Abstract.
Given that the C580Y polymorphism in the Plasmodium falciparum propeller domain of the kelch 13 gene (pfk13) was documented in Guyana, monitoring for mutations associated with antimalarial resistance was undertaken in neighboring Roraima state in Brazil. Polymorphisms in the pfmdr1 and pfk13 genes were investigated in 275 P. falciparum samples. No pfk13 mutations were observed. Triple mutants 184F, 1042D, and 1246Y were observed in 100% of the samples successfully sequenced for the pfmdr1 gene, with 20.1% of these having an additional mutation at codon 1034C. Among them, 2.5% of samples harbored two copies of the pfmdr1 gene. We found no evidence of the spread of C580Y parasites to Roraima state, Brazil. As previously observed, the 184F, 1042D, and 1246Y mutations in the pfmdr1 gene appear to be fixed in this region. Continued molecular surveillance is essential to detect any potential migration or local emergence of artemisinin-resistant mutation.
The introduction of artemisinin-based combination therapies (ACTs) for the treatment of malaria began in the 2000s in Cambodia and extended to South American countries and Africa in the early 2000. The first report of artemisinin resistance, resulting in delayed parasite clearance posttreatment, was reported in 2008 in Cambodia.1 Since then, artemisinin resistance has been detected in several countries in the Greater Mekong Subregion (GMS).2–5 Mutations in the Plasmodium falciparum kelch 13 propeller domain (pfk13) have been identified as molecular markers of artemisinin resistance.6 Currently, nine single nucleotide polymorphisms in the pfk13 gene are validated as molecular markers for artemisinin resistance: F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, and C580Y.7
Historically, resistance to antimalarial drugs, including chloroquine and sulfadoxine–pyrimethamine, has evolved independently in South America and South East Asia and then spread globally (reviewed in ref. 8). Interestingly, a focus of independent emergence of pfk13 C580Y mutation was reported in Guyana based on samples collected in 2010.9 A molecular monitoring study using samples collected from 2017 in different parts of Guyana confirmed the presence of this mutation (Musset et al., in press). Genome analysis of these samples confirmed evolution of C580Y allele in a single genetic background in parasites belonging to the Guyana region (L. Musset, personal communication). The presence of this mutation in Guyana has not been linked to phenotypic association with drug failure; however, a study from Suriname, bordering Guyana and Brazil, reported > 10% of patients with delayed parasite clearance in a therapeutic efficacy study (TES), although this TES found no pfk13 mutations.10 Nevertheless, these findings highlight the importance of continued monitoring for antimalarial drug resistance using TES and molecular monitoring in this region.
In 2018, Roraima reported 23,369 cases of malaria which was a 62% increase compared with 2017. Data from the Brazilian Secretariat of Health Surveillance revealed that 50% of patients seen in the State of Roraima were from Venezuela and Guyana. Given the high rates of migration of people from Venezuela and Guyana to Brazil, there is a concern that pfk13 mutations from Guyana could spread to Brazil. Therefore, we conducted a molecular monitoring study to assess the prevalence of known molecular markers of ACT resistance in Roraima state.
This study was part of a larger study aimed at understanding the epidemiology and characterization of autochthonous and imported malaria in Roraima state and approved by the Federal University of Roraima Ethical Committee (CAAE: 44055315.0.0000.5302) and the CDC. The study was carried out in three municipalities in Roraima state: Boa Vista, Pacaraima, and Rorainapolis, Figure 1.
Figure 1.
Study sites used for samples collection.
All individuals seeking a malaria diagnosis in the health posts within the study sites were eligible to participate in the study. Informed consent was obtained from all enrolled participants from whom venous blood samples were obtained. The blood was used to prepare Giemsa-stained microscopic slides for malaria diagnosis and dried blood spots (Whatman 903 Protein Saver Cards) for molecular studies. All patients who were positive by microscopy for malaria parasites were treated with artemether–lumefantrine, according to the Brazilian national therapeutic guidelines for malaria. The DNA was isolated using a QIAmp DNA Mini Kit (QIAGEN, Valencia, CA) following the manufacturer’s recommendations. Samples were tested for species identification using photo-induced electron transfer PCR (PET-PCR) assay as previously described,11 and all P. falciparum samples were analyzed for polymorphisms in the pfk139 propeller domain and the pfmdr1 gene using the Sanger sequencing method as previously described.12,13 The purified PCR products were sequenced using the BigDye Terminator kit (Applied Biosystems, Foster City, CA), version 3.1, and an Applied Biosystems 3130XL capillary sequencer (Foster City, CA). Sequence analysis was performed using Geneious 7.1.7 software (Biomatters, Auckland, New Zealand). Pfmdr1 Copy number variation (CNV) was determined using TaqMan real-time PCR (Stratagene MX3005P; Agilent Technologies, Santa Clara, CA) as previously described.9
A total of 751 specimens were collected between March 2016 and November 2017: In all, 354 (47.1%) were obtained from Pacaraima, 340 (45.3%) from Boa Vista, and 57 (7.6%) from Rorainapolis. Of these, 275 (36.6%) were P. falciparum positive (age range, 16–89 years), 96 (34.9%) from Boa vista, 178 (64.7%) from Pacaraima, and 1 (0.4%) from Rorainapolis. We did not observe any mutations (associated with artemisinin resistance or otherwise) in the pfk13 gene in the 239 (86.9%) successfully sequenced isolates, Table 1. A total of 224 samples were successfully sequenced for pfmdr1 gene; all the 224 samples had mutations at codons 184F, 1042D, and 1246Y. Of these, 45 (20.1%) also had mutation at codon 1034. No mutations were observed at pfmdr1codon 86. Of the 240 samples successfully investigated for pfmdr1 CNV, 6 (2.5%), all from Pacaraima, were shown to have two copies, Table 1.
