Abstract
Background and Objective
Compared with tissue biopsy, liquid biopsy is the most preferable non-invasive promising method in personalized medicine, although it has many limitations in isolating circulating tumor cells (CTC). Lung cancer associated mortality is drastically increased due to a shortfall of early-stage detection, which remains a challenge. Herein, we aimed to detect lung cancer at an early-stage using CellCollector device.
Methods
39,627 volunteers underwent low-dose computed tomography; 2508 cases with pulmonary nodules and 7080 with no pulmonary nodules were chosen. After follow-up, 24 patients were diagnosed with early-stage non-small cell lung cancer (NSCLC), and subjected to CTC detection using CellCollector, along with 72 healthy volunteers. Immunofluorescence staining for EpCAM/CKs and CD45 were performed for CTC validation.
Results
Fifteen out of twenty-four (stage I, n = 18; stage II, n = 6) early-stage lung cancer patients were found to be CTC-positive, whereas no CTC was found in the control group. Genetic mutation of TP53, ERBB2, PDGFRA, CFS1R and FGFR1 in the CTC revealed 71.6% of the mutation sites similar to the tumor tissues of 13 patients. Molecular characterization revealed higher expression of protein PD-LI in CTC (40%) as compared to tumor tissue (26.7%). Moreover, CTC clusters were detected in 40% of patients.
Conclusion
CTC detection using the CellCollector in early-stage NSCLC had a relative high capture rate. Moreover, CTC analysis is a prospective setting for molecular diagnostic in cases when tumor tissue biopsy is not desirable.
Keywords: CellCollector, in vivo detection, PD-L1, gene mutation, next generation sequencing
Introduction
Lung cancer is a major cause of cancer-related mortality worldwide. According to the GLOBOCAN survey in 2018, approximately 2.09 million new lung cancers reported and 1.76 million deaths occurred worldwide.1 Non-small cell lung cancer (NSCLC) comprising about 80–85% of all lung cancer cases and is a lethal disease with a five-year survival rate ranging from 50% in pathological stage I to 2% in stage IV.2,3 Currently, patients screening using low-dose computed tomography (LDCT) is considered as a promising approach that could increase the rate of early detection which contributes to the ultimate goal of reducing lung cancer mortality.3,4 However, LDCT can produce 20% or more false-positive results in baseline screens and 3% or more in subsequent screens. This is might be due to various disadvantages of LDCT screening, such as low sensitivity and specificity, which may increases patient-stress and misleads diagnosis.5 For lung cancer patients, surgical resection provides the best chance for diagnosis and cure. Tumor tissues for pathology are the gold standard for confirmed diagnosis for cancers. Beside pathological diagnosis, tumor biopsies could also provide additional gene mutation and immunohistochemistry information, but some reasons might cause the tumor tissues unavailable obtained. Hence, biomarkers suitable to search for improving diagnosis sensitivity and providing more gene and immunological information are urgently needed.6–9
Circulating tumor cell (CTC) is metastatic tumor cell that have emerged from the primary tumor site into the bloodstream to form secondary tumors at distinct sites.10 These cells present notable advantages in that they are a noninvasive biomarker and provide both diagnostic and prognostic information.11,12 Several in vitro approaches have been reported to detect CTC in the peripheral blood of cancer patients. However, currently used in vitro techniques have limitations, mainly due to low volumes of blood and millions of white blood cells in the background that limits the yield and sensitivity of using CTC.13 To increase the yield when capturing CTC, the GILUPI CellCollector®, a first innovative in vivo CTC isolation product in the world, has been used in clinical applications.5 This medical device, which was approved with a Conformité Européenne (CE) mark, for application in solid cancers and by the China Food and Drug Administration for breast cancer, offers the opportunity to capture and isolate CTC from the directly from the circulating blood.14 Numerous clinical application studies have demonstrated that this new strategy has high capture rates when used in advanced stage lung cancer and even could detect CTC in ground glass nodule patients.15,16 However, the use of CTC in early-stage lung cancer screening and molecular diagnosis has rarely been reported.
The sensitivity of target-based drug molecules is mainly related to the genetic mutation of individual tumors. Therefore, it is important to identify genetic mutations in tumor cells, which could provide appropriate treatment option for patients. Most genetic variations involving “driver mutations” have been identified in NSCLC. Approximately 50% of NSCLC harbor recurrent somatic oncogenic mutations in genes including EGFR, HER2, and KRAS. Moreover, immunotherapies targeted against programmed death ligand 1 (PD-L1) and its receptor programmed death protein-1 (PD-1) have improved survival in a subset of patients with advanced lung cancer. PD-L1 protein expression has emerged as a biomarker that predicts which patients are more likely to respond to immunotherapy.17 Generally, gene and immunological analyses are performed on tumor tissues, small biopsies or cytological samples obtained from the primary neoplastic site. It is well known that there are many controversies associated with these invasive detection methods, such as puncture sampling may not obtain tumor cells, patients are rarely subjected to re-biopsy, and the invasion of the sampling process has a potential risk of cancer metastasis.18 Coping with these challenges, minimally invasive technologies that capture the genomic contents of tumors in fluids have been combined with sensitive genotyping assays.19–21 Therefore, in this present study, we evaluate the potential role of CTC as an alternative to tissue biopsy.
So far, due to the limitations of in vitro detection methods, CTC has been rarely studied in early-stage lung cancer. Here, we analyzed 24 patients with early-stage NSCLC and 72 individuals with no pulmonary nodules matched with lung cancer group were included in the further analysis to evaluate the use of CTC detection rate and to assist early-stage lung cancer molecular diagnosis. This is the first study aimed to do molecular characterization of CTC focusing the PD-L1 expression and gene mutational sequencing of CTC isolated using CellCollector and tumor tissues obtained from CTC-positive patients of early-stage lung cancer.
Materials and Methods
Study Design
This was a mono-centric population-based screening program for lung cancer patients who underwent LDCT in the Fourth Hospital of Hebei Medical University during the period of Jan 2014 to Mar 2017. The study included 9528 asymptomatic individuals (aged 40–75 years old and resided in heavy air pollution area) from a large population, including 39,627 individuals in Hebei Province. All asymptomatic individuals (9528) were screened for lung cancer detection using LDCT. A total of 2508 subjects were diagnosed with pulmonary nodules. All these individuals were monitored from the date of enrollment to September 2017. Eventually, 29 of the subjects with pulmonary nodules had been diagnosed with suspected lung cancer. Excluding 3 patients who underwent surgical resection in other hospitals, the other 26 of the suspected lung cancer patients were treated in the Fourth Hospital of Hebei Medical University. Before surgery, 26 patients were undergone CTC detection, and tumor tissues were collected from the CTC-positive patients and performed pathological analysis. All isolated CTC and tumor tissue obtained from the CTC-positive patients were analyzed using next-generation sequencing (NSG) and PD-L1 expression to explore the consistency of its expression across patients. The ratio of suspected lung cancer group and healthy control group was 1:3. Seventy-two individuals from the 7020 no pulmonary nodule population were served as control group and matched with lung cancer group for sex, age, family history, smoking status of cancer and other possible risk factors in the study. All the 72 control individuals were subjected to CTC detection. This study was performed in accordance with the Declaration of Helsinki. Approval was obtained from the Forth Hospital of Hebei Medical University ethics committee (Shijiazhuang, Hebei, China), and written informed consent was obtained from all the patients (Figure 1).
