Abstract
Benzylisoquinoline alkaloids (BIAs) are a major class of plant metabolites with many pharmacological benefits. Sacred lotus (Nelumbo nucifera) is an ancient aquatic plant of medicinal value because of antiviral and immunomodulatory activities linked to its constituent BIAs. Although more than 30 BIAs belonging to the 1-benzylisoquinoline, aporphine, and bisbenzylisoquinoline structural subclasses and displaying a predominant R-enantiomeric conformation have been isolated from N. nucifera, its BIA biosynthetic genes and enzymes remain unknown. Herein, we report the isolation and biochemical characterization of two O-methyltransferases (OMTs) involved in BIA biosynthesis in sacred lotus. Five homologous genes, designated NnOMT1–5 and encoding polypeptides sharing >40% amino acid sequence identity, were expressed in Escherichia coli. Functional characterization of the purified recombinant proteins revealed that NnOMT1 is a regiospecific 1-benzylisoquinoline 6-O-methyltransferase (6OMT) accepting both R- and S-substrates, whereas NnOMT5 is mainly a 7-O-methyltransferase (7OMT), with relatively minor 6OMT activity and a strong stereospecific preference for S-enantiomers. Available aporphines were not accepted as substrates by either enzyme, suggesting that O-methylation precedes BIA formation from 1-benzylisoquinoline intermediates. Km values for NnOMT1 and NnOMT5 were 20 and 13 μm for (R,S)-norcoclaurine and (S)-N-methylcoclaurine, respectively, similar to those for OMTs from other BIA-producing plants. Organ-based correlations of alkaloid content, OMT activity in crude extracts, and OMT gene expression supported physiological roles for NnOMT1 and NnOMT5 in BIA metabolism, occurring primarily in young leaves and embryos of sacred lotus. In summary, our work identifies two OMTs involved in BIA metabolism in the medicinal plant N. nucifera.
Keywords: plant biochemistry, plant molecular biology, secondary metabolism, enzyme catalysis, mass spectrometry (MS), natural product biosynthesis, natural product, benzylisoquinoline alkaloids, Nelumbo nucifera, O-methyltransferases, regiospecificity, stereospecific
Introduction
Benzylisoquinoline alkaloids (BIAs)2 are a major class of plant-specialized metabolites, many of which possess significant pharmacological value. The 1-benzylisoquinoline (S)-norcoclaurine has been recognized as the common intermediate to all biosynthetically characterized BIAs (Fig. 1), and is formed via the enantioselective condensation of the tyrosine derivatives, dopamine and 4-hydroxyphenylacetaldehyde, catalyzed by norcoclaurine synthase (NCS) (1). Functional group modifications, especially O- and N-methylations, of the 1-benzylisoquinoline scaffold combined with various C-C and C-O coupling reactions yield additional structural subclasses including aporphines (e.g. magnoflorine), bisbenzylisoquinolines (e.g. dauricine), pavine (e.g. eschscholtzidine), protoberberines (e.g. berberine), morphinans (e.g. morphine), phthalideisoquinolines (e.g. noscapine), and benzo[c]phenanthridines (e.g. sanguinarine) (2).
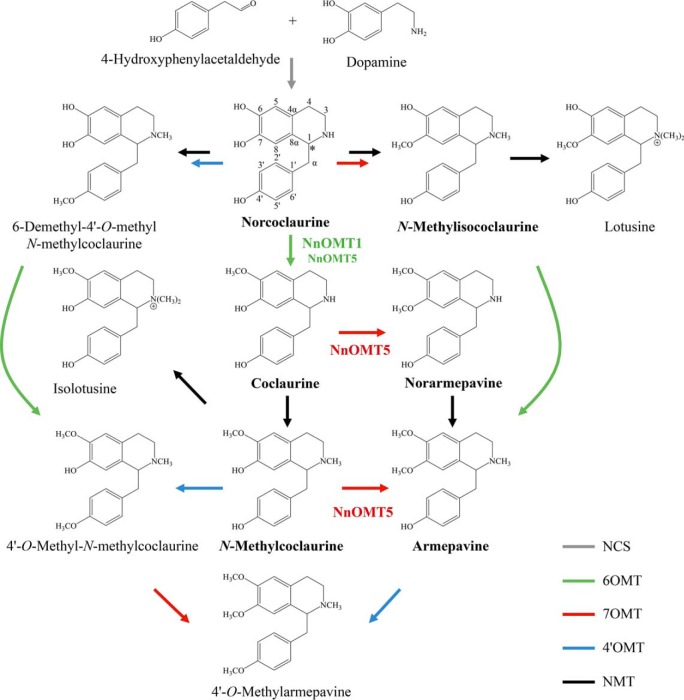
Figure 1.
Schematic representation of the proposed 1-benzylisoquinoline alkaloid biosynthetic pathway in sacred lotus (N. nucifera). The enzymes functionally characterized in this study (NnOMT1 and NnOMT5) are indicated, and detected alkaloids are shown in bold. Asterisk denotes the chiral center in norcoclaurine. Gray arrow, norcoclaurine synthase (NCS); green arrows, 6-O-methyltransferase (6OMT); red arrows, 7-O-methyltransferase (7OMT); blue arrows, 4′-O-methyltransferase (4′OMT); black arrows, N-methyltransferase (NMT).
The natural occurrence of BIAs is restricted to a relatively small number of plant families primarily in the order Ranunculales, which includes the opium poppy (Papaver somniferum), an extensively investigated model species that remains the sole commercial source for the narcotic analgesic morphine and the antitussive and potential anticancer drug noscapine (3). BIAs have also been detected in other plant orders including the Cornales, Laurales, Magnoliales, Piperales, Sapindales, and Proteales, although BIA metabolism in members of these taxa has not yet been empirically determined (4). Sacred lotus (Nelumbo nucifera Gaertn.; Fig. 2a) is an aquatic basal eudicot in the order Proteales that has been cultivated as an edible crop for nearly seven millennia, and is highly valued in both Chinese and Ayurvedic traditional medicine for its healing properties (5). The pharmacological properties of sacred lotus are largely the result of the BIA constituents in the plant. For instance, 1-benzylisoquinolines such as norcoclaurine and its 6-O-methylated derivative coclaurine have exhibited anti-HIV activity (6), whereas the 6,7-O,O- and N-methylated derivative armepavine has displayed promising immunomodulatory effects for the treatment of autoimmune diseases such as systemic lupus erythematosus (7). In addition, major aporphines (e.g. nuciferine) and bisbenzylisoquinolines (e.g. neferine) have shown potential for the treatment of lung, liver, ovarian, and breast cancers (8–12). However, none of the biosynthetic genes and enzymes responsible for the BIA biosynthesis in sacred lotus have been characterized (13).
Figure 2.
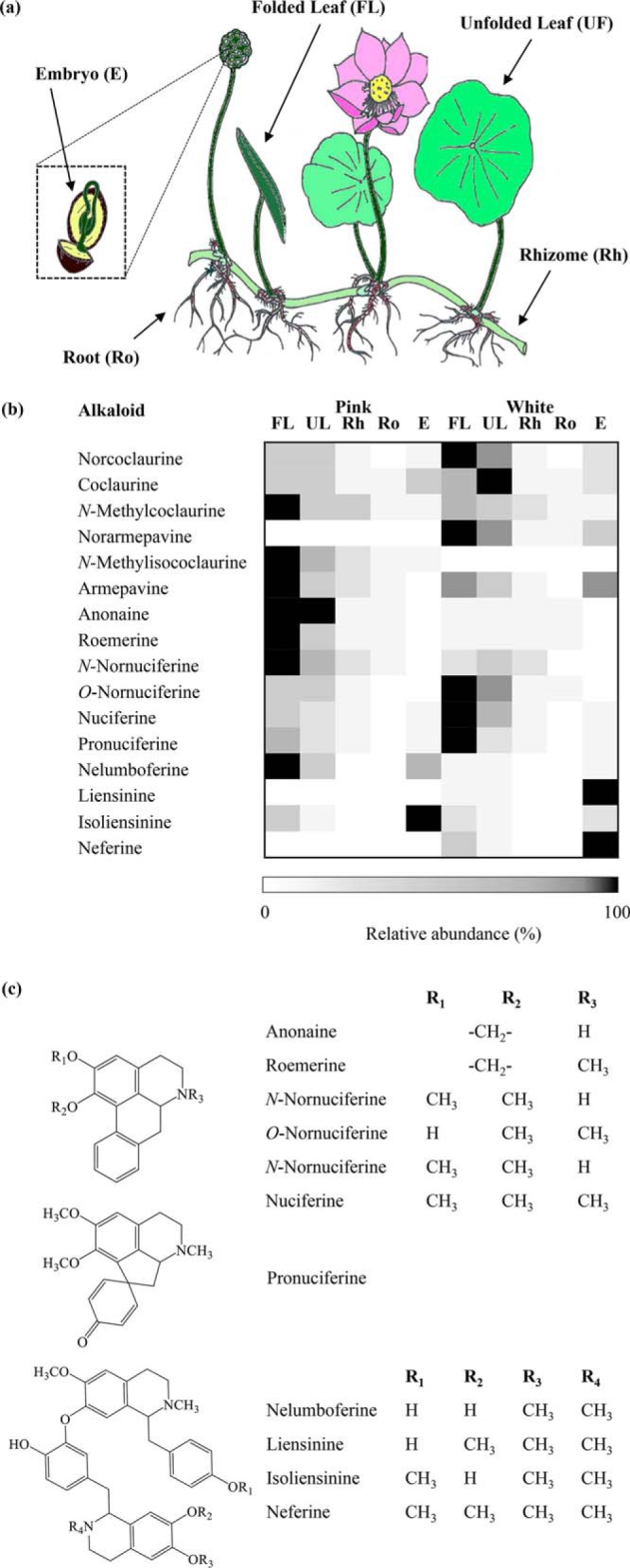
(a) Diagram of sacred lotus. (b) Heat map showing the organ-specific profile of BIAs in two varieties of sacred lotus (Pink and White). Relative abundance corresponds to the mean value of three independent replicates. Values were normalized to the sample with the highest level for each compound. (c) Structures corresponding to detected aporphine, pro-aporphine, and bisbenzylisoquinoline alkaloids isolated from sacred lotus (for 1-benzylisoquinoline alkaloid structures refer to Fig. 1). FL, folded leaf; UL, unfolded leaf; Rh, rhizome; Ro, roots; E, embryos.
Based on the reported alkaloid profile in sacred lotus and well-characterized pathways in opium poppy, BIA metabolism is predicted to begin with the formation of norcoclaurine and proceed via a limited number of enzyme types including one or more O-methyltransferases (OMTs), N-methyltransferases, and cytochrome P450 monooxygenases potentially belonging to CYP80 and CYP719 subfamilies (13). Specifically, the formation of known 1-benzylisoquinoline alkaloids in sacred lotus would involve only O-methylations and N-methylations of norcoclaurine (Fig. 1). Because O-methylation contributes substantially to the diversity of BIAs in sacred lotus, we targeted the isolation and functional characterization of OMTs.
