Summary
Regulatory T cells (Tregs) are specialized in immune suppression and play a dominant role in peripheral immune tolerance. Treg cell lineage development and function maintenance is determined by the forkhead box protein 3 (FoxP3) transcriptional factor, whose activity is fine‐tuned by its post‐translational modifications (PTMs) and interaction partners. In this review, we summarize current studies in the crystal structures, the PTMs and interaction partners of FoxP3 protein, and discuss how these insights may provide a roadmap for new approaches to modulate Treg suppression, and new therapies to enhance immune tolerance in autoimmune diseases.
Keywords: allosteric modifiers, FoxP3, protein interactions, post‐translational modifications, regulatory T cells
FOXP3 is the master transcription factor in regulating Treg cell development and function. The forkhead domain of FOXP3 forms a domain‐swapped dimer which can bridge two long‐range FOXP3‐targeted genes (a). The zinc‐finger and leucine‐zipper domain forms oligomerization through the hydrophobic coiled‐coil surface (b). Acetylation of K250 and K252 located in the coiled‐coil region changes the conformation of FOXP3, which implies a structure‐based regulation of the conformation and activity of FOXP3 by integration of posttranslational modifications may modulate Treg function. Understanding the molecular and structural features of Foxp3 helps to design rational therapeutic strategies against autoimmune diseases.
![]()
Introduction
Regulatory T cells (Tregs) maintain peripheral immune homeostasis and limit inflammatory tissue damage during vigorous and deleterious immune responses. The forkhead box protein 3 (FoxP3) transcription factor is an important regulator of the Treg cell lineage. FoxP3 mutations often leads to ‘Scurfy’ in rodents and to X‐linked autoimmune disorders (XLAAD) in humans. FoxP3 protein contains multiple structural domains, interaction partners and various kinds of post‐translational modifications. Studies of the regulatory mechanisms of Treg cells have been partially elucidated by manipulating and studying changes in FoxP3 protein function.
In this review, we will first introduce current understanding of the FoxP3 protein’s function in Treg cell biology, and then review the known FoxP3 domain crystal structures, FoxP3 interaction partners and post‐translational modifications. We then discuss ways to explore the complexity of FoxP3 by rationally designing compounds to modify FoxP3 protein function based on its structure or the structure of associated molecules that mediate a specific function. This review focuses on the regulation of Treg cell function via targeting the molecule and associated modifiers as well as the atomic structure of the FoxP3 protein.
FoxP3 and Treg cells
Immune tolerance processes enable our immune system to respond to non‐self‐induced damage while limiting collateral damage to self‐tissues. The first studies to define Treg cells as the basis for infectious tolerance were from the Waldmann laboratory 1. CD4+CD25+ Tregs are immune suppressive T cells which play a predominant role in peripheral immune tolerance 2. Treg cells can be classified into two main types: thymus Treg cells (tTreg) and peripheral Treg cells (pTreg), based on current understanding of their origins 3, 4. Although many factors contribute to the stable Treg cell lineage, including surface protein markers 5, 6, 7 and genomic epigenetic modifications 8, 9, accumulating evidence has demonstrated that FoxP3 transcription factor acts as a dominant regulator in Treg cell development and function maintaining 10, 11, 12.
The role of FoxP3 in regulating immune tolerance was determined by analysis of lethal autoimmunity that developed in the Scurfy mice and in the immune dysregulation polyendocrinopathy enteropathy X‐linked syndrome (IPEX) patients, a recessive immune disorder that appears at an early age 13, 14, 15. Targeted deletion of FoxP3 in CD4+ T cells of mice leads to severe autoimmunity 16, while ectopic expression of FoxP3 can re‐program conventional CD4+ T cells into cells which act as anti‐inflammatory Treg cells, and confers certain capacities to suppress autoinflammation in vivo 10, 11. Enforced expression of FoxP3 induces a Treg phenotype in human leukemic Jurkat T cells, including increased expression of Treg‐associated cell surface markers as well as inhibition of cytokine production. FoxP3‐transduced Jurkat T cells suppress the proliferation of human conventional T cells 17.
FoxP3 belongs to a forkhead transcription factor family which mainly localizes in the nucleus 18, 19. Genome‐wide analyses of FoxP3 target genes has revealed that FoxP3 binds to perhaps as many as ~700 genes and miRNA, which are involved in the T cell receptor (TCR) signaling pathway, cell communication and transcriptional regulation 20. This complexity highlights the important role for FoxP3 in modifying transcriptional and receptor signaling networks in Treg cell development and maintenance. FoxP3 directly targeted genes comprise only a small portion of the entire program of Foxp3‐dependent gene expression, suggesting that FoxP3 regulates a substantial part of the FoxP3‐dependent transcriptional program indirectly through association with other transcription factors. FoxP3 plays a dual role as both a transcriptional activator and repressor of its target genes in Treg cells 21. FoxP3 heterodimerize with FoxP1, which enforces FoxP3‐mediated regulation of gene expression and co‐ordinates regulatory T cell function 22.
