Abstract
Introduction:
Proteolysis – targeting chimeras (PROTACs) have emerged as a new modality with the potential to revolutionize drug discovery. PROTACs are heterobifunctional molecules comprising of a ligand targeting a protein of interest, a ligand targeting an E3 ligase and a connecting linker. The aim is instead of inhibiting the target to induce its proteasomal degradation.
Areas covered:
PROTACs, due to their bifunctional design, possess properties that differentiate them from classical inhibitors. A structural analysis, based on published crystal aspects, kinetic features and aspects of selectivity are discussed. Specific types such as homoPROTACs, PROTACs targeting Tau protein and the first PROTACs recently entering clinical trials are examined.
Expert opinion:
PROTACs have shown remarkable biological responses in challenging targets, including an unprecedented selectivity over protein family members and even efficacy starting from weak or unspecific binders. Moreover, PROTACs are standing out from classical pharmacology by inducing the degradation of the target protein and not merely its inhibition. However, there are also challenges in the field, such as the rational structure optimization, the evolution of computational tools, limited structural data and the greatly anticipated clinical data. Despite the remaining hurdles, PROTACs are expected to soon become a new therapeutic category of drugs.
Keywords: Degradation, PROTAC, structural analysis
1. Introduction
Proteolysis-targeting chimeras (PROTACs) were first reported in 2001 [1] as chimeric molecules that artificially target the ubiquitin ligase complex, Skp1-Cullin-F box. In the beginning, PROTACs were considered merely an academic exercise or as Craig Crews stated a ‘cute chemical curiosity’ [2]. Nowadays, almost two decades later, PROTACs are recognized as a new modality in drug discovery and have the potential to become the new blockbuster therapeutics [2].
PROTACs are bifunctional molecules that hijack the ubiquitin proteasome system (UPS) in order to achieve the degradation of a disease-related target protein. The UPS and the autophagy/lysosomal routes are the main pathways for the degradation of intracellular proteins and the maintenance of homeostasis. The proteasome recognizes proteins that are tagged with ubiquitin, a small 76 amino acid residue protein, which is highly conserved among all eukaryotes. Protein ubiquitination is an ATP-dependent enzymatic reaction that comprises three steps with three enzyme classes: ubiquitin-activating enzymes (E1 enzymes), ubiquitin-conjugating enzymes (E2 enzymes) and ubiquitin ligases (E3 enzymes). First, a ubiquitin molecule is activated by an E1 enzyme in an ATP-dependent manner. Then, the activated ubiquitin is transferred to an E2 enzyme and finally, an E3 ligase catalyzes the transfer of the ubiquitin molecule from E2 to a lysine residue on the substrate via the formation of a covalent bond [3]. Appropriately ubiquitin-tagged proteins are recognized by the 26S proteasome and are destroyed by proteolysis. The discovery of the uttermost importance of the ubiquitin-proteaseome homeostasis system was recognized with the Nobel Prize in Chemistry in 2004 to Aaron Ciechanover, Avram Hershko, and Irwin Rose.
1.1. Mode of action and unique features
PROTACs are bifunctional molecules consisting of a ligand that binds to an E3 ligase, connected by a linker to another ligand that binds to the protein of interest (POI). The rationale behind this design is that by bringing the E3 ligase in the vicinity of the protein of interest, ubiquitination by the E3 ligase and subsequent proteasomal degradation will be triggered. Interestingly, PROTACs trigger an artificially induced target degradation, by bringing into close proximity two proteins that normally would not interact. Thus, the successful interaction relies on the bridging molecule and the adequate affinity of the PROTAC toward both the E3 ligase and the POI [4]. In contrast to classical drug pharmacology, no functional activity is necessary for degrading the POI.
The mode of action of PROTACs is considerably changing the paradigm of ‘druggable’ targets. The druggability of a target protein is usually dependent on the inhibition of its activity by designing small molecules that can bind to a cavity or pocket, leading to therapeutic benefit [5]. To date, thousands of protein–protein interactions (PPIs) are known, without deep pockets, with the absence of well-defined binding sites and with flat protein interfaces and thus remain challenging targets for small molecules [6]. PROTACs, on the other hand, have been shown to be suitable for targeting transcription factors that lack an active binding site [7] or for membrane – bound proteins [8].
Regarding their mode of action, in the case of PROTACs, it is event-driven, rather than occupancy-driven [4]. Occupancy-driven modalities are a hallmark of classical receptor pharmacology and require high drug concentrations in order to maintain a level of target occupancy that provides sufficient clinical benefit. However, high drug concentrations are also linked to off-target effects, which can be reduced by drugs with high specificity and favorable pharmacokinetic properties. Conversely, PROTACs show a catalytic behavior in their ability to induce proteasomal degradation at substoichiometric levels [9]. Their efficacy is not limited by equilibrium occupancy. It has been shown by Crews et al. [9] that a reduction in protein levels of more than 90% can be reached at nanomolar concentrations, hitherto impossible to achieve with the occupancy-driven modality. Moreover, in a recent study of PROTAC-mediated degradation of receptor tyrosine kinases (RTK) [8], further advantages of degradation over protein inhibition were demonstrated, including a more sustained reduction in downstream signaling and the maintenance of response duration even after washout of the PROTAC. The long-lasting biological effect and differential downstream signaling of PROTACs represent significant advantages over classical high receptor occupancy dependent drugs.
PROTACs are not consistent with Lipinski’s rule of five, which is a significant indication for cell permeability of small molecules. Although they have relatively large molecular mass, PROTACs can sustain sufficient intracellular concentrations, which in combination with the catalytic mechanism of action, is successfully leading to protein degradation. The exact mechanism of cellular uptake is not fully understood, but the fact that diverse PROTACs chemo-types are showing cell penetration in different cell types is an indication of a passive process [10]. Next, Sun et al. [11] have systematically investigated the potential usage of PROTACs in mice and pigs and rhesus monkeys. They observed that the PROTAC approach could markedly reduce the concentration of the POIs FKBP12 and BTK in vivo. For example, PROTAC-mediated depletion of FKBP12 by oral administration occurred in all organs or tissues except the brain, with a constant effect for about 1 week after a single treatment. These findings suggest the efficiency and reversible potential of PROTAC approach in animals and provide a robust basis for future clinical trials in human patients.
1.2. Types of degraders: cereblon, VHL, MDM2, cIAP1, other degraders
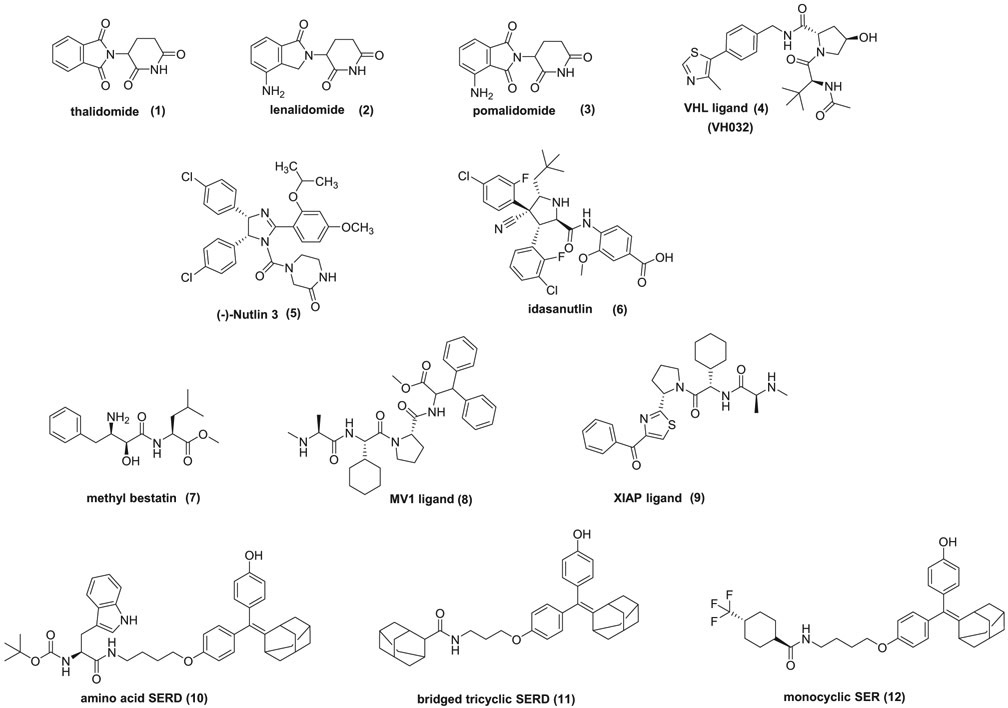
In the proteasome-mediated protein degradation process, the E3 ligases are critical components. In humans, more than 600 E3 ligases are known, but to date only a handful of them have been utilized in PROTACs. PROTACs can be classified according to the E3 ligases used, which most commonly are cereblon, Von Hippel Lindau (VHL), mouse double minute 2 homolog (MDM2), cellular inhibitor of apoptosis protein 1 (cIAP1) and other degraders (Figure 1).
Figure 1.
Structures of E3 ligase degraders.