Figure 1.
Schematic overview of the procedure for molecular analysis of CTC and tumor tissue.
In vivo Application of the CellCollector
The CellCollector (GILUPI GmbH, Potsdam, Germany) is an innovative medical device captures CTC in vivo using EpCAM antibodies. The CellCollector is made of medical stainless steel with a polycarboxylate/hydrogel which is coated on a gold-plated layer coating on one end. This coating (functional area) is 2 cm long, adhered to the coating is an EpCAM antibody used to separate circulating epithelial cell adhesion molecule-positive cells. This antibody is normally used to detect CTC in humans.14
Before using this device, the functional area (0.5 mm diameter) of the device was inserted into the peripheral venous blood through a 20G indwelling needle cannula. Following insertion, the functional field of CellCollector was exposed to 1.5 L blood in the arm vein of patients for 30 min, which increases the chance to capture CTC.14,22
CTC Identification and PD-L1 Expression Analysis
For CTC identification, the CellCollector after its usage was gently washed in washing buffer and then incubated with 2% (w/v in PBS) bovine serum albumin (BSA) for 30 min at room temperature. After incubation, captured CTC was detected by immunocytochemical staining for EpCAM or cytokeratins 8, 18, and 19. CTC attached to the wire were then probed with FITC-conjugated mouse monoclonal antibody directed to EpCAM (Acris, clone HEA125-FITC) and an APC-conjugated rabbit antibody raised against CD45 (Exbio, clone MEM-28-Alexa647). Finally, CTC was counterstained with Hoechst33342 (Sigma). The fluorescence intensity of the stained CTC was viewed and captured using an Axio A1m microscope equipped with AxioCam digital camera system and the AxioVision 4.6 software (Zeiss, Jena, Germany). EpCAM/cytokeratin-positive cells should had additional morphological features such as extensive cell body (diameter 10–50 μm), asymmetric nucleus, and a high imbalance in the nuclear to cytoplasmic ratio. Finally, cells were counted on each CellCollector by an operator blinded to the clinical information of the patients. For CTC PD-L1 identification, PE was coated to PD-L1 antibody to observe PD-L1 (clone 28-8, ab205921, Abcam) at orange channel by fluorescence microscope. The staining protocol for CTC identification was performed referring the previously published method.15
Tumor PD-L1 expression was measured by PD-L1 immunohistochemical (IHC) 22C3 pharmDx (Dako, Inc.). Tumor cells with negative or positive PD-L1 expression were interpreted as a Tumor proportion score (TPS). PD-L1 expression at <1% tumor cells as negative, that is, the percentage of tumor cells with partial or complete cell membrane staining was less than 1%. PD-L1 positivity was defined as the percentage of tumor cells with partial or complete cell membrane staining was more than 1%, among which TPS ≥50% was judged as high expression of PD-L1.
CTC Separation and Whole Genomic Amplification
After identification of CTC with IF staining, the positive cells were relocated and cut with a special cutter under microscope. The cells and CellCollector fragment together were collected into an EP tube to lyses cells and amplify genomic DNA. The whole genomic amplification was performed following the instructions of commercial WGA kit (REPLI-g Single Cell Kit (Qiagen)). Briefly, Buffer D2 was prepared. The cell sample was added to 4 μL Mg2+-free and Ca2+-free PBS followed by the addition of 3μL D2 buffer. The content in the tube were mixed gently by flicking the tube and then centrifuged. Then incubated in a thermal cycler at 65°C for 10 min. Additionally, 3 μL Stop Solution was added, mixed and stored on ice. Then, 40 µL of master mix was added to each prepared 10 µL DNA sample and mixed. Finally incubated at 30°C for 6 h, and then 65°C for 3 min to inactivate REPLI-g SC DNA polymerase.
Library Preparation
Totally 50 ng of genomic DNA (Nanodrop concentration as the standard) was used to construct sequencing libraries using the Ion Ampliseq Library Kit 2.0 (ThermoFisher) and Ion Ampliseq Cancer Hotspot Panel v2 in accordance with the instructions of kit. The library quality was determined according to the Agilent2100 and Agilent DNA High Sensitivity Kit (Agilent Technologies), and the qualified library fragments ranged from 100 bp to 300 bp; Kapa Library Quantification Kit (Roche) was performed to accurately quantify the molar concentration of library, which was not less than 1000 pmol/L. The qualified libraries were used for subsequent sequencing.
Next-Generation Sequencing
Qualified libraries were diluted to 100 pmol/L with nuclease-free water in accurate quantitative concentration by fluorescence quantitative PCR method, and then the samples were mixed in line with the data volume of 2.2 M reads of each sample. Then, 10 µL of the mixed library was taken for template preparation via Ion PI Hi-Q OT2 200 Kit (ThermoFisher) and Ion One Touch2 system. Next, the template quality control was carried out by Ion Sphere quality control kit and Qubit 3.0 (ThermoFisher), and ISPs% of 10–30% was qualified. After enrichment on Ion One Touch ES system, qualified templates were sequenced on Ion Torrent Proton and Ion PI chip using Ion PI Hi-Q sequencing 200 Kit (ThermoFisher). For panel NGS analysis, the Life proton system was used.
Statistical Analysis
Differences between the healthy control and NSCLC patients group were determined by Student’s t test and Pearson’s chi-square test or Fisher’s exact test as appropriate using SPSS 21.0 software. All figures were constructed using GraphPad Prism 5.0 software (GraphPad, San Diego, CA, USA). P value <0.05 was considered to be statistically significant.
Results
Characteristics of the Study Individuals
Twenty-four subjects were diagnosed with lung cancer (stage I, n = 18; stage II, n = 6). In the lung cancer group, the mean age was 61.4 years old, the smoking rate was 45.8% and 20.8% of the patients had a family history of cancer. Histological characterization of the tumor exhibited squamous cell carcinoma (n = 6) and adenocarcinoma (n = 18). No patients were staged for distant overt metastases and local tumor progression (T stage), and all patients showed T1 or T2 tumors (Table S1). Moreover, the 72 healthy subjects matched with the lung cancer group included 42 males and 30 females. There was no significant difference in any of these values including sex, age, smoking status and family history of cancer between the lung cancer patients and the control group (Figure 1).