Table 1.
Observed polymorphisms in selected genes of P. falciparum
| Gene/codons assayed | Number tested successfully | Mutations observed |
|---|---|---|
| pfk13 | 239 (86.9%) | No mutations |
| Codons: F446I, N458Y, M476H, Y493H, R539T, I543T, P553L, R561H, and C580Y | ||
| pfmdr1 | 224 (81.5%) | 86Y (0; 0.0%) |
| Codons: 86, 184, 1034, 1042, and 1246 | 184F (224; 100%) | |
| 1034C (45; 20.1%) | ||
| 1042D (224; 100.0%) | ||
| 1246Y (224; 100.0%) | ||
| pfmdr1 | 240 (87.3%) | With two copies (6; 2.5%) |
| Copy number variation |
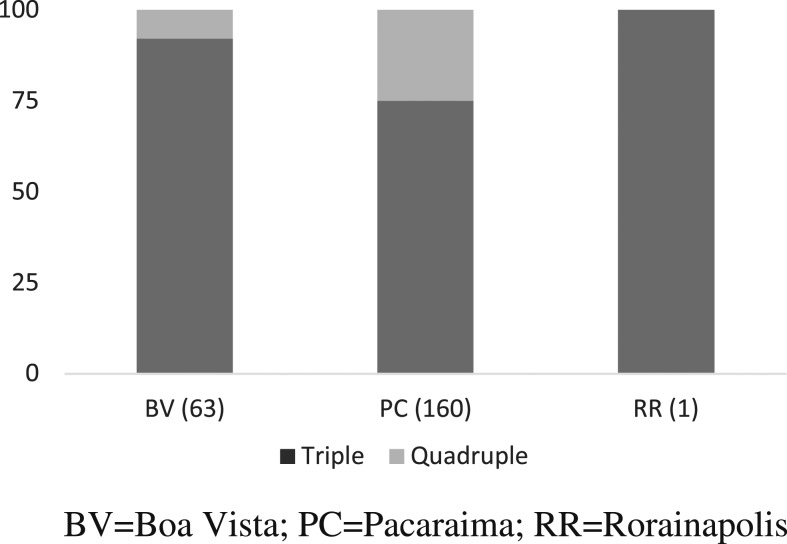
Differences in the proportions of quadruple and triple pfmdr1 mutants in Boa Vista and Pacaraima were analyzed using chi-squared test for proportions. We observed a significantly higher proportion of quadruple pfmdr1 mutants in Pacaraima than Boa vista, X2 (1, N = 223) = 28.1709, P < 0.05, Figure 2.
Figure 2.
Distribution of Pfmdr1 triple and quadruple mutations in the study sites.
Results from our study show no evidence that pfk13 polymorphisms, associated with resistance to artemisinin, have spread to Roraima state of Brazil. Our findings corroborate recent studies conducted in this region of Brazil, which revealed no pfk13 polymorphisms in the parasites investigated.14 In a recent study, none of the tested samples collected in the Brazilian Amazon basin before ACT introduction (1984–2006) and after ACT introduction (2006–2011) and between 2010 and 2017 were found to harbor any pfk13 polymorphisms.14 A search for pfk13 polymorphisms outside the GMS that included testing of 14,037 parasite samples collected in 59 malaria endemic countries, including Brazil, revealed no evidence for the existence of these polymorphisms in countries outside the GMS.15 Recent TES carried out in the Amazon basin in northern Brazil, the region that accounts for 99.8% of cases in the country, demonstrated adequate treatment outcomes using artemether–lumefantrine16 and artesunate–mefloquine.17 Neither studies identified any pfk13 polymorphisms associated with artemisinin resistance, thus supporting the continued use of these ACTs for treatment of malaria in Brazil.