1.2.1. Cereblon (CRBN)-based degraders
In 2010, Ito et al. [12] revealed that the molecular target and the primary cause of the teratogenic activity of thalidomide, a commonly prescribed sedative in pregnant women in the 1950s, was the protein cereblon (CRBN). Thalidomide (1) and its derivatives lenalidomide (2) and pomalidomide (3) are characterized as immunomodulatory drugs (IMiDs) and have received approvals for multiple myeloma. Mechanistically, IMiDs target the E3 ubiquitin ligase CUL4-RBX1-DDB1-CRBN [13], which is also known as CRL4CRBN . The binding of IMiDs to cereblon allows for the recruitment of the transcription factors of the IKAROS family (IKZF1 and IKZF3) and their ubiquitination over the endogenous cereblon substrate. In 2014, the crystal structures of DDB1-CRBN complexes bound to thalidomide [14] and lenalidomide [15] were solved. Since then, PROTACs with IMiD small molecules targeting CRBN and diverse proteins of interest, including the bromodomain and extra-Terminal (BET) proteins (BRD2/3/4) [16-18], FKBP12 [16], BCR-ABL [19], BRD9 [20], Sirt2 [21], CDK9 [22,23], FLT3 [24], BTK [24,25], ALK [26], CDK4/CDK6 [27,28] and HDAC6 [29] have been reported.
1.2.2. Von Hippel–Lindau (VHL)-based degraders
Reports for peptide-based PROTACs targeting the Von Hippel Lindau (VHL) E3 ligase were already described in 2004 [30]. These early, peptide-based PROTACs were constructed on a peptide sequence, deriving from the transcription factor hypoxia-inducible factor 1α (HIF-1α) and thus had the ability to bind to VHL. The presence of a poly-D-arginine tag assisted in cell penetration. Later on, considerable effort resulted in the discovery of small-molecule inhibitors targeting the interaction between HIF-1α and VHL [31-33]. For the highly potent small molecule inhibitors, X-ray structures elucidated the binding mode. Furthermore, regarding VHL-PROTACs, the initial – HIF1-α-derived-peptide was replaced with small molecules bearing the hydroxyproline moiety (4), thus leading to high-affinity and high-specificity binders for the VHL. VHL-PROTACs, based on small molecules in this study, resulted in the effective degradation of estrogen-related receptor (ERRα) and the kinase RIPK2 [9]. Examples of small molecule-based VHL-PROTACs have shown effective degradation of HaloTag fusion proteins [34], oncogenic BCR-ABL [19], BRD4 [35,36], TBK1 [37], several transmembrane tyrosine kinases (EGFR, HER2, and c-Met) [8] and TRIM24 [38].
1.2.3. MDM2-based degraders
PROTACs based on MDM2, the major E3 ligase targeting the tumor suppressor p53, are also reported, although there are considerably fewer reports compared to CRBN and VHL. Nutlins, which are ligands that bind to the p53-binding pocket of MDM2, are used in the construction of these PROTACs and disrupt the interaction of MDM2 with the transcription factor p53, without affecting the E3 ligase activity of MDM2 [39]. In 2008, Crews et al. synthesized PROTACs bearing nutlins (5), as the MDM2 ligands and the non-steroidal androgen receptor ligand (SARM) [40]. The cell-permeable PROTAC, successfully recruited the androgen receptor to MDM2, which as the E3 ligase triggered its ubiquitination and proteasomal degradation. Moreover, Crews et al. [41] showed that A1874, an MDM2-recruiting, BRD4 degrading PROTAC, which consists of idasanutlin (6) as MDM2 ligand and JQ1 as BRD4/BET inhibitor, was able to degrade the target protein by 98% with nanomolar potency. It is noteworthy that this is the first report of synergistic antiproliferative effect deriving from the E3 ligase ligand and the targeting warhead, since the PROTAC was able both to degrade BRD4 and at the same time stabilize p53.
1.2.4. cIAP1-based degraders
The cellular inhibitor of apoptosis protein 1 (cIAP1) has also been utilized in PROTAC design. In 2010, Hashimoto’s group [42] disclosed fully chemical PROTACs consisting of methyl bestatin (MeBS) (7), which selectively binds to the BIR3 domain of cIAP1, the RING domain of which promotes auto-ubiquitination, and all-trans retinoic acid (ATRA), which is the endogenous ligand of retinoic acid receptors and can recruit the intracellular retinoic-acid binding proteins CRABP-1 and CRABP-2. The cIAP1-based PROTAC successfully induced the proteasomal degradation of the target proteins CRABP-1 and CRABP-2. Further improved PROTACs were reported [43], by replacing the MeBS moiety with the MV1 moiety (8), which is a cIAP1/cIAP2/XIAP panligand and in this case, the PROTAC achieved the double protein knockdown of cIAP1and CRABP-2. Other examples of protein degradation utilizing hybrid small molecules named SNIPER (Specific and Non-genetic IAP-dependent Protein Eraser) have been reported for estrogen receptor alpha (ERα) [44,45], TACC3 [46] and BCR-ABL [47]. These SNIPERs were bestatin-based but the use of an improved high-affinity IAP ligand (9) that preferentially recruits X-linked IAP (XIAP) rather than cellular IAP1 led recently to potent SNIPERs against ERα, BCL-ABL, BRD4, and phosphodiesterase-4 (PDE4) [48]. Furthermore, effective degradation of the androgen receptor was also demonstrated [49].
1.2.5. Other degraders
The discovery of novel degrons (degradation – inducing inhibitors) is crucial for expanding the PROTAC toolbox. In 2012, Hedstrom et al. [50] showed that the tert-butyl carbamate-protected arginine (Boc3Arg) moiety can induce the degradation of ligands linked to it and remarkably it was proven that the process was ATP- and ubiquitin-independent. The degradation of glutathione-S-transferase was achieved by linking Boc3Arg with the covalent inactivators ethacrynic acid and thiobenzofurazan, whereas dihydrofolatereductase was degraded by linking Boc3Arg to the non-covalent inhibitor trimethoprim. In those cases, degradation is occurring via the 20S proteasome, however ATP is not necessary and the ubiquitin pathways are not involved [51]. The authors show that the Boc3Arg-linked ligands are localizing target proteins to the 20S proteasome and thus induce degradation. In 2018, Sharma et al. [52] revealed novel scaffolds that act as selective estrogen-receptor degraders (SERDs) and show ER antagonistic properties. Three distinct degron classes were reported with nanomolar potency as ERα degraders and inhibition of ER target gene expression; lipophilic amino acids (Leu, Phe, and Trp) (10), bridged bi- and tri-cyclic systems (11) and monocyclic systems (12).
2. Structural analysis
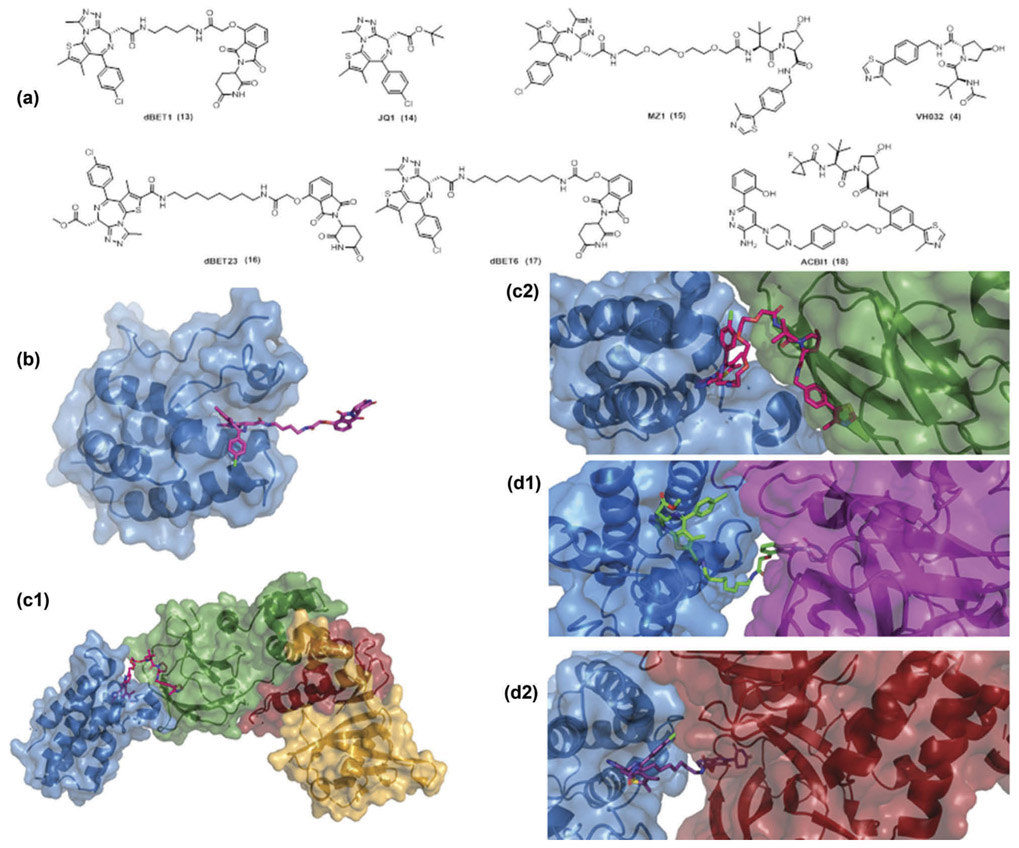
The crucial step in the mechanism of action for PROTACs is the formation of a high affinity, long-lasting ternary complex of an E3 ligase – PROTAC – protein of interest. Until recently there was a lack of structural data for ternary complexes. In 2015, a crystal structure was reported for a PROTAC bound to one protein of interest in a binary complex [16]. The high-resolution structure showed that the PROTAC dBET1(13) was bound to BRD4, proving similar recognition to the inhibitor JQ1 (14) (PDB 4ZC9) (Figure 2).