Overview of CellCollector and CTC Identification
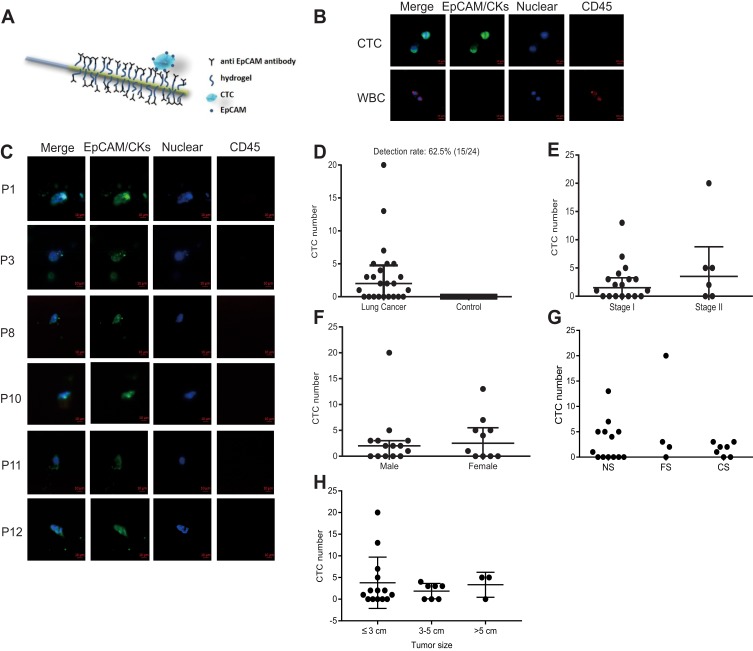
The CellCollector was inserted into the arm vein of each patient. During 30 min application time, EpCAM-positive cancer cells were captured by anti-EpCAM which were covalently bound to the functional domain of the CellCollector (Figure 2A). The isolated tumor cells were stained for EpCAM and/or keratins, and then counterstained with nuclear specific Hoechst-33342. CD45 staining was used to identify false-positive events (white blood cells). EpCAM/CKs-positive, nuclear positive and CD45-negative cells were identified as CTC, and EpCAM/CKs-negative, nuclear positive and CD45-positive cells were identified as leukocytes. Tumor cells were identified as EpCAM- and/or pan-keratin-positive (green) and CD45-negative (red). The representative images of CTC and WBC were shown in Figure 2B. The representative of CTC that captured from enrolled patients were shown in Figure 2C.
Figure 2.
Mechanism of CTC isolation and detection CellCollector. (A) Pictorial representation of the CellCollector wire and its application. (B) Representative pictures of WBCs and CTC (bar = 10 μm). EpCAM antibodies coated wire facilitates binding of EpCAM positive cells when exposed into the arm vein of cancer patients. (C) Identification of EpCAM/CKs positive CTC from patients. Cancer cells were observed for EpCAM- and/or pan-keratin-positive (green) and CD45-negative (red) events. Nuclear counterstaining (blue) was performed by Hoechst33342 (bar = 10 μm). (D) The CTC detection rate in lung cancer patients and healthy control. (E) The CTC detection in stage I and stage II. (F) The CTC detection in male and female. (G) The CTC detection in person with differential smoking history [NS, never smoker; FS, former smoker; CS, current smoker]. (H) The CTC detection rate in three types of tumor size: ≤3 cm, 3–5 cm and >5 cm.
CTC Counts and Correlation to Clinical and Pathological Characteristics
To evaluate clinical correlation between CTC counts and clinical parameters, CTC detection rate and CTC counts were used. In total, 62.5% (15/24) of the patients were positive for ≥1 CTC (range, 1–20 cells). Interestingly, no CTC-like events were observed in the control group (Figure 2D). More importantly, no adverse events were occurred in all subjects. CTC detection rates were 61.1% (11/18) and 66.7% (4/6) in stage I and stage II, respectively (P = 0.603) (Figure 2E). Nine male patients and six female patients were detected as CTC positive, with the detection rates of 64.3% (9/14) and 60.0% (6/10), there was no significant difference in detection rate for gender (P = 0.582) as shown in Figure 2F. CTC detection rates were 53.8% (7/13), 75.0% (3/4) and 71.4% (5/7) in the three groups of never smoker, former smoker and current smoker, respectively (P = 0.625) (Figure 2G). Then, patients were divided into 3 categories according to their tumor size: ≤3 cm, 3–5 cm and >5 cm, with the detection rate of 64.3% (9/14), 57.1% (4/7) and 66.7% (2/3), respectively, and no significant difference in detection rate for tumor size was noticed (P>0.05) (Figure 2H). Detailed clinical information and CTC detection of patients were shown in Table S1.
PD-L1 Expression Analysis in Tumor Tissue, CTC and CTC Cluster
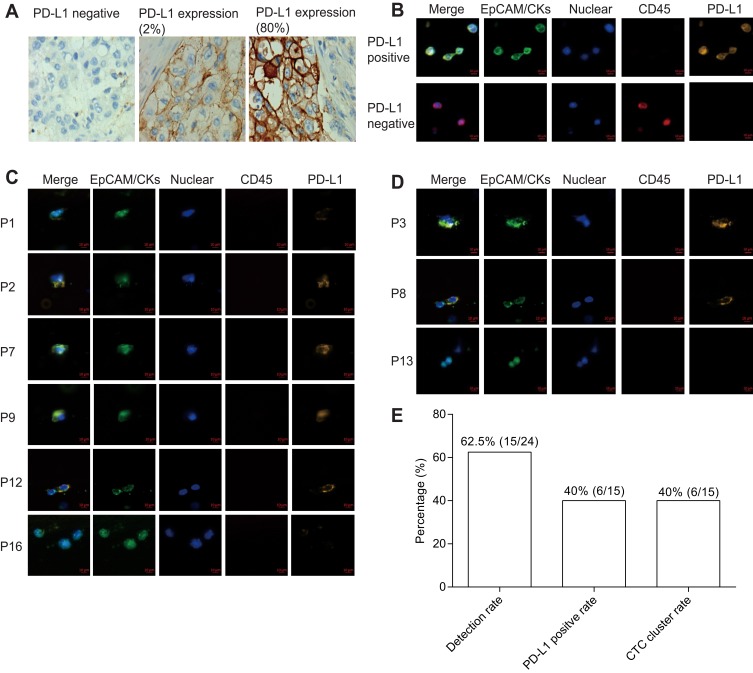
PD-L1 expression in tumor tissue was analyzed by immunohistochemistry. PD-L1 negative expression, weak expression and strong expression of tumor tissue are shown in Figure 3A. SK-BR-3 and NK92 cell lines were used as PD-L1 positive and negative cells, respectively (Figure 3C). In total, six patients were found as CTC PD-L1 positive (Figure 3B). Forty percent patients were detected ≥1 PD-L1+ CTC and the PD-L1 positive expression rate was 26.7% in tumor tissue. Thus, the PD-L1 status of CTC did not correlate with the tumor tissue PD-L1 (P = 0.439). In total, six patients (6/15) were detected CTC clusters in this study (Figure 3D and E).
