of a unified cleavability predictor.
The improved performance of the HLAthena models can be attributed to several factors: (i) our models are trained exclusively on MS data of eluted peptides from mono-allelic cell lines; (ii) we integrate several critical endogenous features, such as peptide cleavage and gene expression; (iii) our rich dataset reliably captures not only allele- but also length-specific motifs, widely covering the space of HLA binding preferences; (iv) we preferentially predict with allele-and-length-specific models for their demonstrated accuracy over pan-allele-pan-length predictors which are employed only for uncharacterized alleles.
Despite improvements in epitope prediction, we recognize that further innovations are required if we are to achieve near perfect accuracy. We offer evidence that allele complexity and motif abundance may partially drive the observed variability in prediction power across alleles. The former implies a benefit in obtaining even larger training datasets, while the latter necessitates techniques to determine non-binders at large scale to collect reliable true negative datasets against which to evaluate model performance. Other innovations that could boost prediction fidelity include increased LC-MS/MS instrument sensitivity and better de novo HLA peptide identification methods. For predicting peptide presentation in tumor cells, better accuracy can be achieved by taking into account the uneven allele utilization and weighting predictions accordingly. While we include expression as a variable in our predictors, it is important to note that RNA-seq of tumors may not be representative of all clones and usually includes non-malignant cells that obfuscate tumor gene expression. Finally, we emphasize that our models do not predict whether the HLA-presented peptides can interact with the T cell receptors in an individual, a problem that remains unsolved.
Online Methods
Generation of HLA-A, B and C single allele cell lines
Single HLA allele-expressing cDNA vectors in a pcDNA-3 backbone were ordered from GenScript@. The HLA class I deficient B721.221 cell line was transfected with the HLA allele expression vectors using lipofectamine, as described previously4. Cell lines with stable surface HLA expression were generated first through selection using 800 μg/ml G418 (Thermo Scientific), followed by enrichment of HLA positive cells through up to 2 serial rounds of fluorescence-activated cell sorting (FACS) and isolation using a pan-HLA antibody (W6/32; Santa Cruz) on a FACSAria II instrument(BD Biosciences). Priority was given to HLA alleles with lack of binding data in public databases or over 1% frequency in the US organ donor registry populations13.
Generation of primary human samples
All human tissues were obtained through DFCI or Partners Healthcare approved IRB protocols. Conditions for growth and in vitro propagation of melanoma and GBM tumor cell lines and of monocyte-derived dendritic cells were described previously15,26. PBMC from patients with chronic lymphocytic leukemia (CLL) were enriched for CD19 positive CLL tumor cells and were used in IP/MS analysis. Tumor specimens from patients with clear cell renal cell carcinoma (ccRCC) were collected following informed consent for enrollment on a tissue collection research protocol approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (IRB). Surgically resected ccRCC tumor tissue was mechanically dissociated with scalpels, and then enzymatically dissociated using a mixture of collagenase D (Roche), Dispase (STEMCELL Technologies), and DNase I (New England BioLabs) at room temperature, and filtered through a 100 micron cell strainer using the sterile plunger of a syringe. Red blood cells were lysed using ammonium-chloride-potassium buffer (Gibco). The cell suspension was stained for viability (Zombie Aqua; BioLegend), anti-CD45 (BV605; BD Biosciences), and anti-carbonic anhydrase IX (PE; R&D Systems). Viable, CD45-negative, CAIX-positive tumor cells were isolated by FACS (BD FACSAria II cell sorter; BD Biosciences). Cells were cultured in a specialized growth medium consisting of OptiMEM GlutaMax media (Gibco), 5% fetal bovine serum, 1mM sodium pyruvate (Gibco), 100 units/mL penicillin and streptomycin, 50 micrograms/mL gentamicin, 5 micrograms/mL insulin (Sigma), and 5 ng/mL epidermal growth factor (Sigma). Following successive passages, CAIX expression was confirmed by flow cytometry (anti-CAIX, PE-conjugated; R&D Systems) and by immunohistochemical analysis of a cell pellet. Ovarian cancer patient-derived cells were propagated within a xenograft model, which was generated by serial passaging of tumor cells from a patient with advanced ovarian cancer. These cells originated from solid tumor or pleural effusion (3 million cells/mouse) that were injected orthotopically in the abdominal cavity in NOD-SCID mice (8-week old, Jackson labs). Tumor growth was monitored weekly by observing mice for signs of abdominal distension. Cells were harvested 4 months after initial injection and banked for future experiments. For interferon stimulation, cultured cells were stimulated with 2000Unit/ml of IFNγ (Peprotech) for 3 days and were used in IP/MS analysis.