Figure 2.
(a) structures of dBET1 PROTAC (13), JQ1 (14) (BET inhibitor), MZ1 PROTAC (15), VH032 (4) (VHL inhibitor), dBET23 PROTAC (16), dBET6 PROTAC (17) and ACBI1 PROTAC (18), (b)Binary complex of dBET1 (purple sticks) with BRD4 (blue surface) [PDB 4ZC9], C1)Ternary complex of MZ1 (magenta sticks) with BRD4 (blue surface), pVHL (green surface), elongin C (red surface), elongin B (gold surface) [PDB 5T35], C2)closeup view BRD4 (blue surface) – MZ1 (magenta sticks) – pVHL (green surface), D1)Ternary complex of dBET23 (green sticks) with BRD4 (blue surface) and CRBN (magenta surface) [PDB 6BN7], D2) Ternary complex of dBET6 (purple sticks) with BRD4 (blue surface) and CRBN (red surface) [PDB 6BOY].The figure was prepared in Pymol (The PyMOLMolecular graphics system, version 2.0 Schrödinger, LLC).
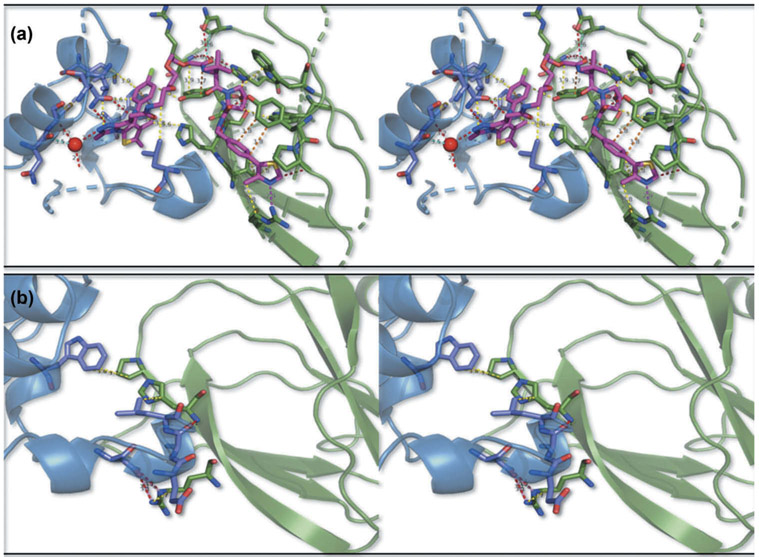
In 2017, Ciulli et al. [53] solved the first crystal structure of a ternary complex with 2.7Å resolution (PDB 5T35). The PROTAC molecule MZ1 (15) is comprised of the pan-BET bromodomain inhibitor JQ1 (14) with the potent and specific VHL ligand VH032 (4) and a three-unit PEG linker. MZ1 is bound to the second bromodomain of Brd4 (Brd4BD2) and pVHL:ElonginC:ElonginB and is inducing extensive new hydrophobic and electrostatic protein – protein interactions and protein – ligand contacts (Figure 3), while maintaining the individual interactions of the respective ligands with the E3 ligase and the bromodomain; the ligand JQ1 binds in the acetyllysine binding pocket of Brd4BD2, whereas VH032 binds to the hydroxyproline binding site of VHL. The PEG linker forms additional protein – ligand interactions, including van der Waals interactions and a hydrogen bond. Moreover, solvent-exposed areas of the JQ1 and VH032 ligands are buried in the interface, as the two proteins come in close proximity. In total, the extended buried surface area of the ternary complex reaches 2,621 Å2, from which 1,933 Å2 refers to the surface buried by the folding of the ligand. The burial of extensive surface areas and the formation of new PPIs are leading to stability of the ternary complex and converts a non-selective pan-BET inhibitor to a selective degrader.
Figure 3.
(a)Stereo view of ligand – protein interactions between MZ1 (magenta sticks) with BRD4 (blue cartoon), pVHL (green cartoon): hydrogen bonds (red dashes), cat_dip (magenta dashes), dipolar (cyan dashes), hdon_pi (yellow dashes), vdW (dark blue dashes), pi_pi (orange dashes). The key aminoacids participating in the interactions are shown in sticks (b)Stereo view of protein – protein interactions between BRD4 (blue cartoon), pVHL (green cartoon):H bonds as red, ionic as pink and van der Waals as yellow dashes. The key aminoacids participating in the interactions are shown in sticks. The figure was prepared by using Scorpion (Desert Scientific Software, http://saas1.desertsci.com/) and Pymol (The PyMOL molecular graphics system, version 2.0 Schrödinger, LLC).
In 2018, Nowak et al. [54] solved multiple X-ray structures of degrader – bound CRL4CRBN – BRD4 complexes and showed that the bound degrader has a unique effect on distinct binding conformations. The degraders consist of the E3-moiety thalidomide (1) that binds to CRL4CRBN, the ligand JQ1 (14) that binds to BRD4BD1 and BRD4BD2 with equal affinities and flexible linkers of varying length and composition. In the obtained X-ray structures of PROTACs dBET23 (16) and dBET6 (17), the linker length and the linkage position resulted in distinct binding conformations in the ternary complex. The observed plasticity in the degrader binding in the same protein (BRD4BD1) provides evidence for the selectivity profiles among the set of degraders that share the same E3 and target moiety. Thus, the features of the linker (type, length, attachment position) can affect which surface residues in the target protein might be involved in the complex formation. Interestingly, the interprotein contacts, even though having little contribution to the binding affinity of the interaction, seem to be the main drivers of selectivity. The plasticity of the binding and the distinct conformations of the degraders can result in effective degradation even in the absence of tight binding of the small molecule to the POI. In particular for relatively short linkers, the conformational constraints give access to only a few interprotein contact conformations and this feature could be a driver of selectivity and at the same time improve the drug-like properties of the degrader. In a study published earlier this year by Smith et al. [55], isoform-selective p38-MAPK targeting PROTACs were designed by using only one warhead and one E3 ligase. In all cases, the kinase inhibitor foretinib was used and VHL inhibitors. The authors show that varying the linker length and linker attachment point induced significant differences in isoform selectivity (p38α or p38δ), even by adding only one extra atom on the linker.
To date, there are limited examples of X-ray structures of ternary complexes. Very recently Ciulli et al. solved high-resolution ternary complex crystal structures and together with biophysical data rationally optimized the structures toward ACBI1 (18), a potent and cooperative degrader of SMARCA2, SMARCA4 and PBRM1 [56].
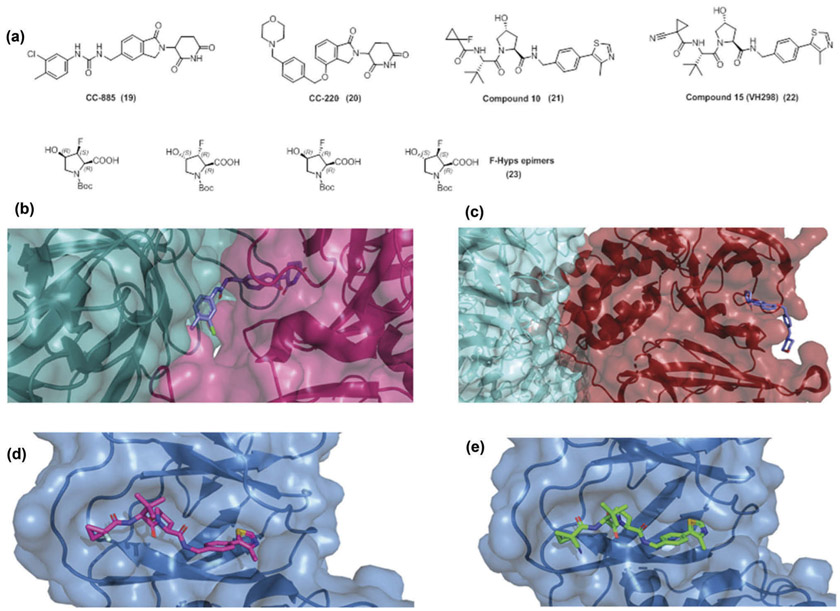
Regarding the design of degraders, valuable structural information is accumulating regarding the binding modes of the E3 ligase ligands. In 2010, Ito et al. [12] identified the molecular target of thalidomide as cereblon and in 2014, Hartmann et al. [57] showed that thalidomide and its derivatives mimic uridine. The nature of the binding pocket resembles an aromatic cage with three tryptophan residues. Thalidomide and its derivatives bind through the glutarimide ring into the aromatic cage, while the rest of the molecule protrudes from the binding pocket. More recently, Boichenko et al. [58] investigated thoroughly the chemical ligand space of cereblon, by solving multiple X-ray structures. The binding of the compounds was determined by a FRET assay and teratogenic effects were evaluated in a zebrafish model. A pharmacophore model for binding to cereblon was established; at least one carbonyl group of the glutarimide ring is necessary for binding, whereas the second carbonyl increases the affinity for 5- and 6-membered rings. In general, 5-membered rings showed higher affinity than 6-membered, whereas 7-membered rings didn’t show detectable binding. Moreover, 4-membered lactams were comparable to 5-membered rings. The presence of a heteroatom in the ring and the effect of substitutions were also explored. In all cases, a non-substituted NH group in the ring is crucial for binding. The structural data indicate the optimized minimal binding moieties needed for cereblon binding and provide valuable insight for future cereblon effectors. Moreover, a new co-crystal structure for the cereblon modulator (CC-885) (19), which demonstrated antitumor effects through the recruitment and degradation of G1 to S phase transition 1 protein (GSPT1), was disclosed [59]. Furthermore, SARs of glutarimide analogues deriving from CC-885 were established for compounds that promote the degradation of Aiolos and/or GSPT1 [60]. A cereblon modulator (CC-220) (20) showed improved cellular degradation of the transcription factors Ikaros and Aiolos [61], which in this case derives from improved affinity between the compound and cereblon. The crystal structure of cereblon – CC-220 and DDB1 (Damage Specific DNA Binding Protein 1) indicates that the increase in potency correlates with increased contacts between CC-220 and cereblon, away from the modeled binding site of Ikaros and Aiolos (Figure 4).
