Abstract
During oxidative stress, inflammation, or environmental exposure, ribo- and deoxyribonucleotides are oxidatively modified. 8-Oxo-7,8-dihydro-2′-guanosine (8-oxo-G) is a common oxidized nucleobase whose deoxyribonucleotide form, 8-oxo-dGTP, has been widely studied and demonstrated to be a mutagenic substrate for DNA polymerases. Guanine ribonucleotides are analogously oxidized to r8-oxo-GTP, which can constitute up to 5% of the rGTP pool. Because ribonucleotides are commonly misinserted into DNA, and 8-oxo-G causes replication errors, we were motivated to investigate how the oxidized ribonucleotide is utilized by DNA polymerases. To do this, here we employed human DNA polymerase β (pol β) and characterized r8-oxo-GTP insertion with DNA substrates containing either a templating cytosine (nonmutagenic) or adenine (mutagenic). Our results show that pol β has a diminished catalytic efficiency for r8-oxo-GTP compared with canonical deoxyribonucleotides but that r8-oxo-GTP is inserted mutagenically at a rate similar to those of other common DNA replication errors (i.e. ribonucleotide and mismatch insertions). Using FRET assays to monitor conformational changes of pol β with r8-oxo-GTP, we demonstrate impaired pol β closure that correlates with a reduced insertion efficiency. X-ray crystallographic analyses revealed that, similar to 8-oxo-dGTP, r8-oxo-GTP adopts an anti conformation opposite a templating cytosine and a syn conformation opposite adenine. However, unlike 8-oxo-dGTP, r8-oxo-GTP did not form a planar base pair with either templating base. These results suggest that r8-oxo-GTP is a potential mutagenic substrate for DNA polymerases and provide structural insights into how r8-oxo-GTP is processed by DNA polymerases.
Keywords: DNA damage, 8-oxoguanine (8-oxo-G), DNA polymerase, DNA repair, structural biology, DNA replication, mutagenic nucleobase, nucleotidyl transferase reaction, oxidized ribonucleotide, r8-oxo-G lesion
Introduction
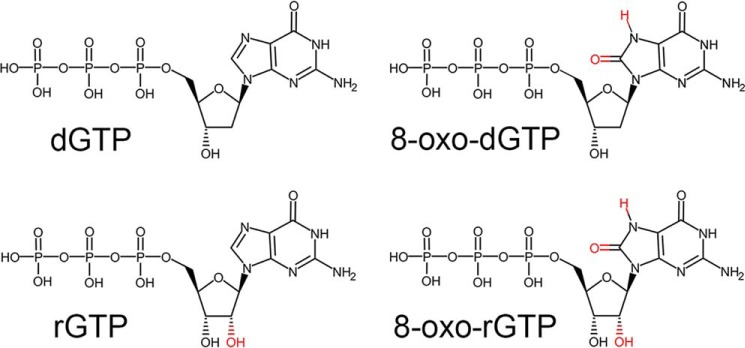
Oxygen radicals produced during oxidative stress can damage bases within both duplex DNA and the nucleotide pool, causing cytotoxicity and mutagenesis (1, 2). Specifically, oxidation of the nucleotide pool has been shown to be a significant contributor to DNA damage (3–6). Guanine is particularly susceptible to oxidation and readily forms 8-oxo-7,8-dihydro-2′-guanosine (8-oxo-G)2 (7). The deoxynucleotide form of 8-oxo-G is a highly mutagenic nucleobase substrate for DNA polymerases and, consequently, has been implicated in several human diseases (8–11). In contrast, the ribonucleotide form of 8-oxo-G (r8-oxo-G) remains to be fully characterized. The r8-oxo-G lesion has two deviations from the canonical nucleotide: an adducted oxygen on the nucleotide base (O8) and an additional oxygen on the ribose sugar (O2′) (Fig. 1). Importantly, the combined effect of both noncanonical oxygens during nucleotide discrimination is not well-understood.
Figure 1.
The chemical structure of various guanine nucleotides. Deviations from the canonical DNA polymerase substrate are highlighted in red.
During DNA replication, polymerases must select the correct base and sugar combination to ensure high fidelity. DNA polymerase β (pol β) is an established mammalian model for studying nucleotide discrimination because of its amenability to structure–function studies and conformational nucleotide selection mechanism akin to replicative polymerases (12, 13). The conformational change occurs upon correct nucleotide binding, where the polymerase N-subdomain closes to facilitate key nucleic acid–protein interactions for efficient catalysis (12–18). Following closure, catalysis is carried out by nucleophilic attack of the primer terminus oxyanion (O3′) on the α-phosphate (αP) of the incoming nucleotide, resulting in deoxynucleoside monophosphate incorporation and generation of a pyrophosphate (19).
There are several key steps involved in pol β closure and nucleotide insertion that discriminate between correct and incorrect nucleotides (20). To discriminate against ribonucleotides, Tyr-271 on α-helix M acts as a steric gate to prevent insertion of incoming ribonucleotides. Specifically, the backbone carbonyl of Tyr-271 clashes with ribose O2′ of the incoming nucleotide (Fig. 2) (21, 22). Although most DNA polymerases have an equivalent steric gate, misincorporation of ribonucleotides is an extremely common replication error that is estimated to occur millions of times per cell division in normal cells (23–25). Moreover, mildly oxidative conditions in human cells results in oxidation of 0.2%–5% of free guanine ribonucleotides (rGTP) into r8-oxo-GTP (4, 24, 26). Recent studies in yeast, bacteria, and humans have shown that r8-oxo-GTP is incorporated into DNA, indicating that r8-oxo-GTP is a potential polymerase substrate (27–29). Furthermore, human DNA polymerase δ is unable to proofread lesions containing either a O2′ or O8, suggesting reduced proofreading of r8-oxo-GMP during replication (24, 30).
Figure 2.
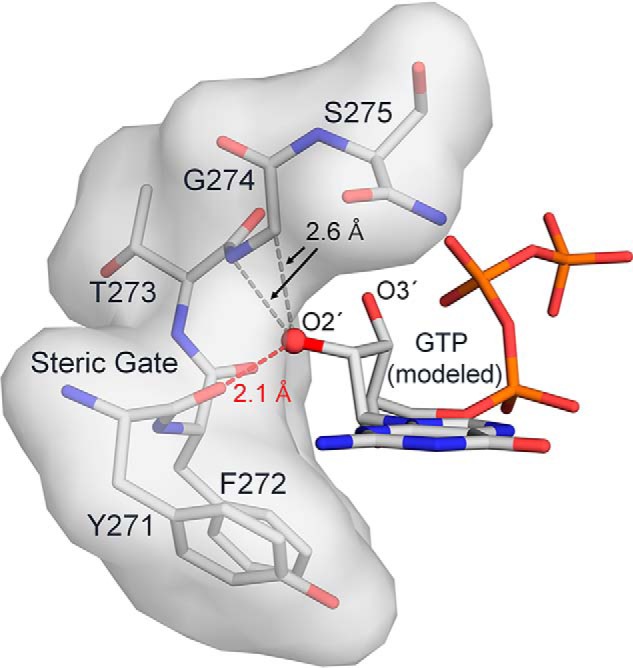
The ribonucleotide steric gate in DNA pol β. Helix M is shown as both surface and stick representation (gray). The ribose sugar O2′ (red sphere) was added to the pol β structure with an incoming dGTP (PDB code 4UB4). Steric clashes between the O2′ and helix M are indicated.
