Abstract
Directing the flow of protein traffic is a critical task faced by all cellular organisms. In Gram-negative bacteria, this traffic includes lipoproteins. Lipoproteins are synthesized as precursors in the cytoplasm and receive their acyl modifications upon export across the inner membrane. The third and final acyl chain is added by Lnt, which until recently was thought to be essential in all Gram-negatives. In this report, we show that Acinetobacter species can also tolerate a complete loss-of-function mutation in lnt. Absence of a fully functional Lnt impairs modification of lipoproteins, increases outer membrane permeability and susceptibility to antibiotics, and alters normal cellular morphology. In addition, we show that loss of lnt triggers a global transcriptional response to this added cellular stress. Taken together, our findings provide new insights on and support the growing revisions to the Gram-negative lipoprotein biogenesis paradigm.
Keywords: Acinetobacter, cell envelope, lipoprotein, Lnt, stress response
Introduction
The Gram-negative cell envelope is composed of an inner membrane (IM) and an outer membrane (OM), with the aqueous periplasm in between [1]. The peptidoglycan cell wall is contained within the periplasm [2]. Biogenesis of the cell envelope relies on the activity of multiple proteins, many of which function as part of multi-subunit molecular machines [3–5]. For example, delivery of lipopolysaccharide (LPS) to the outer leaflet of the OM requires the Lpt machine [3]. Outer membrane protein (OMP) assembly requires the Bam complex [6], while lipoproteins are sorted between the IM and OM by the Lol transporter [7]. In model Gram-negative organisms like Escherichia coli, the functions of these assembly factors are essential under standard laboratory conditions [8]. However, there is increasing evidence that the requirement for these factors varies among different Gram-negative species and under particular conditions [9–12].
Lipoproteins are synthesized as precursors in the cytoplasm and then targeted for export across the IM [7]. These precursors then undergo an ordered series of modifications to install the lipid moieties. First, the diacylglyceryl transferase Lgt adds two acyl chains to a conserved cysteine residue adjacent to the amino-terminal signal sequence [13]. Next, the signal sequence is cleaved by the signal peptidase LspA [14]. Finally, the third acyl chain is added to the amino terminus of the cysteine residue by the apolipoprotein N-acyl transferase Lnt [15]. In E. coli, lipoproteins with an aspartate residue at position +2 will be retained at the IM [16, 17]; in other cases, the +3 or +4 residue guides IM retention [18]. Lipoproteins destined for the OM are routed there through the Lol pathway [19].
The Lol pathway consists of an IM platform composed of the LolCDE proteins, which functions as an ABC transporter [20]. Fully acylated lipoproteins are routed to LolCDE. If destined for the OM, lipoproteins are then delivered to the periplasmic carrier LolA which carries them to the OM receptor LolB for assembly into the OM [21, 22]. Lipoproteins can remain anchored to the inner leaflet of the OM, or in some cases may flip to the outer leaflet of the OM [23]. This flipping mechanism is an area of active research interest [4, 19]. Recently, the classical model of how Lol functions has been challenged [12]. In some mutant strains of E. coli, the need for both LolA and LolB can be bypassed upon activation of an envelope stress response [12]. Grabowicz and Silhavy also propose the existence of a redundant machinery for lipoprotein trafficking, and that the true role of Lol is to ensure that the OM does not become overloaded with lipoproteins [12]. However, the identity of this novel machinery has not yet been revealed.
In E. coli, all three lipoprotein modification factors – Lgt, Lsp, and Lnt – as well as the Lol machinery, are essential for growth under standard laboratory conditions [8]. Until recently, this was thought to be the case for all Gram-negative bacteria. However, this no longer appears to be the case. The first demonstration that lnt is dispensable for growth came from work in Francisella and Neisseria species [10]. Here, we show that Lnt is not essential but important for growth in Acinetobacter baylyi or Acinetobacter baumannii. These two organisms have recently emerged as models for studying complex genetic interactions and pathogenesis of drug-resistant microbial infections, respectively [24, 25]. We also found that the impact of crippling lipoprotein biogenesis by mutating lnt is broad. Without Lnt, the bacterial cell envelope becomes more permeable and modification of OM lipoproteins is impaired. Given the central importance of lipoproteins during cell envelope biogenesis, we reasoned that the absence of Lnt would impair the flow of lipoprotein traffic and thus trigger a cellular stress response. By using RNA-seq and quantitative PCR, we identified nearly 80 genes whose expression is altered in the absence of lnt. Taken together, our findings support a key role for Lnt in lipoprotein maturation, as well as providing a foundation for future studies of stress responses in Acinetobacter species. Defining the lipoprotein biogenesis pathways in A. baylyi and A. baumannii is of fundamental importance to elucidating the biology of these important Gram-negative microbes.