Figure 4.
(a) Chemical structures of CC-885 (19), CC-220 (20), compound 10 (21)and compound 15 (VH298) (22)and F-Hyps epimers (23), (b) Crystal structure of CC-885 (purple sticks) with GSPT1 (light blue surface) and CRBN (magenta surface) [PDB 5HXB], (c) Crystal structure of CC-220 (purple sticks) with DDB1 (light blue surface) and CRBN (red surface) [PDB 5V3O], (d) Crystal structure of compound 10 (magenta sticks) with pVHL:EloB:EloC (light blue surface) [PDB 5NVX], E) Crystal structure of compound 15/VH298 (green sticks) with pVHL:EloB:EloC (light blue surface) [PDB 5LLI].The figure was prepared in Pymol (The PyMOL molecular graphics system, version 2.0 Schrödinger, LLC).
Regarding inhibitors for the Von Hippel Lindau (VHL) E3 ubiquitin ligase, the group of Ciulli [62] has reported extensive SARs and solved X-ray structures in an effort to optimize the series. The best compounds show double-digit nanomolar affinities for binding to VHL and improved cellular activity on the VHL:HIF-1α-PPI (compounds (21), (22)). All known examples of VHL inhibitors contain the moiety of hydroxyproline (Hyp), since VHL features a Hyp recognition site that targets for degradation post-translationally hydroxylated HIF-1α subunits. Recently, Ciulli et al. [63] focused on novel fluorinated hydroxyprolines (F-Hyps). A synthetic route was successfully established for all four diastereoisomers of 3-fluoro-4-hydroxyprolines (F-Hyps) (23), followed by quantum mechanical calculations, NMR spectroscopy and small-molecule X-ray crystallography to delineate the effect of the fluorination on the conformational preferences of the core. The fluorination of Hyp had negligible effects on the hydrogen bond donor capacity of the C4 hydroxyl, however it actually led to the inversion of the natural preference from C4-exo pucker to the C4-endo pucker. Despite this inversion, F-Hyps were still able to bind to the VHL E3 ligase. The observed preferential recognition of the (3R,4S) epimer of F-Hyp could be utilized for expanding the chemical space of degraders. Moreover, despite a weakened affinity, the (3S,4S)-F-Hyp that was incorporated in the PROTAC MZ1 still led to Brd4-selective cellular degradation.
The same group utilized fragment-based screening and computational methods for surface probing in order to identify ligandable pockets on the VHL E3 ligase [64]. Until recently, only the HIF-recognition site was known as a ligandable. Ciulli et al. identified two more ligandable sites and reported crystal structures of the VHL: EloC: EloB E3 ubiquitin ligase with fragment-based hits. Two fragments were bound to a small cavity at the EloC:Cul interface, whereas one more fragment was bound in a cryptic pocket in VHL.
The accumulation of structural data, both for the E3 ligases commonly targeted in proteolysis targeted chimeras, as well as the crystal structures of the ternary complexes are expected to significantly facilitate the design and optimization of the degraders in the future.
3. Computational tools
Computational tools have already been applied in PROTAC design in an effort to rationally design and optimize the different components of the heterobifunctional molecule. A clear overview is provided in the recent work of Drummond and Williams [65]. In general, the multiple possible conformations and the observed plasticity in the recently disclosed ternary complexes' X-ray structures show that there are still challenges to overcome. One of the main concerns is the applicability of the computational approaches in multiple targets, including different E3 ligases and varying linkers that are known to have a significant impact on the possible conformations. In their recent work, Drummond and Williams [65], propose and validate four different methods for generating in silico ternary complexes, covering different ways for the preparation of PROTAC conformations. The one extreme being the sampling of conformations separately from their binding proteins and the other extreme that the whole ternary complex being present during the sampling. In general, the protein-protein docking-based method, in which PROTAC conformations were sampled independently of the proteins, but protein-protein docking was included to provide possible ligase-target arrangements, was considered superior and was accurate enough to complement structural optimization and provide crystal-like ternary complexes. Even so, it is expected that the predictability and accuracy of computational tools for PROTAC will keep evolving as more structural data become available.
4. Ternary complexes and kinetics
Regarding their mode of action, PROTACs differ significantly from classical inhibitors. Until recently, the design of PROTACs mostly considered the formation of the complex with the proteins as two binary interactions, in which the two warheads were optimized separately for individual interactions with the target proteins. However, now it is unambiguously proven that the linkers have a much more active role than keeping the ligands in proximity and can greatly affect the degradation, as well as the isoform specificity [53-55]. In contrast to a typical binary complex that occurs during the interaction of an inhibitor with the target protein, where the inhibitor is required to bind to a functional binding site and block a single protein interaction, PROTACs result in a ternary complex in which recognition is crucial, whereas potency is of reduced significance [66]. There are three different possibilities regarding the formation of ternary complex and the subsequent effective degradation [67]. The first possibility for effective degradation to occur is via the formation of a stable ternary complex, which requires high affinity between the POI and the PROTAC, as well as favorable interactions with the E3 ligase. In the second possibility, even with somehow weak affinity, but with favorable interactions, degradation can be effective if the ternary complex is stable. On the contrary, in the third possibility, high affinity in the absence of favorable interactions results in an unstable ternary complex and thus degradation is ineffective. Therefore, PROTACs are suitable for ‘difficult’ targets, where the known inhibitors are able to interact with the target, but due to weak binding are unsuitable for further clinical development or for protein – protein interactions where the absence of well-defined pockets is a typical feature.
Regarding kinetics, PROTACs also differ from classical inhibitors. In multiple studies with PROTACs, the ‘hook effect’ was frequently observed when high concentrations were used. In case of high concentrations, the binary complexes PROTAC: E3 ligase and PROTAC: POI are hindering the formation of the ternary complex due to saturation, which is required for degradation. Moreover, since there are two proteins involved in the formation of the ternary complex, the binding affinity of the PROTAC to one protein partner may be either enhanced or reduced by the presence of the second protein. The quantification of this effect, which is known as ‘cooperativity’ is possible by defining the ratio of binary and ternary dissociation constants of PROTAC binding to the first protein [53,68]. Positive cooperativity implies that the ternary binding affinity is enhanced compared to binary, whereas in negative cooperativity the ternary complex is destabilized. Neutral cooperativity indicates that there is no change in the presence of the second protein.
The understanding of these phenomena and the kinetic analysis of ternary complexes are crucial for further PROTAC optimization and development. The applicability of biophysical techniques (X-ray, NMR, ITC, AlphaLISA, TR-FRET) to study binary and ternary complexes has been reviewed in detail by Ciulli et al. [66]. More recently, the group of Ciulli [68] developed an SPR-based assay to quantify the stability of PROTAC-induced ternary complexes and to explore the kinetics equilibria between binary and ternary complex formation as a quantitatively label-free technique.
In 2018, a modular live-cell platform utilizing endogenous tagging was disclosed and applied to the monitoring of the PROTAC-mediated degradation of bromodomains [69]. The authors combine CRISPR/Cas9, endogenous tagging and luminescent technology in order to kinetically measure target protein levels. Combination of this technology with optimized bioluminescence resonance energy transfer (NanoBRET) was applied to kinetic measurements of intracellular protein interactions along the degradation pathway, including the ternary complex formation, ubiquitination, and PROTAC-target engagement. Overall, the technique allows for better understanding of the cellular mechanism of action and assesses which stages of the degradation process are impacted by the recruitment of different E3 ligase complexes.