To maintain the Treg lineage specificity under normal physiological conditions, FoxP3 has been suggested to reprogram certain T cell metabolic processes to modify cellular function in stressful environments. FoxP3 has been observed to suppress Myc expression and cell glycolysis. While lactate levels impair effector T cells through lactate dehydrogenase (LDH)‐mediated nicotinamide adenine dinucleotide (NAD) depletion, FoxP3 expression may limit pyruvate to lactate reduction. Ectopic expression of FoxP3 expression increases oxygen consumption rates (OCR) and oxidative phosphorylation (OXPHOS) 23.
Structure of FoxP3 domains
Structurally, FoxP3 contains three major domains: a proline‐rich N‐terminal domain (1–97 aa) responsible for transcriptional activation and repression, a central zinc‐finger leucine‐zipper domain (98–260 aa) implicated in oligomer formation or association with other factors and a conserved C‐terminal forkhead domain (337–423 aa) responsible for DNA binding. These domains operate together to modulate FoxP3 function 24.
A high‐resolution crystal structure of a ternary complex containing the NFAT1 DNA‐binding domain, the FoxP3 FKH domain and the ARRE2 site DNA of the interleukin (IL)‐2 promoter has been both modeled and solved. Although the ternary complex of FoxP3, NFAT and DNA was observed only in the presence of DNA containing the composite ARRE2 element, the FKH domain of FoxP3 tends to form a domain‐swapped dimer through a DNA‐independent process (Fig. 1a). An IPEX mutation (F373A) in the FKH domain may impair domain‐swapping, which might abrogate FoxP3 suppressor function. Furthermore, the FoxP3 FKH dimer can simultaneously bind two distinct FoxP3‐binding sites in solution and bring them into close approximation 25. These structural and biochemical analyses suggest that a unique transcriptional function of FoxP3 is to mediate long‐range interactions of FoxP3‐targeted genes. In fact, another study revealed that FoxP3 is specialized for bridging long‐distance genomic elements and reorganizes the three‐dimensional structure of the T cell genome to stabilize cellular differentiation programs. The circular chromosome conformation capture sequencing (4C‐seq) analysis demonstrated that FoxP3 could reorganize the chromosomal ‘interactome’ at the Ptpn22 locus by domain‐swapping and/or through interactions with other factors 26.
Figure 1.
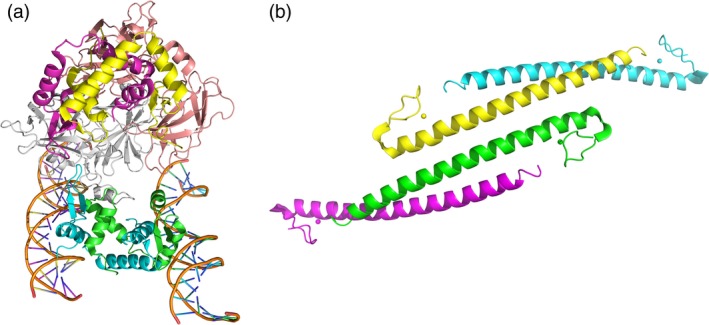
(a) The FKH domain of forkhead box P3 (FoxP3) forms a domain‐swapped dimer. One FKH monomer (green) forms a domain‐swapped dimer with the other FKH monomer (cyan), thereby bridging two molecules of DNA. The picture was generated based on the crystal structure of FoxP3 forkhead domain complexed with DNA and NFAT (PDB code 3QRF) using Pymol. (b) The crystal structure of FoxP3 ZL domain, which tends to form oligomerization through a coiled‐coil motif. The picture was generated based on the structure of FoxP3 ZL domain (PDB code 4I1L) using Pymol.
The crystal structure of the mouse FoxP3 zinc finger and leucine zipper domain (FoxP3‐ZL) from our laboratory provided details about the atomic features of homo‐ and hetero‐association of FoxP3. The mFoxP3‐ZL homodimer features an unusual two‐stranded anti‐parallel α‐helical coiled coil with a twofold symmetry, and crystal packing analysis revealed that the mFoxP3‐ZL coiled‐coil dimer tends to form clusters due to the extended hydrophobic stretch on the coiled coil surface (Fig. 1b). Lysine 251 plays an indispensable role in FoxP3 coiled coil‐mediated dimerization, and the delK251 mutant mFoxP3 fails to repress IL‐2 production. Of note, K249 and K251 of mouse FoxP3, i.e. human FoxP3 K250 and K252 in the coiled coil region, were identified as acetylation sites, which implies that a structure‐based regulation of the conformation and activity of FoxP3 protein by integration of post‐translational modifications may modulate Treg function 27.
The crystal structure of the FoxP3 N‐terminal region and the full‐length FoxP3 are still unavailable, because the N‐terminal region displays intrinsic disorder. We have been successful in developing small crystals of an N‐terminal region that extends to amino acid 262, and we postulate that partner binding and/or post‐translational modification may stabilize this region and help to define the structural features of the FoxP3 N‐terminal domain. The roles of binding partners and post‐translation modifications in regulating FoxP3 function have been defined to some extent.