To gain mechanistic insight into r8-oxo-GTP insertion, we utilized pol β for structure–function studies of the nucleotidyl transferase reaction with an incoming r8-oxo-GTP. Here we report precatalytic ternary (polβ:DNA:rNTP) substrate X-ray crystallographic structures of r8-oxo-GTP opposite templating adenine (mutagenic) or cytosine (nonmutagenic), FRET measurements of pol β closure, and kinetic characterization of r8-oxo-GTP insertion efficiencies. Our results provide molecular insight into how DNA polymerases incorporate r8-oxo-GTP into DNA.
Results
Kinetic characterization of r8-oxo-GTP insertion
The efficiency of r8-oxo-GTP and rGTP insertion into a single-nucleotide gap by pol β was measured with both a templating cytosine (dC) and adenine (dA) (Fig. 3). To contextualize r8-oxo-GTP insertion, these insertion efficiencies were compared with undamaged deoxyribonucleotide (dGTP) and oxidized deoxyribonucleotide (8-oxo-dGTP) insertion (11, 12). Relative to correct dGTP insertion, r8-oxo-GTP insertion efficiency opposite dC and dA is reduced more than 250,000-fold and 4,000-fold, respectively. The 60-fold preference for insertion of r8-oxo-GTP opposite dA, compared with dC, demonstrates the mutagenicity of r8-oxo-GTP. This is consistent with 8-oxo-dGTP, which is also preferentially inserted opposite dA. However, insertion of r8-oxo-GTP is less efficient than 8-oxo-dGTP by 700- and 4,300-fold opposite dA and dC, respectively. This demonstrates that the ribose O2′ hinders insertion of the 8-oxo-G base, but the effect is not strictly additive. For comparison, the O2′ of the undamaged base (rGTP) reduces insertion efficiency by 3,500-fold opposite the correct base, dC. Notably, rGTP is extremely difficult to misinsert opposite the wrong base; rGTP insertion opposite dA could not to be measured in our assay or previously (31). Despite the reduced efficiencies imparted by the O2′, ribonucleotides are still frequently inserted into DNA (25), and we show that mutagenic r8-oxo-GTP insertion has a similar efficiency as ribonucleotide insertion. To provide molecular insight into the basis of these observations, we turned to a FRET polymerase closure assay and X-ray crystallography.
Figure 3.
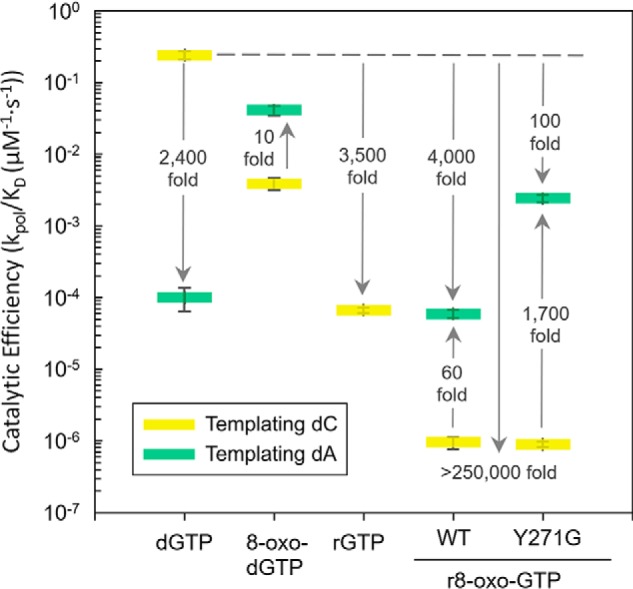
Discrimination plot evaluating r8-oxo-GTP insertion. The catalytic efficiencies (kpol/Kd) for insertion of r8-oxo-GTP opposite dC (yellow) or dA (green) for pol β are shown. The distance between the respective catalytic efficiencies is a measure of discrimination/fidelity. Each short horizontal bar represents the standard deviation of the mean of triplicate independent determinations. The insertion efficiencies for dGTP were reported in Refs. 11, 12 and 8-oxo-dGTP in Ref. 11. The catalytic efficiency values are reported in Fig. S1C and are as follows: 2.4 × 10−1 ± 3.0 × 10−2 μm−1·s−1 (dGTP:dC), 1.0 × 10−4 ± 3.6 × 10−5 μm−1·s−1 (dGTP:dA), 3.9 × 10−3 ± 7.0 × 10−5 μm−1·s−1 (8-oxo-dGTP:dC), 4.1 × 10−2 ± 4.1 × 10−3 μm−1·s−1 (8-oxo-dGTP:dA), 6.7 × 10−5 ± 5.7 × 10−6 μm−1·s−1 (rGTP:dC), 9.5 × 10−7 ± 1.8 × 10−7 μm−1·s−1 (r8-oxo-GTP:dC), 5.9 × 10−5 ± 7.9 × 10−5 μm−1·s−1 (r8-oxo-GTP:dA), 8.9 × 10−7 ± 8.3 × 10−8 μm−1·s−1 (Y271G:r8-oxo-GTP:dC), and 2.4 × 10−3 ± 2.9 × 10−4 μm−1·s−1 (Y271G:r8-oxo-GTP:dA).
Polymerase closure is impaired for ribonucleotide and mismatched substrates
To evaluate how polymerase subdomain closure might contribute to the decrease in kinetic efficiency for r8-oxo-GTP insertion, we utilized a steady-state FRET-based assay with pol β. Closure of the N-subdomain, triggered by binding of the nucleotide, was measured using FRET between the fluorescent AEDANS label, located at position V303C of the N-subdomain, and the quencher (Dabcyl) located at position −8 relative to the templating base of the DNA substrate (Fig. S2). As expected, the correct pairing of dGTP opposite dC results in closure of pol β, indicated by a shift of the N-subdomain ∼12 Å closer to the DNA compared with the binary open complex (Fig. 4). Of note, our calculated distances for the open binary and closed correct ternary associate closely with previous measurements using this assay and those expected from crystallographic data (15). Performing the same experiment with either 8-oxo-dGTP, rGTP, or r8-oxo-GTP insertion opposite dC resulted in a lack of significant N-subdomain closure (shifting only 2.9–4.5 Å). These results indicate that modifications on either the sugar (O2′) or the base (O8) inhibit the enzyme from closing and are consistent with the observed reduction in their insertion efficiency opposite dC (Fig. 3).