For primary tumors and patient cell lines, HLA-peptide complexes were immunoprecipitated from 0.1 to 0.2g tissue or up to 50 million cells. Solid tumor samples were dissociated using tissue homogenizer (Fisher Scientific 150) and HLA complexes were enriched as described above.
HLA peptide enrichment and LC-MS/MS analysis
Soluble lysates from up to 50 million single HLA expressing B721.221 cells and up to 0.2 g from tumor samples were immunoprecipitated with W6/32 antibody (sc-32235, Santa Cruz) as described previously4. 10 mM iodoacetamide was added to the lysis buffer to alkylate cysteines for 71 alleles (Supplementary Table 1c; Supplementary Data 2). Peptides of up to three IPs for single HLA expressing samples and up to four IPs for tumor samples were combined, acid eluted either on StageTips or SepPak cartridges34, and analyzed in technical duplicates using high resolution LC-MS/MS on a QExactive Plus (QE+), QExactive HF (QE-HF) or Fusion Lumos (Thermo Scientific). For acquisition parameters see Supplementary Note 2.
HLA peptide identification using Spectrum Mill
Mass spectra were interpreted using the Spectrum Mill software package v6.1 pre-Release (Agilent Technologies, Santa Clara, CA). MS/MS spectra were excluded from searching if they did not have a precursor MH+ in the range of 600–4000, had a precursor charge >5, or had a minimum of <5 detected peaks. Merging of similar spectra with the same precursor m/z acquired in the same chromatographic peak was disabled. Prior to searches, all MS/MS spectra had to pass the spectral quality filter with a sequence tag length >2 (i.e. minimum of 4 masses separated by the in-chain masses of 3 amino acids). MS/MS spectra were searched against a protein sequence database that contained 98,298 entries, including all UCSC Genome Browser genes with hg19 annotation of the genome and its protein coding transcripts (63,691 entries), common human virus sequences (30,181 entries), recurrently mutated proteins observed in tumors from 26 tissues (4,167 entries), as well as 259 common laboratory contaminants including proteins present in cell culture media and immunoprecipitation reagents. Mutation files for 26 tumor tissue types were obtained from the Broad GDAC portal (gdac.broadinstitute.org). Recurrent mutations in the coding region within each of the 26 tumor types (frequency=3 for stomach adenocarcinoma, uterine corpus endometrial carcinoma; frequency=5 for adrenocortical carcinoma, pancreatic adenocarcinoma, melanoma; frequency=2 for rest) were included. MS/MS search parameters included: no-enzyme specificity; fixed modification: cysteinylation of cysteine; variable modifications: carbamidomethylation of cysteine, oxidation of methionine, and pyroglutamic acid at peptide N-terminal glutamine; precursor mass tolerance of ±10 ppm; product mass tolerance of ± 10 ppm, and a minimum matched peak intensity of 30%. Variable modification of carbamidomethylation of cysteine was only used for HLA alleles that included an alkylation step (performed in 2017 or later). Peptide spectrum matches (PSMs) for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to apply target-decoy based FDR estimation at the PSM level of <1% FDR. Peptide auto-validation was done separately for each HLA allele with an auto thresholds strategy to optimize score and delta Rank1 – Rank2 score thresholds separately for each precursor charge state (1 thru 4) across all LC-MS/MS runs for an HLA allele. Score threshold determination also required that peptides had a minimum sequence length of 7, and PSMs had a minimum backbone cleavage score (BCS) of 5. BCS is a peptide sequence coverage metric and the BCS threshold enforces a uniformly higher minimum sequence coverage for each PSM, at least 4 or 5 residues of unambiguous sequence. The BCS score is a sum after assigning a 1 or 0 between each pair of adjacent AA’s in the sequence (max score is peptide length-1). To receive a score, cleavage of the peptide backbone must be supported by the presence of a primary ion type for HCD: b, y, or internal ion C-terminus (i.e. if the internal ion is for PWN then BCS is credited only for the backbone bond after the N). The BCS metric serves to decrease false-positives associated with spectra having fragmentation in a limited portion of the peptide that yields multiple ion types. PSMs were consolidated to the peptide level to generate lists of confidently observed peptides for each allele using the Spectrum Mill Protein/Peptide summary module’s Peptide-Distinct mode with filtering distinct peptides set to case sensitive. A distinct peptide was the single highest scoring PSM of a peptide detected for each allele. MS/MS spectra for a particular peptide may have been recorded multiple times (e.g. as different precursor charge states, from replicate IPs, from replicate LC-MS/MS injections). Different modification states observed for a peptide were each reported when containing amino acids configured to allow variable modification; a lowercase letter indicates the variable modification (C-cysteinylated, c-carbamidomethylated). These unfiltered peptide lists are provided as Supplementary Data 1.