Importance of FoxP3 interactions
FoxP3 has a proline‐rich N‐terminal domain in comparison with other members of FoxP3 transcriptional factor family 28. This unique N‐terminal domain of the FoxP3 protein leads to interaction with many molecules, including FoxP1 29, Eos 30, AML1/Runx1 31, NFAT 32, RORα 33, TIP60 and HDAC7 34, which are involved in various aspects of Treg function and development, as reviewed elsewhere 35.
Studies have revealed that a green fluorescent protein (GFP) insertion at the FoxP3 N‐terminal without a flexible linker leads to steric hindrance that blocks interaction with Eos, Tip60 and HDAC7, and thus to impaired iTreg development and nTreg stability in an autoimmune or inflammatory environment. Consequently, FoxP3gfp expression on the autoimmune‐prone non‐obese diabetic (NOD) background dramatically accelerated diabetes 36. The N‐terminal insertion of GFP of FoxP3 alleviates arthritis but exacerbates diabetes. Mechanistically, Foxp3fgfp Treg cells display a significant over‐representation of interferon regulatory factor 4 (IRF4)‐dependent transcripts in which GFP insertion at FoxP3 N‐terminal enhances the physical interaction with IRF4, but abrogates the interaction with hypoxia‐inducible factor 1 (HIF‐1). In contrast, the FoxP3 and FoxP3 heterodimer formation thought to occur via the leucine‐zipper region distal from the GFP insertion site remains intact.
These observations imply that perturbations in Treg cell phenotype and functional characteristics can affect autoimmune diseases in a unique but unfortunately unpredictable manner, and that specific modifications of FoxP3 and its interactions could serve as a basis for therapeutically modulating Treg cell function in a qualitative manner 37.
T cells from rheumatoid arthritis patients display down‐regulated TIP60 expression, which impairs Treg development. Ectopic Tip60 expression appears to rescue FoxP3 function and prevents synovial inflammation and immune cell infiltration of the engrafted synovial tissue 38. A missense mutation (A384T) in the FKH domain of FoxP3 impairs the suppressive function of Treg cells while maintaining its ability to repress inflammatory cytokines.
A specific perturbation of FoxP3 interaction with TIP60 accounts for the impaired suppressive function of FoxP3A384T Treg cells. The loss of function could be rescued by using an allosteric modifier to enhance FoxP3–TIP60 interaction pharmacologically (Fig. 2a). Our preclinical results have shown that the allosteric modification of TIP60 improves the FoxP3‐mediated transcriptional program in FoxP3A384T Treg cells, the therapeutic restoration of FoxP3–TIP60 interaction by this small molecule modifier promotes Treg cell function without directly affecting T effector cell responses, and enhances disease protection in mouse models of colitis and collagen‐induced arthritis 39.
Figure 2.
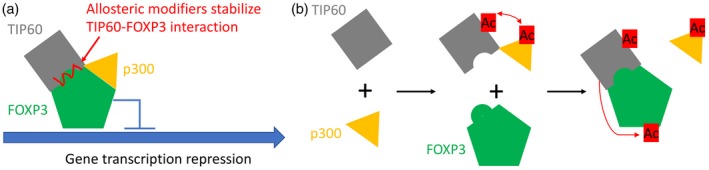
Enhanced regulatory T cell (Treg) suppressive function by targeting TIP60, p300 and forkhead box P3 (FoxP3) interaction. (a) A model of TIP60 allosteric modifiers help to stabilize TIP60–FoxP3 interactions by delaying the release of TIP60 from the TIP60–p300‐FoxP3 complex. (b) TIP60 and p300 promote the acetylation of each other, and then promote FoxP3 acetylation co‐operatively (gray square = TIP60; orange triangle = p300; green pentagon = FoxP3; red Ac = acetylation).
These studies imply that FoxP3 functions as a transcriptional complex defined by multiple FoxP3 partners. In fact, a single amino acid deletion of the zinc finger–leucine zipper domain region of FoxP3 (delE251) that defines an IPEX mutation disrupts the formation of the complex and heteromerization with FoxP1. This disruption limits repression of IL‐2 transcription by FoxP3 upon T cell activation and compromises Treg cell suppressive function, which explains some features of the pathogenesis of a disease syndrome developed in IPEX patients 29. Certain aspects of the super‐complex have also been comprehensively analyzed using biochemical and mass‐spectrometric approaches 40. The efforts focused on a T cell line TCli expressing biotin‐tagged FoxP3 expression protein, and isolated the FoxP3 interaction complex by biotinylation‐based pulldown, and then sequenced molecules by microliquid chromatography. Biotin‐tagged FoxP3 forms multi‐protein complexes of 400–800 kDa and associates with 361 partner proteins, among which ~30% are transcription‐related. Of note, expression of many of these partners may also be influenced by FoxP3‐related processes 40.
Post‐translational modifications of FoxP3
Post‐translational modifications (PTMs) represent a dynamic mechanism to co‐ordinate environmental signals and the functional properties of proteins. There is emerging evidence indicating that the transcriptional activity of FoxP3 can be fine‐tuned by its post‐translational modifications.