Figure 4.
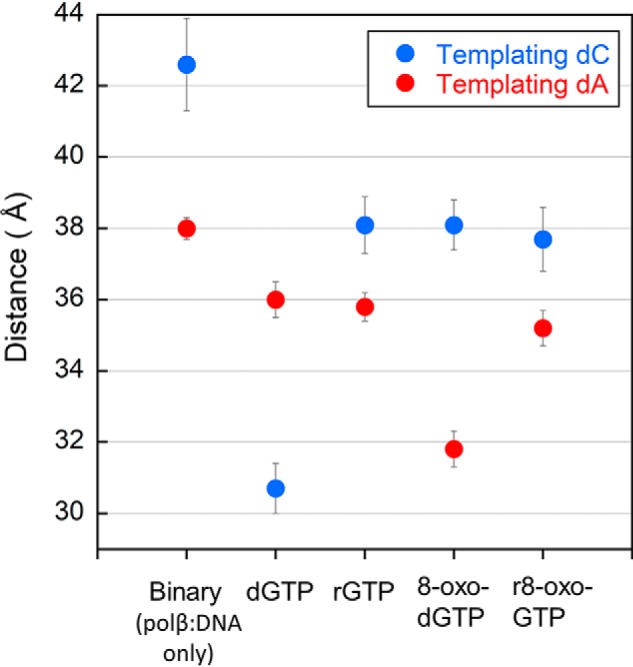
Calculated interprobe distances from FRET efficiencies during steady-state FRET experiments. Efficiencies from triplicate measurements are plotted for the following complexes opposite either a templating cytosine (dark blue) or templating adenine (red): binary; ternary with dGTP, rGTP, or r8-oxo-GTP; with error bars (black) calculated using standard deviation of the mean. Distance values are reported in Fig. S3 and are as follows: 42.6 ± 1.3 Å (binary:dC), 38.0 ± 0.3 Å (binary:dA), 30.7 ± 0.7 Å (dGTP:dC), 36.0 ± 0.5 Å (dGTP:dA), 38.1 ± 0.8 Å (rGTP:dC), 35.8 ± 0.4 Å (rGTP:dA), 38.1 ± 0.7 Å (8-oxo-dGTP:dC), 31.8 ± 0.5 Å (8-oxo-dGTP:dA), 37.7 ± 0.9 Å (r8-oxo-GTP:dC), and 35.2 ± 0.5 Å (r8-oxo-GTP:dA).
The mismatched insertion of dGTP opposite dA results in only a 2.0 Å shift of the N-subdomain, from 38.0 Å in the binary complex to 36.0 Å in the ternary, and is associated with a significant loss in catalytic efficiency (Fig. 3). In contrast, insertion of 8-oxo-dGTP, the oxidized deoxyribonucleotide, opposite dA leads to a 6.2 Å shift in the N-subdomain, from 38.0 Å to 31.8 Å, which is similar to the distance observed in the presence of the correct dTTP:dA pair (30.9 Å) (Fig. S3). The ability of pol β to undergo subdomain closure during 8-oxo-dGTP insertion opposite dA correlates with the high catalytic efficiency of this mutagenic deoxyribonucleotide. The lack of closure measured by FRET during rGTP insertion opposite dA provides a potential explanation for the unmeasurable catalytic activity (Fig. 4). Last, insertion of r8-oxo-GTP opposite dA resulted in only a 2.8 Å shift in the N-subdomain, which indicates an intermediate or partially closed state and correlates with its intermediate insertion efficiency.
Structural snapshots of r8-oxo-GTP insertion opposite dA and dC
To investigate the active-site geometries and base-pairing properties of r8-oxo-GTP, we sought to collect X-ray crystallographic ternary complex structures with WT pol β. Unfortunately, we were unable to obtain a WT pol β:DNA:r8-oxo-GTP ternary complex, likely as a result of the poor binding between r8-oxo-GTP and pol β. A previous study determined that removal of the Tyr-271 side chain did not appreciably alter insertion of dCTP opposite dG and increased nondamaged ribonucleotide insertion efficiency by 12-fold compared with WT pol β (31). Therefore, we utilized a Y271G pol β variant to obtain ternary structures of pol β bound to r8-oxo-GTP. Consistent with this, Y271G insertion of r8-oxo-GTP showed increased catalytic efficiency opposite dA (2,700-fold) but not opposite dC when compared with the WT (Fig. 3). We proceeded to use the Y271G pol β variant to obtain ternary crystal structures of r8-oxo-GTP insertion opposite both dA and dC with MnCl2 and a dideoxy-terminated primer, which allows for binding of r8-oxo-GTP while preventing catalysis (12, 22, 32).