Methylations play a central role in the functionalization of specialized metabolites (14). In particular, OMTs involved in BIA metabolism catalyze methyl transfer to the hydroxyl group of an alkaloid substrate, using S-adenosyl-l-methionine (SAM) as the methyl donor group and proceeding via an SN2-like nucleophilic attack yielding a methylated alkaloid product and S-adenosyl-l-homocysteine (SAH) (15). OMTs are typically homodimeric or, as recently discovered, heterodimeric enzymes (16, 17). Within each monomer, the C-terminal domain adopts a canonical Rossmann-fold (i.e. a β-strand sandwiched by two α-helices) and plays a fundamental role in substrate binding and catalysis, whereas the N-terminal domain is of central importance in dimerization (15, 18–20).
In opium poppy, the methylation of (S)-norcoclaurine to (S)-coclaurine by (R,S)-norcoclaurine 6-O-methyltransferase (Ps6OMT) represents the first tailoring reaction in BIA metabolism (21, 22). Recent crystallographic studies on 6OMT from meadow rue (Thalictrum flavum, Tf6OMT) have revealed key determinants for substrate recognition and turnover (18). In opium poppy, at least six other OMTs involved in BIA biosynthesis have been functionally characterized, including (R,S)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase 2 (Ps4′OMT2), (R,S)-reticuline 7-O-methyltransferase (Ps7OMT) and norreticuline 7-O-methyltransferase (PsN7OMT), all involved in 1-benzylisoquinoline biosynthesis (i.e. reticuline, laudanine and norlaudanine), as well as 4′-O-demethyl-3-O-acetylpapaveroxine 4′-O-methyltransferase (PsOMT2:PsOMT3 and PsOMT2:Ps6OMT heterodimers) and scoulerine 9-O-methyltransferase (PsSOMT1), which function in phthalideisoquinoline (e.g. noscapine) metabolism (16, 17, 19, 21–26). Although the isolation of a specific 3′-OMT has been elusive, low 3′-O-methyltransferase activity has been detected for PsSOMT1, as well as for SOMTs from related species such as yellow horned poppy (Glaucium flavum, GfOMT6) and California poppy (Eschscholzia californica, EcSOMT) (26–28). Finally, a columbamine O-methyltransferase has been isolated from Japanese goldthread (Coptis japonica; CjCoOMT) (29). Although several functionally related OMTs have been investigated, these enzymes have been isolated from fewer than 10 species all from the Ranunculales (20).
Sacred lotus accumulates 1-benzylisoquinolines, and derived aporphines (via C8-C2′ intramolecular coupling) and bisbenzylisoquinolines (via C8-C3′/5′ and C7-O-C3′/5′ intermolecular coupling) (5), most of which are O-methylated at C6, C7, and/or C4′ (13). The availability of a sacred lotus draft genome (30, 31) greatly facilitates gene mining for candidate OMTs based on amino acid sequence similarity with respect to characterized O-methyltransferases (32–34). However, OMT candidates have so far been investigated only in terms of gene expression, with no functional characterization of the encoded proteins. In addition, enzyme stereospecificity is an intriguing feature of BIA metabolism in sacred lotus, because norcoclaurine has been isolated from the plant as both R- and S-enantiomers, and other BIAs, including coclaurine, norarmepavine, and N-methylcoclaurine have been exclusively reported as the R-conformer (13), in contrast with the predominantly S-stereochemistry of most BIAs in opium poppy and related plants (3). Herein we report the identification and characterization of O-methyltransferases involved in BIA metabolism in sacred lotus.
Results
Sacred lotus BIA profile
Alkaloids corresponding to the leaf (folded and unfolded developmental stages), rhizome, root, and embryo (lotus plumule) of two N. nucifera cultivars, referred as Pink and White because of their distinct floral coloration, were extracted and analyzed by mass spectrometry (MS) (Fig. 2, b and c, Table S1). Fifteen alkaloids (i.e. six 1-benzylisoquinolines, four aporphines, one pro-aporphine, and four bisbenzylisoquinolines) were identified based on their retention times and collision-induced dissociation (CID) spectra compared with available authentic standards or previously published data. 1-Benzylisoquinoline and aporphine alkaloids were prevalent in the leaves, whereas bisbenzylisoquinolines were most abundant in the embryos. In general, the rhizome and roots contained only low BIA levels, although no bisbenzylisoquinolines were detected. Notably, the structural isomers N-methylisococlaurine and norarmepavine were exclusively detected in the Pink and White varieties, respectively. The bisbenzylisoquinolines liensinine and neferine were found only in the White variety, whereas nelumboferine and isoliensinine were the only bisbenzylisoquinolines identified in Pink and accumulated at 20- (in folded leaves) and 4-fold (in embryos) higher levels compared with the White variety. In addition, alkaloids were detected in the leaves (norarmepavine, nelumboferine, and isoliensinine), rhizome (norcoclaurine, coclaurine, N-methylcoclaurine, norarmepavine, N-methylisococlaurine, anonaine, roemerine, N-nornuciferine, O-nornuciferine, nuciferine and pronuciferine), roots (coclaurine, N-methylcoclaurine, norarmepavine, N-methylisococlaurine, armepavine, anonaine, roemerine, N-nornuciferine, and O-nornuciferine), and embryo (norarmepavine and O-nornuciferine) that had not been previously reported (5, 13). Overall, the alkaloid profile found for both varieties supports the occurrence of 6-O-, 7-O-, and 4′-O-methyltransferases in sacred lotus.
NnOMT identification and phylogeny
Sacred lotus transcripts encoding five putative OMTs (NnOMT1–NnOMT5) were identified based on amino acid sequence identity with opium poppy OMTs (Table S2). The predicted NnOMT1–NnOMT5 translation products displayed predicted molecular masses of 38.4, 38.7, 38.8, 39.2, and 38.1 kDa and expected isoelectric points of 6.5, 7.8, 6.1, 5.7, and 5.6, respectively. Four sacred lotus OMTs shared ∼80% amino acid sequence identity, whereas NnOMT5 exhibited <50% identity with respect to the others. NnOMT1 showed ∼70% amino acid sequence identity compared with functionally characterized 6OMTs, whereas all candidates shared ∼40–60% sequence identity with previously reported 7OMTs and 4′OMTs (Table S3). Phylogenetic relationships among NnOMTs and functionally characterized BIA O-methyltransferases placed NnOMT1–NnOMT4 in a single clade related to several characterized 6OMTs, whereas NnOMT5 emerged on a distal clade that also contained CjCoOMT (Fig. 3).
Figure 3.
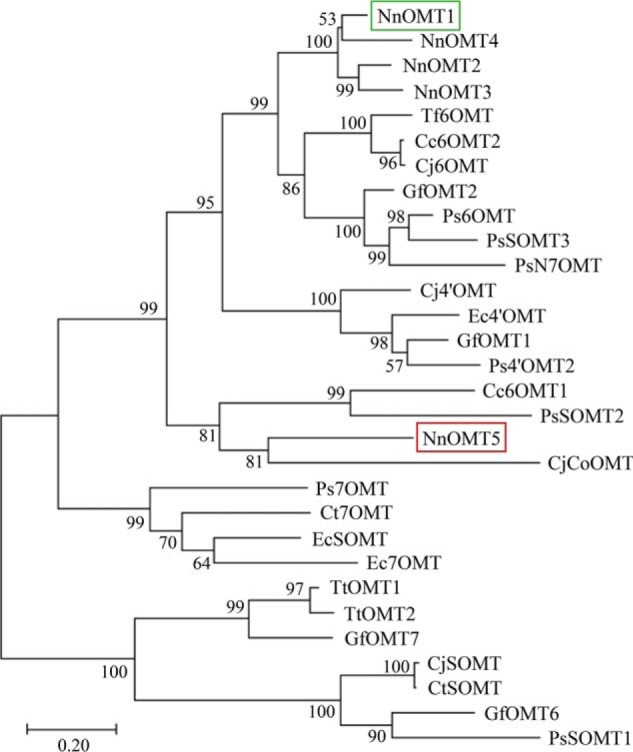
Phylogenetic relationships among sacred lotus O-methyltransferase (NnOMT) candidates and functionally characterized OMTs from BIA-accumulating plants in the Ranunculales. The evolutionary history was inferred using the Maximum Likelihood and the JTT matrix-based model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree with the highest log likelihood is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGAX. Cc, C. chinensis; Cj, C. japonica; Ct, C. teeta; Ec, E. californica; Gf, G. flavum; Nn, N. nucifera; Ps, P. somniferum; Tf, T. flavum; Tt, Thalictrum tuberosum. Sacred lotus NnOMT1 (green box) and NnOMT5 (red box) are indicated.
Amino acid sequence alignment of the functionally characterized Tf6OMT and sacred lotus OMT candidates showed a high conservation of key amino acid residues, including the catalytic determinants His256, Asp257, and Glu315 (18), except for NnOMT4, in which case Asp257 was substituted with a Tyr residue (Fig. 4). Amino acids implicated in BIA substrate binding (Gly165, Asp169, Cys253, and Asp306) were also conserved among sacred lotus OMT candidates except for G165V and C253W substitutions in NnOMT2, G165A in NnOMT3, G165S in NnOMT4, and G165A, D169H, and C253S in NnOMT5. The main residues involved in SAM binding including Thr170, Gly195 (motif I), Asp218 (motif II), Asp238 (motif III), and Lys252 (motif IV) were conserved across sacred lotus OMT candidates, excluding Asp218 (substituted by Gly in NnOMT4) and Asp238 (substituted by Asn in NnOMT3). Two methionine residues, Met166 and Met307, assisting in the isoquinoline placing through sulfur-aromatic interactions were also conserved in lotus OMT candidates. The aromatic residues Trp149 and Phe162, which interact with both SAM and BIA substrates, were mutated, Trp149 to Cys, Ser, and Ile in NnOMT3, NnOMT4, and NnOMT5, respectively, and Phe162 to Leu in NnOMT5. The conserved GXGXGX sequence in motif I, a hallmark of SAM-dependent methyltransferases (14), was fully maintained in all the candidates except NnOMT5, in which the middle Gly residue was mutated to Cys.
Figure 4.
Multiple sequence alignment among NnOMT candidates and T. flavum 6OMT (Tf6OMT). Residues shared by Tf6OMT and sacred lotus OMTs are shaded in gray, and fully conserved residues are shaded in black. Asterisks indicate catalytic (His256, Asp257, and Glu315) and other key residues implicated in BIA (Gly165, Asp169, Cys253, and Asp306) and SAM (Thr170, Gly195, Asp218, Asp238, and Lys252) binding, as reported for Tf6OMT. Conserved motives I–IV are underlined.
