Abstract.
The genus Phlebovirus is a diverse group of globally occurring viruses, including tick-, mosquito-, and sand fly–borne pathogens. Phleboviruses have historically been classified by serological methods. However, molecular methods alone have been used to identify emergent novel and related strains in recent years. This makes reconciling the classification of historically and newly characterized viruses challenging. To address this in part, we describe the characterization of the genomes of the Frijoles and Chilibre species complex phleboviruses, and three unclassified phleboviruses isolated in the Americas: Caimito, Itaporanga, and Rio Grande viruses that had previously only been described at the serological level. With the exception of Itaporanga virus, the phleboviruses sequenced in this study are phylogenetically related to the current species Frijoles phlebovirus, Bujaru phlebovirus, or the Chagres antigenic complex. Unexpectedly, molecular and phylogenetic analysis suggests Chilibre and Caimito viruses are taxonomically related to the family Peribunyaviridae. These viruses have a genomic architecture similar to peribunyaviruses and form monophyletic groups within the genus Pacuvirus. Our data highlight the importance of reconciling serological and molecular taxonomic classification. In addition, we suggest the taxonomy of Chilibre and Caimito viruses should be revised.
INTRODUCTION
The genus Phlebovirus contains about 70 named viruses, distributed widely in Europe, Africa, Central Asia, and the Americas. These viruses are currently classified into 10 species by the International Committee on Taxonomy of Viruses (ICTV): Bujaru phlebovirus, Candiru phlebovirus, Chilibre phlebovirus, Frijoles phlebovirus, Punta Torro phlebovirus, Rift Valley fever phlebovirus, Salehabad phlebovirus, Sandfly fever Naples phlebovirus, Mukawa phlebovirus, and Uukuniemi phlebovirus. In addition, at least 32 unclassified strains are associated with this genus.1 Phleboviruses have a genome comprising three segments of negative sense, single-stranded RNA. The large segment (L) encodes the viral RNA-dependent RNA polymerase, the medium segment (M) encodes the structural glycopolyproteins, and the small segment (S) has an ambisense strategy encoding the nucleoprotein (NP) in the negative sense and the nonstructural small (NSs) protein in the positive sense.2
Phleboviruses are largely transmitted by sand flies; Phlebotomus species in the Old World or Nyssomyia (previously Lutzomyia) species in the New World.3 However, some viruses in this genus are transmitted by mosquitoes, Culicoides, and ticks. Many phleboviruses have been associated with human and animal disease. Pathogenic strains include Rift Valley fever, Toscana, Candiru, Sandfly fever Naples, Sandfly fever Sicilian, and severe fever with thrombocytopenia syndrome viruses.4 Recent discoveries of Sandfly fever Turkey,5 Heartland,6 Hunter Island Group,7 Fermo,8 and Malsoor9 viruses highlight the extent to which the diversity and public or animal health impact of phleboviruses are still unknown and likely underestimated.
Despite their public health impact, some phlebovirus species remain characterized solely by serological techniques. Given the many recent discoveries, genetic characterization of historically identified phleboviruses is important for the development of diagnostic and surveillance tools, and evolutionary insights into the ever-growing breadth of recognized phleboviruses. Here, we describe the complete sequences of two phlebovirus species complexes, and three previously unclassified phleboviruses isolated in the Americas.
MATERIALS AND METHODS
All protocols followed the manufacturer’s recommendations, unless otherwise noted.
Viruses.
Viruses sequenced in the study were obtained from the CDC Arbovirus Reference Collection, Arboviral Diseases Branch, Division of Vector-Borne Diseases. Isolate details are described in Table 1. RNA was extracted from frozen stocks using the QIAmp Viral RNA Mini Kit (Qiagen, Germantown, MD).
Table 1.