Methods
Strains and growth conditions
All strains of Acinetobacter were grown in LB (Lennox, Fisher Scientific); A. baylyi strains were grown at 30 °C, while A. baumannii strains were grown at 37 °C. A. baumannii AB5075-UW wild-type and lnt mutant (lnt153 :: T26) strains were obtained from the University of Washington [26]. To determine colony morphology, overnight cultures of each strain were serially diluted and then aliquots were spread on the surface of LB agar or MacConkey agar plates. Plate were incubated overnight and photographed the next morning. For liquid growth curves, overnight cultures of each strain were diluted in fresh LB to an optical density (OD) at 600 nm of 0.05. Cultures were incubated with intermittent shaking, and OD measurements were taken every 45 min. All strains were grown in triplicate.
Construction of an A. baylyi lnt :: kan insertion-deletion mutant and complementation experiments
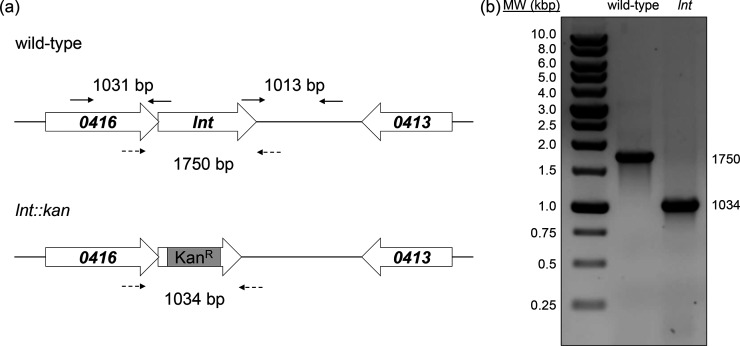
To construct the A. baylyi lnt mutant, we used an overlap-extension PCR strategy using the primers listed in Table S1 (available in the online version of this article) [27]. Briefly, approximately 1 kb of upstream and downstream sequences flanking lnt (ACIAD0415) were amplified by colony PCR from the A. baylyi wild-type reference strain ADP1 (obtained from ATCC). A 795 bp kanamycin-resistance cassette was amplified from plasmid pIM1445 (gift from Ichiro Matsumura) [28]. These three products were joined together in a final PCR using the outermost upstream and downstream flanking primers, resulting in the lnt :: kan insertion-deletion allele. This PCR fragment was purified using a QIAGEN PCR purification kit and then used to transform wild-type A. baylyi, which is naturally competent [25]. Correct incorporation of the insertion-deletion allele was confirmed by PCR analysis (Fig. 1b, c) and DNA sequencing. To construct an lnt complementation strain, the full-length sequence of lnt (Table S2) was cloned into the BamHI site of pWH1266 (ATCC) by custom gene synthesis (GenScript, Piscataway, NJ). The resulting plasmid pLnt was transformed into A. baylyi by electroporation using a Bio-Rad MicroPulser.
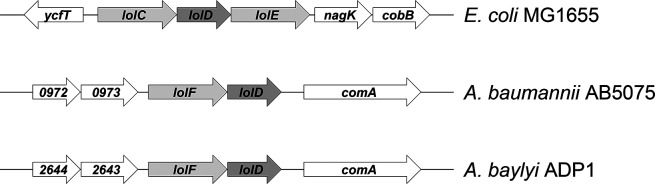
Fig. 1.
Organization of Lol ABC transporter genes in Gram-negative bacteria. In E. coli, the inner membrane (IM) Lol ABC transporter is organized in the standard lolCDE format. In Acinetobacter species, the ABC transporter is organized in a lolFD module. Genes that flank lolFD in A. baumannii AB5075-UW and A. baylyi ADP1 are listed by number.
Immunoblot analysis
Bacterial cultures were grown until mid-exponential phase when 1 ml samples were harvested by centrifugation. Cell pellets were then re-suspended in SDS-PAGE sample buffer in a volume (in ml) equal to the OD600 nm divided by seven. After boiling for 10 min, 10 µl of each sample was resolved by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and probed with polyclonal rabbit antibodies against BamD, RpoD and SecY. Antibodies were generated against His-tagged, purified full-length proteins (BamD and RpoD) or peptides (DNVALARFFKANEGC, SecY) by GenScript (Piscataway, NJ). Blots were probed with antibodies that were diluted 1 : 1000 in Blocking Buffer (LI-COR Biosciences) overnight at 4 °C. Primary antibodies were detected using IRDye 800CW donkey anti-rabbit antibodies (LI-COR Biosciences) prior to imaging with a LI-COR Odyssey Fc imager according to the manufacturer’s protocols.
Scanning electron microscopy
Wild-type and lnt mutant A. baylyi strains were grown overnight on LB plates. Plugs of individual colonies were harvested and then fixed with 3 % glutaraldehyde (in 0.1 M phosphate buffer, pH 7.2) for one hour at room temperature. The glutaraldehyde solution was removed and then samples were washed three times with phosphate buffer. Samples were then dehydrated in an ascending ethanol series (30, 50, 70, 80, 85, 90 and 95 % ethanol for 10 min each and three 100 % ethanol washes for 15 min each). After dehydration, the samples were dried using a Samdri-795 critical-point drier. Samples were secured onto aluminum stubs with double-sided adhesive tabs and were gold-coated using an EMS-550 sputter coater. All samples were visualized using an FEI Quanta 250 scanning electron microscope at 30 kV.