MS/MS data from patient derived cell lines was handled as described above except that they were searched against the database mentioned above with further inclusion of patient specific neoantigen sequences15,26. These peptide lists are provided as Supplementary Data 2.
Filtering of MS-identified peptides
The list of LC-MS/MS identified peptides was filtered to remove potential contaminants in the following ways: (1) peptides observed in negative controls runs (blank beads and blank IPs); (2) peptides originating from the following species: ‘STRSG’, ‘HEVBR’, ‘ANGIO432’, ‘ANGIO394’, ‘ANGIO785’, ‘ANGIO530’, ‘ACHLY’, ‘PIG’, ‘ANGIO523’, ‘RABIT’, ‘STAAU’, ‘CHICK’, ‘Pierce-iRT’, ‘SOYBN’, ‘ARMRU’, ‘SHEEP’ as common laboratory contaminants including proteins present immunoprecipitation reagents. Note that BOVINE peptides derived from cell culture media were not excluded as they appear to have undergone processing and presentation and exhibit anchor residue motifs consistent with the human peptides observed for each allele; (3) peptides which were also identified in a tryptically-digested full proteome Jurkat sample; (4) peptides for which both the preceding and C-term amino acids were tryptic residues (R or K); (5) all possible leader peptides of lengths 8–11 from HLA-A,-B,-C, and -G (first exon, n=410) as they are likely to be presented by HLA-E; (6) peptides with negative deltaFwRevScore as likely falling in the 1% false positive MS identifications; (7) peptides identified for 20 or more of the 95 alleles (n=168); (8) peptides identified as potential C*01:02 contaminants in other alleles due to residual C*01:02 expression in B721.221 (n=383). These peptides were identified by scoring all peptides with the allele-specific C*01:02 model and selecting those with predicted likelihood binding score >0.95 that were also outlier for the allele (mean distance to the nearest 10 peptides >90 percentile). A summary of counts of removed peptides is provided in Supplementary Table 1d. The filtered peptide lists are provided in Supplementary Table 1e (mono-allelic cell lines) and Supplementary Table 6a (patient cell lines).
Immune Epitope Database (IEDB) data access and preparation, related to Figure 1
A curated set of previously identified HLA class I ligand was downloaded from the Immune Epitope Database (IEDB) at http://www.iedb.org/downloader.php?file_name=doc/mhc_ligand_full.zip (accessed on 06/14/2018)17. Records were filtered to MHC allele class=I, Epitope Object Type=Linear peptide, and Allele Name consistent with human HLA class I nomenclature with 4-digit typing (i.e. regex: “ĤLA-[ABCG]\\*[0–9]{2}:[0–9]{2}$”). Peptides with quantitative measurements in units other than nM were removed and so were the following three assay types due to detected inconsistency between predicted (NetMHC 3.0) and actual affinity: “purified MHC/direct/radioactivity/dissociation constant KD”, “purified MHC/direct/fluorescence/half maximal effective concentration (EC50)” and “cellular MHC/direct/fluorescence/ half maximal effective concentration (EC50)”. A peptide was considered a binder if it had a quantitative affinity of <500 nM or qualitative label of “Positive”, “Positive-High”, “Positive-Intermediate”, or “Positive-Low”. In cases where multiple records are available for the same {peptide, allele} pair, we either took the mean affinity or removed the peptide when the difference between the maximum and minimum log-transformed affinities (1 - log(nM)/log(50000)) was >0.2. Similarly, peptides with multiple qualitative records were removed if the same number of positive and negative labels were found or kept otherwise. Our previously published data for 16 HLA-A and B alleles was removed from the analysis of IEDB counts (PubMedID=28228285).