NnOMT purification and characterization
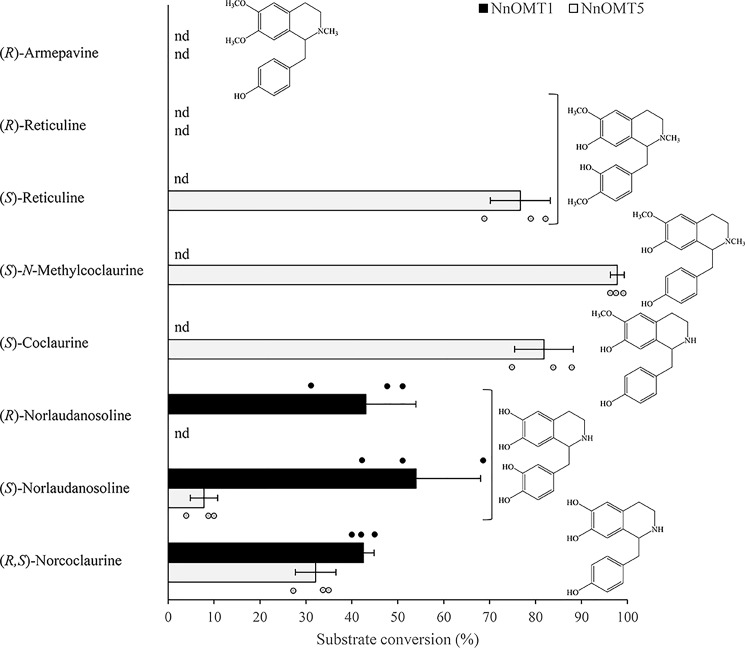
Sacred lotus recombinant His6-tagged OMTs produced in Escherichia coli were purified using cobalt-affinity chromatography and detected on an immunoblot using an anti-His6 antibody (Fig. S1). The molecular weights of the purified proteins and the predicted translation products were similar. Recombinant proteins were initially screened for enzymatic activity using 15 potential BIA substrates representing the 1-benzylisoquinoline, aporphine, protoberberine, and morphinan structural subclasses. The potential 1-benzylisoquinoline substrates differentially contained free hydroxyl groups at C6, C7, C3′, and/or C4′, and both R- and S-enantiomers were in some cases available (Table S4). Reaction products showed an increase of 14 Da with respect to the corresponding substrate and were detected in reactions containing NnOMT1 or NnOMT5 incubated with various 1-benzylisoquinoline substrates (Fig. 5, Fig. S2). (R,S)-Norcoclaurine and (S)-norlaudanosoline were O-methylated by both NnOMT1 and NnOMT5, whereas (R)-norlaudanosoline was only accepted by NnOMT1. In addition, NnOMT5 catalyzed the O-methylation of (S)-coclaurine, (S)-N-methylcoclaurine, and (S)-reticuline with similar efficiencies, but (R)-reticuline was not accepted. Single peaks were detected for all the reactions, indicating the formation of only one product.
Figure 5.
Substrate range for recombinant NnOMT1 and NnOMT5. Values represent the mean ± S.D. of three independent replicates. The structure of the corresponding substrates is shown; nd, not detected.
Reaction products corresponding to the 6-O-methylation of norcoclaurine and norlaudanosoline (coclaurine (m/z 286) and 6-O-methylnorlaudanosoline (m/z 302), respectively) and the 7-O-methylation of N-methylcoclaurine (armepavine (m/z 314)), were unambiguously identified by comparison with available authentic standards. Enzyme assays with coclaurine and reticuline were subjected to positive-mode electrospray ionization (ESI(+)) high performance LC (HPLC)-tandem mass spectrometry (MS/MS) for the accurate characterization of reaction products by CID fragmentation analysis to determine whether O-methylation was associated with the isoquinoline (C7) or benzyl moiety of coclaurine (C4′) and reticuline (C3′) (Fig. S3). ESI(+)-CID of 1-benzylisoquinolines at low ionization energy yielded isoquinoline and/or benzyl moieties as major ion fragments; thus, the addition of 14 Da to either fragment with respect to the parent ion revealed the regiospecificity of the reaction. In this regard, coclaurine showed two main fragments at m/z 178 and 107, corresponding to the isoquinoline and benzyl moieties, respectively. After incubation with NnOMT5, the m/z 107 fragment was retained in the reaction product, indicating that the methyl group was added to the isoquinoline moiety (i.e. 7-O-methylation) resulting in the formation of norarmepavine, an alkaloid found in sacred lotus (the other possible reaction product, 4′-O-methylcoclaurine, has not been reported in this plant). Similarly, the reaction product of NnOMT5 incubated with (S)-reticuline showed a mass increase of the isoquinoline moiety from m/z 192 to 206, corresponding to the 7-O-methylated product laudanine, whereas the mass of the benzyl moiety (m/z 137) remained unchanged. It is notable that neither reticuline nor laudanine have been reported in sacred lotus. 4′OMT activity was not detected with any combination of tested substrates and OMT candidates.
NnOMT1 displayed strict regiospecificity with respect to the C6 hydroxyl and showed similar turnover using racemic norcoclaurine and individual enantiomers of norlaudanosoline, the 3′-hydroxylated derivative of norcoclaurine (Fig. 5). NnOMT5 nonpreferentially methylated the C7 hydroxyl of N-methylated (N-methylcoclaurine and reticuline) or N-demethylated (coclaurine) substrates, and also acted on the C6 hydroxyl of (R,S)-norcoclaurine and (S)-norlaudanosoline. The 6-O-methylation activity of NnOMT5 was not accompanied by secondary methylations (an increase of 28 Da). Contrary to NnOMT1, NnOMT5 showed a strict stereospecificity for the substrates in the S-conformation.
Both enzymes showed a pH optimum of 8.0 using (R,S)-norcoclaurine (NnOMT1) or (S)-N-methylcoclaurine (NnOMT5) as substrates and maximum catalytic activity at 30 and 37 °C, respectively (Fig. S4). NnOMT1 and NnOMT5 displayed Km values of 20 and 152 μm for (R,S)-norcoclaurine, respectively, whereas NnOMT5 exhibited a Km value of 13 μm and a Ki of 870 μm for (S)-N-methylcoclaurine (Table 1, Fig. S5). Catalytic efficiencies (kcat/Km) were similar for the 6OMT activity of NnOMT1 and the 7OMT activity of NnOMT5. The 7OMT activity of NnOMT5 was 50-fold higher than the corresponding 6OMT activity using (R,S)-norcoclaurine as the substrate.
Table 1.
Kinetics parameters for recombinant NnOMT1 (6OMT) and NnOMT5 (6OMT and 7OMT) determined at a fixed saturating concentration of SAM
Values represent the mean ± S.D. of three independent measurements.
| Enzyme | Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|
| μm | nmol min−1 mg−1 protein | s−1 | m−1 s−1 | ||
| NnOMT1 | (R,S)-Norcoclaurine | 20 ± 2 | 52 ± 2 | 0.0336 | 1686 |
| NnOMT5 | (R,S)-Norcoclaurine | 152 ± 36 | 9 ± 1 | 0.0058 | 38 |
| (S)-N-Methylcoclaurine | 13 ± 3 | 30 ± 2 | 0.0191 | 1465 |
NnOMT and BIA correlations in the plant
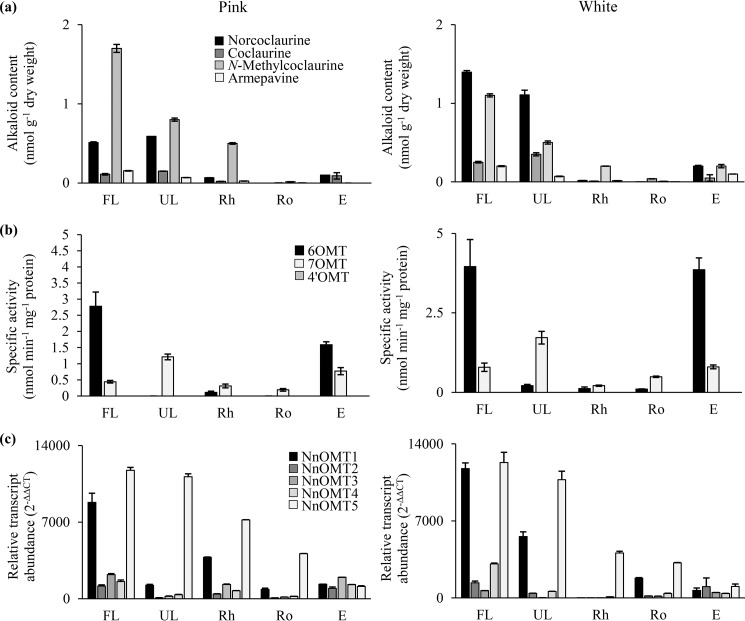
Alkaloid content, O-methyltransferase activity, and OMT gene expression were measured in leaves, rhizome, roots, and embryos for two sacred lotus varieties. Alkaloid content was determined for norcoclaurine, coclaurine, N-methylcoclaurine, and armepavine, for all of which authentic standards were available (Fig. S6). In both the Pink and White varieties, 1-benzylisoquinoline intermediates were predominantly detected in the leaves (Fig. 6a, Table S5). In the White variety, norcoclaurine was the most abundant alkaloid, with folded leaves accumulating 7-, 70-, and 1400-fold higher levels compared with the embryo, rhizome, and root, respectively. The Pink cultivar was richer in N-methylcoclaurine, with the folded leaves containing 3-, 40-, and 90-fold higher levels than the rhizome, root, and embryo, respectively.
Figure 6.
Organ-specific correlational analysis of 1-benzylisoquinoline alkaloids metabolism in two varieties (Pink and White) of sacred lotus. (a) Content of selected 1-benzylisoquinoline alkaloids. (b) 6OMT, 7OMT, and 4′OMT specific activity of plant crude extracts when incubated with (R,S)-norcoclaurine, (S)-N-methylcoclaurine, and (R)-armepavine, respectively. (c) Relative abundance of NnOMT1-NnOMT5 transcripts. Values represent the mean ± S.D. of three independent measurements. FL, folded leaf; UL, unfolded leaf; Rh, rhizome; Ro, roots; E, embryos.
Crude plant protein extracts incubated with (R,S)-norcoclaurine, (S)-N-methylcoclaurine, or (R)-armepavine revealed similarities between the Pink and White varieties (Fig. 6b, Table S6). 6OMT activity was most abundant in folded leaves and embryos, whereas 7OMT activity was predominant in the unfolded leaves. 6OMT activity was 5- and 4-fold higher than 7OMT activity in folded leaves and embryos, respectively. 7OMT activity was 8-fold higher than 6OMT in the White variety, whereas 6OMT activity was not detected in unfolded leaves of the Pink variety. Rhizomes and roots showed only low levels of 6OMT and 7OMT activities. 4′OMT activity was not detected.
Gene expression profiles were similar for both the Pink and White varieties (Fig. 6c, Table S7). NnOMT1 and NnOMT5 transcripts were most abundant in leaves and were 3- to 30-fold higher compared with other OMT genes. The levels of all five NnOMT transcripts were similar in the embryos of both varieties.
Discussion
We have shown that two cDNAs from sacred lotus efficiently catalyze the regiospecific 6-O-methylation of norcoclaurine to coclaurine (NnOMT1) and 7-O-methylation of coclaurine and N-methylcoclaurine to norarmepavine and armepavine, respectively (NnOMT5). NnOMT1 was not stereospecific, whereas NnOMT5 accepted only S-enantiomers. Correlation between transcript levels, enzyme activity, and alkaloid content supported a physiological role for NnOMT1 and NnOMT5 in BIA metabolism.