Viruses sequenced in this study
| Virus | Current classification | Isolate | Collection date | Country | Host | Accession numbers |
|---|---|---|---|---|---|---|
| Cacao | Chilibre phlebovirus | VP-437R | April 12, 1970 | El Aguacate, Panama | Nyssomyia trapidoi* | MK330756-58 |
| Caimito | Unclassified phlebovirus | VP-488A | January 21, 1971 | El Aguacate, Panama | Nyssomyia ylephiletor* | MK330759-61 |
| Chilibre | Chilibre phlebovirus | VP-118D | September 29, 1969 | Canal Zone, Panama | Lutzomyia spp. | MK330762-64 |
| Frijoles | Frijoles phlebovirus | VP-161A | November 24, 1969 | Canal Zone, Panama | Lutzomyia spp. | MK330765-67 |
| Icoaraci | Rift Valley fever phlebovirus | BeAn24262 | October 14, 1960 | Para, Brazil | Rodent | MK330768-70 |
| Itaporanga | Unclassified phlebovirus | original | March 4, 1962 | Itaporanga, Brazil | Sentinel mouse | MK330771-73 |
| Rio Grande | Unclassified phlebovirus | TBM3-204 | May 12, 1973 | Texas, United States | Neotoma micropus | MK330774–75, MN517122 |
* Classified as Lutzomyia species at the time of isolation.
Genome sequencing and assembly.
Complementary DNA was generated from extracted RNA using the Ovation RNA-Seq System V2 Kit (NuGEN, Redwood City, CA). Sequencing libraries were made using the Ion Xpress Plus Fragment Library Kit (Life Technologies, Carlsbad, CA), barcoded with Ion Xpress Barcodes (Life Technologies), and quantified using the Ion library TaqMan Quantitation Kit (Life Technologies). Sequencing templates were prepared using the Ion One Touch 2 System and Ion Hi-Q View OT2 kits (Life Technologies). Whole genome sequencing was performed on the Ion Torrent Personal Genomics Machine system using the Ion Hi-Q View Sequencing Kit and 318 v. 2 chips (Life Technologies), sequencing two libraries per chip.
Genomes were assembled from fastq files with a base Phred quality of Q ≥ 20 in CLC Genomics Workbench v. 11 (Qiagen) using de novo assembly with a bubble size of the average read length. Contigs greater than 500 nt in length were submitted for BLAST analysis through NCBI. Consensus sequences for each segment were extracted from the CLC Genomics Workbench and used for reference-guided assembly in SeqMan NGen (DNASTAR), and putative open-reading frames were identified using EditSeq (DNASTAR, Madison, WI). Terminal ends were determined for all sequences using the 5′ RACE System 2.0 Kit (Life Technologies) and the 3′ Poly(A) Tailing Kit (Ambion-Thermo Fisher Scientific, Waltham, MA) followed by the 3′ RACE Kit (Life Technologies), using gene-specific primers (IDT, Coralville, IA). RACE DNA templates were cloned with TOPO-TA sequencing kits (Life technologies), and five replicates per termini were miniprepped (Qiagen) and sequenced by capillary sequencing on an ABI 3130 instrument (Life Technologies) using the primers provided in the TOPO-TA kits. All sequences generated in this study have been deposited in GenBank (Table 1).
Molecular and phylogenetic analysis.
Conserved domains were identified in each segment by CD-search through NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Predicted proteins were submitted to the TOPCONS server for identification of transmembrane and signal peptide regions.10 Potential glycosylation sites in the glycoproteins were identified using the NetNGlyc server (http://www.cbs.dtu.dk/services/NetNGlyc/). Nucleotide sequences comprising the open-reading frames for each protein were codon-aligned using the ClustalW function of Mega 7.11 Percent sequence identity was calculated in Mega 7 using the p-distance model with G + I frequencies and complete deletion of missing data. Substitution models were determined using the model fit function of Mega 7. Bayesian inference was completed using BEAST v. 1.8.412 executed through the CIPRES Scientific Gateway (www.phylo.org).13 The model GTR + G + I was used for all trees except the orthobunyavirus S tree, which used GTR + G. A lognormal relaxed clock and a coalescent constant tree prior were used with an MCMC of 100 million generations and 10% burn-in. Priors were tested using the generalized stepping-stone sampling method.14 Convergence of parameters was verified using TRACER v. 1.5.15 Maximum clade credibility trees were generated using TreeAnnotator12 and FigTree v. 1.4.3 showing the posterior probability of each branch. Maximum likelihood trees generated in Mega 7 verified Bayesian tree topologies.