Allele similarity analyses, related to Figure 2
To assess which alleles are similar to each other we considered similarity according to the observed binding motifs (peptide space) as well as similarity according to the HLA binding grooves (HLA binding pocket or HLA protein space). Similarity in peptide space was evaluated by tabulating the frequency of each of the 20 amino acids at each position along the peptide sequence (1 through 9) per allele, forming a vector of size 20*9=180. The pairwise correlations of these frequency vectors were used to quantify similarity (Fig. 2a).
To evaluate similarity in HLA binding pocket space, HLA protein sequences were downloaded from IMGT®, the international ImMunoGeneTics information system® http://www.imgt.org (http://hla.alleles.org/alleles/text_index.html, accessed 05/05/2018) and aligned. From the full HLA protein sequences we selected positions which are in contact with the peptide (within a distance of 2 ) or positions that are most frequently mutated across alleles to represent the binding pocket: {7, 9, 13, 24, 31, 45, 59, 62, 63, 65, 66, 67, 69, 70, 71, 73, 74, 76, 77, 80, 81, 84, 95, 97, 99, 110, 114, 116, 118, 138, 143, 147, 150, 152, 156, 158, 159, 163, 167, 171} (Fig. 2b). The residue at each position of the binding pocket was featurized by its amino acid physical properties encoded as 10 Kidera Factors (available from R package Peptides v2.4, data(AAdata))38 and 3 principal components derived from a dimensionality reduction of a large set of physicochemical properties39. The full binding pocket was represented by the concatenated list of positions and allele similarity was assessed by Euclidean distance.
Given the two approaches to evaluating allele similarity (motif space and pocket space), we assessed how well they agree by identifying the closest neighbors for each allele in motif space and the closest neighbors for each allele in pocket space and counting how many of the former are also found in the latter (Supplementary Fig. 2a). The closest neighbor in motif space were considered to be alleles with correlation greater than 97.5% of all pairwise correlations. Analogously, closest neighbor alleles in pocket space were considered to be alleles within distance less than 97.5% of all pairwise distances.
Analysis of submotifs across alleles, related to Figure 2
Grouping the 9-mer peptides identified for each allele into submotifs (see Peptide distance visualization and sub-clustering of binding motifs) identified 1133 submotifs across the 95 alleles supported by at least 20 peptides (Fig. 2d; Supplementary Fig. 2c). To determine whether any of those submotifs are shared by two or more alleles, each submotif was represented as a vector of amino acid frequencies per peptide position (analogously to main motifs representation in allele similarity analysis), projected onto two dimensions (umap() function from R package umap v0.2)40, and clustered (dbscan() function from dbscan R package). This approach identified 101 distinct clusters of submotifs with 1–22 alleles participating in each (Fig. 2c,d; Supplementary Fig. 2b,c).
Evaluation and validation of length-dependent motif differences, related to Figure 3
To compare 8-mer motifs to 9-mer motifs, we generated pseudo 8-mer motifs from 9-mers by dropping middle residues (positions 4, 5, or 6). To compare 10- and 11-mer motifs to 9-mers we generated pseudo 9-mer motifs by dropping middle residues from 10-mers (positions 5 or 6) and 11-mers (positions 5 and 6, or 6 and 7). The pseudo motif which was most similar to the true motif was used to evaluate the change in frequency and entropy at every peptide position. 8-, 10-, or 11-mer motifs with at least 100 identified peptides, that had an absolute difference in residue frequency with the true motif of >0.25 or an absolute difference in entropy of >0.2 at any position were considered as different. For example, if proline is observed at position 5 in 5% of peptides associated with motif1 but 40% of peptides associated with motif2 the absolute difference in frequency is |0.05–0.40|=0.35>0.25, thus deeming motif1 and motif2 different.