Figure 3.
PD-L1 expression and CTC cluster analysis. (A) PD-L1 expression in tumor tissue. (B) PD-L1 expression in CTC of cancer patients (bar = 10 μm). (C) Immuno-staining differentiates PD-L1 expression in positive and negative control (bar = 10 μm). (D) Represent CTC cluster in patients. (E) Detection rate, CTC cluster rate and PD-L1 positive rate (≥1 PD-L1+ CTC) of lung cancer patients.
Comparison of Gene Mutation Between CTC and Tumor Tissue
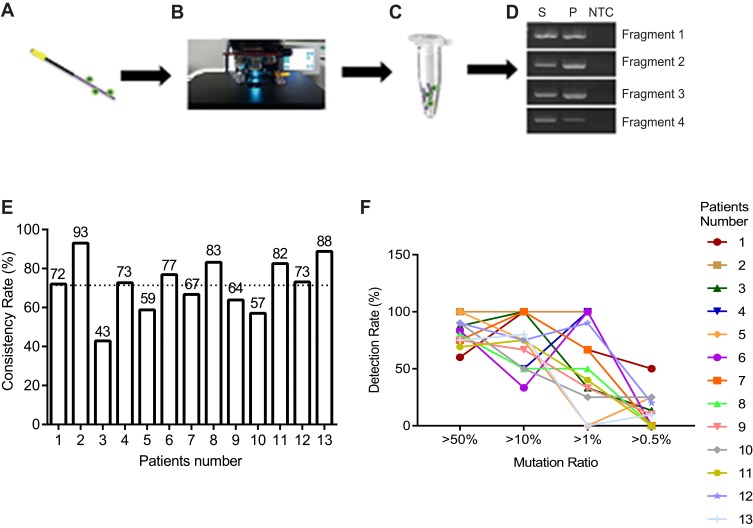
Next, we wanted to determine the genetic information of captured CTC. To test this, we performed gene mutations analysis in CTC using high-throughput NGS methods. Cells isolated with the CellCollector were observed microscopically by means of immunofluorescence staining according to the staining procedure. The functional domain of the CellCollector was fragmented and whole genomic amplification (WGA) was performed. The quality of WGA DNA was analyzed using PCR assay with four primer pairs (Figure 4A–D).
Figure 4.
Genomic mutation analysis between CTC and tumor tissue using NGS. (A–D) Flow chart of CTC genomic DNA extraction. (E) Consistency of mutation between tumor tissue and CTC, mean value was shown as dotted line. (F) Detection rate of mutation in CTC showing different mutation load in tumor tissue.
Gene mutations were analyzed with Hotspot Panel v2 in 13 lung cancer patients’ CTC and tumor tissue. In total, consistency of gene mutation between CTC and tumor tissue was from 43% to 93% (mean 71.6%, SD 13.7) (Figure 4E). Moreover, we analyzed different mutation rate in tumor tissue and CTC. Interestingly, we found that high mutation rates in tumor tissue could be easily found in CTC compared with those with low mutation rates (Figure 4F).
Biofunction of Gene Mutations in the CTC Genome
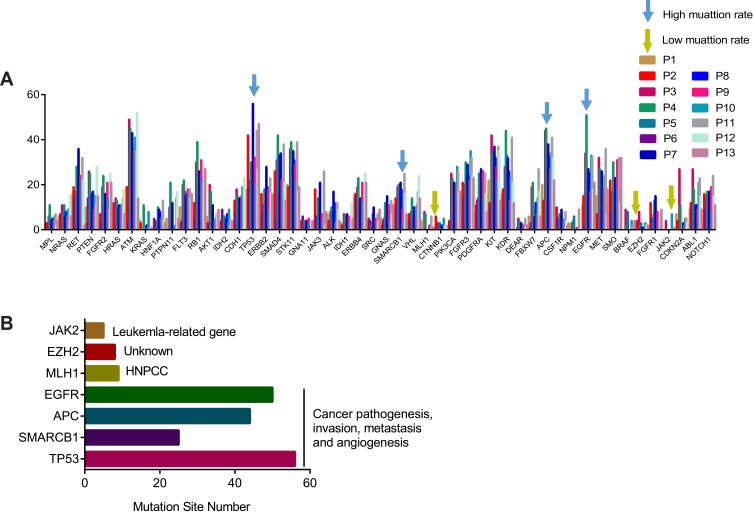
Genomic mutations were analyzed in 13 subjects for understanding the mutational profiles in CTC. Although all 50 analyzed genes were found exist mutation sites, the number of mutation site was different in those genes. However, CTC genomes exhibited similar mutational profiles (Figure 5). All the mutated genes were shown in Table 1. Additionally, we selected seven genes, including three low mutational genes and four high mutational genes to identify the biofunctions and clinical significance of those genes (Figure 6A). The low mutational genes included JAK2, EZH2 and MLH1, were non-lung cancer-related biofunctions. However, the high mutational genes, including EGFR, APC, SMARCB1 and TP53, played roles in cancer pathogenesis, invasion, metastasis and angiogenesis (Figure 6B). Although many patients are found to have EGFR mutations, the clinical significance of many types of mutations is not clear. Moreover, five of the mutation sites, including TP53, ERBB2, PDGFRA, CFS1R and FGFR1, were diagnostic markers that were found in all the tested samples.
Figure 5.
CTC genomic mutation analysis of 13 patients for 50 genes’ panel. The mutation sites number for each gene identified by second-generation sequencing in 13 patients (each bar represented one gene for an individual patient).
Table 1.