Reassortment was evaluated by concatenating the open-reading frames of 47 complete genomes representing at least one virus from each clade. Only reassortment events associated with a full segment of the concatenated genome that had P < 0.05 using the Bonferoni adjustment and detected by three or more models using recombination detection program (RDP) v. 4.916 were considered significant.17
RESULTS
Genome analysis.
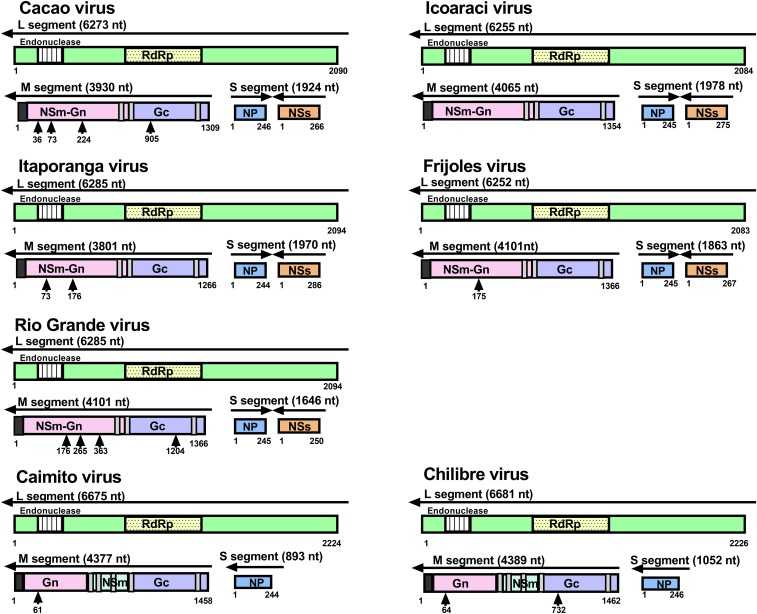
Complete genomes of Cacao, Caimito, Chilibre, Frijoles, Icoaraci, Itaporanga, and Rio Grande viruses were sequenced (Table 2) and found to include three negative-sense RNA molecules consistent with bunyavirus S, M, and L segments (Figure 1).
Table 2.
Next-generation sequencing metrics of viruses in this study
| Virus | Segment | Sequencing depth (reads) | Average coverage (total nt./contig length) |
|---|---|---|---|
| Cacao | Large | 941,866 | 23,107 |
| Medium | 239,818 | 9,789 | |
| Small | 72,084 | 6,089 | |
| Caimito | Large | 164,510 | 3,720 |
| Medium | 89,167 | 2,963 | |
| Small | 15,104 | 2,177 | |
| Chilibre | Large | 119,283 | 3,205 |
| Medium | 459,251 | 15,904 | |
| Small | 82,185 | 11,899 | |
| Frijoles | Large | 430,190 | 9,348 |
| Medium | 269,486 | 8,304 | |
| Small | 165,384 | 11,441 | |
| Icoaraci | Large | 551,243 | 11,244 |
| Medium | 257,597 | 8,380 | |
| Small | 106,093 | 8,045 | |
| Itaporanga | Large | 359,773 | 8,162 |
| Medium | 210,617 | 6,941 | |
| Small | 124,241 | 8,740 | |
| Rio Grande | Large | 330,900 | 6,655 |
| Medium | 390,455 | 11,280 | |
| Small | 71,130 | 5,575 |
Figure 1.
Genomic organization of viruses sequenced in this study. The whole genomes of sequenced viruses were analyzed for conserved domains, transmembrane domains, and glycosylation sites (see Methods). Putative open-reading frames are numbered by amino acid. Dark gray squares are the signal peptide, and light gray squares are transmembrane domains in the glycoprotein. Black arrows denote possible N-linked glycosylation sites. RdRp = RNA-dependent RNA polymerase. This figure appears in color at www.ajtmh.org.
