Abstract
Budding yeast (Saccharomyces cerevisiae) responds to low cytosolic iron by up-regulating the expression of iron import genes; iron import can reflect iron transport into the cytosol or mitochondria. Mmt1 and Mmt2 are nuclearly encoded mitochondrial proteins that export iron from the mitochondria into the cytosol. Here we report that MMT1 and MMT2 expression is transcriptionally regulated by two pathways: the low-iron-sensing transcription factor Aft1 and the oxidant-sensing transcription factor Yap1. We determined that MMT1 and MMT2 expression is increased under low-iron conditions and decreased when mitochondrial iron import is increased through overexpression of the high-affinity mitochondrial iron importer Mrs3. Moreover, loss of iron-sulfur cluster synthesis induced expression of MMT1 and MMT2. We show that exposure to the oxidant H2O2 induced MMT1 expression but not MMT2 expression and identified the transcription factor Yap1 as being involved in oxidant-mediated MMT1 expression. We defined Aft1- and Yap1-dependent transcriptional sites in the MMT1 promoter that are necessary for low-iron- or oxidant-mediated MMT1 expression. We also found that the MMT2 promoter contains domains that are important for regulating its expression under low-iron conditions, including an upstream region that appears to partially repress expression under low-iron conditions. Our findings reveal that MMT1 and MMT2 are induced under low-iron conditions and that the low-iron regulator Aft1 is required for this induction. We further uncover an Aft1-binding site in the MMT1 promoter sufficient for inducing MMT1 transcription and identify an MMT2 promoter region required for low iron induction.
Keywords: iron, iron–sulfur protein, metal homeostasis, mitochondria, transcription, transport metal, Aft1, Yap1
Introduction
Iron is essential for all organisms but may also be highly toxic because of its ability to participate in redox reactions. Organisms have evolved mechanisms to regulate uptake, intracellular transport, and storage of iron to protect cells from iron toxicity and deliver iron to sites of utilization. In particular, mitochondria require iron to make heme and iron–sulfur (Fe–S)2 clusters. Saccharomyces cerevisiae has two high-affinity mitochondrial iron importers, Mrs3 and Mrs4 (1–4), and iron can also be imported through the pyrimidine transporter Rim2 (5, 6). We reported previously that mitochondria can act as an iron storage organelle, protecting cells from cytosolic iron toxicity (4). For mitochondria to act as a reservoir for iron implies that there may be mechanisms to release mitochondrial iron. S. cerevisiae has two mitochondrial iron exporters, Mmt1 and Mmt2 (3, 7), that belong to the family of cation diffusion facilitator transporters (8, 9). Homologs of Mrs3 and Mrs4, the mitoferrins, are found in all eukaryotes, whereas, to date, homologs for Mmt1 and Mmt2 are only found in fungi and plants (3, 7, 10). It has been suggested that mitochondrial iron in mammals can be exported through the ABC transporter Abcb8 (11–13).
In yeast, the expression of many iron transporters is under the regulation of two systems: the low-iron-induced Aft1/Aft2 regulon(s) that turn(s) on induction of 22 genes (14), including the mitochondrial iron importer Mrs4, and the high-iron-induced regulon, which is under control of the transcription factor Yap5 (15–18). Both systems are affected by Fe–S cluster levels. Loss of Fe–S clusters activates Aft1 translocation to the nucleus, resulting in induction of the low-iron regulon for iron acquisition. Under high Fe–S cluster levels, nuclear localized Yap5 binds a 2Fe–2S cluster that induces a conformational change, which activates transcription of Yap5 target genes such as the vacuolar iron exporter CCC1 (17–19). In this study, we determined that the expression of the mitochondrial exporter genes MMT1 and MMT2 is regulated by two different conditions. First, increased transcription of MMT1 and MMT2 occurs when cells are grown in iron-limited medium, and Aft1 is necessary for this induction. We identify an Aft1 binding site on the MMT1 promoter and low-iron-sensing domains on the MMT2 promoter. We also discovered that increased oxidants induced transcription of MMT1 and that the transcription factor Yap1 mediates this oxidant-induced expression, and we identify a putative Yap1 binding site on the MMT1 promoter.
Results
MMT1 and MMT2 expression is increased under low-iron conditions
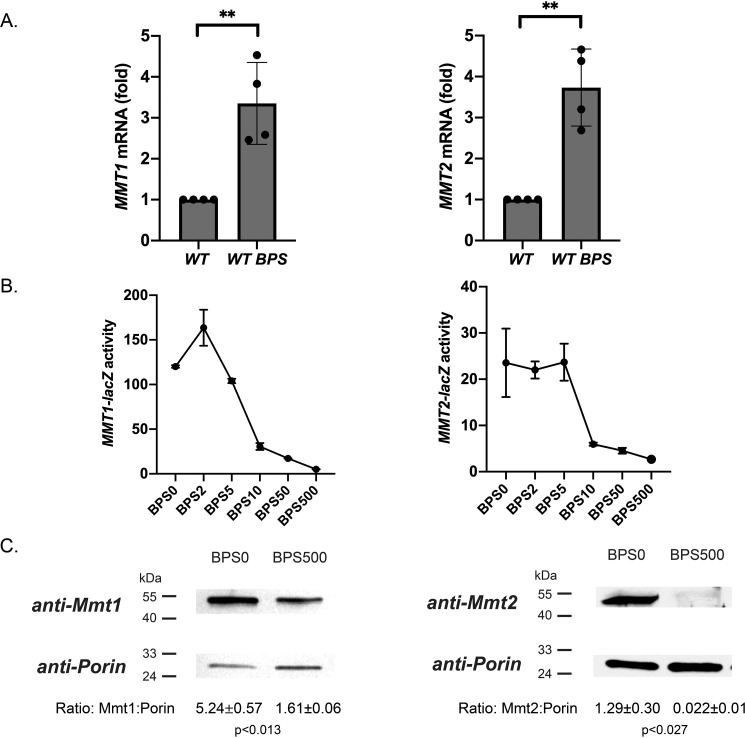
Cytosolic iron levels in yeast are regulated by iron import across the plasma membrane through high- and low-affinity iron transport mechanisms such as Fet3/Fth1 (20, 21) or Fet4 (22) and iron export from organelles such as the vacuole through Fet5/Fth1 (23) or Smf3 (24). We previously provided evidence that Mmt1 and Mmt2 are mitochondrial iron exporters in S. cerevisiae that can alter cytosolic iron levels (3, 25). To determine whether alterations in intracellular iron affect the expression of MMT1 and MMT2, we grew WT cells in complete minimal (CM) medium made low-iron by addition of the iron chelator bathophenanthroline sulfonate (BPS). In previous studies, we determined that the optimal iron level for WT growth was around BPS10-BPS20 (micromolar) and is equivalent to normal medium growth (20). We measured MMT1 and MMT2 transcript changes by RT-qPCR. MMT1 and MMT2 transcripts were increased under low-iron conditions (Fig. 1A, WT BPS). To determine whether iron levels affected transcription, we generated reporter constructs containing the promoter region of either MMT1 or MMT2 fused to the β-gal gene (lacZ). For MMT1, the promoter region we utilized encompasses the complete intergenic region between MMT1 and the 5′-distal gene ECM5. For MMT2, the promoter region we utilized encompasses the complete intergenic region between MMT2 and the 5′-distal gene GRE1. Both MMT1-lacZ and MMT2-lacZ activities were increased under low-iron growth conditions and decreased with increasing iron (Fig. 1B). MMT1-lacZ activity was affected more than MMT2-lacZ activity in response to changes in medium iron. Importantly, increased transcription of MMT1 and MMT2 under low-iron growth conditions equated to increased Mmt1 and Mmt2 protein levels (Fig. 1C).
Figure 1.