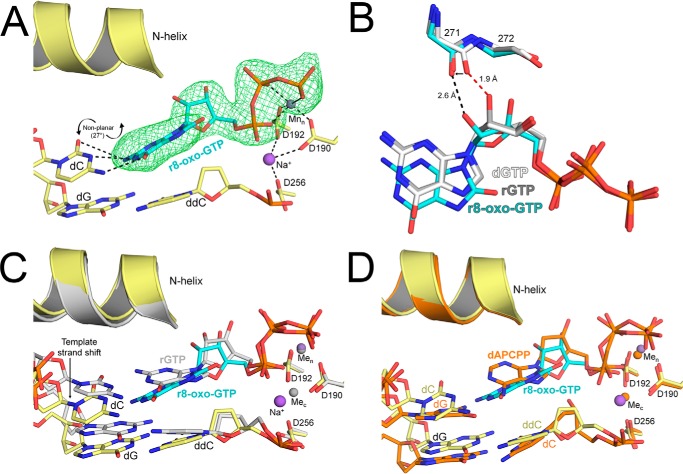
The ternary complex, showing Y271G pol β bound to single-nucleotide gapped DNA with a templating dC and an incoming r8-oxo-GTP, diffracted to 2.55 Å (Table S1). The resulting structure contained two pol β complexes in the asymmetric unit, with one complex in the open conformation and the other in the closed conformation. The density for r8-oxo-GTP in the open active site of pol β was relatively poor, with only clearly defined density for the triphosphate moiety (Fig. S4). In contrast, the density for r8-oxo-GTP in the closed pol β conformation was more defined and is the focus of our analysis (Fig. 5A). In this structure, the r8-oxo-GTP is bound to the pol β active site with a single Mn2+ in the nucleotide metal-binding site, and the catalytic metal-binding site contains a sodium ion. The lack of divalent catalytic metal binding has been observed previously for dideoxy-terminated ternary complexes (33). The incoming r8-oxo-GTP resides in the anti conformation opposite a dC template in a closed complex, but the base-pairing is not planar. Instead, the angle between base planes in the r8-oxo-GTP:dC nascent pair is 27°, positioning the O8 oxygen in the direction of the N-helix (34). This twist results in the adducted O8 of r8-oxo-GTP sitting 0.5 Å farther away from the O5′ compared with the O8 in a previously solved 8-oxo-dGTP:dC complex (35). In the 8-oxo-dGTP:dC complex, a Ca2+ ion interacts with the α-phosphate of the incoming nucleotide to neutralize the close proximity of the O8 and O5′. This metal is not present in the r8-oxo-GTP:dC complex, likely because of the propeller twist accommodating any potential clash between O8 and O5′. The ribose sugar of the incoming nucleotide resides in an O4′-endo conformation, with the O2′ pointed toward the backbone carbonyl of Gly-271 (Fig. 5B). This results in the backbone carbonyl of Gly-271 shifting 0.9 Å, relative to its position in the dGTP:dC complex, to avoid a clash with the O2′ (Fig. 5B). A stabilizing hydrogen bond between the 3′-OH of the incoming nucleotide and the nonbridging β-phosphate oxygen, observed in the rGTP:dC complex, is not present with r8-oxo-GTP because the 3′-OH is 5 Å from the nonbridging β-phosphate oxygen of the nucleotide. In a previously obtained Y271A pol β rCTP:dG complex, a rotameric shift of Phe-272 (Fig. S5) and a shift in the primer terminus toward the major groove were observed (22). These structural features are also present in our r8-oxo-GTP:dC complex as a result of removing the Tyr-271 side chain. In the r8-oxo-GTP:dC complex, we also observed a shift of the templating dC along with the 5′-end of the templating strand upstream compared with an rGTP:dC complex control (Fig. 5C). Ternary mismatches also shift the templating strand upstream; however, the r8-oxo-GTP:dC complex does not shift to the same extent (Fig. 5D) (12, 36). This is likely due to weak interactions between the Watson–Crick faces of r8-oxo-GTP and the templating dC.
Figure 5.
X-ray crystallographic structure of Y271G pol β in complex with r8-oxo-GTP opposite dC. A, precatalytic closed ternary complex of Y271G pol β (yellow) with r8-oxo-GTP (blue sticks) bound across from dC. Mn2+ and Na+ ions are shown in purple, and potential hydrogen bonds are shown as black dashed lines. A polder map (green mesh) contoured at 3.0 σ is shown for the incoming r8-oxo-GTP. B, overlay of the incoming nucleotide and steric gating residues for the r8-oxo-GTP:dC complex (blue), the undamaged rGTP:dC complex (gray), and the dGTP:dC complex (white) (PDB code 4UB4). Potential clashes are highlighted by red dashed lines and potential hydrogen bonds as black dashed lines. C, overlay between the r8-oxo-GTP:dC complex (yellow/blue) and an undamaged rGTP:dC complex (gray). Key residues and DNA are shown as sticks and the N-helix as a cartoon. D, overlay between the r8-oxo-GTP:dC complex (yellow/blue) and a mismatched dAPCPP:dG complex (orange) (PDB code 3C2M). Key residues and DNA are shown as sticks and the N-helix as a cartoon.
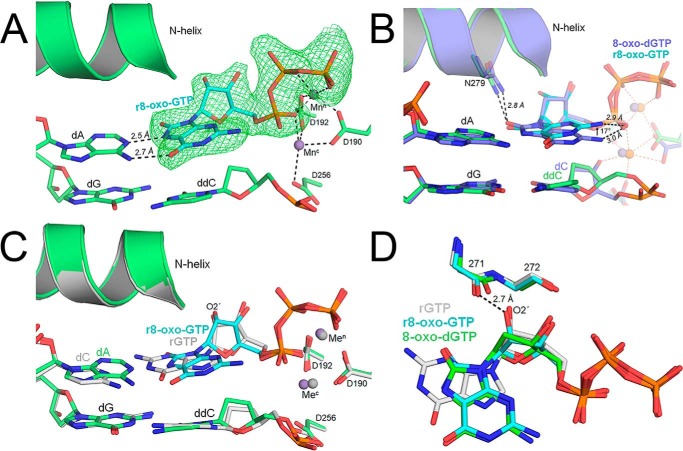
8-oxo-dGTP prefers to insert opposite adenine via a rotation of the glycosidic bond and formation of a Hoogsteen base pair with dA. In our solution studies, r8-oxo-GTP similarly prefers to insert opposite adenine (Fig. 3). To evaluate whether r8-oxo-GTP uses Hoogsteen base-pairing, we obtained a ternary Y271G pol β structure with an incoming r8-oxo-GTP opposite a templating dA. This complex diffracted to 2.05 Å, with only one pol β molecule per asymmetric unit (Table S1). In the resulting structure, pol β assumed a closed conformation with clear density for r8-oxo-GTP in the nucleotide binding pocket (Fig. 6A). r8-oxo-GTP was bound with Mn2+ in both the nucleotide and catalytic metal binding sites, indicating a more optimal active-site geometry compared with r8-oxo-GTP opposite dC (Fig. 5A). Similar to 8-oxo-dGTP, r8-oxo-GTP Hoogsteen base-pairs opposite dA using the syn conformation. This places the adducted O8 oxygen into the minor groove, where it is stabilized by Asn-279, which was also observed with 8-oxo-dGTP (Fig. 6B). However, one clear difference between structures with 8-oxo-dGTP and r8-oxo-GTP is that the nucleoside breaks from planarity by 17° with the ribo form, pointing the N2 away from the α-phosphate and toward the N-helix (Fig. 6B) (37). Despite being nonplanar, there was no significant change to the distance of the hydrogen bond between the N2 and the α-phosphate (2.9 Å), which has been proposed to stabilize the Hoogsteen conformation during 8-oxo-dGTP insertion opposite dA (3.0 Å) (Fig. 6B) (35). The overall organization of the active site during r8-oxo-GTP insertion opposite dA aligns well with our rGTP:dC structure and is consistent with respect to their similar kinetic efficiencies (Fig. 6C). The ribose O2′ was positioned 2.8 Å away from the Gly-271 backbone carbonyl, similar to rGTP:dC (2.6 Å) (Fig. 6D). However, the ribose sugar adopts a C4′-exo conformation as opposed to the more competent 3′-endo in our rGTP:dC complex and other pol β:rNTP structures (Fig. 6D). Additionally, the r8-oxo-GTP 3′-OH and its nonbridging β-phosphate oxygen are within hydrogen bonding distance at 3.2 Å.
Figure 6.