The S segments of Rio Grande, Cacao, Frijoles, Icoaraci, and Itaporanga range in size from 1,646 nt to 1,978 nt in length and have an ambisense coding strategy with the negative-sense NP and the positive-sense NSs, similar to other phleboviruses. The NP proteins range in size from 244 amino acids (Itaporanga) to 246 amino acids (Cacao), whereas the NSs proteins are more variable in size and range from 250 amino acids (Rio Grande) to 286 amino acids (Itaporanga). Surprisingly, the S segments of Caimito and Chilibre viruses are significantly shorter in length (893 and 1,052 nt, respectively) and only encode the NP (244 and 246 amino acids, respectively).
The M segments encode for the polyprotein, which varies in length. Cacao, Frijoles, Icoaraci, Itaporanga, and Rio Grande have smaller polyproteins, ranging in size from 1,266 amino acids (Itaporanga) to 1,366 amino acids (Rio Grande and Frijoles). These polyproteins encode for the NSm upstream of the Gn and Gc proteins and have three transmembrane domains (Figure 1), which is similar to other phleboviruses.18,19 However, Caimito and Chilibre polyproteins are larger in size (1,458 and 1,462 amino acids, respectively) and encode for Gn, NSm, and Gc. The polyprotein possesses five transmembrane domains, similar to viruses in the family Peribunyaviridae.20
The L segments encode for the L protein, ranging in size from 2,083 amino acids (Frijoles) to 2,094 amino acids (Itaporagna and Rio Grande) in length. The Caimito and Chilibre encode for a larger protein, 2,224 and 2,226 amino acids in length, respectively. All L segment open-reading frames encode for the N-terminal endonuclease domain21 and the central polymerase motifs22 common to all viruses in the order Bunyavirales.
The terminal ends for each segment of Cacao, Icoaraci, Frijoles, and Itaporanga are the canonical sequences (in coding sense) 5′-ACACAAAG….CTTTGTGT-3′ typical of all phleboviruses. Whereas the Rio Grande virus has these expected terminal ends for most segments, the 5′ of the M segment is 5′-ACACAAGG and the 3′ of the S segment is CTCTGTGT-3’. The terminal ends of Caimito and Chilibre are consistent with the terminal ends found in orthobunyaviruses (in the coding sense), 5′-AGTAGTGTACT….AGCACACTACT-3′, with the exception of Caimito S segment that has the terminal end AGCAGCTACT-3’. In addition, NCBI BLAST analysis of the proteins encoded by Caimito and Chilibre reveals the highest identity to Tapirape, Pacui, and Rio Preto da Eva, viruses isolated in Brazil from rodents and sand flies.23 Although the terminal ends of pacuviurses have not been derived, the genomic organization, terminal end sequences, and NCBI BLAST analysis of Caimito and Chilibre suggest that these viruses belong to the family Peribunyaviridae, genus Pacuvirus.
Phylogenetic relationships.