MMT1 and MMT2 expression is induced under low iron. A, WT cells in CM medium or CM medium treated with 80 μm BPS medium were grown overnight. Complementary DNA was synthesized from total RNA from cells, and RT-qPCR was performed for MMT1, MMT2, and ACT1 using primers listed in Table 2. Error bars represent S.D.; n = 4. B, WT cells were transformed with MMT1-lacZ or MMT2-lacZ and grown in CM medium treated with BPS containing different concentrations (micromolar) of FeSO4 overnight, and β-gal activity was assessed as described previously (27). A representative iron concentration (BPSX) curve is shown for MMT1-lacZ and MMT2-lacZ. n = 3. C, crude mitochondria were isolated from cells grown in BPS(0) or BPS(500), and Mmt1, Mmt2, and Porin levels were determined using Western blot analysis. Representative blots and their corresponding Mmt/Porin ratios are shown. n = 3. **, p ≤ 0.01.
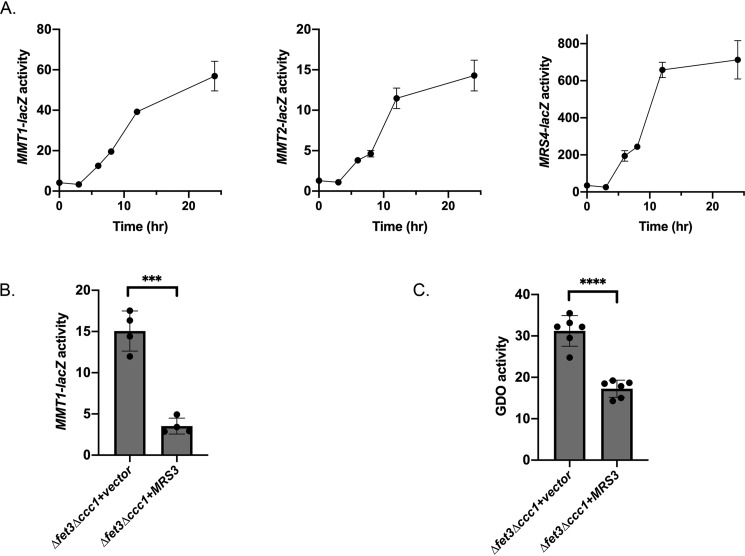
Previous studies demonstrated that transcription of MRS4, a mitochondrial iron importer, was induced under low-iron conditions (26). We examined whether the kinetics of induction of MRS4 were similar or different from mitochondrial iron exporter expression. To determine whether MMT1 and MMT2 expression was coordinated with mitochondrial importer (MRS4) expression, we generated an MRS4-lacZ fusion construct containing the MRS4 promoter and compared the timing of expression of MMT1-lacZ, MMT2-lacZ, and MRS4-lacZ in cells grown in BPS(0) for 0–24 h. MMT1-lacZ, MMT2-lacZ, and MRS4-lacZ showed similar kinetics for induction (Fig. 2A). From these data, we conclude that WT cells grown under low-iron conditions induce the expression of both mitochondrial iron importers and exporters. There was significantly higher relative expression of MRS4-lacZ compared with MMT1-lacZ or MMT2-lacZ.
Figure 2.
Low-iron induction of MMT1 and MMT2 occurs with kinetics similar to MRS4 induction. A, WT cells transformed with MMT1-lacZ, MMT2-lacZ, or MRS4-lacZ were grown overnight in CM medium, washed, and then reinoculated in CM medium with 80 μm BPS and grown for 0–24 h. β-Gal activity and protein levels were determined as described under “Experimental procedures.” A representative time course is shown for MMT1-lacZ, MMT2-lacZ, and MRS4-lacZ. n = 3. B, Δfet3Δccc1 cells transformed with either control vector or the TET-regulated MRS3 plasmid and MMT1-lacZ were grown in CM medium overnight, and β-gal activity was measured. Error bars represent S.D. n = 4. C, cells as in B were transformed with a control plasmid or a plasmid containing GDO and grown in CM medium overnight, and GDO activity was measured. Error bars represent S.D. n = 6. ***, p ≤ 0.001; ****, p ≤ 0.0001.
It is possible that the signal for transcription of MMT1 and MMT2 is either low cytosolic iron or low mitochondrial iron. Under most conditions, low cytosolic iron is indistinguishable from low mitochondrial iron, as mitochondrial iron is derived from cytosolic iron. Thus, it is unclear whether increased MMT1 or MMT2 transcription is responding to low cytosolic iron or low mitochondrial iron. We developed an experimental protocol to differentiate between these two possibilities, using overexpression of the mitochondrial iron transporter Mrs3 to increase mitochondrial iron. Cells with low iron stores (Δccc1) and an impaired high-affinity cell surface iron transporter system (Δfet3) can grow in iron-replete medium because of the activity of low-affinity cell-surface iron transporters (27). We transformed a plasmid containing a tetracycline-regulated MRS3 (pTETMRS3), which is on in the absence of doxycycline (4), into a Δfet3Δccc1 yeast strain to determine whether movement of iron from the cytosol to mitochondria affected the expression of MMT1-lacZ. We utilized MMT1-lacZ as our reporter, as the range of expression was larger compared with MMT2-lacZ. Overexpression of MRS3 reduced the expression of MMT1-lacZ (Fig. 2B). To confirm that changes in cytosolic iron occurred upon expression of Mrs3, we utilized our previously described iron-dependent gentisate 1,2-dioxygenase (GDO) enzyme assay (7), whose activity is sensitive to changes in cytosolic iron. We found that overexpression of the iron importer Mrs3 reduced cytosolic iron and, consequently, reduced MMT1 expression, resulting in lower cytosolic GDO activity (Fig. 2C). Mrs3 expression decreases cytosolic iron and increases mitochondrial iron. If MMT1 expression was responding to cytosolic iron levels, then MMT1-lacZ expression would be predicted to go up when Mrs3 is overexpressed and not down as observed. These results confirm that mitochondrial iron exporter expression is regulated by changes in mitochondrial iron levels.
Low-iron-induced MMT1 and MMT2 expression is mediated through Aft1
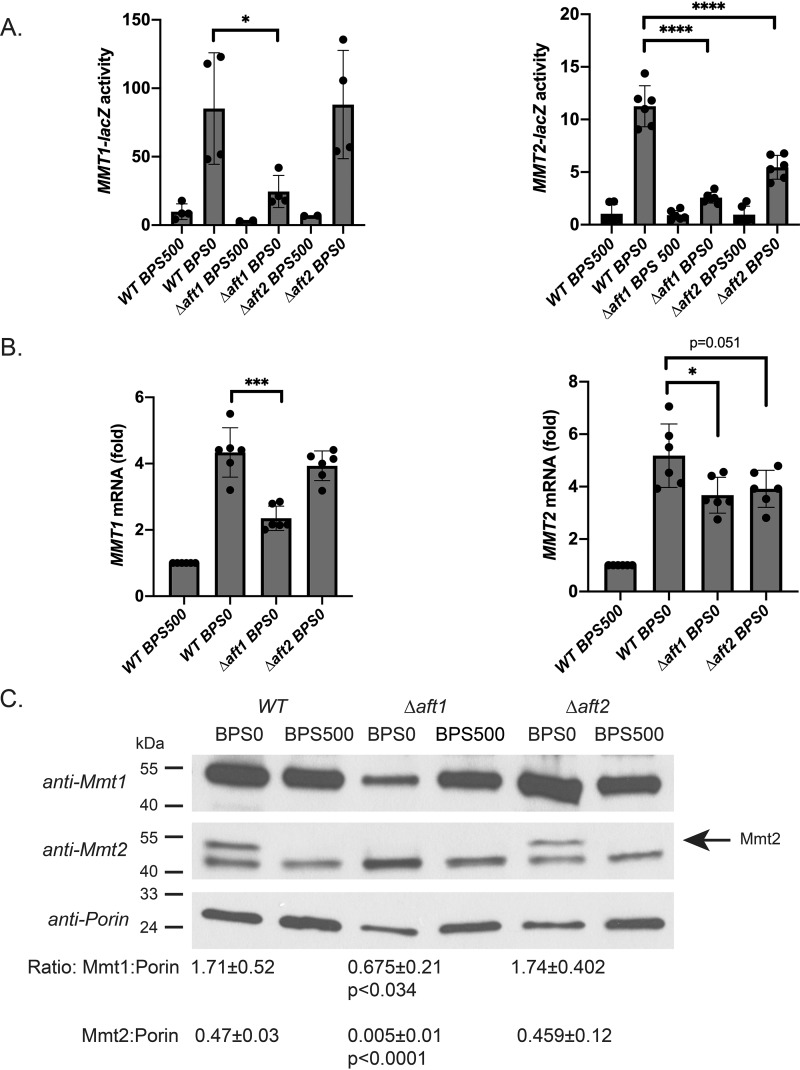
The low-iron regulon is induced by the transcription factors Aft1 and Aft2 (for a review, see Ref. 14). To determine whether MMT1 and MMT2 expression is regulated by Aft1 or Aft2, we utilized our MMT1-lacZ and MMT2-lacZ reporter constructs expressed in WT, Δaft1, or Δaft2 cells grown in low-iron or high-iron medium. Loss of Aft1 greatly reduced MMT1-lacZ and MMT2-lacZ expression, whereas loss of Aft2 had no effect on MMT1-lacZ expression but partially reduced MMT2-lacZ expression (Fig. 3A). These results were validated by RT-qPCR, where loss of Aft1 resulted in decreased MMT1 and MMT2 expression; loss of Aft2 did not alter low-iron induction of MMT1 expression but did have a small effect on MMT2 expression (Fig. 3B). We confirmed that Mmt1 and Mmt2 protein levels were reduced in Δaft1 cells grown in low-iron medium compared with WT controls (Fig. 3C). In contrast, loss of Aft2 did not affect Mmt1 and Mmt2 protein levels. There was still a 2- to 3-fold increase in mRNA for both MMT1 and MMT2 under-low iron conditions in the absence of Aft1, and Mmt1 protein was still detected, whereas no Mmt2 protein was detected. This suggests that there may be other mechanisms that regulate MMT1 and MMT2 expression and protein levels.