Mutation Sites of 50 Genes from 13 Patients
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPL | 2 | 3 | 0 | 6 | 11 | 5 | 4 | 5 | 4 | 6 | 7 | 2 | 4 |
| NRAS | 6 | 7 | 0 | 11 | 7 | 11 | 6 | 8 | 6 | 9 | 4 | 15 | 6 |
| RET | 16 | 19 | 13 | 17 | 28 | 16 | 36 | 15 | 24 | 22 | 32 | 22 | 2 |
| PTEN | 10 | 3 | 14 | 26 | 25 | 5 | 16 | 17 | 9 | 15 | 11 | 28 | 15 |
| FGFR2 | 6 | 7 | 3 | 19 | 24 | 11 | 16 | 10 | 9 | 15 | 9 | 25 | 21 |
| HRAS | 10 | 12 | 0 | 14 | 13 | 11 | 10 | 11 | 10 | 11 | 14 | 18 | 7 |
| ATM | 19 | 19 | 3 | 49 | 45 | 7 | 43 | 32 | 36 | 41 | 35 | 52 | 14 |
| KRAS | 4 | 3 | 0 | 0 | 11 | 0 | 0 | 2 | 1 | 8 | 0 | 1 | 1 |
| HNF1A | 0 | 5 | 4 | 1 | 4 | 10 | 9 | 8 | 5 | 5 | 13 | 4 | 3 |
| PTPN11 | 5 | 0 | 0 | 10 | 21 | 5 | 12 | 0 | 1 | 2 | 14 | 17 | 5 |
| FLT3 | 10 | 3 | 4 | 20 | 22 | 17 | 15 | 16 | 18 | 15 | 18 | 14 | 16 |
| RB1 | 6 | 12 | 5 | 30 | 39 | 14 | 26 | 23 | 31 | 22 | 21 | 22 | 27 |
| AKT1 | 6 | 3 | 3 | 20 | 17 | 0 | 11 | 4 | 4 | 5 | 7 | 8 | 9 |
| IDH2 | 3 | 4 | 9 | 3 | 6 | 3 | 5 | 7 | 3 | 9 | 2 | 5 | 4 |
| CDH1 | 2 | 13 | 7 | 18 | 15 | 6 | 14 | 11 | 15 | 19 | 19 | 23 | 0 |
| TP53 | 18 | 42 | 18 | 22 | 30 | 28 | 56 | 32 | 32 | 20 | 44 | 27 | 47 |
| ERBB2 | 6 | 16 | 8 | 11 | 18 | 13 | 28 | 19 | 18 | 9 | 23 | 16 | 16 |
| SMAD4 | 13 | 26 | 5 | 31 | 42 | 15 | 33 | 34 | 22 | 19 | 38 | 21 | 13 |
| STK11 | 20 | 19 | 0 | 36 | 39 | 28 | 35 | 31 | 29 | 21 | 39 | 17 | 19 |
| GNA11 | 4 | 4 | 6 | 4 | 4 | 2 | 4 | 4 | 5 | 2 | 0 | 4 | 4 |
| JAK3 | 1 | 18 | 6 | 3 | 14 | 14 | 21 | 5 | 7 | 4 | 26 | 2 | 8 |
| ALK | 7 | 4 | 0 | 8 | 10 | 7 | 17 | 12 | 3 | 5 | 12 | 9 | 4 |
| IDH1 | 3 | 2 | 0 | 7 | 6 | 0 | 7 | 0 | 6 | 3 | 5 | 12 | 5 |
| ERBB4 | 15 | 16 | 15 | 19 | 23 | 9 | 14 | 14 | 21 | 12 | 16 | 25 | 21 |
| SRC | 1 | 5 | 4 | 2 | 3 | 7 | 10 | 2 | 5 | 1 | 7 | 4 | 9 |
| GNAS | 5 | 2 | 4 | 5 | 12 | 6 | 15 | 6 | 11 | 4 | 13 | 7 | 11 |
| SMARCB1 | 6 | 14 | 10 | 19 | 20 | 16 | 21 | 18 | 9 | 4 | 25 | 16 | 7 |
| VHL | 3 | 7 | 4 | 8 | 14 | 6 | 10 | 5 | 5 | 11 | 17 | 24 | 14 |
| MLH1 | 4 | 0 | 0 | 8 | 7 | 0 | 0 | 0 | 2 | 0 | 6 | 0 | 1 |
| CTNNB1 | 1 | 6 | 0 | 3 | 0 | 3 | 0 | 2 | 0 | 5 | 2 | 2 | 0 |
| PIK3CA | 2 | 4 | 3 | 25 | 23 | 7 | 21 | 17 | 20 | 28 | 26 | 17 | 17 |
| FGFR3 | 8 | 21 | 14 | 20 | 30 | 24 | 29 | 24 | 22 | 35 | 32 | 24 | 23 |
| PDGFRA | 7 | 11 | 13 | 14 | 25 | 13 | 27 | 11 | 26 | 14 | 25 | 16 | 8 |
| KIT | 22 | 12 | 10 | 42 | 34 | 18 | 37 | 32 | 28 | 22 | 37 | 35 | 13 |
| KDR | 16 | 18 | 9 | 28 | 44 | 33 | 32 | 26 | 13 | 23 | 41 | 21 | 12 |
| DEAR | 0 | 0 | 1 | 5 | 4 | 3 | 3 | 0 | 2 | 1 | 5 | 1 | 2 |
| FBXW7 | 6 | 1 | 3 | 19 | 21 | 4 | 6 | 12 | 7 | 14 | 27 | 17 | 6 |
| APC | 20 | 13 | 0 | 44 | 45 | 12 | 38 | 32 | 27 | 34 | 41 | 32 | 22 |
| CSF1R | 2 | 10 | 5 | 1 | 7 | 6 | 9 | 4 | 4 | 5 | 8 | 4 | 2 |
| NPM1 | 3 | 0 | 0 | 3 | 4 | 6 | 0 | 0 | 1 | 0 | 9 | 0 | 0 |
| EGFR | 11 | 15 | 3 | 34 | 51 | 22 | 27 | 25 | 17 | 33 | 32 | 18 | 21 |
| MET | 15 | 7 | 0 | 32 | 27 | 9 | 26 | 11 | 18 | 24 | 36 | 7 | 18 |
| SMO | 10 | 22 | 7 | 17 | 30 | 16 | 23 | 20 | 31 | 18 | 32 | 12 | 32 |
| BRAF | 0 | 0 | 0 | 9 | 2 | 8 | 0 | 0 | 0 | 4 | 2 | 0 | 4 |
| EZH2 | 2 | 4 | 0 | 8 | 2 | 3 | 1 | 1 | 0 | 3 | 3 | 2 | 2 |
| FGFR1 | 1 | 12 | 5 | 3 | 7 | 13 | 15 | 4 | 8 | 1 | 5 | 7 | 9 |
| JAK2 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 1 |
| CDKN2A | 7 | 4 | 0 | 27 | 11 | 0 | 0 | 3 | 2 | 5 | 5 | 3 | 0 |
| ABL1 | 9 | 12 | 9 | 27 | 18 | 5 | 11 | 11 | 9 | 21 | 23 | 15 | 9 |
| NOTCH1 | 5 | 16 | 9 | 15 | 17 | 14 | 17 | 5 | 19 | 8 | 24 | 15 | 11 |
Figure 6.
Functional analysis of gene mutation in CTC genome. (A) Mutation rate of cancer-driven genes in subjects, blue arrow indicates high mutation rate, yellow arrow indicates low mutation rate. Different color indicates individual subject. (B) Gene annotation of indicated genes including three low mutational genes (JAK2, EZH2, MLH1) and four high mutational genes (EGFR, APC, SMARCB1, TP53).