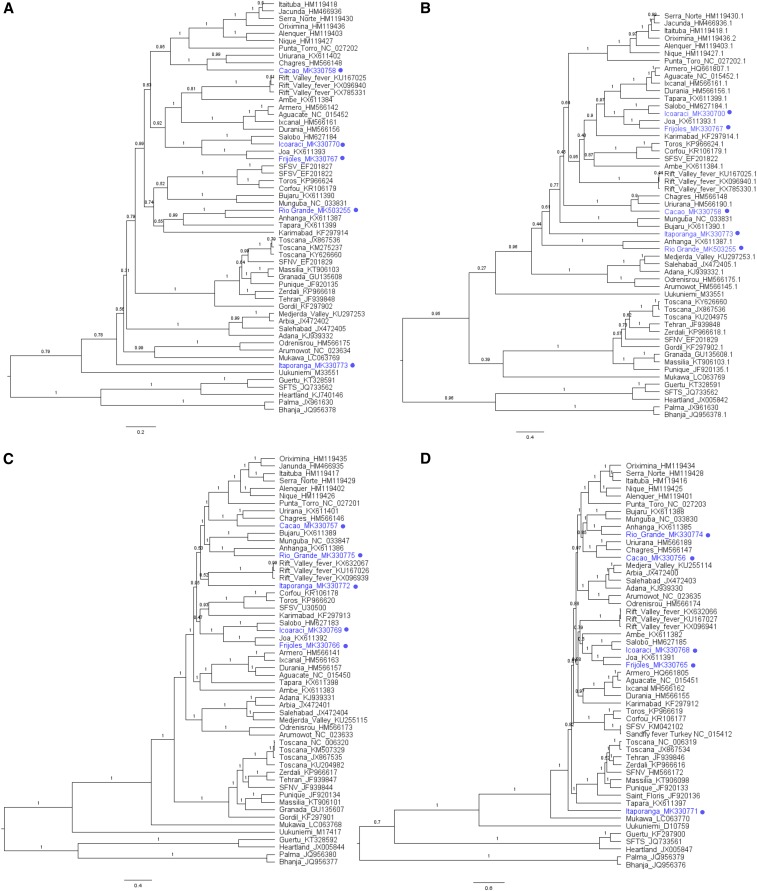
Bayesian phylogenetic inference of the coding sequence of each protein resulted in 18 well-supported clades within the genus Phlebovirus19 (Figure 2); however, higher divergence in the NSs tree resulted in weaker supported nodes. Viruses that cluster together create the same clade regardless of the protein coding sequence analyzed; however, tree topologies are subtly different. Nine of these clades represent nine species complexes described by the ICTV: Candiru, Bujaru, Frijoles, Rift Valley fever, Salehabad, Sandfly fever Naples, Punta Torro, Mukawa, and Uukuniemi.
Figure 2.
Bayesian maximum clade credibility trees of the genus Phlebovirus. Nucleotide coding sequences of phleboviruses depicting phylogeny of the (A) nucleoprotein, (B) nonstructural small (NSs) protein, (C) M segment, and (D) L segment. Taxa are labeled with virus names and GenBank accession numbers. Viruses sequenced in this study are labeled with a circle. Branches are labeled with the posterior probabilities, and scale bar depicts nucleotide substitutions per site. SFNV = Sandfly fever Naples virus; SFSV = Sandfly fever Sicilian virus, SFTS = severe fever with thrombocytopenia virus; TOSV = Toscana virus. This figure appears in color at www.ajtmh.org.
Of the classified viruses sequenced in this study, Frijoles groups with species member Joa virus and Icoaraci consistently groups with Salobo.24 Salobo and Icoaraci consistently form a well-supported monophyletic group with Frijoles and Joa viruses in each of the protein-coding sequences evaluated. Surprisingly, Cacao virus groups with the Chagres antigenic complex viruses. The Cacao and Chagres group viruses form clades with the Punta Toro and Candiru species complexes in the NP and M trees (Figure 2A and C), whereas the NSs and L form clades with the Bujaru species complex (Figure 2B and D).
Of the unclassified viruses sequenced in this study, Rio Grande virus consistently forms a clade with Anhanga virus. Rio Grande and Anhanga appear in a group with Tapara in the NP tree (Figure 2A), Salehabad in the NSs tree (Figure 2B), and Bujaru complex in the M and L trees (Figure 2C and D). Itaporanga virus displays a distinct branch pattern in each of the four protein-coding sequences. Itaporanga forms a single-virus clade in the NP (Figure 2A), NSs (Figure 2B), and L (Figure 2D) trees, branching near the ancestral node of the sand fly/mosquito-borne virus group and Mukawa virus25 in the NP and L coding trees. The M sequence of Itaporanga forms a clade with Rift Valley fever (Figure 2D).