5. Homo-PROTACs
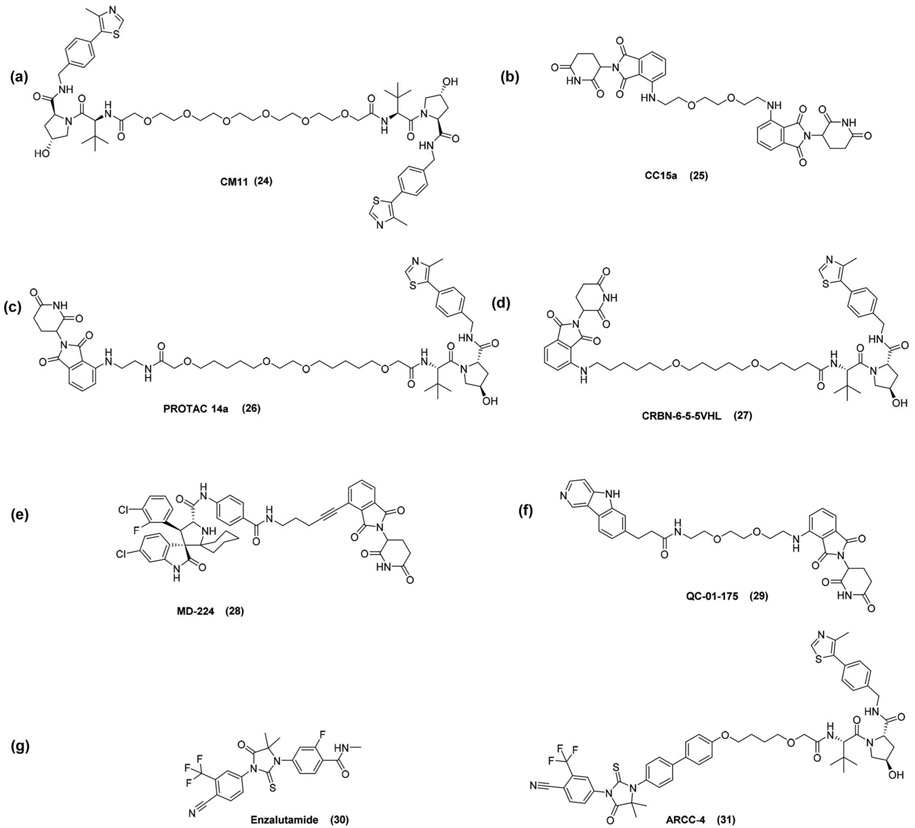
Homo-PROTACs are a unique type of proteolysis-targeting chimeras comprised of two identical molecules linked together. Homo-PROTACs were first reported for the VHL [70], based on the structures of two potent VHL ligands (VH032 and VH298). The linker was attached on different positions and the length of the polyethylene glycol chains varied from 3 to 5 ethylene glycol units. The biological evaluation of the homo-PROTACs revealed that the most active compounds were selectively degrading the long isoform of VHL. The position of the linker and the stereochemistry were crucial for degradation. The trans epimer of Hyp was required for degradation, as expected. Moreover, shorter linker lengths led to decreased degradation. The most active compound (CM11) (24) induced complete deletion of pVHL30 after 4 h at 10nM. In high micromolar concentrations, the ‘hook – effect’ was observed. A competition experiment confirmed that the degradation activity was due to VHL binding. Furthermore, isothermal titration calorimetry (ITC), size-exclusion chromatography (SEC) and AlphaLISA proximity assay were performed to assess the formation of the ternary complex in solution. The binding ratio of CM11 to VHL was proved to be 1:2, in contrast to the 1:1 ratio observed for the inhibitor VH032. Overall, PROTAC CM11 is described as a chemical probe for rapid and selective pVHL30 knockdown, useful for further investigating the biological function of pVHL.
In 2018, homobifunctional PROTACs that utilize cereblon (CRBN) as the hijacked degrader and at the same time, as the protein targeted for degradation, were described, aiming at the chemical-induced CRBN degradation [71]. Two pomalidomide moieties were conjugated via linear linkers with varying length, varying hydrophobicity, and different attachment positions. Western blot analysis was performed for the multiple myeloma cell line MM1S, which expresses endogenous CRBN and its immunomodulatory-induced (IMiD) neo-substrates IKZF1, IKZF3, and casein kinase 1A1 (CK1α). All homo-PROTACs in these series induced a dose-dependent decrease in IKZF1 protein levels, however the impact on IKZF1 degradation varied. The compound with linker length of eight atoms (CC15a) (25) was identified as the most potent CRBN degrader. In high concentrations, the ‘hook effect’ was observed. Co-immunoprecipitation proved that the homo-PROTAC leads to the formation of ternary complexes with 2:1 stoichiometry with two CRBNs and one PROTAC molecule. The most active PROTAC degraded specifically CRBN and showed only weak effects on the neo-substrates, with no effect on other members of the CRL3 ligase family (Figure 5).
Figure 5.
(a) VHL HomoPROTAC, (b) CRBN HomoPROTAC, (c and d) CRBN – VHL PROTACs, (e) MDM2 – CRBN PROTAC, (f) Tau-CRBN PROTAC, (g) structures of enzalutamide and ARCC PROTAC.
In continuation of the homo-PROTAC approach, Ciulli et al. [72] investigated the hypothesis that the E3 ligases themselves could be hijacked against each other using a heterodimerizing PROTAC, leading either to degradation of both E3 ligases or preferential degradation of one E3 ligase. A library of CRBN – VHL PROTACs was designed and synthesized, using pomalidomide as CRBN handle and as for the VHL handle, structural modifications were designed on two known VHL ligands, including different attachment points of the linker. Three series of PROTACs were designed and the degradation was studied by western blot analysis in HeLa cells, including the previously disclosed homo-PROTACs CM11 and CC15a as positive controls for VHL and CRBN degradation, respectively. A few compounds led to significant degradation of CRBN, whereas none of the compounds showed significant degradation of VHL. Testing the compounds at low concentration to rule false-negative results due to the ‘hook effect’, revealed that some compounds showed up to 50% degradation of pVHL30. Thus, the concentration could affect the preferential degradation of one ligase over the other. The most active compound (PROTAC 14a)(26) was further studied in both HeLa and HEK293 cells and induced CRBN degradation rapidly, with high potency and to profound levels. In this ‘double-hijacking’ approach the VHL – CRBN PROTACs resulted in preferential degradation of CRBN over VHL. Further mechanistic studies are expected to illustrate the effect of the different elements that are involved; the conjugation patterns, the linker lengths or the structures of the heterodimerizing PROTACs.
Moreover, Gütschow et al. [73] have also described eight VHL – CRBN PROTACs and investigated the degradation on the myeloma cell line MM1S. In most cases, CRBN levels were decreased, whereas no reduction was observed for VHL protein levels. The most active compound CRBN-6-5-5-VHL (27) showed a negligible effect on the degradation of the neo-substrates IKZF1 and IKZF3 and was superior compared to the homo-PROTAC 15a. Regarding SAR, the linker length and the lipophilicity were considered the crucial factors, whereas the polar surface area was less significant. Overall, only CRBN was degraded by the CRBN – VHL PROTACs. Such compounds have potential as chemical probes to elucidate ligand specificities, as well as potential therapeutic value.
Wang et al. [74] reported MDM2 heterodimer PROTACs, by linking the MDM2 inhibitor MI-1061 either with a CRBN ligand or a VHL ligand. For MDM2 – CRBN PROTACs, SARs were established regarding the length and the type of linker, modifications on the cereblon ligand and attachment point of the linker. The most potent compound, MD-224 (28), was the result of linker rigificiation; two methylene groups were converted into an alkyne group and this significantly improved the potency. The authors investigated also MDM2 – VHL PROTACs with the MDM2 inhibitor MI-1061, however in all cases MDM2 – VHL PROTACs were less potent than the corresponding MDM2 inhibitor. Extensive mechanistic studies were performed for MD-224. The compound was able to achieve complete and durable tumor regression in the xenograft tumor model in mice and was proven to be much more efficacious than the MDM2 inhibitor MI-1061. Interestingly, a ‘no-linker’ version was also active in degrading MDM2, however to a much lesser extent.
6. Tau-PROTACs
Tauopathies belong to neurodegenerative diseases, with characteristic accumulation of aberrant forms of tau protein, which results in neuronal death in focal brain areas. In particular for Alzheimer’s disease (AD), although the exact pathogenesis remains elusive, several hypotheses have been proposed, such as chronic inflammation, oxidative stress, acetylcholine abnormalities, β-amyloid cascade, and pathogenic tau protein. In 2016, Li et al. [75] showed that multifunctional molecules, consisting of a Tau-recognizing peptide moiety, a cell-penetrating peptide, and an E3 ligase-recognizing peptide moiety, can enhance Tau degradation in cells. The multifunctional peptides were tested for their ability to induce Tau degradation in a stable mouse neuroblastoma N2a-based cell line. The compound TH006 appeared to be the most potent and it was further studied in a fluorescence polarization assay. Confocal microscopy data showed that TH006 was able to enter into cells and further analysis by western blots and flow cytometry proved that it induced effective intracellular tau degradation.