Disc diffusion antibiotic sensitivity assays
Aliquots of 100 µl from overnight cultures of each strain were spread onto the surface of LB agar plates and allowed to dry. Antibiotic-containing discs (BD BBL Sensi-Discs) were placed on the agar surface, and zones of growth inhibition around the discs were measured the next day. Discs contained the following amounts of antibiotics: bacitracin (10 units), novobiocin (30 µg), vancomycin (30 µg) and polymyxin B (300 units).
RNA-seq analysis
All RNA-seq experiments were conducted with Cofactor Genomics (St. Louis, MO), using the following workflow.
Ribosome-depletion and library preparation
Triplicate cultures of both wild-type and lnt mutant A. baylyi were grown in LB at 30 °C and harvested during mid-exponential phase. Total RNA was extracted using the RNeasy kit (Qiagen) following the manufacturer’s instructions. Total RNA was processed for library construction by Cofactor Genomics (http://cofactorgenomics.com, St. Louis, MO) as follows. Species-specific rRNA probes were hybridized to total RNA to remove any ribosomal RNA, and the resulting ribosome-depleted RNA was then fragmented. First-strand cDNA synthesis was performed using reverse transcriptase and random primers in the presence of Actinomycin D, followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. Double-stranded cDNA was end-repaired and A-tailed for subsequent adaptor ligation. Indexed adaptors were ligated to the A-tailed cDNA. Enrichment by PCR was performed to generate the final cDNA sequencing library. The library was sequenced as single-end 75 base pair reads on an Illumina NextSeq 500 following the manufacturer’s protocols.
Quality control and data analysis
Initial quality control was performed by Cofactor Genomics (http://cofactorgenomics.com, St. Louis, MO). Raw sequence data in FASTQ format were assessed for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and ribosomal RNA content using sortmeRNA (http://bioinfo.lifl.fr/RNA/sortmerna/). NovoAlign (Novocraft) was used to align reads to a set of transcript sequences. Alignments to the genome were performed using STAR (https://github.com/alexdobin/STAR). Only unique alignments to the genome were allowed. The genome alignment loci from all samples were combined and clustered to generate contiguous read coverage. The reads per kilobase of transcript per million mapped reads (RPKM) expression value was calculated for each sample and used as the basis for expression comparison and statistical analysis. The resulting comparative expression data were visualized in ActiveSite (Cofactor Genomics). All raw data were deposited via SRA (SRP133498).
Quantitative RT-PCR (qPCR)
RNA was extracted as described above. cDNA was synthesized using the iScript reverse transcriptase kit (Bio-Rad). The primers used for expression analysis are listed in Table S2. qPCR was carried out using POWER SYBR Green Supermix on a StepOnePlus thermocycler (Applied Biosystems), with the 16S rRNA gene as the reference. Reactions were set up according to the manufacturer’s protocols using 500 nM primers and 2 µl of the cDNA template (diluted 1 : 10). Relative expression was determined using the comparative cycle threshold (ΔΔCT) method [29]. The cycling conditions used were as follows: amplification stage – 95 °C for 10 min and then 40 amplification cycles of 95 °C for 30 s, 60 °C for 1 min and 72 °C for 30 s; melting curve stage – 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. No-template and no-reverse transcriptase reactions served as the negative controls. All reactions were carried out in triplicate, using cDNA derived from triplicate cultures.
Results and Discussion
Acinetobacter lnt mutants are viable, but exhibit growth defects
Based on the work of LoVullo et al., in organisms where the LolCDE ABC transporter is instead organized as LolFD, lnt is dispensable for growth [10]. Examination of the Acinetobacter baylyi and Acinetobacter baumannii genomes revealed the existence of this LolFD format (Fig. 1 and [10]). Therefore, we hypothesized that lnt mutants would be viable in Acinetobacter spp. To test this hypothesis, we created an insertion-deletion mutant in the A. baylyi lnt gene ACIAD0415 [27]. We obtained kanamycin-resistant colonies and confirmed integration of the insertion-deletion allele into the chromosome by PCR and DNA sequence analysis (Fig. 2). Compared to wild-type A. baylyi, colonies of the lnt mutant were much smaller when grown on LB agar plates (Fig. 3a, upper panels). The lnt mutant also displayed a growth defect when cultured in broth (Fig. 3c). Notably, a previous study reported that lnt is essential in A. baylyi [30]. The mutants constructed in this study were cultured using a defined minimal medium. In the experiments we report here, the A. baylyi lnt mutant was generated on a rich medium and displays a profound growth defect. We have found that growth of the lnt mutant on minimal medium is severely impaired; normal-sized colonies appear only after 3 days of incubation, so the lnt mutant could have been overlooked and incorrectly labelled as essential for growth.