Figure 3.
Low-iron-induced MMT1 and MMT2 expression is mediated through Aft1. A, WT, Δaft1, and Δaft2 cells transformed with MMT1-lacZ or MMT2-lacZ were grown in CM medium treated with 80 μm BPS with or without 500 μm iron overnight, and β-gal activity was measured. Error bars represent S.D. n = 4. B, RT-qPCR was performed for MMT1, MMT2, and CMD1 from cells as in A, using primers listed in Table 2. Error bars represent S.D. n = 6. C, crude mitochondria were isolated from cells as in A, and Mmt1, Mmt2, and Porin levels were determined using Western blot analysis. Representative blots are shown and quantified as described under “Experimental procedures.” n = 3. The arrow identifies the Mmt2-specific band. *, p ≤ 0.05; ***, p ≤ 0.001; ****, p ≤ 0.0001.
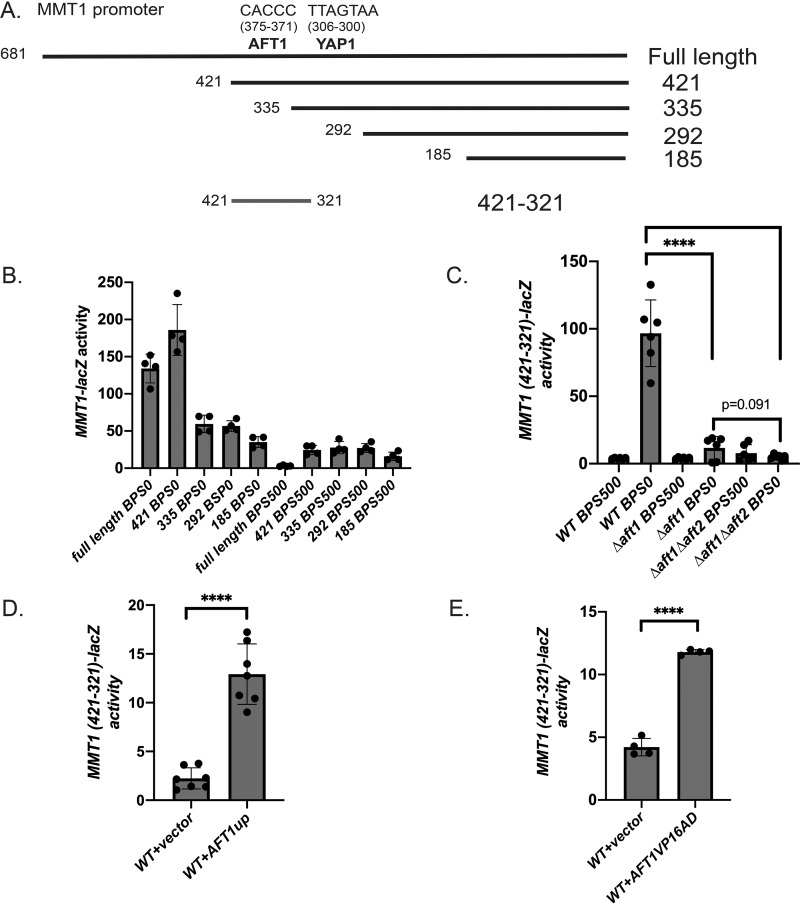
Sequence analysis identified a putative core Aft1-binding site (CACCC) in the MMT1 promoter 375–371 bp upstream of the translational ATG start site. We generated truncation mutants of the MMT1 promoter to test for regions that were responsible for low-iron-mediated expression (Fig. 4A). Removal of the putative Aft1-binding site reduced low-iron induction by more than 50% (Fig. 4B, full-length versus construct 335). Additional truncations further reduced low-iron-mediated expression (Fig. 4B, construct 185), suggesting additional regulatory regions. We cloned a minimal domain of the MMT1 promoter containing the putative Aft1-binding site (421–321, 101 bp, construct 421–321) into the expression vector pYC7, which contains a multiple cloning site upstream of the TATA box of the CYC1 gene fused to the lacZ gene (28), and examined low-iron-dependent expression in the presence or absence of Aft1 and in the absence of Aft1 and Aft2. The minimal MMT1 promoter (421–321) still showed low-iron-dependent induction, whereas loss of Aft1 eliminated most of the low-iron MMT1-lacZ expression (Fig. 4C). Loss of Aft2 in the Δaft1 strain further reduced low-iron MMT1 421–321-lacZ expression, although not to significance, suggesting that Aft2 contributes to inducing expression of MMT1 under low-iron conditions when Aft1 is absent. In addition, a WT strain transformed with construct 421–321 and an empty vector or the iron-independent constitutively active Aft1 (AFT1up), which has been used extensively to study Aft1-mediated expression (26, 29–32), showed increased expression compared with the empty vector control (Fig. 4D). Aft1 has been suggested previously to bind to the MMT1 promoter (33). To further determine whether Aft1 is involved in low-iron MMT1 induction, we expressed the minimal MMT1 421–321–lacZ construct in a yeast strain expressing either an empty vector or a chimera of the Aft1 DNA-binding domain tethered to the VP16 activation domain, which has been shown previously to be sufficient for binding to DNA (26). The presence of the Aft1 DNA binding domain alone was sufficient to induce expression of MMT1 421–321–lacZ (Fig. 4E). Together, our data strongly support that Aft1 is necessary to induce MMT1 expression.
Figure 4.
Aft1 is necessary for low-iron-mediated induction of MMT1. A, diagram of the MMT1 promoter with putative Aft1- and Yap1-binding sites and truncation constructs. B, WT cells transformed with full-length or truncation mutants of the MMT1 promoter fused to lacZ were grown in CM medium with 80 μm BPS (low iron) or high iron (500 μm) FeSO4 overnight. β-Gal activity and protein levels were determined. Error bars represent S.D. n = 4. C, WT, Δaft1, or Δaft1Δaft2 cells expressing MMT1 421–321-lacZ were grown as described in B, and β-gal activity was measured. Error bars represent S.D. n = 6. D, WT cells expressing MMT1 421–321-lacZ were transformed with an empty vector or an AFT1up plasmid. Cells were grown in CM medium overnight, and β-gal activity was measured. Error bars represent S.D. n = 7. E, WT cells expressing MMT1 421–321-lacZ were transformed with an empty vector or an AFT1VP16AD plasmid. Cells were grown in CM medium overnight, and β-gal activity was measured. Error bars represent S.D. n = 4. ****, p ≤ 0.0001.