Despite sharing an apparently common biosynthetic pathway to N-methylcoclaurine (Fig. 1), sacred lotus and opium poppy exhibit substantial differences in their alkaloid profiles. In this regard, whereas reticuline constitutes a key branch point intermediate in the biosynthesis of most BIAs in opium poppy (3), sacred lotus does not accumulate reticuline and other 1-benzylisoquinolines containing a C3′ functional group, or reticuline-derived alkaloids including phthalideisoquinolines, benzo[c]phenanthridines, protoberberines, and morphinans, which occur in members of the Ranunculales (13). In contrast, aporphines and bisbenzylisoquinolines are the major BIAs in sacred lotus, although alkaloid profiles vary among nearly 600 known varieties, and across different plant organs and developmental stages (32–37). In addition, sacred lotus aporphine alkaloids do not display substitutions in the benzyl moiety (13), in contrast with the reticuline-derived aporphines (e.g. magnoflorine, corytuberine and glaucine) found in members of the Ranunculales (27, 38, 39).
The sacred lotus BIA profile showed a preferential accumulation of 1-benzylisoquinolines and aporphines in leaves, and bisbenzylisoquinolines in embryos (Fig. 2b), in agreement with previous investigations (36, 37, 40). Alkaloids containing 6-O-, 7-O-, and/or 4′-O-methylations were detected in Pink and White varieties, supporting the search for specific O-methyltransferase activities in the plant. OMT gene candidates have been predicted in sacred lotus based on sequence similarity to functionally characterized OMTs from BIA-accumulating species (32–34), but the purported catalytic activities have not been assessed. We show that two of these candidates, NnOMT1 and NnOMT5, catalyze the O-methylation of 1-benzylisoquinolines.
Fifteen BIA substrates possessing diverse structural backbones were screened, including 1-benzylisoquinolines with a free hydroxyl at C6, C7, and C4′. As described for several 6OMTs from the Ranunculales (41, 42), NnOMT1 displayed strict C6 regiospecificity and accepted both (R)- and (S)-1-benzylisoquinoline conformers (Fig. 5). NnOMT1 contains all conserved catalytic and substrate-binding residues occurring in Tf6OMT (Fig. 4). In Tf6OMT, the O-methyltransfer reaction occurs via His256 base-assisted deprotonation of the substrate C6 hydroxyl, followed by a nucleophilic attack on the SAM methyl group. Interactions of His256 and Asp257 with the C6 hydroxyl group provide proper orientation and Glu315 contributes to the strong basicity in the His256 NE2 required for catalysis (18). Because NnOMT1 and Tf6OMT possess equivalent key catalytic residues (His256, Asp257, and Glu315) and share ∼70% amino acid sequence identity, a similar catalytic mechanism is expected for NnOMT1.
NnOMT5, the most phylogenetically unique sacred lotus OMT, catalyzed the methyltransfer to the C6 hydroxyl of (R,S)-norcoclaurine and (S)-norlaudanosoline, but more efficiently to the C7 hydroxyl of (S)-coclaurine, (S)-N-methylcoclaurine, and (S)-reticuline (Table 1). Despite the closest phylogenetic relationship between NnOMT5 and CjCoOMT, their substrate ranges are different. NnOMT5 did not accept the tested protoberberines (Table S4), whereas CjCoOMT preferred columbamine and other protoberberines, but did not accept the 1-benzylisoquinolines norlaudanosoline, coclaurine, and reticuline (29). NnOMT5 also showed a close relationship with a sister clade containing PsOMT2, which together with PsOMT3 or Ps6OMT forms the heterodimeric enzyme capable of O-methylate a seco-berbine intermediate in noscapine biosynthesis (16, 17). Notably, this sister clade also included Coptis chinensis 6OMT1 (Cc6OMT1), which was reported to catalyze either 6-O- or 7-O-methylation of (S)-norcoclaurine (43). Dual 6-O- and 7-O-methyltransferase activity has also been reported for Cc6OMT2, along with enzymes from Coptis teeta (Ct7OMT), P. somniferum (Ps6OMT and Ps7OMT), and G. flavum (GfOMT1) (22, 27, 43).
Regardless of a nomenclature suggestive of regiospecific functions, OMTs involved in BIA metabolism are generally promiscuous and exhibit apparent redundancy in most plants (22, 26–28, 43, 44). In Tf6OMT co-crystalized with (S)-norlaudanosoline the C7 hydroxyl group of the BIA substrate was found in close proximity to the catalytic His256, suggesting that minor adjustments in substrate positioning could lead to productive 7-O-methyltransfer (18) and the observed dual 6-O- and 7-O-methyltransferase activity of some OMTs. Based on the available crystal structure for Tf6OMT (Protein Data Bank 5ICE), we constructed a homology model for NnOMT5 (Fig. S7). In the resulting structure, the key catalytic histidine (His251 in NnOMT5) involved in hydrogen bonding with the target C6-OH (3.0 Å), is also near the C7-OH (3.1 Å). Analysis of residues likely involved in BIA substrate binding showed that G165A and C253S mutations did not have a significant impact (i.e. the main chain carbonyl involved in hydrogen bond formation with the BIA substrate C3′ and C6 hydroxyl groups, respectively, remained in a similar position) in agreement with the observed variability of these residues in other 7OMTs (i.e. G165A in Ps7OMT, PsN7OMT and Ct7OMT, C253W in Ps7OMT and Ct7OMT, C253N in PsN7OMT, and C253G in Ec7OMT). Conversely, the D169H mutation disrupted a hydrogen bond with the C4′-OH (2.6 Å), which stabilizes the substrate in the active site. Interestingly, whereas Asp169 is a conserved residue across all 6OMTs, or is replaced by Glu in 4′OMTs, no consensus was found in functionally characterized 7OMTs (i.e. Asp in PsN7OMT, Ser in Ps7OMT and Ec7OMT, and Thr in Ct7OMT); thus, this residue might not play a significant role in 7OMT regiospecificity.
When both C6 and C7 hydroxyl groups were available, NnOMT5 exclusively catalyzed 6-O-methylation (Fig. S2). Similar results were reported for E. californica and Coptis japonica 4′OMTs, which catalyze norcoclaurine 6-O-methyltransfer, although with lower efficiency than 4′-O-methylation on already 6-O-methylated substrates (45), in support of the importance of 6-O-methylation as a purported rate-limiting step in BIA metabolism (21). However, because NnOMT5 catalyzed 7-O-methylation of (S)-N-methylcoclaurine 50-fold more efficiently than 6-O-methylation of (R,S)-norcoclaurine, the physiological relevance of the latter activity is questionable compared with NnOMT1 (Table 1). Although coclaurine and N-methylcoclaurine are almost certainly physiological substrates of NnOMT5 (Fig. 1), the occurrence of 1-benzylisoquinolines such as N-methylisococlaurine and lotusine, which contain 7-O-methyl and C6 hydroxyl moieties, does not correlate with the inability of NnOMT5 to 7-O-methylate norcoclaurine in vitro. In this regard, it is feasible that norcoclaurine N-methylation should occur first for NnOMT5 to catalyze 7-O- rather than 6-O-methylation, however, the required substrate to test this possibility was not available.
NnOMT5 also preferred norcoclaurine over norlaudanosoline (Fig. 5), whereas NnOMT1 showed no substrate preference. Nonetheless, the main catalytic activity detected for NnOMT5 was the 7-O-methylation of several N-methylated and N-demethylated substrates (Fig. 5). Although our understanding of 7OMT substrate recognition and catalysis is limited, it has been suggested that the conserved residue Asp306 in PsN7OMT is able to hydrogen bond with the nitrogen of its only substrate norreticuline, whereas Ps7OMT and Ec7OMT contain smaller uncharged residues at equivalent positions presumably to ease steric hindrance with (R,S)-reticuline and other N-methylated substrates (22, 23, 44), and Ct7OMT, with a Asp306 to Ser substitution, is able to 7-O-methylate (S)-norcoclaurine (43). Notably, NnOMT5 contains a corresponding Asp306 residue (Figs. S7 and S8), but catalyzes similarly efficient 7-O-methylation of N-methylated and N-demethyl substrates (Fig. 5).
The catalytic activity of recombinants NnOMT1 and NnOMT5 with (R,S)-norcoclaurine and (S)-N-methylcoclaurine, respectively, exhibited similar reaction efficiency (kcat/Km), whereas NnOMT5 showed a 50-fold more efficient 7-O- compared with 6-O-methylation (Table 1). The Km of NnOMT1 with (R,S)-norcoclaurine (20 μm) is similar to that reported for Ps6OMT (10 μm) and Tf6OMT (52 μm) (22, 41), whereas the Km of NnOMT5 (152 μm) is substantially higher, suggesting that NnOMT1 is the main 6OMT in sacred lotus. On the other hand, the Km of NnOMT5 with (S)-N-methylcoclaurine (13 μm) is substantially lower compared with that for (R,S)-norcoclaurine (152 μm), but similar to that reported for Ps7OMT with (S)-reticuline (16 μm) (22) and PsN7OMT with (S)-norreticuline (44 μm) (23), suggesting that NnOMT5 acts primarily as a 7OMT in the plant.
The apparent lack of recombinant NnOMT (Fig. 5, Fig. S2) or plant crude extract activity (Fig. 6b, Table S6) on substrates 3′- or 4′-hydroxyl groups partially agrees with the sacred lotus BIA profile, which lacks C3′-substituted alkaloids (13). In contrast, 4′-O-methylated BIAs were detected (Fig. 2b), including isoliensinine and neferine (Fig. 2c). Low 4′OMT activity has been reported for enzymes with primarily 6-O- or 7-O-methyltransferase activity, such as GfOMT2 and Ps7OMT (22, 27). However, NnOMT1 and NnOMT5 did not show 4′-O-methyltransferase activity on any substrate, including (R)-armepavine, although the actual substrate(s) might not have been available. The phylogenetic relationships between NnOMT5 and PsOMT2, and between NnOMT1 and PsOMT3 and Ps6OMT, suggest that 4′OMT activity in sacred lotus could also involve a heterodimeric enzyme. It is notable that in the Ranunculales, a Berberis koetineana 4′OMT (Bk4′OMT), Cj4′OMT, Ec4′OMT, and Ps4′OMT2 act only on 3′-hydroxylated substrates (22, 25, 27, 42, 45, 46), suggesting that sacred lotus 4′-O-methyltransferase activity could involve a unique enzyme.
Assayed aporphine, protoberberine, and morphinan alkaloids were not accepted as substrates (Table S4). OMTs shown to O-methylate 1-benzylisoquinolines and protoberberines occur in species accumulating both alkaloid types (26–28), although specificity for 1-benzylisoquinolines (e.g. Cj6OMT and PsN7OMT) has also been reported (23, 42). Although sacred lotus does not accumulate morphinans or protoberberines (13), aporphines are major alkaloids in the leaves (34, 35). The lack of NnOMT activity on aporphines suggests that the C8–C2′ coupling yielding the aporphine scaffold occurs after the O-methylation of 1-benzylisoquinoline intermediates. However, Dactylicapnos scandens 7OMT was recently reported to O-methylate corytuberine, demonstrating the plausibility of aporphines as OMT substrates (47). It is notable that the aporphines and bisbenzylisoquinolines of sacred lotus were not available; thus, it remains possible that certain intermediates within these BIA structural types are NnOMT substrates.