Fig. 2.
Construction of an A. baylyi lnt mutant. (a) Genomic region of lnt (top panel) and location of primers (solid arrows) used to generate an lnt :: kan insertion-deletion allele (bottom panel) as described in the Methods. Mutant construction was confirmed by PCR analysis of products amplified using primers (dashed lines) flanking lnt or lnt :: kan. (b) Products were amplified from chromosomal DNA of wild-type or lnt mutant cells by colony PCR and resolved by agarose gel electrophoresis.
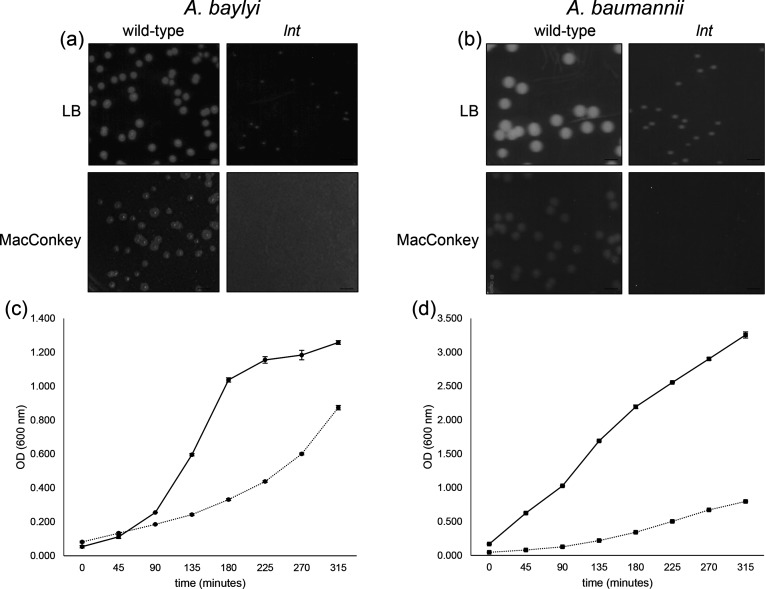
Fig. 3.
Growth of A. baylyi and A. baumannii lnt mutants is impaired relative to wild-type. Overnight cultures of the indicated A. baylyi (a) or A. baumannii (b) strains were serially diluted and plated on either LB (upper panels) or MacConkey (lower panels) agar plates. Scale bar=3.5 mm. Growth of A. baylyi (c) and A. baumannii (d) wild-type (solid line) and lnt mutant (dashed line) strains was monitored by optical density (OD) at 600 nm. Shown is a representative experiment of strains grown in triplicate, where error bars indicate standard deviation from the mean.
To determine whether lnt was dispensable in other species of Acinetobacter that possess a LolFD module, we searched the A. baumannii AB5075-UW transposon mutant library collection browser and found three confirmed insertions within the lnt gene (http://www.gs.washington.edu/labs/manoil/baumannii.htm) [26]. Similar to our observations with A. baylyi, the lnt mutant of A. baumannii AB5075-UW showed a growth defect on LB agar and a diminished generation time in liquid culture (Fig. 3b, d). We also observed that the lnt mutants have a profound growth defect when plated on MacConkey agar (Fig. 3a, b, lower panels). MacConkey agar contains bile salts, a detergent which impairs the growth of Gram-negative bacteria with defective outer membranes [31]. Though wild-type A. baylyi displayed reduced growth on MacConkey, colonies of the lnt mutant failed to appear even after extended incubation. Similarly, the A. baumannii lnt mutant was also unable to grow on MacConkey. These findings suggest that the function of Lnt, while important, is not required to sustain growth of Acinetobacter species under normal laboratory conditions. However, when bacteria are subjected to membrane-disrupting detergents like bile salts, the function of Lnt is vital.