To experimentally validate the observed length-specific motifs we selected 18 peptides from 3 alleles representing 4 length-specific motifs that were predicted to be strong binders by our algorithm (MSi) but weak binders according to NetMHCpan-4.0 and tested them for binding in in vitro binding assays. Peptide affinity measurements were performed at Immunitrack, Copenhagen, Denmark as previously described41.
PPV vs PPV at 40% Recall
To evaluate predictive power, we constructed datasets consisting of the observed allele- and length-specific binders in our MS data (n) along with 999*n random decoys from the human proteome and considered the fraction of correctly predicted binders in the top 0.1% of the dataset (i.e. positive predictive value, ). We advocated for the PPV evaluation metric in Abelin et al.4, over the commonly used AUC, because it is better suited for the HLA presentation prediction problem space where a relatively small number of true binders need to be identified amongst an excess of non-binders. Each HLA allele is expected to present a repertoire of approximately ~10,000 peptides17,42–45 among the 1.1×107 9-mer peptides in the proteome, meaning that approximately only 1 out of a thousand peptides (0.1%) gets presented.
The definition of PPV described above is equivalent to PPV at recall=PPV% since the number of positive calls equals the number of true positives. A different version of this metric used recently to quantify algorithm performance is PPV at recall=40%, that is 7. In addition, Bulik-Sullivan et al. used a dataset with a ratio of 1 hit : 10,000 decoys for the evaluation of single-allele dataset, rather than a ratio of 1:999 used by us, which reduced PPV. Due to these differences, model performance evaluated with PPV is not comparable to model performance evaluated with PPV40%recall, h:d=1:10000 (Fig. 5d).
Development of integrative HLA-binding prediction models
Overview of model training procedure
Machine learning models were created to predict the likelihood that a given peptide will be endogenously presented by a given HLA class I molecule. The positive training set consisted of MS-identified peptides from the set of 95 B721.221 mono-allelic cell lines (hits). The negative set consisted of random peptides drawn from the human proteome which did not overlap with the hits (decoys). Note that one decoys set was used for training and a separate non-overlapping decoy set was used for evaluation. The number of positive and negative training examples were balanced by sampling 10x #hits decoys, and sampling the hits 10 times with replacement (this was done after splitting the data into folds to ensure that each hit is only present in one unique fold). Training was carried out in a standard 5-fold cross validation (CV) procedure: training data was split into 5 equal parts, each part was left out one at a time and a model was trained on the remaining 4 parts, obtaining 5 models each of which is evaluated on its corresponding left out set. The 5-fold CV training was repeated 3 times with different model initialization random seeds. The final predictions score for each peptide was the average of the 3 initializations. Neural network models were trained using the Theano and keras python libraries.
Allele-and-length-specific models
To build models that are both allele- and length-specific only the MS-hits identified for that particular allele and length were used to train the model. This was done for each of the 95 alleles and for lengths 8, 9, 10, and 11 where at least 40 peptides were identified. The MSIntrinsic (or MSi) neural network models were fully-connected with one hidden layer of size 50 and tanh activation. Training was carried out in batches of size 30, for 10 epochs, and early-stopping determined by evaluating on a 20% hold-out partition of the training set to avoid overfitting. Three different models were trained with three different encodings of the peptide sequence: (1) one-hot (a.k.a. Binary or dummy) encoding; (2) similarity encoding using the blosum62 matrix; (3) similarity encoding based on the PMBEC matrix46. In addition to the peptide sequence encoding features, the MSi models included the following features:
Amino acid properties - each peptide residue was represented by the first three principal components derived from a dimensionality reduction of a large set of physicochemical properties39.