The occurrence of most BIAs as R-conformers is another unique feature in sacred lotus compared with members of the Ranunculales (13). The importance of the stereochemistry of 1-benzylisoquinoline substrates is evident in opium poppy, in which reticuline conformation determines the formation of major end point alkaloids, with morphine derived from (R)-reticuline, and other alkaloids such as noscapine and sanguinarine produced from (S)-reticuline. BIA stereochemistry is initially established by the stereoselectivity of NCS. Although all characterized NCS isoforms from the Ranunculales yield exclusively (S)-norcoclaurine (48–50), norcoclaurine in sacred lotus has been detected as both S- and R-enantiomers (6, 51, 52) and consequently, the presence of both enantiomeric forms of norcoclaurine-derived alkaloids depends on the stereospecificity of downstream enzymes. We show that NnOMT1 catalyzed the 6-O-methylation of (R)- and (S)-norlaudanosoline with similar turnover efficiency, whereas NnOMT5 accepted only (S)-norlaudanosoline and (S)-reticuline (Fig. 5, Table S4). Previously characterized OMTs have either shown a lack of stereospecificity (e.g. Cj6OMT, Ps6OMT, and Ps7OMT) or strict S-stereospecificity (Bk4′OMT) (22, 42, 53). Our results support the formation of (R)-coclaurine and (S)-armepavine as previously reported in lotus (6, 54). However, despite the fact that NnOMT5 showed strict stereospecificity toward S-enantiomers, neither norlaudanosoline nor reticuline are physiologically relevant substrates and it is possible that the enzyme catalyzes the turnover of (R)-coclaurine and (R)-N-methylcoclaurine in the formation of the reported compounds (R)-norarmepavine and (R)-armepavine, respectively (55). Unfortunately, none of the relevant R-enantiomers were available as potential substrates.
Previous work on the BIA profile in sacred lotus focused on the quantification of aporphines and bisbenzylisoquinolines in leaves and embryos, respectively, but limited information has been provided for 1-benzylisquinoline intermediates (34, 36). We quantified key alkaloids produced in the early steps of the pathway (i.e. norcoclaurine, coclaurine, N-methylcoclaurine, and armepavine) in different sacred lotus organs (Fig. 6a). Although 1-benzylisoquinolines were predominant in leaves, the detected compounds were likely intermediates leading to aporphine and bisbenzylisoquinoline end products. The low levels of 1-benzylisoquinolines in embryos could also reflect their transformation to predominant bisbenzylisoquinolines (36, 37, 40, 56).
Assays containing crude plant extracts incubated with (R,S)-norcoclaurine, (S)-N-methylcoclaurine, and (R)-armepavine showed 6-O- and 7-O-methyltransferase activity in all organs (Fig. 6b), but mostly in leaves and embryos. OMT transcripts were also expressed in all organs, although NnOMT1 and NnOMT5 showed higher expression levels, mainly in leaves (Fig. 6c). These results are consistent with a recent report on NnOMT expression at different leaf developmental stages, whereby NnOMT1 and NnOMT5 transcript levels were up to 10-fold higher compared with NnOMT2–NnOMT4 (34). A lag between NnOMT expression and aporphine alkaloid accumulation was also noted in this study, which is potentially associated with the involvement of OMTs early in BIA metabolism and a requirement for the expression of additional genes to complete aporphine and bisbenzylisoquinoline biosynthesis. It is notable that because this earlier study did not perform a functional characterization of candidate genes, NnOMT5 was reported as a 4′OMT (34). In agreement with the suggested order of O-methylations in other plants (21, 45), NnOMT1 appears to act earlier than NnOMT5 in the leaves because 6OMT activity decreased in unfolded leaves, whereas 7OMT activity increased (Fig. 6b). Similarly, NnOMT1 transcript levels were higher in folded leaves than unfolded leaves, whereas NnOMT5 transcript levels remained constant (Fig. 6c), consistent with a previous report (34).
Although NnOMT1 and NnOMT5 transcript levels were 5- to 15-fold lower in embryos compared with folded leaves, specific 6- and 7-O-methyltransferase activities were similar in both organs, likely because of the prevalence of photosynthetic leaf proteins. Interestingly, the levels of transcripts encoding CYP80G, purportedly involved in aporphine biosynthesis, in leaves were 5 times higher than levels of transcripts encoding CYP80A, potentially involved in bisbenzylisoquinoline biosynthesis, in embryos (33), suggesting that lower biosynthetic gene transcript levels in embryos might be sufficient in this organ. However, neither CYP80G nor CYP80A orthologs in sacred lotus were functionally characterized, so it is not certain that these transcripts are relevant to BIA metabolism. Bisbenzylisoquinoline biosynthesis has been proposed to occur in leaves, with alkaloids subsequently transported to embryos (36). NCS activity (4) and purported NnNCS transcripts (57) were detected in leaves, suggesting that BIA biosynthesis occurs in this organ. The detection of O-methyltransferase activity indicates that embryos are also involved in alkaloid biosynthesis.
From an evolutionary perspective, it is not known whether OMT recruitment occurred repeatedly or if an ancestrally promiscuous OMT gave rise to functionally distinct enzymes (20). The 6-O-methyltransferase activity of NnOMT1 was correctly predicted based on similarity to Tf6OMT and other 6OMTs. However, the 7-O-methyltransferase activity of NnOMT5 was unexpected because of its relatively low similarity to characterized 7OMTs from the Ranunculales (Fig. 3, Table S3), suggesting a possible independent recruitment in sacred lotus. The characterization of other enzymes (e.g. norcoclaurine synthase, coclaurine N-methyltransferase, and aporphine and bisbenzylisoquinoline synthases) will provide additional key insights into the evolutionary history of BIA biosynthesis in sacred lotus.
Experimental procedures
Plant material
Seeds for two sacred lotus (N. nucifera) varieties, named Pink and White because of their distinct flower colors, were germinated in water according to the instructions of the vendor (Rarexoticseeds). After the emergence of embryogenic leaves, the seedlings were planted in pots containing heavy loam soil and submerged in an artificial pond. Plants were grown at 30 °C and under high-intensity lighting with a 16-h photoperiod. Alternatively, seeds were carefully opened with a bench press and the embryos were isolated. All harvested plant samples were immediately frozen in liquid nitrogen and stored at −80 °C until used.
Chemicals and reagents
(R,S)-Norcoclaurine, (R)-norlaudanosoline, (S)-norlaudanosoline, (S)-coclaurine, and (13S,14R)-1,13-dihydroxy-N-methylcanadine were purchased from Toronto Research Chemicals (ON, Canada); (R)-reticuline was purchased from Santa Cruz Biotechnology (Santa Cruz, CA); (R)-armepavine was purchased from MuseChem (NJ); (S)-scoulerine was purchased from ChromaDex (CA); (+)-bulbocapnine and (+)-isocorydine were purchased from Sequoia Research Products (Pangbourne, UK); and boldine and SAM were purchased from Sigma-Aldrich. (S)-Reticuline was a gift from Tasmanian Alkaloids Pty. Ltd. (Westbury, Australia), and morphine and codeine were gifts from Sanofi-Aventis (Paris, France). (S)-N-Methylcoclaurine was generated from (S)-coclaurine using purified recombinant NMT1 from G. flavum (58). Enzymatic conversion was performed at 30 °C, with 1 mm (S)-coclaurine, 1 mm SAM, and 20 μg of recombinant GfNMT1. Reactions were extracted by ethyl acetate three times and the product identity and purity confirmed by HPLC-MS/MS. All other chemicals were purchased from Sigma-Aldrich or Bioshop Canada (ON, Canada).
Plant alkaloid extraction and LC-MS analysis
N. nucifera samples corresponding to folded and unfolded leaves, rhizomes, roots, and embryos were ground to a fine powder under liquid nitrogen using a mortar and pestle. Alkaloid extraction was performed using a modified version of a previously reported protocol for sacred lotus (36). Briefly, 1 ml of methanol was added to ∼100 mg of plant material, and samples were sonicated in an ultrasonic bath for 30 min at room temperature, centrifuged at 14,000 × g for 10 min, and the supernatant was filtered (0.2-μm Acrodisc® syringe filter, Pall Corporation, NY). The insoluble material was dried and weighed. Samples (5 μl) were fractionated on a Zorbax C18 column, 2.1 × 50 mm, 1.8 μm (Agilent, CA) using a Dionex UltiMate 3000 HPLC system (Thermo Scientific) at a flow rate of 500 μl/min. The mobile phase consisted of solvent A (10 mm ammonium acetate, pH 5.5, 5% (v/v) acetonitrile) and solvent B (100% acetonitrile), starting at 100% (v/v) solvent A, and ramping from 0 to 20% (v/v) solvent B over 5 min, 20 to 50% (v/v) solvent B over 3 min, 50 to 100% (v/v) solvent B over 3 min, remaining isocratic at 100% (v/v) solvent B over 2 min, ramping from 100 to 0% (v/v) solvent B over 0.1 min, and remaining isocratic at 100% (v/v) solvent A over 1.9 min. The total run time was 15 min and data were collected for the first 10 min. Heated ESI source and interface conditions were operated in positive ion mode as follows: vaporizer temperature 400 °C, source voltage 3 kV, sheath gas 60 arbitrary units, auxiliary gas 20 arbitrary units, capillary temperature 380 °C, capillary voltage 6 V, and tube lens 45 V. A LTQ-Orbitrap-XL (Thermo Fisher) mass spectrometer was operated using LTQ Tune Plus version 2.5.5 SP1 and Xcalibur software (version 2.1.0.1140), with additional analyses using the QualBrowser feature of Xcalibur. Internal and external calibration, tuning, and general operations were performed as previously reported (27, 59). Error was maintained at <2 ppm. Exact masses, retention times, and CID spectra of available authentic standards, or previously published data, was used for alkaloid identification (Table S1, Fig. S6). Quantification of norcoclaurine, coclaurine, N-methylcoclaurine, and armepavine was performed using authentic standards.