Phosphorylation of FoxP3
Phosphorylation modification can occur on serine, threonine and tyrosine residues of protein, and these modifications are reversible. Phosphorylation regulates multiple aspects of protein function, including stability, localization and trafficking. Mouse and human FoxP3 sequences contain several conserved serine, threonine and tyrosine residues, which are potential interaction sites of certain kinases. FoxP3 can be phosphorylated on threonine residues 41.
The primary structure of FoxP3 contains four cyclin‐dependent kinase (CDK) motifs (Ser/Thr‐Pro) within the N‐terminal repressor domain. CDK2 can partner with cyclin E to phosphorylate FoxP3 at these sites. Mutation of the N‐terminal CDK motifs (Ser/Thr3>Ala) in FoxP3 elevated protein stability and increased FoxP3 transcriptional activity. Consistently, CD4 T cells expressing the Ser/Thr3>Ala mutant FoxP3 displayed significantly elevated suppressive capacity compared with cells expressing wild‐type FoxP3 42. These results explain the finding that Cdk2‐deficient Treg are more suppressive than wild‐type Treg, as measured by the ability to suppress the proliferation of conventional CD4+ T cells in vitro and to ameliorate colitis in an in‐vivo mouse model of inflammatory bowel disease 43. A role for CDK2 in influencing Treg function through phosphorylation‐dependent regulation of FoxP3 is apparent.
Pim‐1 and Pim‐2 kinases can act as oncogenic serine/threonine kinases, and their over‐expression contributes to lymphoid transformation. Pim‐1 and Pim‐2 kinases promote the rapamycin‐resistant survival, growth and proliferation of lymphocytes, including Treg cells 44, 45. Both Pim‐1 and Pim‐2 kinases have been suggested to phosphorylate FoxP3 protein. Sequence alignment analysis predicts a potential Pim‐1 interaction and phosphorylation motif within the FoxP3 forkhead domain. Pim‐1 phosphorylates human FoxP3 at Ser422 of the forkhead domain and attenuates FoxP3 DNA binding activity in regulation of Treg feature genes 46. In contrast, Pim‐2 kinase phosphorylates at multiple sites in the FoxP3 N‐terminal domain, as identified by mass spectrum analyses. Pim‐2‐deficient mice are more resistant to dextran sulfate sodium (DSS)‐induced acute colitis than their wild‐type counterparts 47. Phosphorylation of FoxP3 by Pim‐1 and Pim‐2 kinase negatively regulates Treg suppressive function. Of note, TCR stimulation down‐regulated Pim‐1 expression but up‐regulated FoxP3 expression. Therefore, at least some FoxP3 phosphorylation mediated by Pim‐1 may be regulated by TCR stimulation.
FoxP3 phosphorylation is enhanced by TCR activation with incubation of anti‐CD3/CD28 antibodies or pharmacological treatment with phorbol 12‐myristate 13‐acetate (PMA) and ionomycin. The TCR‐mediated signaling pathway can regulate FoxP3 phosphorylation through the activation of Nemo‐like kinase (NLK) in a transforming growth factor (TGF)‐β TAK1‐dependent manner. NLK‐mediated phosphorylation of FoxP3 resulted in the stabilization of protein levels by preventing association with the STUB1 E3‐ubiquitin protein ligase and proteasomal degradation. Conditional deletion of NLK in Treg cells results in the loss of in‐vivo suppressive capacity and autoinflammation in aged mice. NLK directly phosphorylates FoxP3 on multiple residues in the same manner as Pim‐2 and CDK kinases 48.
One substantial caveat in these studies is that NLK or Pim‐2 cannot be predicted to phosphorylate specific sites of FoxP3. There is incomplete information concerning the existence of other endogenous kinases involved in FoxP3 phosphorylation. It is clear, however, that FoxP3 undergoes phosphorylation modifications, and phosphorylation of FoxP3 can either positively or negatively modulate Treg function.
Human and mouse FoxP3 possess four evolutionarily conserved tyrosine residues. Lymphocyte‐specific protein tyrosine kinase (LCK), a Src family kinase that regulates tumor progression, co‐localizes with FoxP3 in the MCF‐7 cell nucleus and phosphorylates Tyr‐342 of FoxP3. Tyr‐342 phosphorylation of FoxP3 is involved in the inhibitory regulation of cancer malignancy by SKP2, vascular endothelial growth factor (VEGF)‐A and matrix metallopeptidase 9 (MMP9) expression, while Y342F mutant blocks this inhibition 49. Our preliminary data indicate that comparable FoxP3 tyrosine phosphorylation also occurs in human T cells, although its biological significance in modulating T cell function remains further investigation (unpublished data).
Tumor necrosis factor (TNF)‐α may impair synovial Treg cell function, a process that relates to the pathogenesis of active rheumatoid arthritis. TNF‐α induces protein phosphatase 1 (PP1) to dephosphorylate Ser‐418 in the C‐terminal DNA‐binding domain of FoxP3, and TNF‐α antagonist therapy (for example, treatment with a TNF‐α‐specific antibody: infliximab) may restore Treg cell function 50.