In 2018, Jiang et al. [76] reported a peptide PROTAC targeting Tau by recruiting the Keap1 – Cul3 ubiquitin E3 ligase. The substrate of the Keap1 – Cul3 ubiquitin E3 ligase is the transcription factor NF-E2-related factor-2 (Nrf2), which is involved in the regulation of oxidative stress. The dysregulation of Keap1-Nrf2 signaling is affecting both oxidative stress and inflammatory-related diseases. The authors focused on peptide PROTACs, hijacking the Keap1 and thus leading to ubiquitination and degradation of Tau, as an alternative to inhibiting Nrf2. One of the peptide PROTACs was able to interact effectively with Keap1 and Tau, showed cell penetration and induced Tau degradation in different cell lines overexpressing Tau. The data from this study clearly show the potential of PROTAC degradation in neurodegenerative diseases. Likely, however, peptide-based PROTACs and even small molecule-based PROTACs will need considerable optimization to pass the blood-brain-barrier.
Recently, an application of PROTAC combined with position emission tomography (PET) tracers, which are useful tools for the diagnosis of tauopathies, was reported [77]. The most clinically advanced tau PET tracer (18F-T807 or 18F-AV-1451) was coupled via linkers to pomalidomide. A library of 25 hetero-bifunctional molecules with various linker sizes and attachment chemistry was synthesized. The lead PROTAC QC-01–175 (29) was evaluated in a biolayer interferometry (BLI) assay for degradation against the wild-type and the two variant forms of recombinant human tau: A152T and P301L. Although the PET tracer 18F-T807 shows off-target activity against the monoamine oxidases A and B (MAO-A, MAO-B), the off-target MAO binding was significantly reduced with the PROTAC QC-01–175. Moreover, the PROTAC was evaluated in a human neuronal cell model of tauopathy and promoted tau clearance in a concentration-dependent manner, thus achieving the rescue of tau-mediated neuronal stress vulnerability. It is noteworthy that the PROTAC had minimal effects on tau from wild-type control neurons and targeted preferentially tau species from FTD (frontotemporal dementia) neurons, expressing tau-152T or tau-P301L, indicating specificity for disease-relevant forms. The mechanism of action of QC-01–175 was also investigated by targeting each component that is expected to be involved in proteasomal degradation. The data showed that the degradation depends on CRBN and tau binding, as well as neddylation and proteasome function, whereas autophagy is not involved. Further optimization of the compound would be necessary for clinical development, since it is a relatively large and flexible molecule and might suffer from poor brain penetration or fast metabolism. Although the PROTAC showed improved off-target effects in the case of MAO-A and MAO-B, mass spectrometry global proteome analysis showed that members of the C2H2 zinc finger protein family were also downregulated, due to CRBN-binding. Optimization to remove these IMiD off-target effects would be necessary toward a more selective tau degrader. Nevertheless, QC-01–175 will significantly contribute to studies in human tauopathies.
The PROTAC-mediated Tau degradation is also being considered for the Alzheimer’s disease treatment. In a recent patent highlight [78], compounds with tau binding properties were linked to thalidomide or lenalidomide with varying linker length and composition. The ability of these compounds to induce tau degradation was demonstrated in assays with human cells. Further in vivo assays were performed and the blood-brain barrier permeability was examined.
7. Toward clinical trials
PROTAC protein degraders entered the patent literature for the first time by the biotech Proteinix in 1999 but they never followed up with their patent [79]. Two years later, Craig Crews from Yale University started publishing on targeted degraders [1]. By 2008, Crews and coworkers reported the first non-peptidic PROTACs, based entirely on small molecules and degrading androgen-receptor (AR) by recruiting MDM2 as the E3 ligase [40]. In 2013, Crews founded the biotechnology start-up Arvinas (New Haven, CT) to develop PROTAC technology to the clinic. In 2017, Arvinas selected AR PROTAC for prostate cancer and estrogen-receptor (ER) PROTAC for breast cancer as the first clinical trial candidates.
ARV-110 is the first PROTAC protein degrader which is clinically evaluated by Arvinas. Currently, a phase 1 clinical trial is ongoing with ARV-110 in patients with metastatic castration-resistant prostate cancer (mCRPC) [, https://clinicaltrials.gov/ct2/show/NCT03888612?spons=Arvinas&rank=1].
The structure of ARV-110 is not disclosed. ARV-110 is an orally bioavailable AR PROTAC which shows consistent activity and potency in various in vitro and in vivo systems and shows efficacy in enzalutamide-resistant prostate cancer. ARV-110 degrades AR in all tested cell lines (for example LNCaP and MCF7) with a half-maximal degradation concentration (DC50) of ~1nM. AR degradation by ARV-110 leads to the suppression of the AR-target gene PSA expression, inhibition of AR-dependent cell proliferation and induction of potent apoptosis in VCaP cells. DMPK and exploratory toxicology studies show robust oral, dose-proportional drug exposure in rodent and non-rodent species. In mouse models, ARV-110 degrades clinically relevant mutant AR. It shows activity in a high androgen environment. In mouse xenograft studies, more than 90% AR degradation is observed at a 1 mg/kg PO QD dose. Significant inhibition of tumor growth and AR signaling can be achieved in both an intact and castrate setting. Further ARV-110 demonstrates in vivo efficacy and reduction of downstream oncogenic Erg protein in a long term, castrate, enzalutamide-resistant VCaP tumor model [80].
An interesting head-to-head comparison between enzalutamide (30), and a similar PROTAC derivative to ARV-110, ARCC-4 (31), across different cellular models of prostate cancer drug resistance, revealed ≥95% of cellular androgen receptor depletion, inhibition of prostate tumor cell proliferation, degradation of clinically relevant androgen receptor point mutants and unlike enzalutamide, retainment of antiproliferative effect in a high androgen environment. Thus, AR PROTACs have the potential to overcome drug resistance of direct AR inhibitors [81].
8. Conclusion
PROTACs are not following the classical drug discovery rules and in a lot of aspects, such as the mechanism of action, the kinetics, the formation of the ternary complex, the catalytic mode-of-action, the obvious deviation from Lipinsky’s rule of five, there is still a lot to be established. However, the first crystal structures of ternary complexes, as well as kinetic studies and biophysical assays have significantly improved our understanding for this new modality. Impressive preclinical data have been accumulated for numerous challenging targets, including the Tau protein, the androgen, and the estrogen receptor. Here, we provide an overview of the types of degraders, examples of PROTACs for various targets, a structural analysis based on crystal structures and aspects of PROTACs kinetics. The special cases of homoPROTACs and tau-PROTACs are analyzed further. Overall, PROTACs represent a new modality with game-changing potential in drug discovery.
9. Expert opinion
PROTACs differ significantly from small-molecule inhibitors and in a lot of cases, unanticipated findings were revealed. For example, in 2018 [82], in the case of BET-VHL PROTACs, it was proven that the most potent inhibitor does not necessarily generate the most potent degrader. In the case of BCL6 [83], selective BCL6 inhibitors were developed and subsequently a BCL6-PROTAC, that even though was able to degrade the target, failed to induce a significant phenotypic response, despite achieving cellular concentrations. The latter was the consequence of a residual undegradable BCL6 population. On the contrary, in some cases, such as the targeting of focal adhesion kinase (Fak) [84], where kinase inhibitors have a low success rate in clinical trials, the optimized clinical candidate defactinib was outperformed by the designed PROTAC. PROTACs, apart from applications in difficult or undruggable targets, could also be a suitable strategy to overcome resistance. In the case of Bruton’s tyrosine kinase (BTK) in chronic lymphocytic leukemia (CLL), resistance can occur after the mutation of a cysteine to a serine. Therefore, the irreversible covalent inhibitor ibrutinib is unable to bind to the target. In an interesting approach [25] ibrutinib was converted into a PROTAC, by removing the covalent warhead and was able to even degrade the mutated target, in contrast to the inhibitor. It is noteworthy, that very recently it was shown [85] that resistance in cancer cell lines following chronic PROTAC treatment can occur. In this study, resistance was observed both for VHL- and CRBN-based BET PROTACs, as a result of genomic alterations that compromised core components of the E3 ligase complexes. On the contrary, secondary mutations affecting the compound binding to the target were not observed. It should be noted that the observed resistance was specific for the individual PROTAC, meaning that cells that became resistant to the VHL-recruiting PROTAC remained sensitive to CRBN-based PROTAC and vice versa.
The application of PROTACs against numerous targets and the impressive responses clearly show that they are a powerful new modality. However, they are still aspects that are not fully elucidated. Therefore, the accumulation of more structural data regarding the ternary complex and the different steps involved in the degradation process would significantly facilitate the rational design of PROTACs as well as the optimization of the crucial components.
Moreover, PROTACs are mostly described for four members of the E3 ligase family. However, in humans, there are more than 600 of them. So far, only a very limited part of them has been utilized and the choice of the E3 ligase to target is not always rational. Studies exploring the different E3 ligases suitable for PROTACs are still underdeveloped. This would require the detailed structural analysis of the ligases, their recognition requirements, and their druggability. A recent example in this direction focused on the structural basis for recruitment of DAPK1 to the KLHL20 E3 ligase and the possibility of using this E3 ligase in PROTACs [86]. Moreover, PROTACs are usually designed having as starting point a known, potent ligand for the protein of interest, which is appropriately modified in order to attach the linker and at the same time maintain the features necessary for binding. This is an important limitation, in cases of protein – protein interactions, where there might be lack of small molecules or absence of exact structural data to guide the modification of the structure. Better understanding of the recognition phenomena of PROTACs would be necessary to be able – ideally – to design PROTACs even for targets lacking known inhibitors.