MMT1 and MMT2 expression is regulated by Fe–S cluster synthesis
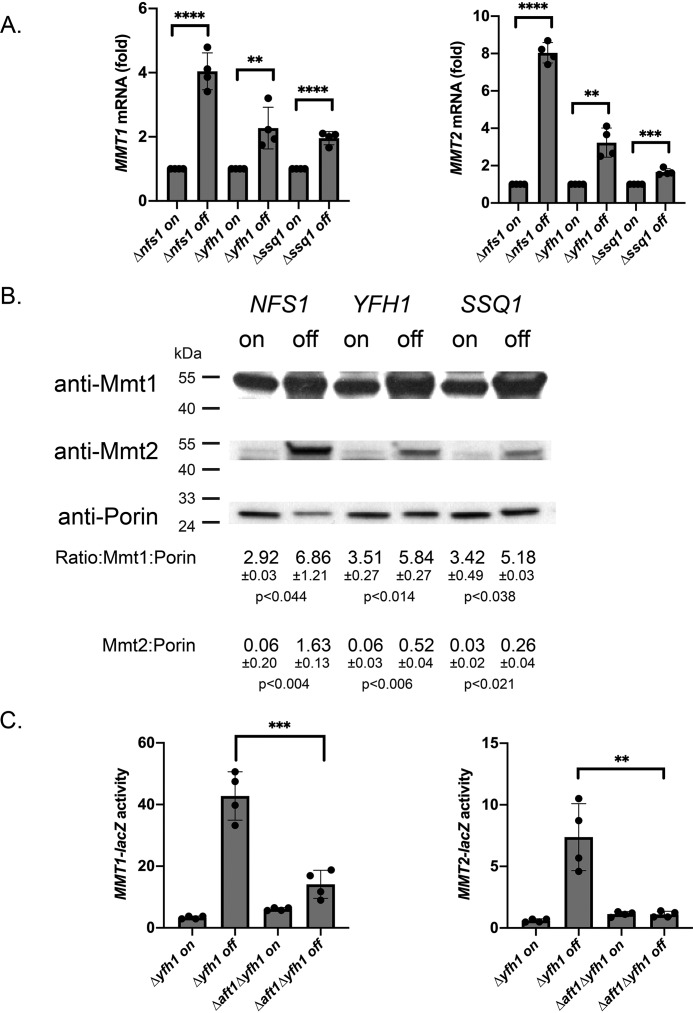
Fe–S cluster biosynthesis in mitochondria has long been known as a signal of cellular iron status and regulates the activity of Aft1/Aft2. To determine whether MMT1 and MMT2 expression is affected by changes in Fe–S cluster synthesis, we took advantage of the ability to shut off Fe–S cluster synthesis using methionine-regulated expression (MET3 promoter). In the absence of methionine, expression is turned on and expression is turned off when cells are grown in 10× methionine (20 mm). We utilized MET3-driven expression of NFS1, YFH1, or SSQ1, genes important in Fe–S cluster synthesis (34, 35), in the corresponding deletion strains Δnfs1, Δyfh1, and Δssq1. MMT1 and MMT2 transcripts were dramatically increased when Fe–S cluster synthesis protein expression was turned off (Fig. 5A). Western blot analysis confirmed that shutoff of Nfs1, Yfh1, or Ssq1 resulted in increased levels of Mmt1 and Mmt2 (Fig. 5B). We determined that sensing of Fe–S cluster synthesis depended on Aft1, as Δaft1 dramatically reduced MMT1-lacZ and MMT2-lacZ expression when YFH1 expression was turned off (Fig. 5C). These results support the hypothesis that Aft1-mediated MMT1 and MMT2 expression is regulated by Fe–S cluster status.
Figure 5.
Loss of Fe–S cluster synthesis induced MMT1 and MMT2 expression. A, Δnfs1pMET3NFS1, Δyfh1pMET3YFH1, and Δssq1pMETSSQ1 cells were grown without methionine (on) or with 10× (20 mm) methionine (off) overnight, and RT-qPCR for MMT1, MMT2, and ACT1 was performed. Error bars represent S.D. n = 4. B, mitochondria were isolated from cells grown as in A, and the levels of Mmt1, Mmt2, and Porin were determined by Western blot analysis. n = 3. C, Δyfh1pMET3YFH1 or Δaft1Δyfh1pMET3YFH1 cells transformed with MMT1-lacZ or MMT2-lacZ were grown as in A, and β-gal activity was measured. Error bars represent S.D. n = 4. **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
The MMT2 promoter contains a repressive domain and a low-iron-responsive domain that regulate expression
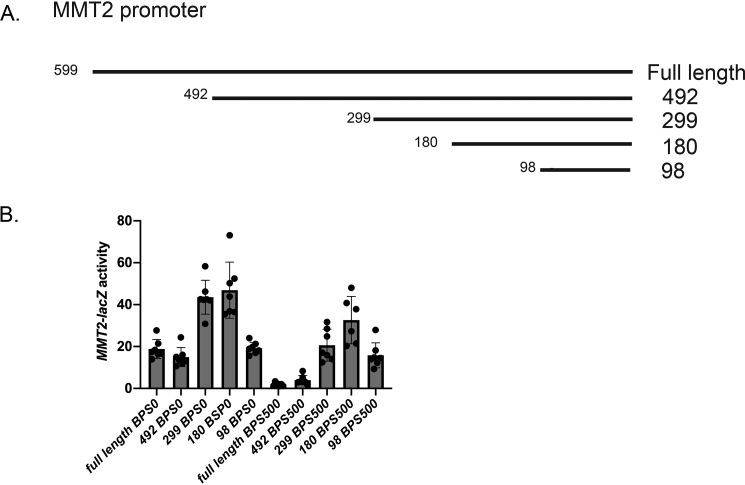
The MMT2 promoter did not have a defined Aft1-binding site. To determine regions of the MMT2 promoter important for low-iron-induced expression, we generated truncation mutants of the MMT2 promoter fused to lacZ (Fig. 6A). Both the full length and construct 492 (492 bp to ATG) showed low-iron-mediated induction of MMT2-lacZ compared with high iron (BPS500) (Fig. 6B). Constructs 299 and 180, which removed additional base pairs upstream of the ATG (492 through 180 bp) showed increased induction of MMT2-lacZ compared with the full-length promoter (Fig. 6B). This was surprising and revealed that there may be sites in the promoter that mediate repression of MMT2 expression. Even under high iron, the truncated version of MMT2-lacZ (299 and 180) showed expression. Loss of an additional 82 bp (construct 98) reduced low-iron induction back to full-length MMT2-lacZ expression. This 82-bp region contains a putative Med8-binding site that is a transcription factor associated with positive and negative regulation of expression (36, 37). We did not examine the exact function of this region of the MMT2 promoter; however, our results suggest that this region is important in regulating MMT2 expression. We were unable to determine an exact region of the MMT2 promoter that is responsible for low-iron induction.
Figure 6.
MMT2 promoter truncations reveal both repression and activation domains in response to low iron. A, a map of the promoter region of MMT2 and truncations mutants. B, WT cells transformed with full-length or truncation mutants fused to lacZ were grown in BPS treated with low-iron medium, and β-gal activity was measured. Error bars represent S.D. n = 6.