The OM permeability barrier is compromised in the absence of lnt
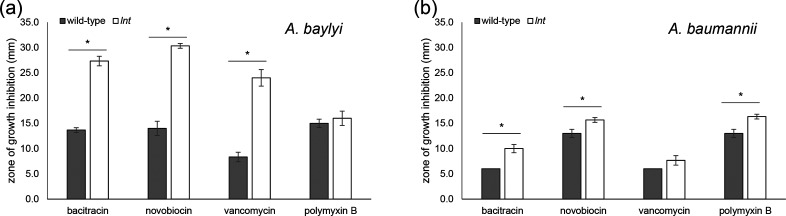
The growth defects associated with the loss of lnt under standard laboratory conditions are consistent with disruption of an important cellular process – namely, the final acylation step of lipoproteins prior to their sorting between the IM and OM. It has already been shown in Francisella that without Lnt, trafficking of diacylated lipoproteins to the OM is diminished [10]. Many OM lipoproteins are critical for establishing the barrier function of the OM. Indeed, this barrier function is compromised in both Francisella and Neisseria lnt mutants as evidenced by slightly increased sensitivity to different antibiotics [10]. Given these findings, we expected the OM to be severely compromised in the Acinetobacter lnt mutants. Indeed, we found that both Acinetobacter lnt mutants were more sensitive to antibiotics than their wild-type strains (Fig. 4). The disparity was particularly notable for A. baylyi. The lnt mutant was extremely sensitive to both bacitracin and vancomycin, which disrupt cell wall synthesis, and novobiocin, which targets DNA gyrase (Fig. 4a P≤0.05) [32–34]. In the case of A. baumannii, the difference in antibiotic sensitivity between wild-type and lnt mutant strains was considerably less pronounced; however, the lnt mutant was significantly more sensitive to bacitracin and novobiocin (P≤0.05) and vancomycin (P≤0.067) (Fig. 4b). Interestingly, only the A. baumannii lnt mutant displayed significantly increased sensitivity to polymyxin B (P≤0.05), which is a membrane-disrupting agent [35]. We do not fully understand the differential in sensitivities between the A. baylyi and A. baumannii strains. The A. baumannii lnt mutant contains a transposon insertion at nucleotide position 929 (of 1560). This mutation should disrupt the active site of Lnt based on previous studies [36, 37]. The A. baylyi lnt mutant is disrupted by an antibiotic resistance cassette, replacing nucleotides 17–1554 (of 1560). However, A. baumannii AB5075-UW is a multidrug-resistant clinical that produces a capsule which likely contributes to the increased antibiotic resistance profile shown in Fig. 4 [38]. A similarly diverse spectrum of antibiotic sensitivities was also noted among lnt mutants in Francisella and Neisseria species [10]. One of the most important functions of the Gram-negative OM is to serve as a barrier against large antibiotics. The fact that the lnt mutants displayed increased sensitivity to various antibiotics indicates a defective OM, particularly in A. baylyi. Given the stronger sensitivity phenotypes associated with the A. baylyi lnt mutant, we conducted our additional analyses with this mutant.
Fig. 4.
In the absence of lnt, the integrity of the outer membrane (OM) is compromised. Overnight cultures of wild-type and lnt mutant for A. baylyi (a) and A. baumannii (b) strains were spread onto the surface of an LB agar plate. After allowing the cultures to dry, filter discs impregnated with the indicated antibiotics [bacitracin (10 units), novobiocin (30 µg), vancomycin (30 µg) and polymyxin B (300 units)] were placed on the agar surface. Zones of growth inhibition (in mm) were measured after overnight growth at 30 °C. Bar charts represent the mean of triplicate measurements, with error bars indicating standard deviation from the mean. *, P≤0.05 by Student’s t-test.
Proper acylation of OM lipoprotein BamD requires Lnt in A. baylyi
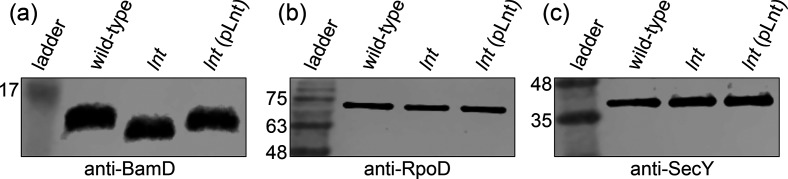
To confirm that the A. baylyi lnt mutant cannot properly acylate precursor lipoproteins, we investigated the OM lipoprotein BamD (Fig. 5). BamD is the only essential lipoprotein component of the Bam complex, which is required for assembly of OMPs in the Gram-negative OM [39]. Without functional BamD, OMP assembly is impaired which destabilizes the OM [40]. We probed whole-cell extracts of wild-type and lnt mutant A. baylyi with antibodies raised against BamD. In the absence of lnt, BamD migrated at a lower molecular weight than in the wild-type parent (Fig. 5a). This is consistent with failure to add the third and final acyl chain to the diacylated BamD precursor. To complement the lnt mutant, we used the Acinetobacter shuttle vector pWH1266 to express full-length lnt. We found that migration of BamD to a higher molecular weight was restored (Fig. 5a). The larger BamD species in the wild-type and complemented mutant runs at about 16 kDa, while the smaller BamD species in the lnt mutant runs at about 15 kDa. We also noticed that the overall level of BamD was slightly decreased in the lnt mutant. Given the central importance of BamD, this finding could partially explain the growth and permeability defects exhibited by the lnt mutant. For comparison, we blotted for the cytoplasmic protein RpoD (Fig. 5b) and the IM protein SecY, demonstrating no migration or expression differences (Fig. 5c).
Fig. 5.
Lnt is required for full maturation of OM lipoprotein BamD. Whole-cell protein extracts derived from an equal amount of wild-type, lnt mutant or complemented mutant (lnt (pLnt)) A. baylyi cells were resolved by SDS-PAGE on a 12 % resolving gel and then probed using the indicated antibodies during an immunoblot. (a) BamD migrates at a lower molecular weight in the lnt mutant as compared to wild-type. (b) Cytoplasmic protein RpoD and (c) IM protein SecY displayed similar gel migration in wild-type and lnt mutant strains. Numbers indicate molecular weight in kDa.