Acetylation/ubiquitination of FoxP3
Acetylation and ubiquitination are post‐translational modifications of FoxP3, which have attracted extensive studies. Acetylation and ubiquitination are often mutually exclusive. Acetylation modifications may prevent polyubiquitination that occurs on the same lysine residue through a competition‐based mechanism.
Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes responsible for histone acetylation, but are also involved in acetylation of protein substrates. There are three major subsets of HATs: the Gcn5/PCAF family, the p300/CBP family and the MYST family 51. TIP60 is a member of the MYST family, and was the first identified acetyltransferase that associates with and promotes FoxP3 acetylation. As discussed above, TIP60 functions as an essential subunit of the FoxP3 repression complex. The N‐terminal 106–190 aa of FoxP3 is required for TIP60–FoxP3 interactions. Knock‐down of endogenous TIP60 by targeted short hairpin RNAs (shRNA) relieves FoxP3‐mediated repression 34. p300, a member of p300/CBP family, was also suggested to acetylate FoxP3. Acetylation of FoxP3 by p300 prevents FoxP3 protein from proteasome‐mediated degradation and treatment with histone deacetylase SIRT inhibitor (NAM) results in significantly increased numbers of functional Treg cells 52.
TIP60 and p300 regulate FoxP3 acetylation and activity in a co‐operative manner. p300 interacts with TIP60 to promote TIP60 autoacetylation and protein stability. The acetylation of TIP60 functions by acting with p300 as a molecular switch to allow TIP60 to change binding partners. Subsequently, p300 is released from this complex, and TIP60 interacts with and acetylates FoxP3. Reciprocally, TIP60 also promotes p300 acetylation that is critical for HAT activity of p300. Transcriptional activity of FoxP3 is thus promoted by TIP60 and p300 co‐operative interactions (Fig. 2b). Conditional deletion of p300 (p300fl/flFoxP3YFP‐Cre) in Treg cells reveals that p300 has a modest effect on the suppressive function of Treg cells. In contrast to p300fl/flFoxP3YFP‐Cre mice, Tip60fl/flFoxP3YFP‐Cre and p300fl/flTIP60fl/flFoxP3YFP‐Cre mice developed severe and fatally autoimmune diseases at an early age, indicating that TIP60 plays a dominant role for the development and function of Treg cells 53.
Loss of abundance of Treg cells accounts for certain autoimmune diseases. SIRT1, a member of the lysine deacetylase Sirtuin (SIRT) family, may decrease FoxP3 acetylation. SIRT1 inhibition may affect FoxP3 acetylation and alter FoxP3 protein levels 52. Sirt1‐deficient iTreg cells display increased FoxP3 stability and demonstrate restrained iTreg conversion into pathogenic T cells. Administration of a pharmacological Sirt‐1 inhibitor, Ex‐527, can attenuate graft‐versus‐host disease (GVHD) while preserving the graft‐versus‐leukemia effect 54. Treatment of mice with the pan‐HDAC inhibitor trichostatin‐A (TSA) was found to increase the proportions and absolute numbers of FoxP3+CD4+ T cells in peripheral lymphoid tissues. HDAC9–/– mice develop less DSS‐induced colitis than wild‐type mice. HDAC inhibitor therapy increases acetylation of both histone and the forkhead domain of FoxP3, as well as FoxP3 protein expression and DNA binding. These changes lead to enhanced Treg suppression and anergy by promoting the association of FoxP3 with target genes. HDAC6, HDAC9 and Sirt1 all deacetylated FoxP3. However, each protein mediates specific effects on discrete transcription factors that control expression of the gene encoding FoxP3, which suggests that combinations of HDAC inhibitors may be used to augment the therapeutic benefits for treating autoimmunity and organ transplantation 55.
Butyrate, a short‐chain fatty acid (SCFA), produced by commensal microorganisms during starch fermentation, increases FoxP3 protein acetylation and facilitates extrathymic generation of Treg cells 56. Treatment with sodium butyrate up‐regulates FoxP3 and IL‐10 expression, and mitigates inflammatory skin reactions in part by induction of functionally active FoxP3+ Treg cells 57.
HDAC inhibitor therapy has been evaluated to modify Treg function in vivo and shows promising effects on allograft survival and autoimmune diseases 58. The administration of the HDAC inhibitor vorinostat in combination with standard GVHD prophylaxis is associated with a lower than expected incidence of severe acute GVHD. Vorinostat‐treated patients who received allogeneic hematopoietic cell transplant (allo‐HCT) experienced significant reductions in the plasma levels of proinflammatory cytokines and promotion of CD25+CD127– Treg frequency of peripheral blood mononuclear cells (PBMCs) with greater suppressive function 59.
FoxP3 protein levels and turnover rate may affect Treg cell lineage stability. The Deubiquitinase USP7 was found to be up‐regulated in Treg cells. Ectopic expression of USP7 decreased FoxP3 polyubiquitination and increased FoxP3 expression. Treg cells pretreated with deubiquitinase inhibitor altered their function in prevention of adoptive transfer‐induced colitis 60. It is thought that FoxP3 undergoes polyubiquitination and proteasome‐based degradation in certain inflammatory conditions.