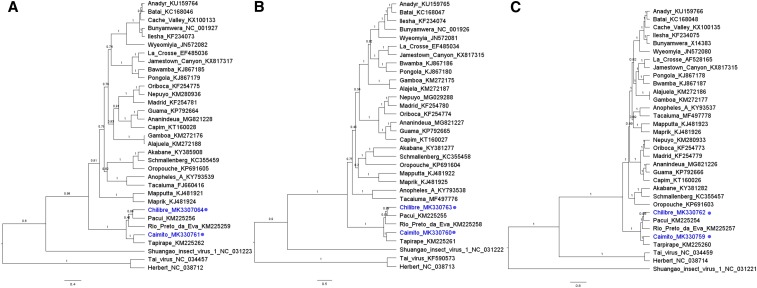
Chilibre and Caimito viruses do not group with any virus in the genus Phlebovirus; however, these viruses appear in well-supported clades with the viruses in the genus Pacuvirus, family Peribunyaviridae (Figure 3A–C). Chilibre is most closely related to Pacui virus, whereas Caimito is most closely related to Tapirape virus in the S, M, and L segment protein-coding sequences. These data further support the classification of Chilibre and Caimito viruses in the genus Pacuvirus.
Figure 3.
Bayesian maximum clade credibility trees of Chilibre and Caimito viruses. Nucleotide coding sequences of viruses in the family Peribunyaviridae depicting phylogeny of the (A) S segment, (B) M segment, and (C) L segment. Taxa are labeled with virus names and GenBank accession numbers. Viruses sequenced in this study are labeled with a circle. Branches are labeled with the posterior probabilities, and scale bar depicts nucleotide substitutions per site. This figure appears in color at www.ajtmh.org.
Reassortment evaluation.
Given the discrepant branching pattern observed from the phylogenetic analyses, we performed RDP analysis on concatenated genomes for at least one named virus in every clade with full genome information available. Our analysis did identify and confirm Aguacate as a reassortant of Armero and Ixcanal.26 However, despite the incongruence in branching patterns, segment reassortment was not identified in any of the viruses sequenced in this study. In addition, segment reassortment was investigated in Chilibre and Caimito viruses as a possible explanation for the antigenic relatedness to other phleboviruses; however, segment reassortment was not identified.
DISCUSSION
The genus Phlebovirus is a vast, geographically and ecologically diverse group. Current understanding of the diversity and classification of phleboviruses has mainly relied on serological relationships. However, thorough serological testing of phleboviruses is not feasible because of increasing numbers and diversity of recognized viruses, limiting serological testing relative to molecular techniques.19,27,28 Although recent efforts to describe the genomes and phylogeny of phleboviruses have advanced our understanding, many viruses remain unclassified and their genomes undescribed.
The goal of this study was to help improve the understanding and classification of the genus Phlebovirus by genetically describing the Frijoles and Chilibre species complexes, and three additional unclassified viruses. Our phylogenetic analysis suggests the classification of Frijoles, Joa, Icoaraci, and Salobo viruses into an expanded Frijoles phlebovirus species complex. These viruses consistently form groups in each tree, and previous studies of partial genomes and serological relationships are supportive of this expanded species.19,24 In addition, Rio Grande virus, isolated from North America, groups with Anhanga virus in all phylogenetic trees and shares serological relatedness, suggesting these viruses may form an additional species complex.29–31 Interestingly, the branching patterns of Itaporanga virus, isolated from mosquitoes,3 near the recently identified Ixodes-associated Mukawa virus,25 suggests the phleboviruses evolved first from tick-associated viruses, to mosquito-associated viruses, and finally sand fly–associated viruses. Future analysis of more Itaporanga and Mukawa isolates will help clarify these relationships and improve our understanding of phlebovirus evolution.
Cacao virus is currently classified as a member of the Chilibre phlebovirus species, based on serological relationships.31 However, our data suggest that Cacao virus could be classified in a complex with Chagres and Uriurana. This is surprising, given the lack of serological relatedness of Cacao virus to Chagres or Uriurana.19,31 In addition, the clustering of Cacao virus with several pathogenic phleboviruses, including Chagres,32 Candiru,28 and Punta Torro,33 and serological evidence of Cacao infection in humans34,35 suggest Cacao virus may be associated with human disease.