NnOMT identification and phylogeny
A previously reported N. nucifera draft genome (30, 31) was searched to identify sequences encoding potential O-methyltransferase involved in BIA biosynthesis using the tBLASTn algorithm and functionally characterized OMTs from P. somniferum as queries (Ps6OMT, Ps7OMT, PsN7OMT, and Ps4′OMT2). Five candidates (NnOMT1–NnOMT5) sharing >40% amino acid sequence identity with query sequences were isolated. The GenBankTM accession codes were as follows: XM_010245752, XM_010249599, XM_010249600, XM_010273389, and XM_010277761) (Table S2). Protein molecular weight and pI predictions were made using the Geneious software package (Biomatters, NJ). Amino acid sequence alignments were performed using the default parameters of the MUSCLE algorithm (60) implemented in MEGA X (61). Evolutionary history was inferred using the Maximum Likelihood method established on the JTT matrix-based model (62). The tree with the highest log likelihood was drawn, with branch lengths proportional to the number of substitutions per site, and nodes labeled according to the percentage of trees in which the associated taxa clustered together in the bootstrap test based on 1000 replicates. The amino acid percent identity matrix among N. nucifera (Nn) and other OMTs was performed using Clustal Omega (63) The GenBankTM accession codes were as follows: Cc6OMT1 (MH165875); Cc6OMT2 (MH165876); Cj4′OMT (D29812); Cj6OMT (D29811); CjCoOMT (AB073908); CjSOMT (D29809); Ct7OMT (MH165877); CtSOMT (MH165874); Ec4′OMT (AB745041); Ec7OMT (AB232153); EcSOMT (LC171865); GfOMT1 (KP176693); GfOMT2 (KP176694); GfOMT6 (KP176698); GfOMT7 (KP176699); Ps4′OMT2 (AY217334); Ps6OMT (AY217335); Ps7OMT (AY268893); PsN7OMT (FJ156103); PsSOMT1 (JN185323); PsSOMT2 (MH029292); PsSOMT3 (MH029294); Tf6OMT (AY610507); Tt6OMT1 (AF064693); Tt6OMT2 (AF064694). Abbreviations were as follows: Cc, Coptis chinensis; Cj, Coptis japonica; Ct, Coptis teeta; Ec, Eschscholzia californica; Gf, Glaucium flavum; Nn, Nelumbo nucifera; Ps, Papaver somniferum; Tf, Thalictrum flavum; Tt, Thalictrum tuberosum; 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; 6OMT, norcoclaurine 6-O-methyltransferase; 7OMT, reticuline 7-O-methyltransferase; CoOMT, columbamine O-methyltransferase; N7OMT, norreticuline 7-O-methyltransferase; SOMT, scoulerine 9-O-methyltransferase. The amino acid sequences of functionally characterized OMTs involved in BIAs biosynthesis used in the present work are provided in Table S9.
NnOMT cDNA isolation and expression
Plant tissues were ground with a mortar and pestle to a fine powder under liquid nitrogen. Total RNA was extracted using the cetyltrimethylammonium bromide method, as previously reported for N. nucifera tissues (64). RNA quality was confirmed by A260/280 and A260/230 absorbance measurements using a NanoDrop ND-1000 (Thermo Fisher Scientific) spectrophotometer, and visualization by agarose gel electrophoresis. After genomic DNA removal (AccuRT Genomics DNA Removal kit, Applied Biological Materials, BC, Canada), first-strand cDNA synthesis was performed on 1 μg of RNA using 5× All-in-One RT MasterMix kit, according to the instructions of the manufacturer (Applied Biological Materials, BC, Canada). NnOMT open reading frames were amplified from cDNA using Q5 High Fidelity DNA polymerase (New England Biolabs, MA) and sequence-specific primers (Table S8a) under the following conditions: 98 °C for 30 s; 35 cycles of 98 °C for 10 s, 52–65 °C (optimized for each NnOMT) for 30 s, and 72 °C for 30 s; 72 °C for 2 min. Amplicons were cloned in a pMiniT 2.0 vector, according to the instructions of the manufacturer (New England Biolabs, MA) and used to transform the E. coli TOP 10 strain for colony PCR screening. Plasmids from positive colonies were purified (GeneJet Plasmid Miniprep kit, Thermo-Fisher Scientific) and sequenced. PCR products were purified (GeneJet Gel Extraction Kit, Thermo-Fisher Scientific) and ligated into pRSET-A (Invitrogen) to construct the expression vectors using one-step sequence- and ligation-independent cloning (65). Full-length NnOMT coding regions were cloned in-frame with sequences encoding an N-terminal His6 tag using specific primers (Table S8a).
Recombinant protein purification
The pRSET-A-NnOMT expression plasmids were used to transform the E. coli Rosetta (DE3) pLysS (EMD Millipore, MA) strain and single colonies were used to inoculate 50 ml of LB medium supplemented with 100 μg/ml of ampicillin and 35 μg/ml of chloramphenicol. Cultures were grown at 30 °C with orbital shaking at 200 rpm for 16 h and used to inoculate 1 liter of LB media (100 μg/ml of ampicillin and 35 μg/ml of chloramphenicol) to a starting OD600 of 0.1. Cultures were grown at 30 °C until OD600 ∼0.6, cooled to 16 °C and the production of recombinant proteins was induced by the addition of isopropyl β-d-thiogalactoside to a final concentration of 0.1 mm. Cultures were kept at 16 °C with shaking at 200 rpm for 16 h and cells were harvested by centrifugation at 5,000 × g for 20 min at 4 °C. Cell pellets were resuspended in 40 ml of protein extraction buffer (100 mm Tris-HCl, pH 7.5, 300 mm NaCl, 10% (v/v) glycerol) supplemented with 1 mg/ml of lysozyme and sonicated on ice for 3 min (10-s on, 30-s off). The crude lysate was centrifuged at 16,000 × g for 20 min at 4 °C to remove cellular debris. The cleared supernatant was loaded onto 1 ml of equilibrated Talon cobalt affinity resin (Clontech) and incubated at 4 °C for 30 min with gentle shaking. The resin was washed with 20 ml of protein extraction buffer, followed by 5 ml of protein extraction buffer containing 20 mm imidazole. Purified protein was eluted using 5 ml of protein extraction buffer containing 200 mm imidazole, and subsequently concentrated and desalted by repeated ultrafiltration on an Amicon Ultra 30K column (EMD Millipore, MA) in storage buffer (100 mm Tris-HCl, pH 7.5, 10% (v/v) glycerol, 1 mm β-mercaptoethanol). Purified His6-tagged protein concentration was determined using the Bradford reagent according to the instructions of the manufacturer (Thermo Fisher) and using BSA as the standard. Protein purity was assessed by SDS-PAGE using a 10% (w/v) gel, which was subsequently transferred to a nitrocellulose membrane. Protein blots were blocked overnight with 5% (w/v) skim milk and incubated for 1 h with a 1:1,000 dilution of His6 antibody. After washing with TBST buffer (200 mm Tris-HCl, pH 7.6, 1.5 m NaCl, 0.1% (v/v) Tween 20), the blots were incubated for 1 h with a 1:10,000 dilution of goat anti-mouse IgG antibodies conjugated with horseradish peroxidize (Bio-Rad). Immunoblots were washed and developed using SuperSignal West Pico chemiluminescent substrate, according to the instructions of the manufacturer (Thermo Scientific) and the bands were visualized using an Amersham Biosciences Imager 600 (GE Healthcare).
Plant protein extraction
Plant tissues were ground to a fine powder under liquid nitrogen using a mortar and pestle. Total proteins were extracted by adding 5 ml of ice-cold extraction buffer A containing 100 mm Tris-HCl, pH 7.5, 10% (v/v) glycerol, 1% (w/v) polyvinylpyrrolidone 40, 5 mm dithiothreitol and 1 mm phenylmethylsulfonyl fluoride to ∼1 g of plant material, and sonicated in an ultrasonic bath for 5 min at 4 °C. Plant debris was eliminated by centrifugation at 14,000 × g for 10 min, and the supernatant was desalted in a PD-10 column (GE Healthcare) and eluted in extraction buffer without dithiothreitol and phenylmethylsulfonyl fluoride. The solution was concentrated by repeated ultrafiltration using an Amicon Ultra 30K column (EMD Millipore) in storage buffer (100 mm Tris-HCl, pH 7.5, 10% glycerol, 1 mm β-mercaptoethanol). Crude plant protein concentration was determined using the Bradford reagent according to the instructions of the manufacturer (Thermo Fisher) and using BSA as the standard.
Enzyme assays
Substrate-acceptance assays were performed in 100 mm Tris-HCl, pH 7.5, using 100 μm alkaloid, 200 μm SAM, and 5 μg of purified recombinant protein in a reaction volume of 50 μl. Negative control assays were performed using purified recombinant protein denatured in boiling water for 10 min. Assays were incubated at 30 °C and quenched after 4 h with 100 μl of acetonitrile, centrifuged at 17,000 × g for 40 min to precipitate proteins, and the supernatant analyzed by LC-MS/MS. Product formation, determined as an increase of 14 Da with respect to the substrate, was confirmed by comparison of retention times and CID spectra with respect to authentic standards, or data reported in the literature. Substrate conversion rates were calculated based on substrate loss. NnOMT1 and NnOMT5 pH optimum assays were conducted in 25 mm sodium citrate, pH 5.0–6.0, 50 mm sodium phosphate, pH 6.0–7.5, and 100 mm Tris-HCl, pH 7.5–9.0, using 100 μm (R,S)-norcoclaurine or 100 μm (S)-N-methylcoclaurine, respectively, 200 μm SAM and 1 μg of purified recombinant protein in a reaction volume of 50 μl. Assays were incubated within the linear range for product formation at 30 °C for 5 min and stopped with 100 μl of acetonitrile. Temperature optima were determined under identical conditions except that 100 mm Tris-HCl, pH 8, and incubation temperatures between 4 and 60 °C were used. Kinetic parameters were measured for 5 min under the same conditions except that NnOMT1 was incubated with (R,S)-norcoclaurine at 30 °C and NnOMT5 with either (R,S)-norcoclaurine or (S)-N-methylcoclaurine at 37 °C, using alkaloid concentrations from 5 to 400 μm and a fixed SAM concentration of 400 μm. Product formation for NnOMT1 and NnOMT5 was quantified by comparison with authentic standards of coclaurine and armepavine, respectively. All assays were performed in triplicates. Saturation curves and kinetic parameters were determined based on the Michaelis-Menten equation (NnOMT1 and NnOMT5 incubated with (R,S)-norcoclaurine) or a substrate-inhibition equation (NnOMT5 incubated with (S)-N-methylcoclaurine), using Prism 5 (GraphPad).
Plant protein assays were performed using 50 μg of crude protein in 100 mm Tris-HCl, pH 7.5, using 200 μm alkaloid and 400 μm SAM in a 100-μl reaction. Negative control assays were performed using crude plant protein extracts denatured in boiling water for 10 min. Reactions were incubated overnight at 30 °C, quenched with 200 μl of acetonitrile and analyzed by LC-MS/MS. Product formation, determined as an increase of 14 Da with respect to the substrate, was confirmed by comparison of retention times and CID spectra with respect to authentic standards.
HPLC-MS/MS
HPLC-MS/MS was performed using a 1200 HPLC instrument coupled to a 6410 triple quadrupole MS (Agilent, CA). Samples (5 μl) were injected onto a Poroshell 120 SB-C18 HPLC column, 2.1 × 50 mm, 2.7-μm particle size (Agilent). Analytes were eluted using a mobile phase gradient of solvent A (10 mm ammonium acetate, pH 5.5, 5% (v/v) acetonitrile) and solvent B (100% acetonitrile) at a flow rate of 600 μl/min. The gradient started at 100% (v/v) solvent A, ramped linearly to 60% (v/v) solvent B by 8 min, further increased linearly to 99% (v/v) solvent B over 2 min, remained isocratic at 99% (v/v) solvent B from 10 to 11 min, and returned to 100% (v/v) solvent A at 11.1 min for a 3-min re-equilibration period. Analytes were applied to the mass analyzer using an electrospray ionization probe operating in positive mode with the following conditions: capillary voltage 4 000 V, fragmentor voltage 110 V, source temperature 350 °C, nebulizer pressure 50 p.s.i., gas flow 10 liters/min. For full-scan analysis, quadrupole 1 and 2 were set to radio frequency only, whereas the third quadrupole scanned from m/z 200 to 700. ESI(+) and CID spectra were analyzed, the precursor m/z was selected in quadrupole 1 and collision energy of 25 eV and an argon collision gas pressure of 1.8 × 10−3 torr were applied in quadrupole 2. The resulting MS2 fragments were resolved by quadrupole 3 scanning from 30 to 5 m/z greater than the precursor ion m/z. Compounds were identified based on retention times and ESI(+)-CID spectra compared with authentic standards or published spectral data.