After lipopolysaccharide (LPS) stimulation, FoxP3 interacts with the stress‐indicator protein heat shock protein (Hsp)70 and the E3 ubiquitin ligase Stub1. As a result, Stub1 mediates K48‐linked polyubiquitination of multiple lysine residues of FoxP3, resulting in proteasome‐based degradation. Knock‐down of Stub1 by shRNA results in the dramatic accumulation of FoxP3 protein without changing the FoxP3 gene transcript, and promotes Treg suppressive function in vitro and in vivo 61.
The E3 ubiquitin ligase Cbl‐b was found to account for the defective development of thymic Treg cells in Cd28‐deficient mice. Upon TCR stimulation, Cbl‐b, together with Stub1, targets FoxP3 for ubiquitination and subsequently degradation in the proteasome. The loss of Cbl‐b rescues the defective development of tTreg in Cd28‐deficient mice 62. Phosphorylation of FoxP3 by NLK increases FoxP3 protein stabilization by preventing association with the STUB1 E3‐ubiquitin protein ligase 48. A recent study reported that the E3 ligase TNF receptor‐associated factor 6 (TRAF6) mediated FoxP3 K63‐linked ubiquitination at the lysine 262 residue. K63‐linked ubiquitination by TRAF6 ensures proper localization of FoxP3 and facilitates FoxP3 transcriptional activity in Tregs 63. The E3 ubiquitin ligase ring finger protein 31 (RNF31) catalyzes the conjugation of atypical ubiquitin chains to the FoxP3 protein. This interaction positively regulates both FoxP3 stability and Treg cell function. Moreover, RNF31 expression is correlated with intratumoral Treg cells in gastric cancer, implying a potential role of RNF31 in mediating tumor immunity 64.
FoxP3 can methylate modification by the arginine methyl transferase protein PRMT5. Conditional knock‐out of the PRMT5 gene in Tregs causes severe Scurfy‐like autoimmunity. Consistently, pharmacological ablation of PRMT5 activity by DS‐437 also reduces human Treg functions, which enhance the anti‐tumor effects of anti‐erbB2/neu monoclonal antibody‐targeted therapy in mice bearing CT26 human epidermal growth factor receptor (HER2) tumors 65.
FoxP3 acts as an important transcriptional factor for Treg cell development and suppressive function in normal physiological and stressed biological environments. Understanding FoxP3 protein features and modulation mechanisms may help the design of rational therapies for immune disorders.
FoxP3 protein has various post‐translational modifications, such as phosphorylation, acetylation, ubiquitination and methylation. Several relevant enzymes have been determined to involve in these modifications. In addition, accumulating evidence suggests that these post‐translational modifications may affect FoxP3 protein conformation and interactions with other partners, which ultimately alters FoxP3 protein activity to modulate Treg suppressive features. Based on the high‐resolution crystal structure of FoxP3, combination therapy by targeting FoxP3 post‐translational modifications with other therapies, may enhance therapeutic approaches in a variety of diseases.
Disclosures
None.
Acknowledgements
The authors thank Drs Yan Xiao and Zheng Cai for their helpful discussion. This work is supported by Start Funding of Peking University Health Science Center (BMU2018YJ011), and by the non‐profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT31039).
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
Regulatory T cells: exploring mechanisms for future therapies. Clinical and Experimental Immunology 2019, 197: 11–13.
From stability to dynamics: understanding molecular mechanisms of regulatory T cells through Foxp3 transcriptional dynamics. Clinical and Experimental Immunology 2019, 197: 14–23.
The role of FOXP3+ regulatory T cells in human autoimmune and inflammatory diseases. Clinical and Experimental Immunology 2019, 197: 24–35.
Mechanisms of human FoxP3+ Treg cell development and function in health and disease. Clinical and Experimental Immunology 2019, 197: 36–51.
Methods to manufacture regulatory T cells for cell therapy. Clinical and Experimental Immunology 2019, 197: 52–63.