X-ray crystallographic structure of Y271G pol β in complex with r8-oxo-GTP opposite dA. A, precatalytic closed ternary complex of Y271G pol β (green) with r8-oxo-GTP (blue sticks) bound across from dA. Mn2+ ions are shown in purple, and potential hydrogen bonds are shown as black dashed lines. A polder map (green mesh) contoured at 3.0 σ is shown for the incoming r8-oxo-GTP. B, overlay between the r8-oxo-GTP:dA complex (green/blue) and the 8-oxo-dGTP:dA complex (purple) (PDB code 4UAW). Key residues and DNA are shown as sticks. Potential hydrogen bonds are shown as black dashed lines and the N-helix as a cartoon. C, overlay between the r8-oxo-GTP:dA complex (green/blue) and a rGTP:dC complex (gray). Key residues and DNA are shown as sticks and the N-helix as a cartoon. D, overlay of the incoming nucleotide and steric gating residues for the r8-oxo-GTP:dA complex (green), the undamaged rGTP:dC complex (gray), and the 8-oxo-dGTP:dA complex (blue) (PDB code 4UAW). Potential hydrogen bonds are shown as black dashed lines.
Discussion
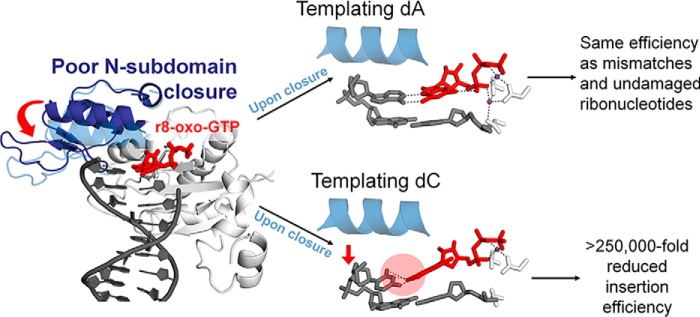
In this study, we characterized r8-oxo-GTP insertion with DNA polymerase β using single-turnover kinetics, a steady-state FRET assay, and X-ray crystallography. Our results show that polymerase subdomain closure opposite both dA and dC is reduced, but differences in active site geometries upon closure lead to the preference for mutagenic (dA) insertion of r8-oxo-GTP (Fig. 7). We also showed that mutagenic insertion of r8-oxo-GTP opposite dA occurs at a similar catalytic efficiency as that of a mismatched deoxyribonucleotide or correctly matched undamaged ribonucleotide. Based on the error rate of misinsertion and measured levels of r8-oxo-GTP, this suggests that r8-oxo-GTP could be a potential mutagenic substrate for DNA polymerases under oxidizing conditions (4, 25). Our findings also demonstrate that the mechanistic barriers that hinder r8-oxo-GTP insertion are manifested by effects arising from the O8 and O2′ that closely resemble their independent effects (11, 22, 31, 35).
Figure 7.
Schematic of r8-oxo-GTP processing by human DNA polymerase β.
The adducted O8 promotes mutagenic r8-oxo-GTP insertion by pol β
It is well characterized that 8-oxo-dGTP is inserted by polymerases across from dA more efficiently than dC, and r8-oxo-GTP continues this trend. 8-Oxo-dGTP and r8-oxo-GTP, respectively, have a 10-fold and 60-fold increase in insertion efficiency opposite dA compared with dC (Fig. 3). However, the ribose sugar of r8-oxo-GTP reduces the insertion efficiency compared with 8-oxo-dGTP. Moreover, when the steric gate residue Tyr-271 is mutated, r8-oxo-GTP:dA insertion increases 40-fold, which is only 17-fold less than 8-oxo-dGTP:dA. Mutagenic r8-oxo-GTP insertion by WT pol β is only 1.7-fold less than its nondamaged mismatch insertion (dGTP:dA), demonstrating the potential frequency of r8-oxo-GTP, given that mismatches are a common replication error. In our FRET experiments, we observed a similar distance between the r8-oxo-GTP:dA and dGTP:dA complexes (35.2 Å and 36.0 Å, respectively) that corroborates the similar catalytic efficiencies (Fig. 3). Conversely, ribonucleotide insertion (rGTP:dC), another frequent replication error, is also within 1.3-fold of r8-oxo-GTP:dA but has a more open conformation (38.1 Å), indicating that other steps in the enzymatic pathway account for the observed differences in catalytic efficiency.
In our X-ray crystal structures, we see similarities between 8-oxo-dGTP and r8-oxo-GTP insertion. Particularly, the adducted O8 oxygen imparts the same general base-pairing properties in the deoxyribo- and ribonucleotide forms. When base-pairing opposite cytosine, there is a retention of the anti conformation that forms Watson–Crick base pairs. Opposite adenine, both r8-oxo-GTP and 8-oxo-dGTP rotate about their glycosidic bond to a syn conformation and Hoogsteen base-pair with dA. Retention of the base pairing properties between 8-oxo-dGTP and r8-oxo-GTP supports the retained mutagenic behavior of the adducted O8.
Mechanistic insight into r8-oxo-GTP discrimination by pol β
The induced-fit model posits that, after initial correct dNTP binding, conformational changes of the ternary complex induce catalytic residues to properly align for catalysis (38). The induced-fit model would also assert that mismatch complexes do not properly align the active site for catalysis and, instead, promote dissociation (38). In a mismatched complex (dG:dAPCPP; PDB code 3C2M), the templating strand is shifted out of coding position, and pol β is positioned in an open conformation (12, 33, 39–41). Similarly, we observed these features in our r8-oxo-GTP:dC complex (Figs. 5D and 4), demonstrating a similar method of discrimination between mismatches and r8-oxo-GTP:dC. Furthermore, we observed an open pol β complex within the asymmetric unit with poorly defined density for the nucleoside and only moderately defined triphosphate density (Fig. S4). Retention of triphosphate density without nucleobase density is consistent with previous findings demonstrating that triphosphate binding occurs prior to Watson–Crick sampling (42, 43). The observance of poor density and reduced occupancy of the r8-oxo-GTP:dC open complex structurally supports our solution experiments showing both a decrease in catalytic efficiency and reduction in pol β closure for this complex. Another feature of the mismatch complex is the absence of Watson–Crick hydrogen bonds in the nascent pair, which led to the hypothesis that optimal base-pairing interactions during nucleotide binding facilitate proper active-site assembly (12). However, we found that r8-oxo-GTP does come within hydrogen-bonding distance with the templating dC despite the template strand shift and propeller twist of the nascent pair. Of note, a similar degree of propeller shift was observed during rCTP:dG insertion with polymerase η (27°) compared with our r8-oxo-GTP:dC twist (27°) (44). Overall, pol β strongly deters r8-oxo-GTP insertion opposite dC via poor r8-oxo-GTP binding, impaired subdomain closure, and significant active-site geometry perturbations should closure occur.