Peptide-level characteristic - 8 peptide-level features were computed with the R package peptides: “boman”, “hmoment”, “hydrophobicity”, “helixbend”, “sidechain”, “xstr”, “partspec”, “pkc”
The logit-transformed output scores from MSi models were used as input features to train logistic regression models that integrate endogenous signals (Supplementary Note 6,7): MSiC models were trained with two features (MSi scores, and cleavability), MSiCE were trained with three input features (MSi scores, cleavability, and expression), and MSiCEB were trained with four input features (MSi scores, cleavability, expression, and presentation bias). Note that despite expression having a larger predictive contribution over cleavability, the cleavability feature is incorporated first to the integrative models (i.e. MSiC, MSiCE instead of MSiE, MSiEC) to allow for samples which lack expression data to utilize the cleavability since upstream and downstream peptide context sequences are readily available from the source protein.
Pan-allele-pan-length models
Pan models were trained similarly with some differences. A single panMSi model was trained with all 8–11-mer peptides identified across the 95 alleles. The panMSi neural networks had additional input features to describe the binding pocket of the HLA protein - each binding pocket residue was represented with 10 Kidera factors and 3 PCs (see Allele similarity analyses, related to Figure 2). The size of the hidden layer was 250, batch size was 5000, and the hidden layer activation function was rectified linear unit (ReLU). Training proceeded for 15 epochs with early-stopping determined by lack of improvement in 4 consecutive epochs.
To construct integrative pan models we considered the four HLA genes (HLA-A, HLA-B, HLA-C, and HLA-G) and the four lengths (8, 9, 10, 11) separately: linear panMSiC, panMSiCE, and panMSiCEB models were trained for all HLA-A alleles and peptides of length 8, all HLA-A alleles and peptides of length 9, all HLA-A alleles and peptides of length 10, and all HLA-A alleles and peptides of length 10, and analogously for HLA-B, C, and G.
Modelling PPV variability, related to Figure 5
A linear regression model was trained to predict the achieved PPV (MSi, 9-mer models) given the following variables:
The total number of 9-mer hits observed for the allele
The sum of entropies at positions 1 through 9 (main motif entropy)
The number of identified submotifs for the allele
The sum of submotif entropies
Estimated abundance of the binding motif calculated by weighting the frequency of each residue at each peptide position by the natural abundance frequency of each of the 20 amino acids.
Evaluation of model performance in multi-allelic samples, related to Figure 6
To evaluate model performance in multi-allelic patient samples, each tumor-presented peptide was scored for binding to each of the sample-specific HLA alleles. To compare scores for different alleles, each score was converted to percentile rank. To this end, empirical cumulative distribution functions (R package stats, function ecdf()) were computed for each model (including benchmark algorithms) from the scores of a background set of 1e6 random decoys. Each decoy set was constructed such that it contains proportions of 8, 9, 10, and 11-mers that are equal to the observed length distribution for the allele (or the HLA gene in the case of pan models). A peptide was considered to be correctly identified as a binder if the predicted binding score for at least one of the alleles in the sample was better than 99.9% of the scores in the corresponding decoy set (0.1 percentile rank; evolution was also performed at different rank thresholds, Supplementary Fig. 5). This approach is very similar to the %Ranks introduced by NetMHC8). A peptide was assigned to the allele(s) for which it had %rank score <0.1, where assignment to more than one allele was allowed.
Data availability
The original mass spectra for 79 newly described mono-allelic datasets, the protein sequence database, and tables of peptide spectrum matches for all 95 alleles have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000084172/. Mass spectrometry data for the 16 previously published mono-allelic datasets in MassIVE can be downloaded at ftp://massive.ucsd.edu/MSV000080527. Datasets for the patient samples are accessible at ftp://massive.ucsd.edu/MSV000084442/. B721.221 RNA seq data for HLA-C (C*04:01, C*07:01) is deposited under GEO: GSE131267. Melanoma RNA-seq data are deposited in dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001451.v1.p115). Glioblastoma bulk RNA-seq data are available through dbGaP (https://www.ncbi.nlm.nih.gov/gap) with accession number phs001519.v1.p126. All other data are available from the corresponding authors upon reasonable request.
Code availability
Code used to generate plots characterizing allele-specific preferences (e.g. logo plots, entropy plots, peptide projection and clustering, overlap with IEDB data, etc.) as well as code to build a sample neural network prediction model is provided as Supplementary code. The HLAthena predictors are available to use online for research purposes only at http://mhc.tools. For commercial usage inquiries please contact the authors or the Broad Institute.