Cells of the lnt mutant have structural defects
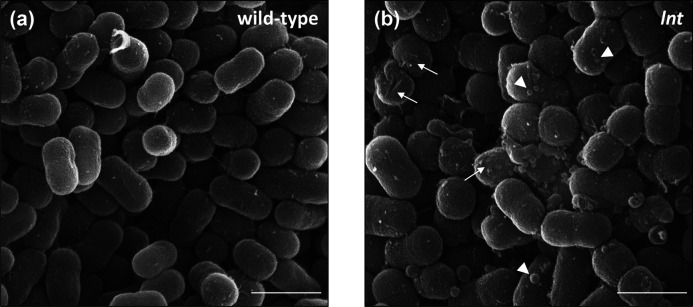
To gain a better picture of the cell envelope defects observed in the lnt mutants, we examined wild-type and mutant cells using scanning electron microscopy (Fig. 6). Wild-type cells appeared as short rods with a smooth surface, typical of A. baylyi (Fig. 6a). We found that the overall shape and size of the lnt mutant cells was similar to wild-type (Fig. 6b). However, the surface of the lnt mutant cells evinced a rougher surface and the presence of outer membrane vesicles (OMVs). These OMVs varied in size, ranging from 70 to 200 nm in diameter, and were consistently observed across multiple fields of view. Notably, increased production of OMVs has been associated with a general response to cell envelope stress [41]. For SEM analysis, we examined microbial cells that were grown under optimal conditions, devoid of major stress stimuli. However, given the important role played by lipoproteins in cell envelope biogenesis, it stands to reason that the lnt mutant cells would exhibit signs of stress even under the most favourable conditions. Accordingly, we postulated that the lnt mutant could be used as a tool to investigate cellular stress responses. In this case, the stress response in question would be triggered by a specific defect in lipoprotein biogenesis. We therefore assessed whether or not the loss of Lnt is sufficient to alter normal patterns of gene expression.
Fig. 6.
Surface roughness and OMV production are increased in lnt mutant cells. Scanning electron microscopy images of wild-type and lnt mutant A. baylyi cells grown on LB agar plates. Arrows indicate rough surface, and arrowheads denote OMVs. Scale bar=1 µm. Shown are representative fields from at least 6 different sample preparations.
The gene expression profile of A. baylyi changes in response to lnt mutation
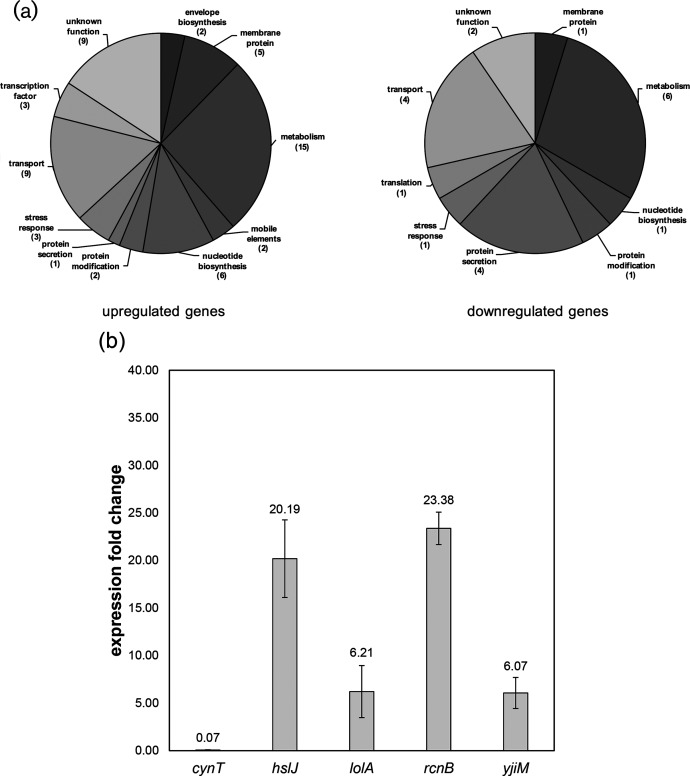
We examined global changes in gene expression using RNA-seq (Fig. 7). Total RNA was collected from cultures of wild-type and lnt mutant strains of A. baylyi grown under stress-free conditions (30 °C in LB broth). We considered genes with a greater than five-fold increase in expression as being induced, while genes with expression levels reduced by five-fold as being repressed (P≤0.01 as determined by Welch’s t-test [42]). By this metric, 57 genes were upregulated in the lnt mutant relative to the wild-type, while 21 were repressed (Table S3). Many of these genes encode proteins with predicted roles in central metabolism, and thus their connection to envelope stress is not immediately obvious (Fig. 7a). Eleven genes have no predicted function. However, there are 17 genes associated with cell envelope biosynthesis (2), protein secretion (5), membrane proteins (6) or stress responses (4).