The r8-oxo-GTP lesion is distinctly nonplanar opposite both dC and dA. However, the O8 and O2′ by themselves do not significantly alter base-pair planarity, as observed during 8-oxo-dGTP and rNTP insertion (22, 35). This suggests that the combined steric burden of O2′ and O8 prevent formation of a planar base pair with either the templating dA or dC. In a previous study, nonplanarity within the nascent pair was suggested to impair catalytic metal binding (42). Our nonplanar r8-oxo-GTP insertion complexes indeed show reduced catalytic metal binding opposite both dA and dC. The r8-oxo-dGTP:dA complex had a reduced occupancy of the catalytic metal (0.8), and r8-oxo-GTP:dC had no divalent metal binding corresponding with its more severe nonplanarity. Therefore, our structures suggest that lack of planarity in the nascent pair impairs proper active-site assembly, including catalytic metal binding. Our data also support the previous suggestion that nonplanar geometry in the nascent base pair prohibits DNA stacking interactions within the DNA polymerase active site that are important for nucleotide binding (36). As determined from our fluorescence measurements, pol β adopts a predominantly open position with r8-oxo-GTP that supports inefficient r8-oxo-GTP binding (Fig. 4). Inefficient binding can also be inferred by the inability to obtain r8-oxo-GTP structures with WT pol β. Overall, the nonplanarity of r8-oxo-GTP contributes to the reduction in catalytic efficiency in addition to poor nucleotide binding, improper organization of the polymerase active site, and inefficient polymerase closure.
Consequences of r8-oxo-GTP insertion
Our results show that the ribose O2′ of r8-oxo-GTP heavily diminishes the rate of insertion opposite adenine and cytosine. Despite this, r8-oxo-GTP can still be mutagenically inserted by pol β at an efficiency similar to undamaged ribonucleotides. If replicative polymerases were to have a similar discrimination for r8-oxo-GTP as pol β, then we can speculate on the amount of mutagenic r8-oxo-GTP insertion events during DNA replication. Using replicative DNA polymerase ϵ as an example, the discrimination between rGTP and dGTP is 12 times worse than pol β, indicating that r8-oxo-GTP may be inserted more efficiently by pol ϵ than by pol β (46). Replicative polymerases are estimated to misinsert more than 1 million ribonucleotides (∼250,000 rGTP) per round of replication (26), and levels of r8-oxo-GTP have been measured to be between 0.2%–5% of the rGTP pool during mild oxidative stress (4, 47). If 0.2% of the ∼250,000 inserted rGTP are oxidized to r8-oxo-GTP, then ∼500 r8-oxo-GTP mutagenic insertion events could potentially occur per cell division under mildly oxidative conditions. The subsequent cellular ramifications of these lesions in duplex DNA are still unclear. Many DNA damage–processing enzymes have been investigated, but most are unable to process r8-oxo-GTP or r8-oxo-G, including 8-oxoguanine glycosylase (OGG1), RNase H2, and ribonucleotide reductase (29, 48–50). MutT-homolog 1 (MTH1) and AP endonuclease (APE1) only have weak activity on r8-oxo-GTP and r8-oxo-G processing, making it uncertain whether the reduced activities are sufficient to protect the cell from r8-oxo-G(TP) (50–52). In cellular extracts subjected to mildly oxidized conditions, r8-oxo-GTP does accumulate (0.3 nmol/106 cells), whereas 8-oxo-dGTP was not detected (4). This suggests that MTH1 is not able to sufficiently remove r8-oxo-GTP from the nucleotide pool, in contrast to its efficient removal of 8-oxo-dGTP. MutY DNA Glycosylase (MUTYH) was found to have r8-oxo-GMP activity and remove the opposing mutagenic dA. However, to our knowledge, the efficiency for inserting the correct base opposite r8-oxo-GMP currently remains unknown.
Experimental procedures
DNA sequences
The DNA sequences used in crystallization studies (16-mer), kinetic studies (34-mer), and FRET studies (45-mer) are provided in Table S2. Each oligonucleotide was suspended in 10 mm Tris-HCl (pH 7.4) and 1 mm EDTA, and the concentration was determined from their UV absorbance at 260 nm. DNA substrates were prepared by annealing three purified oligonucleotides. The annealing reactions were performed by incubating a solution of primer with downstream and template oligonucleotides (1:1.2:1.2 molar ratio, respectively) at 95 °C for 5 min, followed by 65 °C for 30 min, and finally cooling 1 °C min−1 to 10 °C in a PCR thermocycler.
Pol β expression, purification, and crystallization
Human WT pol β was subjected to site-directed mutagenesis to generate the Y271G mutant enzyme using the QuikChange II site-directed mutagenesis protocol and kit. The Y271G mutation was confirmed via sequencing and was overexpressed in BL21-CodonPlus (DE3)-RP Escherichia coli and purified as described previously (53). This mutant was rationally designed to provide flexibility to the steric gate backbone carboxyl of Tyr-271 in pol β. Y271G pol β was incubated with 1-nt gapped/dideoxy-terminated DNA (1:1.2) containing either a templating dA or dC for 30 min. Binary complex crystals were grown via sitting drop vapor diffusion using 2 μl of protein/DNA mixture combined with 2 μl of mother liquor (50 mm imidazole (pH 7.5), 16%–20% PEG3350, and 350 mm sodium acetate) as described previously (33). Binary Y271G pol β:DNA complex crystals were then soaked in a cryosolution containing 20% ethylene glycol, 50 mm imidazole (pH 7.5), 16%–19% PEG3350, 70 mm sodium acetate, 5 mm r8-oxo-GTP, and 50 mm MnCl2 for 1–3 h. This resulted in ternary pol β:DNA:r8-oxo-GTP crystals for X-ray crystallography.
Data collection and refinement
Data were collected at 100 K on a MicroMax-007HF rotating anode generator at a wavelength of 1.54 Å. Diffraction images were collected using a Dectris Pilatus3R 200K-A detector, and the HKL3000 software package was used for processing and scaling the data (54). Initial models were determined using molecular replacement with the previously determined closed ternary (PDB code 2FMS) structure of pol β, and Rfree flags were taken from the starting model, except for PDB code 6UOK, which had two pol β in the asymmetric unit. The metal–ligand coordination restraints were generated by ReadySet (PHENIX). Refinement was performed using PHENIX and model building using Coot (55, 56). Density maps in structure figures (green mesh) were generated using Polder Maps in PHENIX. Polder OMIT maps are reduced-bias σ-A weighted difference density maps (mFO-DFC) that exclude bulk solvent within a 5 Å radius when calculating OMIT maps to improve visualization of weak densities (57). All polder maps were scaled to 3.0 σ, except for Fig. S4, where it was scaled to 4.0σ. Ramachandran analysis determined that 100% of nonglycine residues lie in the allowed regions and at least 96% in favored regions. Local base-pair parameters were calculated using 3DNA, and local interbase angles were corrected for both buckle and propeller contributions (37). The figures were prepared in PyMOL (Schrödinger LLC).