Selectivity so far has proven to be a great advantage of PROTACs, including isoform selectivity [53-55] and the use of non-selective inhibitors that can be converted into selective PROTACs [53]. However, regarding the underlying biology for the E3 ligases aspect, there are still missing data. In particular for IMiDs that bind to cereblon, the neo-substrates include seemingly unrelated proteins (IKZF1, IKZF3, CK1α and GSPT1). IKZF1 and IKZF3 belong to C2H2 zinc finger proteins, which are the largest class of putative transcription factors in the human proteome with approximately 800 members. It was shown recently, that IMiDs indeed are capable of inducing the degradation of a large number of proteins through a C2H2 degron [87]. On the one hand, this observation indicates the potential of CRBN-binding small molecules in targeting transcription factors, but on the other hand raises the question of possible off-target effects deriving from PROTAC-mediated degradation of the C2H2 zinc finger proteins.
With the first PROTACs reaching clinical trials, the results are greatly anticipated to establish their clinical significance. Evidence of their long-term effects or potent toxicity is still missing.
In comparison to other promising techniques, such as CRISPR and RNAi for cancer treatment, PROTACs have the advantage of a catalytic mechanism of action and the reversible binding to the target. The effect is not permanent, thus in contrast to CRISPR, fewer off-target effects are anticipated [88]. It is expected that the highly anticipated clinical data for those techniques will further elucidate their strengths and limitations.
PROTACs are relying on the ubiquitin proteasome system to degrade intracellular targets. A novel expansion to the field of targeted protein degradation, which shows the tremendous potential of this approach, is the heterodimeric molecules called ENDTAC (endosome targeting chimera) [89]. In this very first report, Crews et al. show that extracellular targets can be internalized and degraded via the receptor – mediated endolysosomal pathway. The approach was applied to an extracellular recombinant fusion protein, which was internalized and degraded by hijacking the decoy GPCR receptor, CXCR7. This method could potentially overcome the limitation of intracellular protein targeting by PROTACs and could be applied to secreted, extracellular proteins. Overall, the degradation of protein targets is a highly exciting field and will likely revolutionize drug discovery.
Article highlights.
PROTACs with different degraders have shown impressive biological responses.
The first crystal structures of ternary complexes show plasticity and the great impact of the linker in bringing the two proteins in close proximity.
The stability of a ternary complex is more significant than the high affinity of the ligand.
HomoPROTACs are expected to be valuable probes for investigating biological functions and substrates of the E3 ligases.
PET ligands, such as in the case of Tau protein, can also be utilized in highly effective PROTACs.
PROTACs for androgen and estrogen receptor are the first ones to reach clinical trials.
This box summarizes key points contained in the article.
Acknowledgments
Funding
This research has been supported to (AD) by the National Institute of Health (NIH) (2R01GM097082-05), the European Lead Factory (IMI) under grant agreement number 115489, the Qatar National Research Foundation (NPRP6-065-3-012). Moreover, funding was received through ITN ‘Accelerated Early stage drug dIScovery’ (AEGIS, grant agreement No 675555) and, COFUND ALERT (grant agreement No 665250) and KWF Kankerbestrijding grant (grant agreement No 10504).
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Sakamoto KM, Kim KB, Kumagai A, et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci. USA. 2001;98 (15):8554–8559. DOI: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scudellari M Protein-slaying drugs could be the next blockbuster therapies. Nature. 2019;567(7748):298–300. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67(1):425–479. [DOI] [PubMed] [Google Scholar]
- 4.Salami J, Crews CM. Waste disposal-an attractive strategy for cancer therapy. Science. 2017;355(6330):1163–1167. [DOI] [PubMed] [Google Scholar]
- 5.Nero TL, Morton CJ, Holien JK, et al. Oncogenic protein interafaces: small molecules, big challenges. Nat Rev Cancer. 2014;14 (4):248–262. [DOI] [PubMed] [Google Scholar]
- 6.Zhang QC, Petrey D, Deng L, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chessum NEA, Sharp SY, Caldwell JJ, et al. Demonstrating in-cell target engagement using a pirin protein degradation probe (CCT367766). J Med Chem. 2018;61(3):918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burslem GM, Smith BE, Lai AC, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;25(1):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bondeson DP, Mares A, Smith IE, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol. 2015;11 (8):611–617.• First report of PROTAC inducing degradation with a catalytic mechanism of action.
- 10.Churcher I Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J Med Chem. 2018;61 (2):444–452. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Wang J, Yao X, et al. A chemical approach for global protein knockdown from mice to non-human primates. Cell Discov. 2019;5:10.eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito T, Ando H, Suzuki T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327(5971):1345–1350. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Girona A, Mendy D, Ito T, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26 (11):2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer ES, Böhm K, Lydeard JR, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512 (7512):49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamberlain PP, Lopez-Girona A, Miller K, et al. Structure of the human cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21(9):803–809. [DOI] [PubMed] [Google Scholar]
- 16.Winter GE, Buckley DL, Paulk J, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348 (6241):1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Qian Y, Altieri M, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. ChemBiol. 2015;22(6):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, Hu J, Xu F, et al. Discovery of a small molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem. 2018;61(2):462–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai AC, Toure M, Hellerschmied D, et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl. 2016;55(2):807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remillard D, Buckley DL, Paulk J, et al. Degradation of the BAF complex factor BRD9 by heterobifunctional ligands. Angew Chem Int Ed Engl. 2017;56(21):5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiedel M, Herp D, Hammelmann S. et al. Chemically induced degradation of Sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuinrearranging ligands (SirReals). J Med Chem. 2018;61(2):482–491. [DOI] [PubMed] [Google Scholar]
- 22.Olson CM, Jiang B, Erb MA, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14(2):163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robb CM, Contreras JI, Kour S, et al. Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC). Chem Commun. 2017;53(54):7577–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang HT, Dobrovolsky D, Paulk J, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol. 2018;25(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buhimschi AD, Armstrong HA, Toure M, et al. the C481S ibrutinib-resistance mutation in Bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry. 2018;57(26):3564–3575. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Han XR, Yang X, et al. Proteolysis targeting chimeras (PROTACs) of anaplastic lymphoma kinase. (ALK) Eur J Med Chem. 2018;151:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang B, Wang ES, Donovan KA, et al. Development of dual and selective degraders of cyclin-dependent kinases 4 and 6. Angew Chem Int Ed Engl. 2019;58(19):6321–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rana S, Bendjennat M, Kour S, et al. Selective degradation of CDK6 by a palbociclib based PROTAC. Bioorg Med Chem Lett. 2019;29 (11):1375–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang K, Song Y, Xie H. et al. Development of the first small molecule histone deacetylase 6 (HDAC6) degraders. Bioorg Med Chem Lett. 2018;28(14):2493–2497. [DOI] [PubMed] [Google Scholar]
- 30.Schneekloth JS Jr, Fonseca FN, Koldobskiy M, et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126(12):3748–3754. [DOI] [PubMed] [Google Scholar]
- 31.Buckley DL, Van Molle I, Gareiss PC. et al. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1α interaction. J Am Chem Soc. 2012;134(10):4465–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buckley DL, Gustafson JL, Van Molle I, et al. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1α. Angew Chem Int Ed Engl. 2012;51(46):11463–11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galdeano C, Gadd MS, Soares P, et al. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J Med Chem. 2014;57(20):8657–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley DL, Raina K, Darricarrere N, et al. HaloPROTACS: use of small molecule PROTACs to induce degradation of HaloTag fusion proteins.ACS. Chem Biol. 2015;10(8):1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina K, Lu J, Qian Y, et al. BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113(26):7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10(8):1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crew AP, Raina K, Dong H. et al. Identification and characterization of von Hippel-Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J Med Chem. 2018;61 (2):583–598. [DOI] [PubMed] [Google Scholar]
- 38.Gechijian LN, Buckley DL, Lawlor MA. et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat ChemBiol. 2018;14(4):405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilev LT, Vu BT, Graves B, et al. in vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. [DOI] [PubMed] [Google Scholar]
- 40.Schneekloth AR, Pucheault M, Tae HS, et al. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett. 2008;18(22):5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hines J, Lartigue S, Dong H, et al. MDM2-recruiting PROTAC offers superior, synergistic antiproliferative activity via s simultaneous degradation of BRD4 and stabilization of p53. Cancer Res. 2019;79(1):251–262.••First report of a synergistic, antiproliferative effect from the E3 ligase ligand and the targeting warhead.