The MMT1 promoter contains a Yap1-binding site that is necessary for oxidant-mediated expression
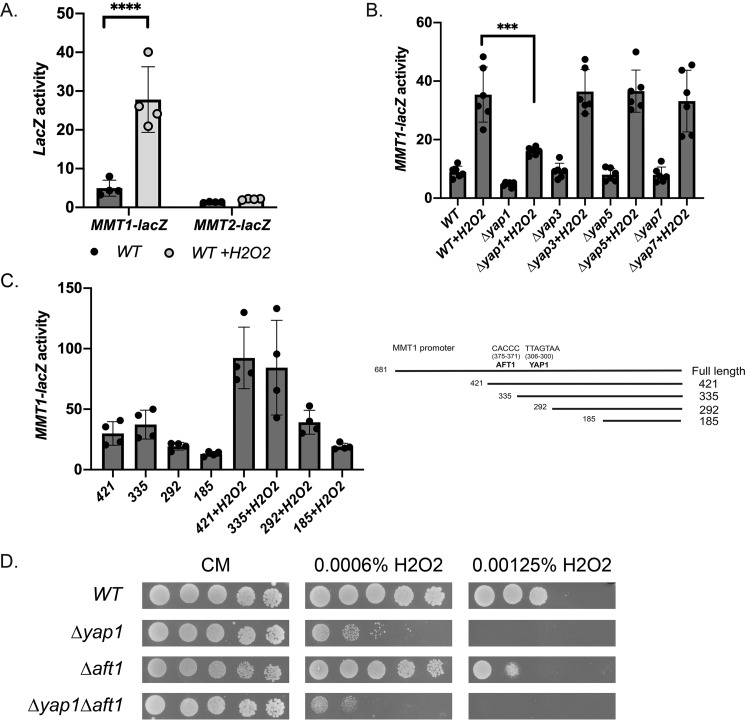
The MMT1 promoter contains a putative Yap1-binding site (Fig. 4A). The Yap family of transcription factors has been shown to be involved in stress response (38), and Yap1 is activated under oxidative stress (39). To determine whether MMT1 and MMT2 expression is responsive to oxidants, we grew cells in the absence or presence of H2O2. MMT1-lacZ expression increased in the presence of H2O2; however, MMT2-lacZ expression was unaltered (Fig. 7A). We confirmed that Yap1 was necessary for H2O2-mediated MMT1-lacZ induction, whereas loss of Yap3, Yap5, and Yap7 had no effect on induction (Fig. 7B). We utilized our MMT1 promoter truncation mutants to examine the minimal domain required for oxidant-mediated induction of MMT1. Loss of the putative Yap1-binding site trended toward reduced H2O2-induced expression of MMT1-lacZ (Fig. 7C, 335 versus 292), supporting the hypothesis that this site is important for oxidant-induced MMT1 expression. However, there is still a 2-fold increase in expression compared with cells that are not exposed to H2O2. This is true for the full-length MMT1 construct (Fig. 7B) as well as our truncation MMT1-lacZ constructs (Fig. 7C). This suggests that there may be other regions in the promoter that contribute to MMT1 expression under oxidative stress. Loss of YAP1 did resulted in increased sensitivity to H2O2, and we determined that loss of AFT1 increased Δyap1 sensitivity to H2O2-mediated oxidative stress (Fig. 7D). This suggests that regulated expression of MMT1 through Yap1 and Aft1 can be protective during times of oxidative stress.
Figure 7.
Loss of YAP1 affects oxidant-mediated induction of MMT1 but not MMT2 expression. A, WT cell transformed with MMT1-lacZ or MMT2-lacZ were grown in the presence or absence of 0.005% H2O2 for 2 h, and β-gal activity was measured. Error bars represent S.D. n = 4. B, WT, Δyap1, Δyap3, Δyap5, and Δyap7 cells transformed with MMT1-lacZ were grown with 0.005% H2O2 for 2 h, and β-gal activity was measured. Error bars represent S.D. n = 6. C, cells expressing truncation mutants as in Fig. 4 were grown in the presence or absence of H2O2, and β-gal activity was measured. Error bars represent S.D. n = 4. D, WT, Δyap1, Δaft1, and Δyap1Δaft1 cells serially diluted onto CM plates in the presence or absence of 0.0006% or 0. 00125% H2O2 were grown for 2 days, and images were captured. ***, p ≤ 0.001; ****, p ≤ 0.0001.
Discussion
Iron import into mitochondria is necessary for heme and Fe–S cluster synthesis, essential processes in most organisms. In yeast, there are two identified high-affinity mitochondrial iron transporters, Mrs3 and Mrs4 (1, 27, 40). Paralogs of these exist in all other eukaryotes, although some species have one instead of two transporters (41, 42). We previously identified two highly homologous mitochondrial cation diffusion facilitators, Mmt1 and Mmt2, that function as mitochondrial iron exporters (3). Most notably, deletion of MMT1 and MMT2 results in phenotypes that are identical to overexpression of the iron importers MRS3/MRS4, whereas overexpression of MMT1 and MMT2 phenocopies deletion of MRS3/MRS4 (7). Indeed, measurements of cytosolic iron levels confirm that overexpression of Mmt1 and Mmt2 results in increased cytosolic iron, whereas overexpression of Mrs3 and Mrs4 reduces cytosolic iron. Based on these observations, we were surprised to discover that transcription of MMT1/MMT2, like transcription of MRS4, is regulated by the low-iron-sensing transcription factors Aft1/Aft2 (14, 43). We determined that both MMT1 and MMT2 are induced under low-iron conditions and that the low-iron regulon transcription factor Aft1 was necessary for MMT1 and MMT2 induction. We identified a minimal domain of the MMT1 promoter containing a putative Aft1-binding site that was sufficient to induce MMT1 transcription and noted that Aft2 may also contribute to the low-iron-mediated increase in MMT1 expression. We were unable to identify any putative Aft1-binding site on MMT2 but did identify a minimal domain of the MMT2 promoter that was necessary for low-iron induction. We determined that both MMT1-lacZ and MMT2-lacZ were induced under low-iron conditions with kinetics similar to those of MRS4-lacZ. This suggests that there is a general response to low iron that induces both mitochondrial iron importers and exporters.
It seems counterintuitive that both mitochondrial iron exporters and iron importers would respond to the same signal. Consideration, however, of the mechanism of induction suggests a possible explanation. The low-iron regulon is induced by movement of Aft1 or Aft2 into the nucleus. The factor that determines the movement of Aft1 and Aft2 is modification by mitochondrially produced Fe–S clusters (31, 44). In the absence of Fe–S modification, Aft1 and Aft2 translocate from the cytosol to nucleus and activate about 20 genes that encompass the low-iron response, which includes the mitochondrial iron importer gene MRS4. An adequate supply of mitochondrial iron is required for mitochondrial Fe–S synthesis. Thus, it makes sense that decreases in cytosolic or mitochondrial iron levels will induce the low-iron response, resulting in increased cytosolic and mitochondrial iron. There is, however, an alternative mechanism that leads to the low-iron response, which is increased oxidant activity, as Fe–S clusters are highly susceptible to oxidant damage. In the presence of oxidants, the low-iron transcriptional response can be induced by compromising Fe–S cluster synthesis. In fact, many of the enzymes in the Fe–S pathway are themselves Fe–S cluster–containing proteins. We posit that the role of Mmt1 and Mmt2 is to protect mitochondria from oxidant damage by reducing the level of mitochondrial “free” iron. Evidence supporting this hypothesis includes the following. First, reducing mitochondrial and cellular iron can protect cells from the consequences of loss of Yfh1, a mitochondrial protein involved in Fe–S cluster synthesis (45–48). Cells deleted for Yfh1 or impaired in Yfh1 activity show increased mitochondrial iron and increased mitochondrial oxidants. Reducing mitochondrial iron by limiting cellular iron or by overexpression of MMT1 or MMT2 protects cells from oxidant damage. Second, yeast with a defect in cytosolic superoxide dismutase 1 show induction of the low-iron regulon (49). Increased cytosolic iron protects these cells from oxidant damage by “soaking up” superoxide. The increase in cytosolic iron can come from increasing plasma membrane iron transport. These cells can also be protected by increased expression of MMT1 and MMT2, as iron exported from mitochondria also reduces superoxide anion (50). Third, support for a role of Mmt1 in suppressing oxidant damage is found in the observation that MMT1 transcription is induced by Yap1, a major regulator of antioxidant defenses (51). We considered that MMT1 and MMT2 play a role in regulating mitochondrial iron levels. The finding that increasing mitochondrial iron levels by overexpressing MRS3 did not, however, induce MMT1 and MMT2 transcription casts doubt on that interpretation. The finding that oxidants (through Yap1) increase transcription of MMT1 supports the view that Mmt1's role is in reducing mitochondrial free iron in the face of oxidants. We expect that increased Mmt1 and Mmt2 levels can reduce mitochondrial free iron to the point where oxidant damage is reduced but not to the point where heme or Fe–S cluster synthesis is affected. This could be mediated by the affinity of these transporters. We hypothesize that the affinity of Mmt1 and Mmt2 for iron is less than the affinity of Mrs3 and Mrs4 for iron and less than the affinity of iron-consuming enzymes within the mitochondria. Thus, only “surplus iron” will be affected by Mmt expression. An alternative hypothesis is that induction of MMT1 and MMT2 may be a protective response needed to tolerate rapid rises in mitochondrial iron after iron deficiency is corrected. This model predicts that these exporters are not normally active in iron-deficient cells but active upon iron replenishment. These models are not mutually exclusive. The low-iron-mediated expression of the MMTs, as measured by β-gal activity, is less than that of MRS4. MRS4-lacZ activity was 10- to 50-fold higher compared with MMT1-lacZ and MMT2-lacZ activity, respectively. This suggests a stronger induction of iron import as well. We recognize that transcript levels do not always equate to protein levels or transporter activity. To our knowledge, exact transporter transcript numbers and the resulting transporter molecules per cell under varying iron conditions have not been determined.