Single-nucleotide gap-filling DNA synthesis
Catalytic efficiencies (kpol/Kd) for single-nucleotide gap-filling reactions were determined by single-turnover analysis (i.e. enzyme ≫ DNA). At least seven time points were gathered for each single-exponential time course determined with three subsaturating concentrations of r8-oxo-GTP. The standard reaction mixture contained 50 mm Tris-HCl (pH 7.4), 100 mm KCl, 5 mm MgCl2, 1 mm DTT, 100 μg/ml BSA, 10% glycerol, 100 nm single-nucleotide gapped DNA, and 1 μm pol β. Concentrations of r8-oxo-GTP varied depending on the identity of the templating base to achieve a wide range of product concentration while avoiding multiple insertions and/or product inhibition. Templating dA kinetics were performed with 10 μm, 25 μm, and 50 μm r8-oxo-GTP during a 4-h time course, with samples taken at 5, 15, 30, 45, 60, 90, 132, 170, 230, and 330 min. Templating dC kinetics were performed with 100 μm, 250 μm, and 500 μm r8-oxo-GTP during a time course of 6.5 h, with samples taken at 15, 30, 60, 95, 132, 180, 230, 275, and 400 min. For rGTP insertion kinetics, 10 μm, 25 μm, and 50 μm rGTP were used in our reactions over a time course of 2.5 h, with samples taken at 2, 5, 15, 30, 45, 60, 90, 120, and 150 min. With the Y271G pol β mutant, kinetic experiments with a templating adenine used 50 nm, 100 nm, and 250 nm r8-oxo-GTP over a 4-h time course, with samples taken at 1, 3, 10, 30, 60, 90, 120, 180, and 240 min. Y271G pol β mutant experiments with a templating cytosine used 200 μm, 400 μm, and 600 μm r8-oxo-GTP over a 6-h time course, with samples taken at 5, 15, 30, 45, 60, 90, 132, 170, 230, and 330 min. Each reaction time course was performed at 37 °C. Reactions were stopped with addition of a quench solution containing 100 mm EDTA, 10 m urea, and formamide dye. The substrates and products were separated on 22% denaturing (8 m urea) polyacrylamide gels. Because a 6-Carboxyfluorescein 5′-labeled primer was used in these assays, the gels were imaged using the GE Typhoon PhosphorImager in fluorescence mode. Substrate and product bands were quantified using ImageJ (58) and then plotted using Kaleidagraph (45). For each concentration of r8-oxo-GTP, the data were fit to a single exponential curve (Fig. S1A). Because subsaturating concentrations of r8-oxo-GTP (<Kd) were used, the data were fit to an alternate form of the Michaelis equation to extract apparent catalytic efficiencies (kpol/Kd):kobs = ((kpol/Kd) × S)/(1 + (S/Kd)), where S refers to the concentration of r8-oxo-GTP (Fig. S1B). Catalytic efficiencies were calculated as an average of three independent experiments ± S.D. (Fig. S1C).
Steady-state FRET
Human pol β containing mutations C239S, C267S, and V303C was used to fluorescently label the N-subdomain with the thiol-reactive IAEDANS, as described previously (15). Fluorescence emission of AEDANS-labeled pol β was measured in either the apoenzyme form (400 nm AEDANS-labeled pol β in 50 mm Tris (pH 8.0), 20 mm NaCl, and 10 mm MgCl2), the binary complex (400 nm AEDANS-labeled pol β mixed with 400 nm single-nucleotide gapped DNA substrate with a dideoxy-terminated primer terminus containing Dabcyl dT at the −8 position relative the templating base), or the ternary complex (formed by adding 1 mm dGTP, 8-oxo-dGTP, rGTP, or r8-oxo-GTP to the binary complex) (Fig. S2). AEDANS was excited at 336 nm, and emission was collected at 400–550 nm using a QuantaMaster 800 fluorometer at room temperature (Fig. S3, A and B) (15). Distances were estimated from the FRET efficiency between AEDANS and Dabcyl and by using a Förster radius of 37.76 Å. Experiments were performed in triplicate, and the error was calculated as a standard deviation of the mean. A table with the calculated distances is provided (Fig. S3C).
Author contributions
M. R. S., K. S. A., J. B. S., and B. D. F. formal analysis; M. R. S. and K. S. A. validation; M. R. S., K. S. A., and N. M. H. investigation; M. R. S. visualization; M. R. S. and K. S. A. methodology; M. R. S. writing-original draft; M. R. S., K. S. A., N. M. H., J. B. S., and B. D. F. writing-review and editing; J. B. S. and B. D. F. supervision; J. B. S. and B. D. F. funding acquisition; B. D. F. conceptualization; B. D. F. project administration.
Supplementary Material
Acknowledgments
We thank Drs. William A. Beard and Amy M. Whitaker for providing additional feedback during manuscript preparation.
This work was supported by National Institutes of Health Grants R01-ES027558 and R35-GM128562 (to B. D. F., M. R. S., and N. M. H) and R01-CA080830 (to J. B. S. and K. S. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5 and Tables S1 and S2.
- 8-oxo-G
- 8-oxo-7, 8-dihydro-2′-guanosine
- pol β
- DNA polymerase β
- AEDANS
- N-(acetylaminoethyl)-5-naphthylamine-1-sulfonic acid
- IAEDANS
- 5((((2-iodoacetyl)amino)ethyl)amino)-naphthalene-1-sulfonic acid.