Fig. 7.
Differential gene expression is induced in the lnt mutant. (a) Transcriptional profile of lnt mutants shows gene upregulation (57 genes) and downregulation (21 genes). Pie charts summarize the data found in Table S3, using a differential expression threshold of five-fold, P≤0.01. (b) qPCR confirmation of a subset of genes with altered expression when lnt function is disrupted. Changes in expression of the indicated genes in A. baylyi are represented as fold change in the lnt mutant as compared to wild-type, using the 16S rRNA gene as the housekeeping control gene. Numbers above the bars are the average values of biological triplicates, with error bars indicating standard deviation from the mean.
We selected a subset of these differentially regulated genes for confirmation by qPCR, some of which have previously been implicated in transcriptional responses to interference with lipoprotein transport [43]. This includes hslJ, lolA and rcnB. We confirmed that expression of lolA was upregulated in the lnt mutant (Fig. 7b). Given that LolA is thought to shuttle lipoproteins between the IM and OM, it is not surprising to see its expression increased. If tri-acylation is required for efficient delivery of lipoproteins to the OM, perhaps an overproduction of LolA increases delivery of di-acylated lipoproteins. Indeed, it has already been shown that lolA expression increases in response to defective lipoprotein sorting via the Rcs stress response and upon depletion of lolCDE [44, 45]. Additionally we observed that expression of hslJ, which encodes a putative periplasmic heat shock chaperone, is increased in the lnt mutant. Interestingly, a previous study also found that in E. coli expression of hslJ was upregulated upon treatment with the LolCDE inhibitor known as compound 2 [43]. This study also discovered that treatment with compound 2 induced expression of lolA and lolB, as well as stress response factors degP, rcsA, and cpxP [43]. Thus, interference with the machinery of lipoprotein biogenesis, by either chemical or genetic means, can induce transcription of stress response genes. The response to defects in the early stages of lipoprotein maturation is an important component of how Gram-negatives counter disruptions in OM biogenesis by adjusting expression of a similar suite of genes [46–48].
We also identified differentially regulated genes with no obvious connection to lipoprotein biogenesis. For example, both yjiM and rcnB were strongly induced in the lnt mutant, while cynT, a predicted carbonic anhydrase, was repressed. Additional studies are required to fully understand the contribution of such a diverse subset of genes toward mitigating defects in lipoprotein biogenesis, particularly so that directly related factors can be separated from indirect effects. Given this wide range of targets, it seems that bacteria have a suite of genes that are used to mitigate stress caused by alteration to the typical lipoprotein composition. It is possible that this suite of genes is also activated during other conditions that trigger cell envelope stress, such as the use of membrane and cell wall-targeting antibiotics. Toward that end, it will be important to monitor gene expression when Acinetobacter is challenged with a different stress to the cell envelope, such as interference with OMP biogenesis or LPS assembly. Further studies will be needed to characterize the specifics of such transcriptional responses; in the meantime, lnt disruption may serve as an important tool to fully classify stress responses that are activated to compensate for defects in lipoprotein maturation.
Supplementary Data
Funding information
This work was supported in part by the National Science Foundation under Grant No. 1615822 and a Faculty Research and Development Grant from Hofstra University to NWR. LRM was partially supported by the National Institute of General Medical Sciences of the US National Institutes of Health (NIH) under award number 1R15GM117501-01A1. LRM is partially funded and has an appointment in the Infectious Diseases and Immunology cluster of the Border Biomedical Research Center (BBRC; National Institute on Minority Health and Health Disparities award number 2G12MD007592), UTEP’s Research Centers in Minority Institutions Program. LRM serves as a Leshner Leadership Institute Public Engagement Fellow in infectious diseases with the American Association for the Advancement of Science.
Acknowledgements
We would like to thank Kathleen Lynch and AnnMarie Gaglio for advice on gene expression analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ABC, ATP-binding cassette; IM, Inner membrane; LPS, lipopolysaccharide; OM, outer membrane; PCR, polymerase chain reaction.
Three supplementary tables are available with the online version of this article.