Discussion
CTC, also known as “liquid biopsy” is used as a surrogate biomarker in several cancers, including lung cancer.8 CTC analysis is a simple noninvasive method causing lesser pain or discomfort than tissue biopsy.23 Although CTC has been extensively studied in stage III and IV lung cancers using in vitro isolation methods, it has rarely been reported in early-stage lung cancer.16 Most of the patients with lung cancer are diagnosed at later-stages with the 5-year survival rate of 18% or less.24 This drastically decreased 5-year survival statistics associated with advanced stages of lung cancer demands early-stage diagnosis to improve patient survival and clinical outcomes. In order to explore the early-stage diagnosis strategy, CTC isolation was performed using the world’s first in vivo CTC isolation product the CellCollector. Exposing this device to approximately 1.5 L of peripheral blood, showed a higher capture rate than the other in vitro assay16 and also has a high detection rate in the early-stages of cancer.
Out of 26 NSCLC patients analyzed for CTC, 24 patients were diagnosed with stage I/II after surgery. Among these patients, 15 were found to be CTC-positive, resulting in a detection rate of 62.5%, which is higher than the detection rate of the FDA-cleared CellSearch system (30–40% detection in advanced lung cancer),16 and there were no false-positive CTC findings in the healthy group and in the two benign patients. The CTC-positive rate detected by CellCollector in advanced stage patients was 73%, and no CTC-like cells were found in the healthy control as reported in our previous study.15 This demonstrates that CellCollector possesses high sensitivity and specificity. To our best knowledge, CTC invasion is one of the typical factors resulting in distant metastasis of malignant tumors.25 Invasion and migration of tumor cells can be induced by changes in adhesion affinity, which causes increased survival rate of CTC in peripheral blood and easier access to other tissues and organs, leading to establishment of metastatic lesions.26 As mentioned above, this in vivo technique was highly sensitive to CTC in patients with either stage I or stage II lung cancer, so the presence of CTC in the blood might therefore serve as a surrogate marker for the purpose of prognosis and diagnosis. Meanwhile, a study conducted by Xie et al have also indicated that the early diagnosis of metastasis and recurrence of lung cancer can be monitored via the changes of CTC before and after surgery or chemotherapy, which is helpful to improve the follow-up treatment regimen and thus enhance the survival rate.27
Molecularly characterization of CTC is important to correlate gene mutation with tumor progression and to identify therapeutic targets for personalized therapies. A new research that assessed the degree of genomic similarity between pulmonary venous CTC (PV-CTC) in surgical resection, primary tumor and metastatic tumor has reported that CTC as a micrometastase is associated with tumor recurrence.28 In our study, comparing gene mutation status between tumor tissues and CTC revealed that approximately 71.6% of the mutation sites were similar in tumor tissues and CTC, indicating that CTC DNA assay could act as a supplemental method for analyzing gene mutation status. The high mutation rate observed in tumor tissue as well as rare mutations that are difficult to identify in tumor tissues could easily be identified in CTC genomic DNA. Besides, in a clinical research conducted by Gorges et al, mutations of KRAS and EGFR genes in CTC-positive cells of two lung cancer patients who had captured CTC using CellCollector were detected by Whole genome amplification and digital PCR technology, which were confirmed in primary tumors.16 Thus, molecular characterization of CTC mimics tumor tissue in terms of genetic alterations and enable real-time monitoring of gene mutations for targeted therapies.
Moreover, we also found several mutation sites that commonly exist in all tested samples, including TP53, ERBB2, PDGFRA, CFS1R and FGFR1. TP53 is a tumor-inhibitor gene that always present in early-stage cancer. Although in-frame insertion mutations in exon 20 of ERBB2 were previously identified in approximately 2% of NSCLC,29 we observed other ERBB2 mutation sites as potential markers due to their occurrence in all the CTC genomic DNA samples. PDGF/PDFGRA signaling has shown to play a vital role in tumor cell proliferation and progression,30 whereas a functional polymorphism in the CSF1R gene was identified as a novel susceptibility marker for lung cancer in never-smoking females,31 Amplification of FGFR1, a receptor tyrosine kinase that binds ligands belonging to the fibroblast growth factor family. The amplification of FGFR1 has been extensively documented in lung cancer for its effects on drug resistance and prognosis.32,33 Although the mutation sites of these genes were found in all the samples, the biofunctions of those mutations remain unclear. The clinical value of these mutations requires a study that uses a large number of samples to demonstrate the sensitivity and specificity of these markers for early-stage lung cancer. They might represent potential markers for cancer screening and individual therapy in the future. The mutation profiles of 13 patients were analyzed in this study and the number of mutation sites in these cancer-driver genes was calculated to build mutational profiles. All the 13 subjects showed a similar mutational profile indicating that the mutation of these cancer-driver genes represents a specific mutational bias. Moreover, we further analyzed the biofunctions of genes with different mutation rates to evaluate their biological significance. Our study showed that genes with a low mutation rate had no connection to lung carcinogenesis, whereas genes that showed high mutation rates were involved in cancer pathogenesis, invasion, metastasis and angiogenesis. Since all these processes are related to CTC hematogenous metastasis, our findings suggest that profiling CTC gene mutation is a potential biomarker for screening individuals at their early-stage of lung cancer and also for monitoring cancer metastasis and invasion.
CTC clusters could be considered as a crucial factor in the prognosis and diagnosis of metastatic process. However, coexistence of CTC cluster with single CTC in the blood stream of cancer patients warrants a superior method over the existing one to specifically isolate CTC clusters for sensitive metastasis detection.34,35 Generally, transiting of CTC cluster through narrow blood vessels is made possible by the cleavage of intercellular adhesions that result in reversible unfolding of clusters into single file chains. This unfolding process is rapid and reversible that enables the CTC to transit successfully as an individual entity instead of cohesive resistive units, and this significantly reduces their overall resistance to flow.36 Aceto et al have indicated that CTC clusters are held together by intercellular adhesion dependent on plakoglobin, which greatly promotes the metastasis of cancer.34 Liu et al have reported that CTC clusters express CD44, and CD44 can mediate cancer cell aggregation, leading to enhanced tumorigenic fitness in different contexts. Moreover, they have also indicated that CD44-CD44 signaling in CTC clusters can augment PAK2 activation, which in turn supports cancer cell stemness and metastatic fitness. Taken together, the high frequency of CTC clusters may have been an effect of CD44-mediated binding and enhanced PAK2 activation.37,38 Previous clinical study has suggested that EpCAM positive CTCs are an independent prognostic factor. Wang et al have showed that CTC clusters were detected in 41.7% (5/12) of CTC-positive samples using anti-EpCAM microbubbles and anti-EpCAM/EGFR microbubbles. Therefore, EpCAM plays a role in the high frequency of CTC clusters.39 Several methods have shown to be capable of capturing CTC clusters, including size-based isolation, microarrays, optical-imaging techniques and negative isolation methods.35,40,41 Here, we used an in vivo detection method to capture CTC clusters and found that these CTC clusters were present in 40% of the detected patients which is in consistent with the previous reports.42 Unfolding the clinical relationship between CTC clusters and prognosis and improvisation of this in vivo isolation technique will ensure new avenues in the clinical application of CTC clusters for early-stage diagnosis of lung cancer.