Of unique interest, our study suggests Chilibre and Caimito viruses are genetically similar to viruses in the family Peribunyaviridae, specifically viruses in the newly recognized genus Pacuvirus. These data were surprising, given Chilibre and Cacao viruses comprise the species complex Chilibre phlebovirus and share reproducible two-way serological relationship in complement fixation.19,30 Reconciling serological and molecular characterizations of rare and underrecognized viruses is a continual challenge to accurate classification.20,36 Although outside the scope of this study, additional independent serological and molecular testing of varied Chilibre and Cacao virus isolates will aid in correlating and confirming recently derived molecular and historically determined serological data. For diagnosis, neutralization tests have identified Cacao virus antibody in humans and can distinguish between Chilibre and Cacao viruses, suggesting neutralization tests remain the gold standard for serological diagnostic confirmation of suspect phlebovirus infection.31
Here, we have described the molecular and phylogenetic relationships of seven viruses recognized within the genus Phlebovirus and provide evidence suggesting two of these viruses, Chilibre and Caimito, are taxonomically related to the family Peribunyaviridae. In light of these findings and recent reports describing the serological misclassification of Estero Real virus,20 serological and genetic methods of classification should be considered in conjunction, and with independent confirmation of multiple strains when possible.
REFERENCES
- 1.Dietzgen R, Calisher CH, Kurath G, Kuzman RLL, IV, Stone DM, et al. 2012. Virus Taxonomy King A, Adams MJ, Carstens EB, Lefkowitz EJ, eds. Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier, 654–681. [Google Scholar]
- 2.Elliott RM, 1990. Molecular biology of the Bunyaviridae. J Gen Virol 71: 501–522. [DOI] [PubMed] [Google Scholar]
- 3.Tesh RB, 1988. The genus Phlebovirus and its vectors. Annu Rev Entomol 33: 169–181. [DOI] [PubMed] [Google Scholar]
- 4.Elliott RM, Brennan B, 2014. Emerging phleboviruses. Curr Opin Virol 5: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carhan A, Uyar Y, Ozkaya E, Ertek M, Dobler G, Dilcher M, Wang Y, Spiegel M, Hufert F, Weidmann M, 2010. Characterization of a sandfly fever Sicilian virus isolated during a sandfly fever epidemic in Turkey. J Clin Virol 48: 264–269. [DOI] [PubMed] [Google Scholar]
- 6.McMullan LK, et al. 2012. A new Phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367: 834–841. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, et al. 2014. Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg Infect Dis 20: 1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remoli ME, Fortuna C, Marchi A, Bucci P, Argentini C, Bongiorno G, Maroli M, Gradoni L, Gramiccia M, Ciufolini MG, 2014. Viral isolates of a novel putative phlebovirus in the Marche region of Italy. Am J Trop Med Hyg 90: 760–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mourya DT, et al. 2014. Malsoor virus, a novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and heartland virus. J Virol 88: 3605–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernsel A, Viklund H, Hennerdal A, Elofsson A, 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37: W465–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummond AJ, Suchard MA, Xie D, Rambaut A, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller MA, Pfeiffer W, Schwartz T, 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, November 14, 2010, 1–8. [Google Scholar]
- 14.Baele G, Lemey P, Suchard MA, 2016. Genealogical working distributions for bayesian model testing with phylogenetic uncertainty. Syst Biol 65: 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond AJ, Rambaut A, 2007. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maljkovic Berry I, et al. 2016. Frequency of influenza H3N2 intra-subtype reassortment: attributes and implications of reassortant spread. BMC Biol 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halldorsson S, Behrens AJ, Harlos K, Huiskonen JT, Elliott RM, Crispin M, Brennan B, Bowden TA, 2016. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc Natl Acad Sci USA 113: 7154–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunes-Neto JP, et al. 2017. Characterization of the Bujaru, Frijoles and Tapara antigenic complexes into the sandfly fever group and two unclassified phleboviruses from Brazil. J Gen Virol 98: 585–594. [DOI] [PubMed] [Google Scholar]