Contributor Information
G. Deng, Email: gdeng@pku.edu.cn.
M. I. Greene, Email: greene@reo.med.upenn.edu.
References
- 1. Qin S, Cobbold SP, Pope H et al “Infectious” transplantation tolerance. Science. 1993; 259:974–7. [DOI] [PubMed] [Google Scholar]
- 2. Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self‐tolerance maintained by activated T cells expressing IL‐2 receptor alpha‐chains (CD25). Breakdown of a single mechanism of self‐tolerance causes various autoimmune diseases. J Immunol 1995; 155:1151–64. [PubMed] [Google Scholar]
- 3. Sakaguchi S. The origin of FOXP3‐expressing CD4+ regulatory T cells: thymus or periphery. J Clin Invest 2003; 112:1310–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee HM, Bautista JL, Hsieh CS. Thymic and peripheral differentiation of regulatory T cells. Adv Immunol 2011; 112:25–71. [DOI] [PubMed] [Google Scholar]
- 5. Chen X, Subleski JJ, Hamano R, Howard OM, Wiltrout RH, Oppenheim JJ. Co‐expression of TNFR2 and CD25 identifies more of the functional CD4+FOXP3+ regulatory T cells in human peripheral blood. Eur J Immunol 2010; 40:1099–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bianchini R, Bistoni O, Alunno A et al CD4(+) CD25(low) GITR(+) cells: a novel human CD4(+) T‐cell population with regulatory activity. Eur J Immunol 2011; 41:2269–78. [DOI] [PubMed] [Google Scholar]
- 7. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity 2019; 50:302–16. [DOI] [PubMed] [Google Scholar]
- 8. Ohkura N, Hamaguchi M, Morikawa H et al T cell receptor stimulation‐induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 2012; 37:785–99. [DOI] [PubMed] [Google Scholar]
- 9. Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 2014; 158:749–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003; 4:330–6. [DOI] [PubMed] [Google Scholar]
- 11. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003; 299:1057–61. [DOI] [PubMed] [Google Scholar]
- 12. Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol 2006; 24:209–26. [DOI] [PubMed] [Google Scholar]
- 13. Chatila TA, Blaeser F, Ho N et al JM2, encoding a fork head‐related protein, is mutated in X‐linked autoimmunity‐allergic disregulation syndrome. J Clin Invest 2000; 106:R75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baud O, Goulet O, Canioni D et al Treatment of the immune dysregulation, polyendocrinopathy, enteropathy, X‐linked syndrome (IPEX) by allogeneic bone marrow transplantation. N Engl J Med 2001; 344:1758–62. [DOI] [PubMed] [Google Scholar]
- 15. Wildin RS, Ramsdell F, Peake J et al X‐linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 2001; 27:18–20. [DOI] [PubMed] [Google Scholar]
- 16. Williams LM, Rudensky AY. Maintenance of the Foxp3‐dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol 2007; 8:277–84. [DOI] [PubMed] [Google Scholar]
- 17. Kim JY, Kim HJ, Hurt EM, Chen X, Howard OMZ, Farrar WL. Functional and genomic analyses of FOXP3‐transduced Jurkat‐T cells as regulatory T (Treg)‐like cells. Biochem Biophys Res Commun 2007; 362:44–50. [DOI] [PubMed] [Google Scholar]
- 18. Lopes JE, Torgerson TR, Schubert LA et al Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol 2006; 177:3133–42. [DOI] [PubMed] [Google Scholar]
- 19. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013; 13:482–95. [DOI] [PubMed] [Google Scholar]
- 20. Marson A, Kretschmer K, Frampton GM et al Foxp3 occupancy and regulation of key target genes during T‐cell stimulation. Nature 2007; 445:931–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome‐wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 2007; 445:936–40. [DOI] [PubMed] [Google Scholar]
- 22. Konopacki C, Pritykin Y, Rubtsov Y, Leslie CS, Rudensky AY. Transcription factor Foxp1 regulates Foxp3 chromatin binding and coordinates regulatory T cell function. Nat Immunol 2019; 20:232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angelin A, Gil‐de‐Gomez L, Dahiya S et al Foxp3 reprograms T cell metabolism to function in low‐glucose, high‐lactate environments. Cell Metab 2017; 25:1282‐93.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hancock WW, Ozkaynak E. Three distinct domains contribute to nuclear transport of murine Foxp3. PLOS ONE 2009; 4:e7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bandukwala HS, Wu Y, Feuerer M et al Structure of a domain‐swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity 2011; 34:479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Chen C, Zhang Z et al DNA binding by FOXP3 domain‐swapped dimer suggests mechanisms of long‐range chromosomal interactions. Nucleic Acids Res 2015; 43:1268–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song X, Li B, Xiao Y et al Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep 2012; 1:665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deng G, Xiao Y, Zhou Z et al Molecular and biological role of the FOXP3 N‐terminal domain in immune regulation by T regulatory/suppressor cells. Exp Mol Pathol 2012; 93:334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B, Samanta A, Song X et al FOXP3 is a homo‐oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol 2007; 19:825–35. [DOI] [PubMed] [Google Scholar]
- 30. Pan F, Yu H, Dang EV et al Eos mediates Foxp3‐dependent gene silencing in CD4+ regulatory T cells. Science 2009; 325:1142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ono M, Yaguchi H, Ohkura N et al Foxp3 controls regulatory T‐cell function by interacting with AML1/Runx1. Nature 2007; 446:685–9. [DOI] [PubMed] [Google Scholar]
- 32. Wu Y, Borde M, Heissmeyer V et al FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 2006; 126:375–87. [DOI] [PubMed] [Google Scholar]
- 33. Du J, Huang C, Zhou B, Ziegler SF. Isoform‐specific inhibition of ROR alpha‐mediated transcriptional activation by human FOXP3. J Immunol 2008; 180:4785–92. [DOI] [PubMed] [Google Scholar]