Quantitative real-time PCR
RNA extraction and cDNA synthesis were performed as described above. Quantitative RT-PCR analysis was performed using PowerUp SYBR Green Master Mix and a QuantStudio-3 Real-Time PCR System (Applied Biosystems). Reactions (10 μl) contained diluted cDNA (∼8 ng), 5 μl of SYBR Green Master Mix (2 times) (Applied Biosystems), and 500 nm of each primer (Table S8b). Thermal conditions were 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 1 s, and 60 °C for 30 s. Amplification specificity was confirmed by melt-curve analysis, generated at a ramp rate of 1.6 °C/s to 95 °C maintained for 15 s, 1.6 °C/s to 60 °C maintained for 1 min, and 0.15 °C/s to 95 °C, which was maintained for 15 s. Primer efficiency was verified using LinRegPCR software (66) and samples with values between 1.8 and 2 were selected for analysis. Relative transcript abundance was calculated by the 2−ΔΔCt method (67), using β-actin as the endogenous reference gene (68). Transcript levels were normalized with respect to the gene showing the lowest expression level.
Homology modeling
The previously reported crystal structure from T. flavum norcoclaurine 6-O-methyltransferase (Tf6OMT) in complex with (S)-norlaudanosoline and S-adenosyl-l-homocysteine (PDB 5ICE) (18) was used as template to generate a NnOMT5 homology model using SWISS-MODEL (69). The resulting NnOMT5 model and Tf6OMT crystal structure were visualized, and figures were generated, using the PyMOL Molecular Graphic System (version 2.3.1, Schrödinger, LLC).
Author contributions
I. M. M.-P. and P. J. F. conceptualization; I. M. M.-P. data curation; I. M. M.-P. formal analysis; I. M. M.-P. and P. J. F. validation; I. M. M.-P. investigation; I. M. M.-P. visualization; I. M. M.-P. methodology; I. M. M.-P. writing-original draft; I. M. M.-P. and P. J. F. writing-review and editing; P. J. F. supervision; P. J. F. funding acquisition; P. J. F. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Jillian Hagel for assistance with LTQ-Orbitrap data analysis and for creating original artwork, and Dr. Jeremy Morris and Dr. Xue Chen for guidance on the experimental work and critical comments.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (to P. J. F.) and a Alberta Innovates Technology Futures Scholarship (to I. M. M-P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S9.
- BIA
- benzylisoquinoline alkaloid
- 4′OMT
- 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase
- 6OMT
- norcoclaurine 6-O-methyltransferase
- 7OMT
- reticuline 7-O-methyltransferase
- CID
- collision-induced dissociation
- CoOMT
- columbamine O-methyltransferase
- CYP
- cytochrome P450 monooxygenase
- ESI
- electrospray ionization
- N7OMT
- norreticuline 7-O-methyltransferase
- NCS
- norcoclaurine synthase
- NMT
- N-methyltransferase
- OD
- optical density
- OMT
- O-methyltransferase
- SOMT
- scoulerine 9-O-methyltransferase.
References
- 1. Dastmalchi M., Park M. R., Morris J. S., and Facchini P. J. (2018) Family portraits: the enzymes behind benzylisoquinoline alkaloid diversity. Phytochem. Rev. 17, 249–277 10.1007/s11101-017-9519-z [DOI] [Google Scholar]
- 2. Hagel J. M., and Facchini P. J. (2013) Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 10.1093/pcp/pct020 [DOI] [PubMed] [Google Scholar]
- 3. Singh A., Menéndez-Perdomo I. M., and Facchini P. J. (2019) Benzylisoquinoline alkaloid biosynthesis in opium poppy: and update. Phytochem. Rev. 18, 1457–1482 10.1007/s11101-019-09644-w [DOI] [Google Scholar]
- 4. Liscombe D. K., MacLeod B. P., Loukanina N., Nandi O. I., and Facchini P. J. (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66, 2501–2520 [DOI] [PubMed] [Google Scholar]
- 5. Chen G., Zhu M., and Guo M. (2019) Research advances in traditional and modern use of Nelumbo nucifera: phytochemicals, health promoting activities and beyond. Crit. Rev. Food Sci. Nutr. 59, 189–209 [DOI] [PubMed] [Google Scholar]
- 6. Kashiwada Y., Aoshima A., Ikeshiro Y., Chen Y. P., Furukawa H., Itoigawa M., Fujioka T., Mihashi K., Cosentino L. M., Morris-Natschke S. L., and Lee K. H. (2005) Anti-HIV benzylisoquinoline alkaloids and flavonoids from the leaves of Nelumbo nucifera, and structure-activity correlations with related alkaloids. Bioorg. Med. Chem. 13, 443–448 10.1016/j.bmc.2004.10.020 [DOI] [PubMed] [Google Scholar]
- 7. Liu C. P., Tsai W. J., Shen C. C., Lin Y. L., Liao J. F., Chen C. F., and Kuo Y. C. (2006) Inhibition of (S)-armepavine from Nelumbo nucifera on autoimmune disease of MRL/MpJ-lpr/lpr mice. Eur. J. Pharmacol. 531, 270–279 10.1016/j.ejphar.2005.11.062 [DOI] [PubMed] [Google Scholar]
- 8. Liu W., Yi D. D., Guo J. L., Xiang Z. X., Deng L. F., and He L. (2015) Nuciferine, extracted from Nelumbo nucifera Gaertn., inhibits tumor-promoting effect of nicotine involving Wnt/β-catenin signaling in non-small cell lung cancer. J. Ethnopharmacol. 165, 83–93 10.1016/j.jep.2015.02.015 [DOI] [PubMed] [Google Scholar]
- 9. Kang E. J., Lee S. K., Park K. K., Son S. H., Kim K. R., and Chung W. Y. (2017) Liensinine and nuciferine, bioactive components of Nelumbo nucifera, inhibit the growth of breast cancer cells and breast cancer-associated bone loss. Evid. Based Complement Alternat. Med. 2017, 1583185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poornima P., Weng C. F., and Padma V. V. (2014) Neferine, an alkaloid from lotus seed embryo, inhibits human lung cancer cell growth by MAPK activation and cell cycle arrest. Biofactors 40, 121–131 10.1002/biof.1115 [DOI] [PubMed] [Google Scholar]
- 11. Yoon J. S., Kim H. M., Yadunandam A. K., Kim N. H., Jung H. A., Choi J. S., Kim C. Y., and Kim G. D. (2013) Neferine isolated from Nelumbo nucifera enhances anti-cancer activities in Hep3B cells: molecular mechanisms of cell cycle arrest, ER stress induced apoptosis and anti-angiogenic response. Phytomedicine 20, 1013–1022 10.1016/j.phymed.2013.03.024 [DOI] [PubMed] [Google Scholar]
- 12. Xu L., Zhang X., Li Y., Lu S., Lu S., Li J., Wang Y., Tian X., Wei J. J., Shao C., and Liu Z. (2016) Neferine induces autophagy of human ovarian cancer cells via p38 MAPK/ JNK activation. Tumour Biol. 37, 8721–8729 10.1007/s13277-015-4737-8 [DOI] [PubMed] [Google Scholar]
- 13. Menéndez-Perdomo I. M., and Facchini P. J. (2018) Benzylisoquinoline alkaloids biosynthesis in sacred lotus. Molecules 23, e2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liscombe D. K., Louie G. V., and Noel J. P. (2012) Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 29, 1238–1250 10.1039/c2np20029e [DOI] [PubMed] [Google Scholar]
- 15. Lam K. C., Ibrahim R. K., Behdad B., and Dayanandan S. (2007) Structure, function, and evolution of plant O-methyltransferases. Genome 50, 1001–1013 10.1139/G07-077 [DOI] [PubMed] [Google Scholar]
- 16. Park M. R., Chen X., Lang D. E., Ng K. K. S., and Facchini P. J. (2018) Heterodimeric O-methyltransferases involved in the biosynthesis of noscapine in opium poppy. Plant J. 95, 252–267 10.1111/tpj.13947 [DOI] [PubMed] [Google Scholar]
- 17. Li Y., and Smolke C. D. (2016) Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 7, 12137 10.1038/ncomms12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robin A. Y., Giustini C., Graindorge M., Matringe M., and Dumas R. (2016) Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 87, 641–653 10.1111/tpj.13225 [DOI] [PubMed] [Google Scholar]
- 19. Cabry M., Offen W. A., Saleh P., Li Y., Winzer T., Graham I. A., and Davies G. J. (2019) Structure of Papaver somniferum O-Methyltransferase 1 reveals initiation of noscapine biosynthesis with implications for plant natural product methylation. ACS Catal. 9, 3840–3848 10.1021/acscatal.9b01038 [DOI] [Google Scholar]
- 20. Desgagné-Penix I., and Facchini P. J. (2012) Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72, 331–344 10.1111/j.1365-313X.2012.05084.x [DOI] [PubMed] [Google Scholar]
- 21. Ounaroon A., Decker G., Schmidt J., Lottspeich F., and Kutchan T. M. (2003) (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum: cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 36, 808–819 10.1046/j.1365-313X.2003.01928.x [DOI] [PubMed] [Google Scholar]
- 22. Pienkny S., Brandt W., Schmidt J., Kramell R., and Ziegler J. (2009) Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J. 60, 56–67 10.1111/j.1365-313X.2009.03937.x [DOI] [PubMed] [Google Scholar]
- 23. Facchini P. J., and Park S. U. (2003) Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 64, 177–186 10.1016/S0031-9422(03)00292-9 [DOI] [PubMed] [Google Scholar]
- 24. Ziegler J., Diaz-Chávez M. L., Kramell R., Ammer C., and Kutchan T. M. (2005) Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta 222, 458–471 10.1007/s00425-005-1550-4 [DOI] [PubMed] [Google Scholar]
- 25. Dang T. T., and Facchini P. J. (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631 10.1104/pp.112.194886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang L., Hagel J. M., and Facchini P. J. (2015) Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol. 169, 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purwanto R., Hori K., Yamada Y., and Sato F. (2017) Unraveling additional O-methylation steps in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica). Plant Cell Physiol. 58, 1528–1540 10.1093/pcp/pcx093 [DOI] [PubMed] [Google Scholar]
- 28. Morishige T., Dubouzet E., Choi K. B., Yazaki K., and Sato F. (2002) Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 269, 5659–5667 10.1046/j.1432-1033.2002.03275.x [DOI] [PubMed] [Google Scholar]
- 29. Morris J. S. and Facchini P. J. (2019) Molecular origins of functional diversity in benzylisoquinoline alkaloid methyltransferases. Front Plant Sci. 10, 1058 10.3389/fpls.2019.01058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gui S., Peng J., Wang X., Wu Z., Cao R., Salse J., Zhang H., Zhu Z., Xia Q., Quan Z., Shu L., Ke W., and Ding Y. (2018) Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 94, 721–734 10.1111/tpj.13894 [DOI] [PubMed] [Google Scholar]