Supplementary Material
Acknowledgements
We acknowledge superb technical assistance from Kristine Pelton, Sandro Santagata, Oliver Spiro, Liudmila Elagina, Binyamin Knisbacher, Sachet Shukla, Joan Brugge, and Annie Apffel. We further express gratitude for constructive input from Michael Rooney, Jenn Abelin and Zhuting Hu. We acknowledge support from National Institutes of Health: NCI-1RO1CA155010-02 (to C.J.W.), NHLBI-5R01HL103532-03 (to C.J.W.), NIH/NCI U24 CA224331 (to C.J.W.), NIH/NCI R21 CA216772-01A1 (to D.B.K.), NCI-SPORE-2P50CA101942-11A1 (to D.B.K), NHGRI T32HG002295 (to S.S.), NCI 5T32CA009172-41 (to D.A.B.), NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (to S.A.C). This work was supported in part by The G. Harold and Leila Y. Mathers Foundation and the Bridge Project, a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center. D.A.B. is supported in part by the John R. Svenson Fellowship. C.J.W. is a scholar of the Leukemia and Lymphoma Society, and is supported in part by the Parker Institute for Cancer Immunotherapy. S.K. is a Cancer Research Institute/Hearst Foundation fellow.
Footnotes
Competing Interests
D.B.K. has previously advised Neon Therapeutics, and owns equity in Aduro Biotech, Agenus Inc., Armata pharmaceuticals, Biomarin Pharmaceutical Inc., Bristol Myers Squibb Com., Celldex Therapeutics Inc., Editas Medicine Inc., Exelixis Inc., Gilead Sciences Inc., IMV Inc., Lexicon Pharmaceuticals Inc., and Stemline Therapeutics Inc. D.A.B. has received consulting fees from Octane Global, Defined Health, Dedham Group, Adept Field Solutions, Slingshot Insights, Blueprint Partnership, Charles River Associates, Trinity Group, Insight Strategy, and is a member of the RCC translational medicine advisory broad of Bristol-Myers Squibb. K.L.L. owns equity and is a founder of Travera LLC and is an advisor to Bristol Myers Squibb Com. and Rarecyte. S.A.C is a member of the scientific advisory boards of Kymera, PTM BioLabs and BioAnalytix and a scientific advisor to Pfizer and Biogen. C.J.W. and N.H. are founders of Neon Therapeutics and members of its scientific advisory board. N.H. is also an advisor for IFM therapeutics. W.J.L is a member of the scientific advisory board of CareDx. All other authors have no competing interests.
Patent applications have been filed on aspects of the described work entitled as follows: “HLA single allele lines”, “Methods for identifying neoantigens”.
References
- 1.Lefranc M-P et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43, D413–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson J et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jurtz V et al. NetMHCpan-4.0: Improved Peptide–MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. The Journal of Immunology 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abelin JG et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 46, 315–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell TJ et al. MHCflurry: Open-Source Class I MHC Binding Affinity Prediction. Cell Syst 7, 129–132.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Gfeller D et al. The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J. Immunol 201, 3705–3716 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Bulik-Sullivan B et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol (2018). doi: 10.1038/nbt.4313 [DOI] [PubMed] [Google Scholar]
- 8.Nielsen M & Andreatta M NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajasagi M et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124, 453–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kruijf EM et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol 185, 7452–7459 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Zhang R-L et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget 8, 107441–107451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson DV, Ozgur M, Sari K, Ghanayem M & Kostyu DD Ramifications of HLA class I polymorphism and population genetics for vaccine development. Genet. Epidemiol 20, 87–106 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Gragert L, Madbouly A, Freeman J & Maiers M Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol 74, 1313–1320 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Solberg OD et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol 69, 443–464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson H et al. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest 126, 4690–4701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vita R et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43, D405–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sette A & Sidney J HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr. Opin. Immunol 10, 478–482 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Robinson J, Malik A, Parham P, Bodmer JG & Marsh SGE IMGT/HLA Database - a sequence database for the human major histocompatibility complex. Tissue Antigens 55, 280–287 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Parham P & Moffett A Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol 13, 133–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen M et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2, e796 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rist MJ et al. HLA peptide length preferences control CD8+ T cell responses. J. Immunol 191, 561–571 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Maenaka K et al. Nonstandard Peptide Binding Revealed by Crystal Structures of HLA-B*5101 Complexed with HIV Immunodominant Epitopes. The Journal of Immunology 165, 3260–3267 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Kaur G et al. Structural and regulatory diversity shape HLA-C protein expression levels. Nat. Commun 8, 15924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celik AA, Simper GS, Hiemisch W, Blasczyk R & Bade-Döding C HLA-G peptide preferences change in transformed cells: impact on the binding motif. Immunogenetics 70, 485–494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keskin DB et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javitt A et al. Pro-inflammatory Cytokines Alter the Immunopeptidome Landscape by Modulation of HLA-B Expression. Front. Immunol 10, 141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Marco M et al. Unveiling the Peptide Motifs of HLA-C and HLA-G from Naturally Presented Peptides and Generation of Binding Prediction Matrices. J. Immunol 199, 2639–2651 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Liepe J et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Faridi P et al. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci Immunol 3, (2018). [DOI] [PubMed] [Google Scholar]
- 31.Mylonas R et al. Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol. Cell. Proteomics 17, 2347–2357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolfs Z, Solntsev SK, Shortreed MR, Frey BL & Smith LM Global Identification of Post-Translationally Spliced Peptides with Neo-Fusion. J. Proteome Res (2018). doi: 10.1021/acs.jproteome.8b00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolfs Z, Müller M, Shortreed MR, Smith LM & Bassani-Sternberg M Comment on ‘A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands’. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassani-Sternberg M et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun 7, 13404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster H et al. The immunopeptidomic landscape of ovarian carcinomas. Proc. Natl. Acad. Sci. U. S. A 114, E9942–E9951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girdlestone J Regulation of HLA Class I Loci by Interferons. Immunobiology 193, 229–237 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Chong C et al. High-throughput and Sensitive Immunopeptidomics Platform Reveals Profound Interferonγ-Mediated Remodeling of the Human Leukocyte Antigen (HLA) Ligandome. Mol. Cell. Proteomics 17, 533–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods-only references
- 38.Kidera A, Konishi Y, Oka M, Ooi T & Scheraga HA Statistical analysis of the physical properties of the 20 naturally occurring amino acids. J. Protein Chem 4, 23–55 (1985). [Google Scholar]
- 39.Bremel RD & Homan EJ An integrated approach to epitope analysis I: Dimensional reduction, visualization and prediction of MHC binding using amino acid principal components and regression approaches. Immunome Res. 6, 7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInnes L, Healy J & Melville J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML] (2018). [Google Scholar]
- 41.Harndahl M et al. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J. Biomol. Screen 14, 173–180 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ & Mann M Mass Spectrometry of Human Leukocyte Antigen Class I Peptidomes Reveals Strong Effects of Protein Abundance and Turnover on Antigen Presentation. Molecular & Cellular Proteomics 14, 658–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt DF et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Rammensee HG, Friede T & Stevanoviíc S MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178–228 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Rammensee H, Bachmann J, Emmerich NP, Bachor OA & Stevanović S SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Kim Y, Sidney J, Pinilla C, Sette A & Peters B Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinformatics 10, 394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra for 79 newly described mono-allelic datasets, the protein sequence database, and tables of peptide spectrum matches for all 95 alleles have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000084172/. Mass spectrometry data for the 16 previously published mono-allelic datasets in MassIVE can be downloaded at ftp://massive.ucsd.edu/MSV000080527. Datasets for the patient samples are accessible at ftp://massive.ucsd.edu/MSV000084442/. B721.221 RNA seq data for HLA-C (C*04:01, C*07:01) is deposited under GEO: GSE131267. Melanoma RNA-seq data are deposited in dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001451.v1.p115). Glioblastoma bulk RNA-seq data are available through dbGaP (https://www.ncbi.nlm.nih.gov/gap) with accession number phs001519.v1.p126. All other data are available from the corresponding authors upon reasonable request.