- 34. Li B, Samanta A, Song X et al FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA 2007; 104:4571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Z, Song X, Li B, Greene MI. FOXP3 and its partners: structural and biochemical insights into the regulation of FOXP3 activity. Immunol Res 2008; 42:19–28. [DOI] [PubMed] [Google Scholar]
- 36. Bettini ML, Pan F, Bettini M et al Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity 2012; 36:717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darce J, Rudra D, Li L et al An N‐terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity 2012; 36:731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su Q, Jing J, Li W et al Impaired Tip60‐mediated Foxp3 acetylation attenuates regulatory T cell development in rheumatoid arthritis. J Autoimmun 2019; 100:27–39. [DOI] [PubMed] [Google Scholar]
- 39. Bin Dhuban K, d'Hennezel E, Nagai Y et al Suppression by human FOXP3(+) regulatory T cells requires FOXP3–TIP60 interactions. Sci Immunol 2017; 2:eaai9297. [DOI] [PubMed] [Google Scholar]
- 40. Rudra D, deRoos P, Chaudhry A et al Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol 2012; 13:1010–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Samanta A, Li B, Song X et al TGF‐beta and IL‐6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci USA 2008; 105:14023–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morawski PA, Mehra P, Chen C, Bhatti T, Wells AD. Foxp3 protein stability is regulated by cyclin‐dependent kinase 2. J Biol Chem 2013; 288:24494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chunder N, Wang L, Chen C, Hancock WW, Wells AD. Cyclin‐dependent kinase 2 controls peripheral immune tolerance. J Immunol 2012; 189:5659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycin‐resistant T cell survival and activation. J Exp Med 2005; 201:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Cutting edge: Foxp3‐mediated induction of pim 2 allows human T regulatory cells to preferentially expand in rapamycin. J Immunol 2008; 180:5794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Z, Lin F, Zhuo C et al PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J Biol Chem 2014; 289:26872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng G, Nagai Y, Xiao Y et al Pim‐2 kinase influences regulatory T cell function and stability by mediating foxp3 protein N‐terminal phosphorylation. J Biol Chem 2015; 290:20211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fleskens V, Minutti CM, Wu X et al Nemo‐like kinase drives Foxp3 stability and is critical for maintenance of immune tolerance by regulatory T cells. Cell Rep 2019; 26:3600–12.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakahira K, Morita A, Kim NS, Yanagihara I. Phosphorylation of FOXP3 by LCK downregulates MMP9 expression and represses cell invasion. PLOS ONE 2013; 8:e77099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nie H, Zheng Y, Li R et al Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF‐alpha in rheumatoid arthritis. Nat Med 2013; 19:322–8. [DOI] [PubMed] [Google Scholar]
- 51. Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res 2004; 32:959–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Loosdregt J, Vercoulen Y, Guichelaar T et al Regulation of Treg functionality by acetylation‐mediated Foxp3 protein stabilization. Blood 2010; 115:965–74. [DOI] [PubMed] [Google Scholar]
- 53. Xiao Y, Nagai Y, Deng G et al Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep 2014; 7:1471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Daenthanasanmak A, Iamsawat S, Chakraborty P et al Targeting Sirt‐1 controls GVHD by inhibiting T‐cell allo‐response and promoting Treg stability in mice. Blood 2019; 133:266–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin‐1 control Foxp3+ regulatory T cell function through shared and isoform‐specific mechanisms. Sci Signal 2012; 5:ra45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arpaia N, Campbell C, Fan X et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Furusawa Y, Obata Y, Fukuda S et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 58. Beier UH, Akimova T, Liu Y, Wang L, Hancock WW. Histone/protein deacetylases control Foxp3 expression and the heat shock response of T‐regulatory cells. Curr Opin Immunol 2011; 23:670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choi SW, Braun T, Chang L et al Vorinostat plus tacrolimus and mycophenolate to prevent graft‐versus‐host disease after related‐donor reduced‐intensity conditioning allogeneic haemopoietic stem‐cell transplantation: a phase 1/2 trial. Lancet Oncol 2014; 15:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van Loosdregt J, Fleskens V, Fu J et al Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg‐cell‐suppressive capacity. Immunity 2013; 39:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen Z, Barbi J, Bu S et al The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 2013; 39:272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao Y, Guo H, Qiao G, Zucker M, Langdon WY, Zhang J. E3 ubiquitin ligase Cbl‐b regulates thymic‐derived CD4+CD25+ regulatory T cell development by targeting Foxp3 for ubiquitination. J Immunol 2015; 194:1639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ni X, Kou W, Gu J et al TRAF6 directs FOXP3 localization and facilitates regulatory T‐cell function through K63‐linked ubiquitination. EMBO J 2019; 38:e99766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu F, Yi G, Liu X et al Ring finger protein 31‐mediated atypical ubiquitination stabilizes forkhead box P3 and thereby stimulates regulatory T‐cell function. J Biol Chem 2018; 293:20099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nagai Y, Ji MQ, Zhu F et al PRMT5 associates with the FOXP3 homomer and when disabled enhances targeted p185(erbB2/neu) tumor immunotherapy. Front Immunol 2019; 10:174. [DOI] [PMC free article] [PubMed] [Google Scholar]