- 31. Ming R., VanBuren R., Liu Y., Yang M., Han Y., Li L. T., Zhang Q., Kim M. J., Schatz M. C., Campbell M., Li J., Bowers J. E., Tang H., Lyons E., Ferguson A. A., et al. (2013) Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14, R41 10.1186/gb-2013-14-5-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meelaph T., Kobtrakul K., Chansilpa N. N., Han Y., Rani D., De-Eknamkul W., and Vimolmangkang S. (2018) Coregulation of biosynthetic genes and transcription factors for aporphine-type alkaloid production in wounded lotus provides insight into the biosynthetic pathway of nuciferine. ACS Omega 3, 8794–8802 10.1021/acsomega.8b00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng X., Zhao L., Fang T., Xiong Y., Ogutu C., Yang D., Vimolmangkang S., Liu Y., and Han Y. (2018) Investigation of benzylisoquinoline alkaloid biosynthetic pathway and its transcriptional regulation in lotus. Hortic. Res. 5, 29 10.1038/s41438-018-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang M., Zhu L., Li L., Li J., Xu L., Feng J., and Liu Y. (2017) Digital gene expression analysis provides insight into the transcript profile of the genes involved in aporphine alkaloid biosynthesis in lotus (Nelumbo nucifera). Front Plant Sci. 8, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen S., Zhang H., Liu Y., Fang J., and Li S. (2013) Determination of lotus leaf alkaloids by solid phase extraction combined with high performance liquid chromatography with diode array and tandem mass spectrometry. Anal Lett. 46, 2846–2859 10.1080/00032719.2013.816960 [DOI] [Google Scholar]
- 36. Deng X., Zhu L., Fang T., Vimolmangkang S., Yang D., Ogutu C., Liu Y., and Han Y. (2016) Analysis of isoquinoline alkaloid composition and wound-induced variation in Nelumbo using HPLC-MS/MS. J. Agric. Food Chem. 64, 1130–1136 10.1021/acs.jafc.5b06099 [DOI] [PubMed] [Google Scholar]
- 37. Zhou M., Jiang M., Ying X., Cui Q., Han Y., Hou Y., Gao J., Bai G., and Luo G. (2013) Identification and comparison of anti-inflammatory ingredients from different organs of lotus Nelumbo by UPLC/Q-TOF and PCA coupled with a NF-κB reporter gene assay. PLoS ONE 8, e81971 10.1371/journal.pone.0081971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris J. S., and Facchini P. J. (2016) Isolation and characterization of reticuline N-methyltransferase involved in biosynthesis of the aporphine alkaloid magnoflorine in opium poppy. J. Biol. Chem. 291, 23416–23427 10.1074/jbc.M116.750893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikezawa N., Iwasa K., and Sato F. (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821 10.1074/jbc.M705082200 [DOI] [PubMed] [Google Scholar]
- 40. Lin Z., Yang R., Guan Z., Chen A., and Li W. (2014) Ultra-performance LC separation and quadrupole time-of-flight MS identification of major alkaloids in Plumula nelumbinis. Phytochem. Anal. 25, 485–494 10.1002/pca.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Do T. C., Nguyen T. D., Tran H., Stuppner H., and Ganzera M. (2013) Analysis of alkaloids in lotus (Nelumbo nucifera Gaertn.) leaves by non-aqueous capillary electrophoresis using ultraviolet and mass spectrometric detection. J. Chromatogr. A 1302, 174–180 10.1016/j.chroma.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 42. Morikawa T., Kitagawa N., Tanabe G., Ninomiya K., Okugawa S., Motai C., Kamei I., Yoshikawa M., Lee I. J., and Muraoka O. (2016) Quantitative determination of alkaloids in lotus flower (flower buds of Nelumbo nucifera) and their melanogenesis inhibitory activity. Molecules 21, e930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hara M., Yazaki K., Tanaka S., and Tabata M. (1995) S-adenosil-l-methionine:norcoclaurine 6-O-methyltransferase from Thalictrum minus cell cultures. Phytochemistry 38, 1131–1135 10.1016/0031-9422(94)00802-Z [DOI] [Google Scholar]
- 44. Morishige T., Tsujita T., Yamada Y., and Sato F. (2000) Molecular characterization of the S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275, 23398–23405 [DOI] [PubMed] [Google Scholar]
- 45. He S. M., Liang Y. L., Cong K., Chen G., Zhao X., Zhao Q. M., Zhang J. J., Wang X., Dong Y., Yang J. L., Zhang G. H., Qian Z. L., Fan W., and Yang S. C. (2018) Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in Coptis species. Front. Plant Sci. 9, 731 10.3389/fpls.2018.00731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujii N., Inui T., Iwasa K., Morishige T., and Sato F. (2007) Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 16, 363–375 10.1007/s11248-006-9040-4 [DOI] [PubMed] [Google Scholar]
- 47. Inui T., Tamura K., Fujii N., Morishige T., and Sato F. (2007) Overexpression of Coptis japonica norcoclaurine 6-O-methyltransferase overcomes the rate-limiting step in benzylisoquinoline alkaloid biosynthesis in cultured Eschscholzia californica. Plant Cell Physiol. 48, 252–262 10.1093/pcp/pcl062 [DOI] [PubMed] [Google Scholar]
- 48. Frenzel T., and Zenk M. H. (1990) S-Adenosyl-l-methionine:3′-hydroxy-N-methyl-(S)-coclaurine-4′-O-methyltransferase, a regio- and stereoselective enzyme of the (S)-reticuline pathway. Phytochemistry 29, 3505–3511 10.1016/0031-9422(90)85265-H [DOI] [Google Scholar]
- 49. He S. M., Song W. L., Cong K., Wang X., Dong Y., Cai J., Zhang J. J., Zhang G. H., Yang J. L., Yang S. C., and Fan W. (2017) Identification of candidate genes involved in isoquinoline alkaloids biosynthesis in Dactylicapnos scandens by transcriptome analysis. Sci. Rep. 7, 9119 10.1038/s41598-017-08672-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee E. J., and Facchini P. (2010) Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22, 3489–3503 10.1105/tpc.110.077958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li J., Lee E. J., Chang L., and Facchini P. J. (2016) Genes encoding norcoclaurine synthase occur as tandem fusions in the Papaveraceae. Sci. Rep. 6, 39256 10.1038/srep39256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sheng X., and Himo F. (2019) Enzymatic Pictet-Spengler reaction: computational study of the mechanism and enantioselectivity of norcoclaurine synthase. J. Am. Chem. Soc. 141, 11230–11238 10.1021/jacs.9b04591 [DOI] [PubMed] [Google Scholar]
- 53. Hong H., Lee Y. I., and Jin D. (2010) Determination of R-(+)-higenamine enantiomer in Nelumbo nucifera by high-performance liquid chromatography with a fluorescent chiral tagging reagent. Microchem. J. 96, 374–379 10.1016/j.microc.2010.06.011 [DOI] [Google Scholar]
- 54. Kato E., Iwata R., and Kawabata J. (2017) Synthesis and detailed examination of spectral properties of (S)- and (R)-higenamine 4′-O-β-d-glucoside and HPLC analytical conditions to distinguish the diastereomers. Molecules 22, e1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sato F., Tsujita T., Katagiri Y., Yoshida S., and Yamada Y. (1994) Purification and characterization of S-adenosyl-l-methionine: norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 225, 125–131 10.1111/j.1432-1033.1994.00125.x [DOI] [PubMed] [Google Scholar]
- 56. Ka S. M., Kuo Y. C., Ho P. J., Tsai P. Y., Hsu Y. J., Tsai W. J., Lin Y. L., Shen C. C., and Chen A. (2010) (S)-Armepavine from Chinese medicine improves experimental autoimmune crescentic glomerulonephritis. Rheumatology (Oxford) 49, 1840–1851 10.1093/rheumatology/keq164 [DOI] [PubMed] [Google Scholar]
- 57. Itoh A., Saitoh T., Tani K., Uchigaki M., Sugimoto Y., Yamada J., Nakajima H., Ohshiro H., Sun S., and Tanahashi T. (2011) Bisbenzylisoquinoline alkaloids from Nelumbo nucifera. Chem. Pharm. Bull. (Tokyo) 59, 947–951 10.1248/cpb.59.947 [DOI] [PubMed] [Google Scholar]
- 58. Vimolmangkang S., Deng X., Owiti A., Meelaph T., Ogutu C., and Han Y. (2016) Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants. Sci. Rep. 6, 26323 10.1038/srep26323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hagel J. M., Morris J. S., Lee E. J., Desgagné-Penix I., Bross C. D., Chang L., Chen X., Farrow S. C., Zhang Y., Soh J., Sensen C. W., and Facchini P. J. (2015) Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 15, 227 10.1186/s12870-015-0596-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morris J. S., Dastmalchi M., Li J., Chang L., Chen X., Hagel J. M., and Facchini P. J. (2016) Plug-and-play benzylisoquinoline alkaloid biosynthetic gene discovery in engineered yeast. Methods Enzymol. 575, 143–178 10.1016/bs.mie.2016.03.023 [DOI] [PubMed] [Google Scholar]
- 61. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kumar S., Stecher G., Li M., Knyaz C., and Tamura K. (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jones D. T., Taylor W. R., and Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 64. Sievers F., and Higgins D. G. (2018) Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang Y. J., Hao X. Y., Liang Z. S., Ke W. D., and Guo H. B. (2013) Efficient isolation of high-quality RNA from lotus Nelumbo nucifera ssp nucifera tissues. Genet. Mol. Res. 12, 223–229 10.4238/2013.January.24.14 [DOI] [PubMed] [Google Scholar]
- 66. Jeong J. Y., Yim H. S., Ryu J. Y., Lee H. S., Lee J. H., Seen D. S., and Kang S. G. (2012) One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 78, 5440–5443 10.1128/AEM.00844-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ruijter J. M., Pfaffl M. W., Zhao S., Spiess A. N., Boggy G., Blom J., Rutledge R. G., Sisti D., Lievens A., De Preter K., Derveaux S., Hellemans J., and Vandesompele J. (2013) Evaluation of qPCR curve analysis methods for reliable biomarker discovery: bias, resolution, precision, and implications. Methods 59, 32–46 10.1016/j.ymeth.2012.08.011 [DOI] [PubMed] [Google Scholar]
- 68. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 69. Dong C., Li G., Li Z., Zhu H., Zhou M., and Hu Z. (2009) Molecular cloning and expression analysis of an Mn-SOD gene from Nelumbo nucifera. Appl. Biochem. Biotechnol. 158, 605–614 10.1007/s12010-008-8410-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.