- 20.Aguilar PV, Marciel de Souza W, Silvas JA, Wood T, Widen S, Fumagalli MJ, Nunes MRT, 2018. Genetic characterization of the patois serogroup (genus Orthobunyavirus; family Peribunyaviridae) and evidence that Estero real virus is a member of the Orthonairovirus. Am J Trop Med Hyg 99: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reguera J, Weber F, Cusack S, 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog 6: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller R, Poch O, Delarue M, Bishop DH, Bouloy M, 1994. Rift valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75: 1345–1352. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues DS, et al. 2014. Pacui virus, Rio Preto da Eva virus, and Tapirape virus, three distinct viruses within the family Bunyaviridae. Genome Announc 2: e00923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu F, Liu D, Nunes MR, DA Rosa AP, Tesh RB, Xiao SY, 2007. Antigenic and genetic relationships among Rift Valley fever virus and other selected members of the genus Phlebovirus (Bunyaviridae). Am J Trop Med Hyg 76: 1194–1200. [PubMed] [Google Scholar]
- 25.Matsuno K, et al. 2018. The unique phylogenetic position of a novel tick-borne phlebovirus ensures an ixodid origin of the genus Phlebovirus. mSphere 3: e00239-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios G, da Rosa AT, Savji N, Sze W, Wick I, Guzman H, Hutchison S, Tesh R, Lipkin WI, 2011. Aguacate virus, a new antigenic complex of the genus Phlebovirus (family Bunyaviridae). J Gen Virol 92: 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palacios G, Tesh RB, Savji N, Travassos da Rosa AP, Guzman H, Bussetti AV, Desai A, Ladner J, Sanchez-Seco M, Lipkin WI, 2014. Characterization of the Sandfly fever Naples species complex and description of a new Karimabad species complex (genus Phlebovirus, family Bunyaviridae). J Gen Virol 95: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palacios G, et al. 2011. Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol 85: 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calisher CH, McLean RG, Smith GC, Szmyd DM, Muth DJ, Lazuick JS, 1977. Rio Grande–a new phlebotomus fever group virus from south Texas. Am J Trop Med Hyg 26: 997–1002. [DOI] [PubMed] [Google Scholar]
- 30.Travassos da Rosa AP, Tesh RB, Pinheiro FP, Travassos da Rosa JF, Peterson NE, 1983. Characterization of eight new phlebotomus fever serogroup arboviruses (Bunyaviridae: Phlebovirus) from the Amazon region of Brazil. Am J Trop Med Hyg 32: 1164–1171. [DOI] [PubMed] [Google Scholar]
- 31.Tesh RB, Peters CJ, Meegan JM, 1982. Studies on the antigenic relationship among phleboviruses. Am J Trop Med Hyg 31: 149–155. [DOI] [PubMed] [Google Scholar]
- 32.Peralta PH, Shelokov A, Brody JA, 1965. Chagres virus: a new human isolate from Panama. Am J Trop Med Hyg 14: 146–151. [DOI] [PubMed] [Google Scholar]
- 33.Gundacker ND, Carrera JP, Castillo M, Diaz Y, Valenzuela J, Tamhane A, Moreno B, Pascale JM, Tesh RB, Lopez-Verges S, 2017. Clinical manifestations of Punta Toro virus species complex infections, Panama, 2009. Emerg Infect Dis 23: 872–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tesh RB, Chaniotis BN, Peralta PH, Johnson KM, 1974. Ecology of viruses isolated from Panamanian phlebotomine sandflies. Am J Trop Med Hyg 23: 258–269. [DOI] [PubMed] [Google Scholar]
- 35.Tesh RB, Peralta PH, Shope RE, Chaniotis BN, Johnson KM, 1975. Antigenic relationships among phlebotomus fever group arboviruses and their implication for the epidemiology of sandfly fever. Am J Trop Med Hyg 24: 135–144. [DOI] [PubMed] [Google Scholar]
- 36.Lima JA, et al. 2019. Characterization of Triniti virus supports its reclassification in the family Peribunyaviridae. J Gen Virol 100: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]