PD-1 and PD-L1 is a pair of immune co-inhibitory molecules that are over expressed in a variety of cancers and mediates tumor immune suppression by inhibiting T cell cytotoxicity.42,43 Recently, FDA has approved drugs targeting PD-1 or PD-L1 and are being used in the treatment of relapsed/refractory melanoma, bladder cancer and NSCLC.44–46 PD-1/PD-L1 inhibitors block the negative regulatory signal by inhibiting the binding of PD-1 to PD-L1, resulting in T cell recovery activity, thereby enhancing immune response. However, the percentage of PD-L1 positive patient respond to PD-L1 inhibitor treatment is reported to be less than 50%. Hence it is necessary to screen the appropriate population of tumor PD-1/PD-L1 immunotherapy, use it in high-selection population, carry out personalized treatment, and vigorously promote precision medicine.47 Isolation of tumor tissue itself is an invasive procedure that poses risk to the patient, as well as a technically challenging procedure. Further heterogeneity of tumor and the dynamic status of PD-L1 expression which modulate in response to various factors especially the interferon levels limit the analysis of PDL-1 expression in tumor tissue taken at one time point. These limitations could be overcome by the non-invasive procedure of isolating CTC from peripheral blood samples that could improve the identification of molecular diagnostic biomarkers and monitoring treatment response. While tissue biopsy provides information about PD-L1 expression at a given time, and site and liquid biopsy provide the whole molecular landscape of tumors. By virtue of its dynamic and non-invasive nature PD-L1 expression in CTC is preferred over tumor biopsy and has been investigated and reported in NSCLC.48–50 In line with the recent findings51 the present study also demonstrates that the liquid biopsy showed higher rate of PD-L1 positivity than tumor biopsy in NSCLC patients.
The outcome of this investigation hypothesized that higher levels of PD-L1 expression observed in CTC might be a reflection of tumor immunosuppression. Detection of PD-L1 expression in CTC of patients whose PD-L1 was negative in tissue biopsy highlights the sensitivity of the assay method. Further, the discrepancies in the level of PD-L1 expression that exist between CTC and tumor biopsy is of paramount importance as it holds promise for an improved diagnostic approach in future. Our data provide a basis for the development of CTC PD-L1 expression as a molecular biomarker for the diagnosis of early-stage NSCLC and warrant an extensive investigation to develop CTC PD-L1 as a marker to measure prognostic, diagnostic and treatment outcome in lung cancer patients.
There are several limitations in this study. Although the outcome of this study provided a valuable information on PD-L1 expression with respect to NSCLC diagnosis, inclusion of small sample size questions the wide applicability of this diagnostic tool. Hence, an in-depth investigation enrolling larger patient population is required to study the molecular characterization of CTC in NSCLC diagnosis with a proper follow-up procedure. Besides, postoperative CTC detection will be conducted in the future to evaluate the number and detection rate of CTC after treatment.
Conclusion
The data described here show that CTC detection using CellCollector in early-stage lung cancer had a relative high capture rate and the CTC could represent promising strategy for lung cancer molecular diagnosis. By comparing gene mutation status between tumor tissues and CTC, we provide a critical clue in support of the clinical significance of liquid biopsy as it provides complementary information about the tumor that might enable clinicians to obtain important diagnostic information. More importantly, in this study, we confirmed that CTC analysis is a prospective setting for molecular diagnostic in cases when tumor tissue is not desirable. In summary, CellCollector is indeed a foolproof device with high sensitivity to isolate CTC where tissue biopsy is not desirable and has a high detection rate in early-stage NSCLC.
Acknowledgments
This work was supported by the Financial Department of Hebei Province (No. [2016]361006 and No. 2017043367-2).
Author Contributions
All authors have made contributions to the concept and design, data acquisition and analysis, and interpretation of the study, as well as drafting and revision of the manuscript. All authors have approved the final manuscript and have agreed to be responsible for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Rabe KF, Longo DL. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377(9):849–861. doi: 10.1056/NEJMra1703413 [DOI] [PubMed] [Google Scholar]
- 3.Shlomi D, Ben-Avi R, Balmor GR, Onn A, Peled N. Screening for lung cancer: time for large-scale screening by chest computed tomography. Eur Respir J. 2014;44(1):217–238. doi: 10.1183/09031936.00164513 [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Qian GS, Bai CX. Lung Cancer Study Group of Chinese Thoracic S, Chinese Alliance Against Lung Cancer Expert G. Chinese consensus on early diagnosis of primary lung cancer (2014 version). Cancer. 2015;121(Suppl 17):3157–3164. doi: 10.1002/cncr.29571 [DOI] [PubMed] [Google Scholar]
- 5.Tammemagi MC, Lam S. Screening for lung cancer using low dose computed tomography. BMJ. 2014;348:g2253. doi: 10.1136/bmj.g2253 [DOI] [PubMed] [Google Scholar]
- 6.He Y, Zhang X, Wang L, et al. Detection of cancer specific mutations in early-stage non-small cell lung cancer using cell-free DNA by targeted sequencing. Int J Oncol. 2016;49(6):2351–2358. doi: 10.3892/ijo.2016.3731 [DOI] [PubMed] [Google Scholar]
- 7.Birse CE, Lagier RJ, FitzHugh W, et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clin Proteomics. 2015;12(1):18. doi: 10.1186/s12014-015-9090-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matikas A, Syrigos KN, Agelaki S. Circulating biomarkers in non-small-cell lung cancer: current status and future challenges. Clin Lung Cancer. 2016;17(6):507–516. doi: 10.1016/j.cllc.2016.05.021 [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt KE, Feinglass JM, DeCamp MM, Bilimoria KY, Odell DD. Treatment trends in early-stage lung cancer in the United States, 2004 to 2013: a time-trend analysis of the National Cancer Data Base. J Thorac Cardiovasc Surg. 2018;156(3):1233–1246 e1231. doi: 10.1016/j.jtcvs.2018.03.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiorelli A, Accardo M, Carelli E, Angioletti D, Santini M, Di Domenico M. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg. 2015;99(6):1899–1905. doi: 10.1016/j.athoracsur.2014.11.049 [DOI] [PubMed] [Google Scholar]