- 42.Itoh Y, Ishikawa M, Naito M, et al. Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc. 2010;132(16):5820–5826. [DOI] [PubMed] [Google Scholar]
- 43.Itoh Y, Ishikawa M, Kitaguchi R, et al. Double protein knockdown of cIAP1 and CRABP-II using a hybrid molecule consisting of ATRA and IAPs antagonist. Bioorg Med Chem Lett. 2012;22(13):4453–4457. [DOI] [PubMed] [Google Scholar]
- 44.Okuhira K, Demizu Y, Hattori T, et al. Development of hybrid small molecules that induce degradation of estrogen receptor-alpha and necrotic cell death in breast cancer cells. Cancer Sci. 2013;104 (11):1492–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demizu Y, Okuhira K, Motoi H, et al. Design and synthesis of estrogen receptor degradation inducer based on a protein knockdown strategy. Bioorg Med Chem Lett. 2012;22(4):1793–1796. [DOI] [PubMed] [Google Scholar]
- 46.Ohoka N, Nagai K, Hattori T, et al. Cancer cell death induced by novel small molecules degrading the TACC3 protein via the ubiquitin-proteasome pathway. Cell Death Dis. 2014;5:e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demizu Y, Shibata N, Hattori T, et al. Development of BCR-ABL degradation inducers via the conjugation of an imatinib derivative and a cIAP1 ligand. Bioorg Med Chem Lett. 2016;26(20):4865–4869. [DOI] [PubMed] [Google Scholar]
- 48.Ohoka N, Okuhira K, Ito M, et al. in vivo knockdown of pathogenic proteins via specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs). J Biol Chem. 2017;292 (11):4556–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibata N, Nagai K, Morita Y, et al. Development of protein degradation inducers of androgen receptor by conjugation of androgen receptor ligands and inhibitor of apoptosis protein ligands. J Med Chem. 2018;61(2):543–575. [DOI] [PubMed] [Google Scholar]
- 50.Long MJ, Gollapalli DR, Hedstrom L. Inhibitor mediated protein degradation. Chem Biol. 2012;19(5):629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi Y, Long MJ, Rosenberg MM, et al. Boc3Arg-linked ligands induce degradation by localizing target proteins to the 20S proteasome. ACS ChemBiol. 2016;11(12):3328–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Guillen VS, Sharma N, et al. New class of selective estrogen receptor degraders (SERDs): expanding the toolbox of PROTAC degrons. ACS Med Chem Lett. 2018;9(8):803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gadd MS, Testa A, Lucas X, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017; 13(5): 514–521.••First report of a ternary complex X ray.
- 54.Nowak RP, DeAngelo SL, Buckley D, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol. 2018; 14(7): 706–714.••The authors show the effect of the linker in the formation of ternary complexes and the observed plasticity in crystal structures.
- 55.Smith BE, Wang SL, Jaime-Figueroa S, et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun. 2019;10(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farnaby W, Koegl M, Roy MJ, et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol. 2019;15(7):672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartmann MD, Boichenko I, Coles M, et al. Thalidomide mimics uridine binding to an aromatic cage in cereblon. J Struct Biol. 2014;188(3):225–232. [DOI] [PubMed] [Google Scholar]
- 58.Boichenko I, Bär K, Deiss S, et al. Chemical ligand space of cereblon. ACS Omega. 2018;3(9):11163–11171.••The authors thoroughly investigate the structural requirements for cereblon binding.
- 59.Matyskiela ME, Lu G, Ito T, et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535(7611):252–257. [DOI] [PubMed] [Google Scholar]
- 60.Hansen JD, Condroski K, Correa M, et al. Protein degradation via CRL4CRBN ubiquitin ligase: discovery and structure-activity relationships of novel glutarimideanalogs that promote degradation of Aiolos and/or GSPT1. J Med Chem. 2018;61(2):492–503. [DOI] [PubMed] [Google Scholar]
- 61.Matyskiela ME, Zhang W, Man HW, et al. A cereblon modulator (CC-220) with improved degradation of Ikaros and Aiolos. J Med Chem. 2018;61(2):535–542. [DOI] [PubMed] [Google Scholar]
- 62.Soares P, Gadd MS, Frost J, et al. Group-based optimization of potent and cell-active inhibitors of the von Hippel–Lindau (VHL) E3 ubiquitin ligase: structure–activity relationships leading to the chemical probe (2S,4R)-1-((S)-2-(1-cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298). J Med Chem. 2018;61(2):599–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Testa A, Lucas X, Castro GV, et al. 3-Fluoro-4-hydroxyprolines: synthesis, conformational analysis, and stereoselectiverecognition by the VHL E3 ubiquitin ligase for targeted protein degradation. J Am Chem Soc. 2018;140(29):9299–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lucas X, Van Molle I, Ciulli A. Surface probing by fragment-based screening and computational methods identifies ligandablepockets on the von Hippel-Lindau (VHL) E3 ubiquitin ligase. J Med Chem. 2018;61 (16):7387–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drummond ML, Williams CI. In silico modeling of PROTAC-mediated ternary complexes: validation and application. J Chem Inf Model. 2019;59(4): 1634–1644 and references therein. [DOI] [PubMed] [Google Scholar]
- 66.Hughes SJ, Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essay Biochem. 2017;61(5):505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.An S, Fu L. Small-molecule PROTACs: an emerging and promising approach for the development of targeted therapy drugs. EBioMedicne. 2018;36:553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy MJ, Winkler S, Hughes SJ, et al. SPR-measured dissociation kinetic of PROTAC ternary complexes influence target degradation rate. ACS Chem Biol. 2019;14(3):361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riching KM, Mahan S, Corona CR, et al. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem Biol. 2018;13(9):2758–2770.••First report of real-time monitoring of every step involved in the PROTAC-mediated protein degradation.
- 70.Maniaci C, Hughes SJ, Testa A, et al. Homo-PROTACs: bivalent small molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun. 2017;8(1):830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steinebach C, Lindner S, Udeshi ND, et al. Homo-PROTACs for the chemical knockdown of cereblon. ACS Chem Biol. 2018;13 (9):2771–2782. [DOI] [PubMed] [Google Scholar]
- 72.Girardini M, Maniaci C, Hughes SJ, et al. cereblon versus VHL: Hijacking E3 ligases against each other using PROTACs. Bioorg Med Chem. 2019;pii: S0968-0896(19):30172–30175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steinebach C, Kehm H, Lindner S, et al. PROTAC-mediated crosstalk between E3 ligases. Chem Commun. 2019;55(12):1821–1824. [DOI] [PubMed] [Google Scholar]
- 74.Li Y, Yang J, Aguilar A, et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J Med Chem. 2019;62(2):448–466.•First report of an MDM2-CRBN-highly potent PROTAC.
- 75.Chu TT, Gao N, Li QQ, et al. Specific knockdown of endogenous Tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chem Biol. 2016;23(4):453–461. [DOI] [PubMed] [Google Scholar]
- 76.Lu M, Liu T, Jiao Q, et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur J Med Chem. 2018;146:251–259. [DOI] [PubMed] [Google Scholar]
- 77.Silva MC, Ferguson FM, Cai Q, et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. ELIFE. 2019;8:e45457.•First report of a PROTAC based on the structure of a PET tracer.
- 78.Kargbo RB. Treatment of Alzheimer’s by PROTAC-Tau protein degradation. ACS Med Chem Lett. 2019;10(5):699–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kenten JH, Roberts SF, (PROTEINEX INC). Controlling protein levels in eucaryotic organisms. US6306663B1 (1999). [Google Scholar]
- 80. a).http://arvinas.com/wp-content/uploads/2018/02/AR-GUASCO2018-final.pdf, Neklesa T, Snyder LB, Willard RR et al. ARV-110: an oral androgen receptor PROTAC degrader for prostate cancer. J Clin Oncol. 2019; 37(7)_suppl:259–259. DOI: 10.1200/JCO.2019.37.7_suppl.259.
- 81.Salami J, Alabi S, Willard RR, et al. Androgen receptor degradation by the proteolysis targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol. 2018;1:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan KH, Zengerle M, Testa A, et al. Impact of target warhead and linkage vector on inducing protein degradation: comparison of bromodomain an extra-terminal (BET) degraders derived from thiazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J Med Chem. 2018;61(2):504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McCoull W, Cheung T, Anderson E, et al. Development of a novel B-cell lymphoma 6 (BCL6) PROTAC to provide insight into small molecule targeting of BCL6. ACS Chem Biol. 2018;13(11):3131–3141. [DOI] [PubMed] [Google Scholar]
- 84.Cromm PM, Samarasinghe KTG, Hines J, et al. Addressing kinase-independent functions of Fak via PROTAC-mediated degradation. J Am Chem Soc. 2018;140(49):17019–17026. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L, Rilery-Gillis B, Vijay P, et al. Acquired resistance to BET-PROTACs (proteolysis targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther. 2019;pii: molcanther:1129. 2018. [DOI] [PubMed] [Google Scholar]
- 86.Chen Z, Picaud S, Filippakopoulos, et al. Structural basis for recruitment of DAPK1 to the KLHL20 E3 ligase. DOI: 10.1101/414250. [DOI] [PMC free article] [PubMed]
- 87.Sievers QL, Petzold G, Bunker RD, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362(6414):pii: eaat0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo J, Liu J, Wei W. Degrading proteins in animals: “PROTAC’tion goes in vivo. Cell Res. 2019;29:179–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nalawansha DA, Paiva S-L, Rafizadeh DN, et al. Targeted protein internalization and degradation by ENDosome Targeting Chimeras (ENDTACs). ACS Cent Sci. 2019; 5:1079–1084 article ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]