There are plant homologs of Mmt1 and Mmt2, most notably the Cucumis sativus homolog MTP6. MTP6 has been shown to efflux iron and manganese from mitochondria, and its expression is up-regulated under iron deficiency and iron excess conditions (10). The only known mitochondrial iron exporter in mammals is Abcb8 (11), a member of a different family of transporters that hydrolyze ATP to move substrates across membranes (52), whereas Mmts are members of the cation diffusion facilitator family (8). That these transporters have different mechanisms for metal transport suggests that they may have evolved separately. The fact that overexpression of Mmts in yeast or Abcb8 in mammals helps protect cells from the toxic effects of defective Fe–S cluster synthesis highlights their role in antioxidant protection and suggests that they play an important role in mitochondrial protection.
Experimental procedures
Yeast, plasmids, and growth medium
Genotypes of strains employed in this study are listed in Table 1. The WT strains employed for these experiments were from the W303 background. Most deletion strains were created by either PCR-amplifying the KanMX cassette from the homozygous diploid deletion collection (Research Genetics) or fusion PCR (53). Cells were grown in CM medium (0.67% yeast nitrogen base, 0.12% dropout amino acid mixture, and 2% dextrose) or CM medium with 10× or 20 mm methionine to turn off expression of NFS1, YFH1, and SSQ1. Media were made iron-deficient by addition of 80 μm BPS, and specific concentrations of ferrous sulfate (micromolar) were added back. The concentration of added iron in micromolar is denoted as BPS(x). Plasmids used in these studies included lacZ reporters generated as described below; MRS4-lacZ, AFT1up, and AFT1VP16AD (all generous gifts from the laboratory of Dr. Dennis Winge, University of Utah); and MMT1, MMT2, TET OFF-MRS3, and cytosolic GDO (4, 7).
Table 1.
Yeast strains
| Yeast Strain | Genotype | Source |
|---|---|---|
| DY150 WT | W303 MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) | 3 |
| Δfet3Δccc1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δccc1::LEU2, Δfet3::HIS3 | This study |
| Δaft1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 lys2 Δaft1:TRP1 | 54 |
| Δaft2 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δaft2::KanMx | 26, 55 |
| Δaft1Δaft2 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δaft1::TRP1 Δaft2::KanMx | Khalimonchuk laboratory |
| Δnfs1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δnfs1::HIS3, MET3-NFS1 | 56 |
| Δyfh1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyfh1::HIS3, MET3-YFH1 | 56 |
| Δssq1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δssq1::KanMx, MET3-SSQ1 | 17 |
| Δyap1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyap1::KanMx | This study |
| Δyap1Δaft1 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyap1::KanMx, Δaft1::TRP1 | This study |
| Δyap3 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyap3::KanMx | This study |
| Δyap5 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyap5::KanMx | 17 |
| Δyap7 | MATa ade2–1 his3–11 leu2–3 112 trp1–1 ura3–52 can1–100(oc) Δyap7::KanMx | This study |
RT-qPCR
Total RNA was isolated using the Agilent Technologies Mini Kit. The SuperScript III kit from Thermo Fisher Invitrogen was used to synthesize first-strand complementary DNA from total RNA. Quantitative PCR was performed using a Roche LightCycler (Idaho Technology) or Bio-Rad iQ5 real-time PCR detection system. RT-qPCR primers for MMT1, MMT2, CMD1, and ACT1 were as described in Table 2.
Table 2.
Primers
| Primer Name | Sequence |
|---|---|
| MMT1 RT-qPCR | F: 5′-gagaccgagcaaaacgacat-3′ |
| R: 5′-tcttacgcctgcatttttcc-3′ | |
| MMT2 RT-qPCR | F: 5′-ggagggccataagaacatca-3′ |
| R: 5′-gaagccttgccttccttctt-3′ | |
| CMD1 RT-qPCR | F: 5′-tctttcgcccagtgaagcagaagt-3′ |
| R: 5′-tcagcggcggagattaaaccatca-3′ | |
| ACT1 RT-qPCR | F: 5′-tgtcaccaactgggacgata-3′ |
| R: 5′-ggcttggatggaaacgtaga-3′ | |
| MMT1-lacZ | F: 5′-cgcggatcccttaaggaaacattgtcagg-3′ |
| R: 5′-cccaagcttcattgtcaaaaatgcgaaaaag-3′ | |
| 421 MMT1-lacZ | F: 5′-cgcggatccccagtattgggtgtttccca-3′ |
| 335 MMT1-lacZ | F: 5′-cgcggatccgcgcgttttgctgtcaatat-3′ |
| 292 MMT1-lacZ | F: 5′-cgcggatccccgtttcttgatagctttcc-3′ |
| 185 MMT1-lacZ | F: 5′-cgcggatccgctcatttccatgacaactt-3′ |
| 421–321 MMT1-lacZ | R: 5′-cctctcgaggacagcaaaacgcgcttttc-3′ |
| F: 5′-cggggtaccccagtattgggtgtttccca-3′ | |
| MMT2-lacZ | F: 5′-ccccccgggctgggtcaggaaacgatgaa-3′ |
| R: 5′-aaactgcaggcatatctttattgcgtttgc-3′ | |
| 492 MMT2-lacZ | F: 5′-ccccccggggagcagcaagtgagaaattt-3′ |
| 299 MMT2-lacZ | F: 5′-ccccccggggtcgaggtatgccgtttcac-3′ |
| 180 MMT2-lacZ | F: 5′-ccccccgggcgttattgtgctgggcaaca-3′ |
| 98 MMT2-lacZ | F: 5′-ccccccgggcagattggtgccttttcaag-3′ |
LacZ reporter constructs
To make lacZ reporter constructs, a PCR product of the MMT1 promoter 681 bp upstream of the ATG start site was digested with BamHI/HindIII and cloned into a Yep354 vector. For MMT2, a PCR product 599 bp upstream of the MMT2 ATG start site was digested with XmaI/PstI and cloned into a Yep354 vector. Other truncations were generated using the same approach. The 695 lacZ construct of the MMT1 promoter was generated by PCR, digested with XhoI/KpnI, and cloned into M2238 CYC1-lacZ (pYC7) (a generous gift from the laboratory of Dr. Stillman, University of Utah). Primers used in cloning are listed in Table 2.