Edited by: T. Schneiders and M. Whiteley
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz N. Filling holes in peptidoglycan biogenesis of Escherichia coli . Curr Opin Microbiol. 2016;34:1–6. doi: 10.1016/j.mib.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci. 2015;370:20150030. doi: 10.1098/rstb.2015.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botos I, Noinaj N, Buchanan SK. Insertion of proteins and lipopolysaccharide into the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160224. doi: 10.1098/rstb.2016.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plummer AM, Fleming KG. From chaperones to the membrane with a BAM! Trends Biochem Sci. 2016;41:872–882. doi: 10.1016/j.tibs.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narita SI, Tokuda H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim Biophys Acta. 2017;1862:1414–1423. doi: 10.1016/j.bbalip.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narita S, Tokuda H. Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J Bacteriol. 2011;193:4832–4840. doi: 10.1128/JB.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS. Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol. 2015;197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabowicz M, Silhavy TJ. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci USA. 2017;114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 14.Yamagata H, Taguchi N, Daishima K, Mizushima S. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli . Mol Gen Genet. 1983;192:10–14. doi: 10.1007/BF00327640. [DOI] [PubMed] [Google Scholar]
- 15.Gupta SD, Wu HC. Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli . FEMS Microbiol Lett. 1991;62:37–41. doi: 10.1016/0378-1097(91)90251-5. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli . Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 17.Seydel A, Gounon P, Pugsley AP. Testing the '+2 rule' for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol. 1999;34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 18.Narita S, Tokuda H. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem. 2007;282:13372–13378. doi: 10.1074/jbc.M611839200. [DOI] [PubMed] [Google Scholar]
- 19.Szewczyk J, Collet JF. The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv Microb Physiol. 2016;69:1–50. doi: 10.1016/bs.ampbs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Narita S, Tokuda H. An ABC transporter mediating the membrane detachment of bacterial lipoproteins depending on their sorting signals. FEBS Lett. 2006;580:1164–1170. doi: 10.1016/j.febslet.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi N, Tokuda H. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J Biol Chem. 2008;283:8538–8544. doi: 10.1074/jbc.M800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci USA. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konovalova A, Mitchell AM, Silhavy TJ. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife. 2016;5:e15276. doi: 10.7554/eLife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott KT, Neidle EL. Acinetobacter baylyi ADP1: transforming the choice of model organism. IUBMB Life. 2011;63:1075–1080. doi: 10.1002/iub.530. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, et al. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii . J Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, et al. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii . BMC Microbiol. 2010;10:279. doi: 10.1186/1471-2180-10-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murin CD, Segal K, Bryksin A, Matsumura I. Expression vectors for Acinetobacter baylyi ADP1. Appl Environ Microbiol. 2012;78:280–283. doi: 10.1128/AEM.05597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Stone KJ, Strominger JL. Mechanism of action of bacitracin: complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc Natl Acad Sci USA. 1971;68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanakunakorn C. Mode of action and in-vitro activity of vancomycin. J Antimicrob Chemother. 1984;14:7–18. doi: 10.1093/jac/14.suppl_D.7. [DOI] [PubMed] [Google Scholar]
- 34.Cozzarelli NR. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977;46:641–668. doi: 10.1146/annurev.bi.46.070177.003233. [DOI] [PubMed] [Google Scholar]
- 35.Hsuchen CC, Feingold DS. The mechanism of polymyxin B action and selectivity toward biologic membranes. Biochemistry. 1973;12:2105–2111. doi: 10.1021/bi00735a014. [DOI] [PubMed] [Google Scholar]
- 36.Gélis-Jeanvoine S, Lory S, Oberto J, Buddelmeijer N. Residues located on membrane-embedded flexible loops are essential for the second step of the apolipoprotein N-acyltransferase reaction. Mol Microbiol. 2015;95:692–705. doi: 10.1111/mmi.12897. [DOI] [PubMed] [Google Scholar]
- 37.Vidal-Ingigliardi D, Lewenza S, Buddelmeijer N. Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J Bacteriol. 2007;189:4456–4464. doi: 10.1128/JB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senchenkova SN, Shashkov AS, Popova AV, Shneider MM, Arbatsky NP, et al. Structure elucidation of the capsular polysaccharide of Acinetobacter baumannii AB5075 having the KL25 capsule biosynthesis locus. Carbohydr Res. 2015;408:8–11. doi: 10.1016/j.carres.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Ricci DP, Silhavy TJ. The Bam machine: a molecular cooper. Biochim Biophys Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli . Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa . J Bacteriol. 2013;195:2971–2981. doi: 10.1128/JB.02267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25:60–83. doi: 10.3102/10769986025001060. [DOI] [Google Scholar]
- 43.Lorenz C, Dougherty TJ, Lory S. Transcriptional responses of Escherichia coli to a small-molecule inhibitor of LolCDE, an essential component of the lipoprotein transport pathway. J Bacteriol. 2016;198:3162–3175. doi: 10.1128/JB.00502-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao K, Narita S, Tokuda H. Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J Bacteriol. 2012;194:3643–3650. doi: 10.1128/JB.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narita S, Tanaka K, Matsuyama S, Tokuda H. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J Bacteriol. 2002;184:1417–1422. doi: 10.1128/JB.184.5.1417-1422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grabowicz M, Silhavy TJ. Envelope stress responses: an interconnected safety net. Trends Biochem Sci. 2017;42:232–242. doi: 10.1016/j.tibs.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raivio TL. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Barchinger SE, Ades SE. Regulated proteolysis: control of the Escherichia coli σ(E)-dependent cell envelope stress response. Subcell Biochem. 2013;66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.