Monitoring all relevant partner drug resistance markers is equally important. In Brazil, artemether–lumefantrine is the first-line antimalarial treatment, commonly used in the Amazon region, and artesunate–mefloquine is used in the other regions. As previously observed, every successfully sequenced parasite harbored the 184F, 1042D, and 1246Y mutations in the pfmdr1 gene, suggesting fixation of these resistance alleles. Some isolates had the additional 1034C polymorphism (20.1%). The presence of mutant pfmdr1 alleles, especially the mutant 184F, that have been associated with tolerance to lumefantrine,18 suggests the vulnerability of partner drugs to resistance pressure in this region. The increase in the copy number of the pfmdr1 gene is associated with mefloquine resistance. Several studies conducted in the Americas reported an absence or limited pfmdr1 gene duplication in P. falciparum–isolated studies (reviewed in ref. 19). In keeping with these findings, we observed only six isolates (2.5%) with two copies of this gene. This contrasts with results obtained in South East Asia in which majority of P. falciparum isolates harbor more than one copy of the pfmdr1 gene.
Overall, we found no evidence of pfk13 C580Y, which supports the continued use of ACTs in this area. However, continued monitoring for potential emergence of resistance parasites is essential.
Acknowledgment:
We would like to acknowledge the Centers for Disease Control and Prevention (CDC) Antimicrobial Resistance Working Group for funding support. J. O. F. is a recipient of a Research Productivity Fellowship from the National Council of Scientific and Technological Development (CNPq) and support from PAEF (IOC-023-FIO-18-2-47).
REFERENCES
- 1.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM; Artemisinin Resistance in Cambodia 1 Study C , 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359: 2619–2620. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S, 2012. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan province, China. Malar J 11: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyaw MP, et al. 2013. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One 8: e57689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phyo AP, et al. 2012. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379: 1960–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariey F, et al. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization , 2018. Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy. Geneva, Switzerland: WHO. [Google Scholar]
- 8.Conrad MD, Rosenthal PJ, 2019. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis 19: e338–e351. [DOI] [PubMed] [Google Scholar]
- 9.Chenet SM, et al. 2016. Independent emergence of the Plasmodium falciparum kelch propeller domain mutant allele C580Y in Guyana. J Infect Dis 213: 1472–1475. [DOI] [PubMed] [Google Scholar]
- 10.Vreden SG, Bansie RD, Jitan JK, Adhin MR, 2016. Assessing parasite clearance during uncomplicated Plasmodium falciparum infection treated with artesunate monotherapy in Suriname. Infect Drug Resist 9: 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucchi NW, et al. 2014. PET-PCR method for the molecular detection of malaria parasites in a national malaria surveillance study in Haiti, 2011. Malar J 13: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffing S, Syphard L, Sridaran S, McCollum AM, Mixson-Hayden T, Vinayak S, Villegas L, Barnwell JW, Escalante AA, Udhayakumar V, 2010. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother 54: 1572–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinayak S, et al. 2010. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis 201: 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue J, Jovel I, Morris U, Aydin-Schmidt B, Islam A, Segurado AC, Bjorkman A, Di Santi S, Martensson A, 2018. Absence of Plasmodium falciparum K13 propeller domain polymorphisms among field isolates collected from the Brazilian Amazon basin between 1984 and 2011. Am J Trop Med Hyg 99: 1504–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menard D, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374: 2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh M, et al. 2018. Efficacy of artemether-lumefantrine for uncomplicated Plasmodium falciparum malaria in Cruzeiro do Sul, Brazil, 2016. Am J Trop Med Hyg 98: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladeia-Andrade S, de Melo GN, de Souza-Lima Rde C, Salla LC, Bastos MS, Rodrigues PT, Luz F, Ferreira MU, 2016. No Clinical or molecular evidence of Plasmodium falciparum resistance to artesunate-mefloquine in northwestern Brazil. Am J Trop Med Hyg 95: 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sisowath C, Ferreira PE, Bustamante LY, Dahlstrom S, Martensson A, Bjorkman A, Krishna S, Gil JP, 2007. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health 12: 736–742. [DOI] [PubMed] [Google Scholar]
- 19.Costa GL, Amaral LC, Fontes CJF, Carvalho LH, de Brito CFA, de Sousa TN, 2017. Assessment of copy number variation in genes related to drug resistance in Plasmodium vivax and Plasmodium falciparum isolates from the Brazilian Amazon and a systematic review of the literature. Malar J 16: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]