β-galactosidase assay
The lacZ reporter constructs were generated, and β-gal activity was assayed as described previously (3). β-Gal–specific activity is reported as nanomoles per minute per milligram of protein.
H2O2 spot assay
Freshly cultured cells were washed. 1:3 serial dilutions were made, spotted onto plates, and incubated at 30 °C for 2 days, followed by plate imaging (25).
Cytosolic gentisate 1,2-dioxygenase assay
Cells were broken by glass beads. Lysates were collected, and GDO activity was assayed as described previously (7).
Other procedures and reagents
Protein concentrations were determined using bicinchoninic acid assay (Pierce) detection reagent from Thermo Fisher Scientific. Proteins were analyzed by 4%–20% SDS-PAGE Tris/glycine, followed by Western blot analysis using Western Lightning (PerkinElmer Life Sciences). Antisera used for probing Western blots included rabbit anti Mmt1 (1:1000), rabbit anti-Mmt2 (1:1000), or mouse anti-Porin (1:1000, Thermo Fisher). Secondary antibodies were either peroxidase-conjugated goat anti-rabbit IgG or peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1:5000). Western blots were quantified using Bio-Rad ImageLabTM software.
Statistics
Statistics were calculated using a two-tailed Student's t test with significance set at p ≤ 0.05: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Author contributions
L. L., J. K., and D. M. W. conceptualization; L. L., S. B., and X. J. data curation; L. L. and D. M. W. formal analysis; L. L., J. K., and D. M. W. writing-review and editing; D. M. W. supervision; D. M. W. funding acquisition; D. M. W. investigation; D. M. W. writing-original draft; D. M. W. project administration.
Acknowledgments
We thank the Ward laboratory members for critically reading the manuscript and the University of Utah Metals Interest Group and Centers for Iron and Heme Disorders for critical feedback. We also thank Dr. Oleh Khalimonchuk (University of Nebraska) for the Δaft1Δaft2 yeast strain.
This study was supported by NIDDK, National Institutes of Health Grants DK052380 and DK030534 and Friedreich Ataxia Research Alliance Grant 10047373 (to D. M. W). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Fe–S
- iron–sulfur
- CM
- complete minimal
- BPS
- bathophenanthroline sulfonate
- GDO
- gentisate 1,2-dioxygenase
- RT-qPCR
- quantitative RT-PCR.
References
- 1. Foury F., and Roganti T. (2002) Deletion of the mitochondrial carrier genes MRS3 and MRS4 suppresses mitochondrial iron accumulation in a yeast frataxin-deficient strain. J. Biol. Chem. 277, 24475–24483 10.1074/jbc.M111789200 [DOI] [PubMed] [Google Scholar]
- 2. Mühlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., Schweyen R. J., Lill R., and Wiesenberger G. (2003) A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278, 40612–40620 10.1074/jbc.M307847200 [DOI] [PubMed] [Google Scholar]
- 3. Li L., and Kaplan J. (1997) Characterization of two homologous yeast genes that encode mitochondrial iron transporters. J. Biol. Chem. 272, 28485–28493 10.1074/jbc.272.45.28485 [DOI] [PubMed] [Google Scholar]
- 4. Lin H., Li L., Jia X., Ward D. M., and Kaplan J. (2011) Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J. Biol. Chem. 286, 3851–3862 10.1074/jbc.M110.190959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Froschauer E. M., Rietzschel N., Hassler M. R., Binder M., Schweyen R. J., Lill R., Mühlenhoff U., and Wiesenberger G. (2013) The mitochondrial carrier Rim2 co-imports pyrimidine nucleotides and iron. Biochem. J. 455, 57–65 10.1042/BJ20130144 [DOI] [PubMed] [Google Scholar]
- 6. Yoon H., Zhang Y., Pain J., Lyver E. R., Lesuisse E., Pain D., and Dancis A. (2011) Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J. 440, 137–146 10.1042/BJ20111036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li L., Miao R., Jia X., Ward D. M., and Kaplan J. (2014) Expression of the yeast cation diffusion facilitators Mmt1 and Mmt2 affects mitochondrial and cellular iron homeostasis: evidence for mitochondrial iron export. J. Biol. Chem. 289, 17132–17141 10.1074/jbc.M114.574723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolaj-Robin O., Russell D., Hayes K. A., Pembroke J. T., and Soulimane T. (2015) Cation diffusion facilitator family: structure and function. FEBS Lett. 589, 1283–1295 10.1016/j.febslet.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 9. Montanini B., Blaudez D., Jeandroz S., Sanders D., and Chalot M. (2007) Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8, 107 10.1186/1471-2164-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Migocka M., Maciaszczyk-Dziubinska E., Malas K., Posyniak E., and Garbiec A. (2019) Metal tolerance protein MTP6 affects mitochondrial iron and manganese homeostasis in cucumber. J. Exp. Bot. 70, 285–300 10.1093/jxb/ery342 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Ichikawa Y., Bayeva M., Ghanefar M., Potini V., Sun L., Mutharasan R. K., Wu R., Khechaduri A., Jairaj Naik T., and Ardehali H. (2012) Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. Proc. Natl. Acad. Sci. U.S.A. 109, 4152–4157 10.1073/pnas.1119338109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ichikawa Y., Ghanefar M., Bayeva M., Wu R., Khechaduri A., Naga Prasad S. V., Mutharasan R. K., Naik T. J., and Ardehali H. (2014) Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Invest. 124, 617–630 10.1172/JCI72931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Issitt T., Bosseboeuf E., De Winter N., Dufton N., Gestri G., Senatore V., Chikh A., Randi A. M., and Raimondi C. (2019) Neuropilin-1 controls endothelial homeostasis by regulating mitochondrial function and iron-dependent oxidative stress. iScience 11, 205–223 10.1016/j.isci.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaguchi-Iwai Y., Dancis A., and Klausner R. D. (1995) AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14, 1231–1239 10.1002/j.1460-2075.1995.tb07106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li L., Bagley D., Ward D. M., and Kaplan J. (2008) Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 28, 1326–1337 10.1128/MCB.01219-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L., Jia X., Ward D. M., and Kaplan J. (2011) Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J. Biol. Chem. 286, 38488–38497 10.1074/jbc.M111.286666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li L., Miao R., Bertram S., Jia X., Ward D. M., and Kaplan J. (2012) A role for iron-sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J. Biol. Chem. 287, 35709–35721 10.1074/jbc.M112.395533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pimentel C., Vicente C., Menezes R. A., Caetano S., Carreto L., and Rodrigues-Pousada C. (2012) The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS ONE 7, e37434 10.1371/journal.pone.0037434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rietzschel N., Pierik A. J., Bill E., Lill R., and Mühlenhoff U. (2015) The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol. Cell. Biol. 35, 370–378 10.1128/MCB.01033-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Askwith C., Eide D., Van Ho A., Bernard P. S., Li L., Davis-Kaplan S., Sipe D. M., and Kaplan J. (1994) The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76, 403–410 10.1016/0092-8674(94)90346-8 [DOI] [PubMed] [Google Scholar]
- 21. Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., and Dancis A. (1996) A permease-oxidase complex involved in high-affinity iron uptake in yeast [see comments]. Science 271, 1552–1557 10.1126/science.271.5255.1552 [DOI] [PubMed] [Google Scholar]
- 22. Dix D. R., Bridgham J. T., Broderius M. A., Byersdorfer C. A., and Eide D. J. (1994) The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269, 26092–26099 [PubMed] [Google Scholar]