- 11.Yu N, Zhou J, Cui F, Tang X. Circulating tumor cells in lung cancer: detection methods and clinical applications. Lung. 2015;193(2):157–171. doi: 10.1007/s00408-015-9697-7 [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41(2):755–768. doi: 10.1159/000458736 [DOI] [PubMed] [Google Scholar]
- 13.Coumans FA, Ligthart ST, Uhr JW, Terstappen LW. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res. 2012;18(20):5711–5718. doi: 10.1158/1078-0432.CCR-12-1585 [DOI] [PubMed] [Google Scholar]
- 14.Saucedo-Zeni N, Mewes S, Niestroj R, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol. 2012;41(4):1241–1250. doi: 10.3892/ijo.2012.1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Shi J, Shi G, et al. Using the new cellcollector to capture circulating tumor cells from blood in different groups of pulmonary disease: a Cohort study. Sci Rep. 2017;7(1):9542. doi: 10.1038/s41598-017-09284-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorges TM, Penkalla N, Schalk T, et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin Cancer Res. 2016;22(9):2197–2206. doi: 10.1158/1078-0432.CCR-15-1416 [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Boyle TA, Zhou C, Rimm DL, Hirsch FR. PD-L1 Expression in Lung Cancer. J Thorac Oncol. 2016;11(7):964–975. doi: 10.1016/j.jtho.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9(8):e103883. doi: 10.1371/journal.pone.0103883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 20.He J, Tan W, Ma J. Circulating tumor cells and DNA for real-time EGFR detection and monitoring of non-small-cell lung cancer. Future Oncol. 2017;13(9):787–797. doi: 10.2217/fon-2016-0427 [DOI] [PubMed] [Google Scholar]
- 21.Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One. 2014;9(1):e85350. doi: 10.1371/journal.pone.0085350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortune JB, Feustel P. Effect of patient position on size and location of the subclavian vein for percutaneous puncture. Arch Surg. 2003;138(9):996–1000; discussion 1001. doi: 10.1001/archsurg.138.9.996 [DOI] [PubMed] [Google Scholar]
- 23.Hanssen A, Loges S, Pantel K, Wikman H. Detection of circulating tumor cells in non-small cell lung cancer. Front Oncol. 2015;5:207. doi: 10.3389/fonc.2015.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan MS, Kirkwood A, Tsigani T, et al. Circulating tumor cells as prognostic markers in neuroendocrine tumors. J Clin Oncol. 2013;31(3):365–372. doi: 10.1200/JCO.2012.44.2905 [DOI] [PubMed] [Google Scholar]
- 26.Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Z, Gao X, Cheng K, Yu L. Correlation between the presence of circulating tumor cells and the pathologic type and staging of non-small cell lung cancer during the early postoperative period. Oncol Lett. 2017;14(5):5825–5830. doi: 10.3892/ol.2017.6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chemi F, Rothwell DG, McGranahan N, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat Med. 2019;25(10):1534–1539. doi: 10.1038/s41591-019-0593-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095 [DOI] [PubMed] [Google Scholar]
- 30.Gialeli C, Nikitovic D, Kletsas D, Theocharis AD, Tzanakakis GN, Karamanos NK. PDGF/PDGFR signaling and targeting in cancer growth and progression: focus on tumor microenvironment and cancer-associated fibroblasts. Curr Pharm Des. 2014;20(17):2843–2848. doi: 10.2174/13816128113199990592 [DOI] [PubMed] [Google Scholar]
- 31.Kang HG, Lee SY, Jeon HS, et al. A functional polymorphism in CSF1R gene is a novel susceptibility marker for lung cancer among never-smoking females. J Thorac Oncol. 2014;9(11):1647–1655. doi: 10.1097/JTO.0000000000000310 [DOI] [PubMed] [Google Scholar]
- 32.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran TN, Selinger CI, Kohonen-Corish MR, et al. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer. 2013;81(3):462–467. doi: 10.1016/j.lungcan.2013.05.015 [DOI] [PubMed] [Google Scholar]
- 34.Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: what we know and what we expect (Review). Int J Oncol. 2016;49(6):2206–2216. doi: 10.3892/ijo.2016.3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63(3):442–449. doi: 10.4149/314_150825N45 [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues P, Vanharanta S. Circulating tumor cells: come together, right now, over metastasis. Cancer Discov. 2019;9(1):22–24. doi: 10.1158/2159-8290.CD-18-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Taftaf R, Kawaguchi M, et al. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov. 2019;9(1):96–113. doi: 10.1158/2159-8290.CD-18-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G, Benasutti H, Jones JF, et al. Isolation of breast cancer CTCs with multitargeted buoyant immunomicrobubbles. Colloids Surf B Biointerfaces. 2018;161:200–209. doi: 10.1016/j.colsurfb.2017.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi JW, Kim JK, Yang YJ, Kim P, Yoon KH, Yun SH. Urokinase exerts antimetastatic effects by dissociating clusters of circulating tumor cells. Cancer Res. 2015;75(21):4474–4482. doi: 10.1158/0008-5472.CAN-15-0684 [DOI] [PubMed] [Google Scholar]
- 41.Divella R, Daniele A, Abbate I, et al. The presence of clustered circulating tumor cells (CTCs) and circulating cytokines define an aggressive phenotype in metastatic colorectal cancer. Cancer Causes Control. 2014;25(11):1531–1541. doi: 10.1007/s10552-014-0457-4 [DOI] [PubMed] [Google Scholar]
- 42.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 43.Tang Y, Fang W, Zhang Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget. 2015;6(16):14209–14219. doi: 10.18632/oncotarget.v6i16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Momtaz P, Postow MA. Immunologic checkpoints in cancer therapy: focus on the programmed death-1 (PD-1) receptor pathway. Pharmgenomics Pers Med. 2014;7:357–365. doi: 10.2147/PGPM.S53163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 46.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philips GK, Atkins M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int Immunol. 2015;27(1):39–46. doi: 10.1093/intimm/dxu095 [DOI] [PubMed] [Google Scholar]
- 48.Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep. 2016;6:31726. doi: 10.1038/srep31726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boffa DJ, Graf RP, Salazar MC, et al. Cellular Expression of PD-L1 in the peripheral blood of lung cancer patients is associated with worse survival. Cancer Epidemiol Biomarkers Prev. 2017;26(7):1139–1145. doi: 10.1158/1055-9965.EPI-17-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ilie M, Szafer-Glusman E, Hofman V, et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann Oncol. 2018;29(1):193–199. doi: 10.1093/annonc/mdx636 [DOI] [PubMed] [Google Scholar]
- 51.Guibert N, Delaunay M, Lusque A, et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer. 2018;120:108–112. doi: 10.1016/j.lungcan.2018.04.001 [DOI] [PubMed] [Google Scholar]