- 23. Urbanowski J. L., and Piper R. C. (1999) The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274, 38061–38070 10.1074/jbc.274.53.38061 [DOI] [PubMed] [Google Scholar]
- 24. Cohen A., Nelson H., and Nelson N. (2000) The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 275, 33388–33394 10.1074/jbc.M004611200 [DOI] [PubMed] [Google Scholar]
- 25. Ward D. M., Chen O. S., Li L., Kaplan J., Bhuiyan S. A., Natarajan S. K., Bard M., and Cox J. E. (2018) Altered sterol metabolism in budding yeast affects mitochondrial iron-sulfur (Fe-S) cluster synthesis. J. Biol. Chem. 293, 10782–10795 10.1074/jbc.RA118.001781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rutherford J. C., Jaron S., and Winge D. R. (2003) Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278, 27636–27643 10.1074/jbc.M300076200 [DOI] [PubMed] [Google Scholar]
- 27. Li L., and Kaplan J. (2004) A mitochondrial-vacuolar signaling pathway in yeast that affects iron and copper metabolism. J. Biol. Chem. 279, 33653–33661 10.1074/jbc.M403146200 [DOI] [PubMed] [Google Scholar]
- 28. Chang Y. C., and Timberlake W. E. (1993) Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamaguchi-Iwai Y., Stearman R., Dancis A., and Klausner R. D. (1996) Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15, 3377–3384 10.1002/j.1460-2075.1996.tb00703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., and Kaplan J. (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278, 45499–45506 10.1074/jbc.M307229200 [DOI] [PubMed] [Google Scholar]
- 31. Rutherford J. C., Ojeda L., Balk J., Mühlenhoff U., Lill R., and Winge D. R. (2005) Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280, 10135–10140 10.1074/jbc.M413731200 [DOI] [PubMed] [Google Scholar]
- 32. Yamaguchi-Iwai Y., Ueta R., Fukunaka A., and Sasaki R. (2002) Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J. Biol. Chem. 277, 18914–18918 10.1074/jbc.M200949200 [DOI] [PubMed] [Google Scholar]
- 33. MacIsaac K. D., Wang T., Gordon D. B., Gifford D. K., Stormo G. D., and Fraenkel E. (2006) An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7, 113 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lill R., and Mühlenhoff U. (2005) Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30, 133–141 10.1016/j.tibs.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 35. Mühlenhoff U., and Lill R. (2000) Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta 1459, 370–382 10.1016/S0005-2728(00)00174-2 [DOI] [PubMed] [Google Scholar]
- 36. Chaves R. S., Herrero P., and Moreno F. (1999) Med8, a subunit of the mediator CTD complex of RNA polymerase II, directly binds to regulatory elements of SUC2 and HXK2 genes. Biochem. Biophys. Res. Commun. 254, 345–350 10.1006/bbrc.1998.9954 [DOI] [PubMed] [Google Scholar]
- 37. Moreno-Herrero F., Herrero P., Colchero J., Baró A. M., and Moreno F. (1999) Analysis by atomic force microscopy of Med8 binding to cis-acting regulatory elements of the SUC2 and HXK2 genes of Saccharomyces cerevisiae. FEBS Lett. 459, 427–432 10.1016/S0014-5793(99)01289-2 [DOI] [PubMed] [Google Scholar]
- 38. Rodrigues-Pousada C., Devaux F., Caetano S. M., Pimentel C., da Silva S., Cordeiro A. C., and Amaral C. (2019) Yeast AP-1 like transcription factors (Yap) and stress response: a current overview. Microb. Cell 6, 267–285 10.15698/mic2019.06.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuge S., Jones N., and Nomoto A. (1997) Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16, 1710–1720 10.1093/emboj/16.7.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y., Lyver E. R., Knight S. A., Pain D., Lesuisse E., and Dancis A. (2006) Mrs3p, Mrs4p, and frataxin provide iron for Fe-S cluster synthesis in mitochondria. J. Biol. Chem. 281, 22493–22502 10.1074/jbc.M604246200 [DOI] [PubMed] [Google Scholar]
- 41. Metzendorf C., Wu W., and Lind M. I. (2009) Overexpression of Drosophila mitoferrin in l(2)mbn cells results in dysregulation of Fer1HCH expression. Biochem. J. 421, 463–471 10.1042/BJ20082231 [DOI] [PubMed] [Google Scholar]
- 42. Shaw G. C., Cope J. J., Li L., Corson K., Hersey C., Ackermann G. E., Gwynn B., Lambert A. J., Wingert R. A., Traver D., Trede N. S., Barut B. A., Zhou Y., Minet E., Donovan A., et al. (2006) Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100 10.1038/nature04512 [DOI] [PubMed] [Google Scholar]
- 43. Blaiseau P. L., Lesuisse E., and Camadro J. M. (2001) Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J. Biol. Chem. 276, 34221–34226 10.1074/jbc.M104987200 [DOI] [PubMed] [Google Scholar]
- 44. Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., and Kaplan J. (2004) Transcription of the yeast iron regulon responds not directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279, 29513–29518 10.1074/jbc.M403209200 [DOI] [PubMed] [Google Scholar]
- 45. Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., and Kaplan J. (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712 10.1126/science.276.5319.1709 [DOI] [PubMed] [Google Scholar]
- 46. Chen O. S., and Kaplan J. (2001) YFH1-mediated iron homeostasis is independent of mitochondrial respiration. FEBS Lett. 509, 131–134 10.1016/S0014-5793(01)03137-4 [DOI] [PubMed] [Google Scholar]
- 47. Santos R., Dancis A., Eide D., Camadro J. M., and Lesuisse E. (2003) Zinc suppresses the iron accumulation phenotype of Saccharomyces cerevisiae lacking the yeast frataxin homologue (Yfh1). Biochem. J. 375, 247–254 10.1042/bj20030835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen O. S., and Kaplan J. (2000) CCC1 suppresses mitochondrial damage in the yeast model of Friedreich's ataxia by limiting mitochondrial iron accumulation. J. Biol. Chem. 275, 7626–7632 10.1074/jbc.275.11.7626 [DOI] [PubMed] [Google Scholar]
- 49. De Freitas J. M., Liba A., Meneghini R., Valentine J. S., and Gralla E. B. (2000) Yeast lacking Cu-Zn superoxide dismutase show altered iron homeostasis: role of oxidative stress in iron metabolism. J. Biol. Chem. 275, 11645–11649 10.1074/jbc.275.16.11645 [DOI] [PubMed] [Google Scholar]
- 50. Jensen L. T., Sanchez R. J., Srinivasan C., Valentine J. S., and Culotta V. C. (2004) Mutations in Saccharomyces cerevisiae iron-sulfur cluster assembly genes and oxidative stress relevant to Cu,Zn superoxide dismutase. J. Biol. Chem. 279, 29938–29943 10.1074/jbc.M402795200 [DOI] [PubMed] [Google Scholar]
- 51. Kuge S., and Jones N. (1994) YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13, 655–664 10.1002/j.1460-2075.1994.tb06304.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilkens S. (2015) Structure and mechanism of ABC transporters. F1000Prime Rep 7, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amberg D. C., Botstein D., and Beasley E. M. (1995) Precise gene disruption in Saccharomyces cerevisiae by double fusion polymerase chain reaction. Yeast 11, 1275–1280 10.1002/yea.320111307 [DOI] [PubMed] [Google Scholar]
- 54. Yun C. W., Ferea T., Rashford J., Ardon O., Brown P. O., Botstein D., Kaplan J., and Philpott C. C. (2000) Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae: evidence for two pathways of iron uptake. J. Biol. Chem. 275, 10709–10715 10.1074/jbc.275.14.10709 [DOI] [PubMed] [Google Scholar]
- 55. Rutherford J. C., Jaron S., Ray E., Brown P. O., and Winge D. R. (2001) A second iron-regulatory system in yeast independent of Aft1p. Proc. Natl. Acad. Sci. U.S.A. 98, 14322–14327 10.1073/pnas.261381198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen O. S., Hemenway S., and Kaplan J. (2002) Inhibition of Fe-S cluster biosynthesis decreases mitochondrial iron-export: evidence that Yfh1p affects Fe-S cluster synthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 16922–16927 10.1073/pnas.232392299 [DOI] [PMC free article] [PubMed] [Google Scholar]