References
- 1. Cadet J., and Wagner J. R. (2013) DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 5, a012559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jena N. R. (2012) DNA damage by reactive species: mechanisms, mutation and repair. J. Biosci. 37, 503–517 10.1007/s12038-012-9218-2 [DOI] [PubMed] [Google Scholar]
- 3. Topal M. D., and Baker M. S. (1982) DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells. Proc. Natl. Acad. Sci. U.S.A. 79, 2211–2215 10.1073/pnas.79.7.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolin C., and Cardozo-Pelaez F. (2007) Assessing biomarkers of oxidative stress: analysis of guanosine and oxidized guanosine nucleotide triphosphates by high performance liquid chromatography with electrochemical detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 856, 121–130 10.1016/j.jchromb.2007.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerchiaro G., Bolin C., and Cardozo-Pelaez F. (2009) Hydroxyl radical oxidation of guanosine 5′-triphosphate (GTP): requirement for a GTP-Cu(II) complex. Redox Report 14, 82–92 10.1179/135100009X392520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haghdoost S., Sjölander L., Czene S., and Harms-Ringdahl M. (2006) The nucleotide pool is a significant target for oxidative stress. Free Radic. Biol. Med. 41, 620–626 10.1016/j.freeradbiomed.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 7. Jovanovic S. V., and Simic M. G. (1986) One-electron redox potentials of purines and pyrimidines. J. Phys. Chem. 90, 974–978 10.1021/j100277a053 [DOI] [Google Scholar]
- 8. Beard W. A., Batra V. K., and Wilson S. H. (2010) DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat. Res. 703, 18–23 10.1016/j.mrgentox.2010.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kulkarni A., and Wilson D. M. 3rd (2008) The involvement of DNA-damage and -repair defects in neurological dysfunction. Am. J. Hum. Genet. 82, 539–566 10.1016/j.ajhg.2008.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakabeppu Y. (2014) Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int. J. Mol. Sci. 15, 12543–12557 10.3390/ijms150712543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown J. A., Duym W. W., Fowler J. D., and Suo Z. (2007) Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2′-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases λ and β. J. Mol. Biol. 367, 1258–1269 10.1016/j.jmb.2007.01.069 [DOI] [PubMed] [Google Scholar]
- 12. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., and Wilson S. H. (2008) Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 10.1016/j.molcel.2008.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beard W. A., Osheroff W. P., Prasad R., Sawaya M. R., Jaju M., Wood T. G., Kraut J., Kunkel T. A., and Wilson S. H. (1996) Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase β. J. Biol. Chem. 271, 12141–12144 10.1074/jbc.271.21.12141 [DOI] [PubMed] [Google Scholar]
- 14. Genna V., Vidossich P., Ippoliti E., Carloni P., and De Vivo M. (2016) A self-activated mechanism for nucleic acid polymerization catalyzed by DNA/RNA polymerases. J. Am. Chem. Soc. 138, 14592–14598 10.1021/jacs.6b05475 [DOI] [PubMed] [Google Scholar]
- 15. Towle-Weicksel J. B., Dalal S., Sohl C. D., Doublié S., Anderson K. S., and Sweasy J. B. (2014) Fluorescence resonance energy transfer studies of DNA polymerase β: the critical role of fingers domain movements and a novel non-covalent step during nucleotide selection. J. Biol. Chem. 289, 16541–16550 10.1074/jbc.M114.561878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang J., Alnajjar K. S., Mahmoud M. M., Eckenroth B., Doublié S., and Sweasy J. B. (2018) The nature of the DNA substrate influences pre-catalytic conformational changes of DNA polymerase β. J. Biol. Chem. 293, 15084–15094 10.1074/jbc.RA118.004564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Werneburg B. G., Ahn J., Zhong X., Hondal R. J., Kraynov V. S., and Tsai M. D. (1996) DNA polymerase β: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry 35, 7041–7050 10.1021/bi9527202 [DOI] [PubMed] [Google Scholar]
- 18. Vande Berg B. J., Beard W. A., and Wilson S. H. (2001) DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β: implication for the identity of the rate-limiting conformational change. J. Biol. Chem. 276, 3408–3416 10.1074/jbc.M002884200 [DOI] [PubMed] [Google Scholar]
- 19. Beard W. A., and Wilson S. H. (2014) Structure and mechanism of DNA polymerase β. Biochemistry 53, 2768–2780 10.1021/bi500139h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Batra V. K., Beard W. A., Pedersen L. C., and Wilson S. H. (2016) Structures of DNA polymerase mispaired DNA termini transitioning to pre-catalytic complexes support an induced-fit fidelity mechanism. Structure 24, 1863–1875 10.1016/j.str.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown J. A., Fiala K. A., Fowler J. D., Sherrer S. M., Newmister S. A., Duym W. W., and Suo Z. (2010) A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 395, 282–290 10.1016/j.jmb.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cavanaugh N. A., Beard W. A., Batra V. K., Perera L., Pedersen L. G., and Wilson S. H. (2011) Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J. Biol. Chem. 286, 31650–31660 10.1074/jbc.M111.253401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Potenski C. J., and Klein H. L. (2014) How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 42, 10226–10234 10.1093/nar/gku773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clausen A. R., Zhang S., Burgers P. M., Lee M. Y., and Kunkel T. A. (2013) Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair 12, 121–127 10.1016/j.dnarep.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E.-B., Burgers P. M., Johansson E., Chabes A., and Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 10.1073/pnas.0914857107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reijns M. A., Rabe B., Rigby R. E., Mill P., Astell K. R., Lettice L. A., Boyle S., Leitch A., Keighren M., Kilanowski F., Devenney P. S., Sexton D., Grimes G., Holt I. J., Hill R. E., et al. (2012) Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 10.1016/j.cell.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ordonez H., and Shuman S. (2014) Mycobacterium smegmatis DinB2 misincorporates deoxyribonucleotides and ribonucleotides during templated synthesis and lesion bypass. Nucleic Acids Res. 42, 12722–12734 10.1093/nar/gku1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sastre-Moreno G., Sánchez A., Esteban V., and Blanco L. (2014) ATP insertion opposite 8-oxo-deoxyguanosine by Pol4 mediates error-free tolerance in Schizosaccharomyces pombe. Nucleic Acids Res. 42, 9821–9837 10.1093/nar/gku711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cilli P., Minoprio A., Bossa C., Bignami M., and Mazzei F. (2015) Formation and repair of mismatches containing ribonucleotides and oxidized bases at repeated DNA sequences. J. Biol. Chem. 290, 26259–26269 10.1074/jbc.M115.679209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flood C. L., Rodriguez G. P., Bao G., Shockley A. H., Kow Y. W., and Crouse G. F. (2015) Replicative DNA polymerase δ but not ϵ proofreads errors in cis and in trans. PLoS Genet. 11, e1005049 10.1371/journal.pgen.1005049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cavanaugh N. A., Beard W. A., and Wilson S. H. (2010) DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 285, 24457–24465 10.1074/jbc.M110.132407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang L., Beard W. A., Wilson S. H., Broyde S., and Schlick T. (2004) Highly organized but pliant active site of DNA polymerase β: compensatory mechanisms in mutant enzymes revealed by dynamics simulations and energy analyses. Biophys. J. 86, 3392–3408 10.1529/biophysj.103.036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., and Wilson S. H. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 10.1016/j.str.2006.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu X.-J., and Olson W. K. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 10.1093/nar/gkg680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freudenthal B. D., Beard W. A., Perera L., Shock D. D., Kim T., Schlick T., and Wilson S. H. (2015) Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature 517, 635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krahn J. M., Beard W. A., and Wilson S. H. (2004) Structural insights into DNA polymerase β deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure 12, 1823–1832 10.1016/j.str.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 37. Lu X.-J., and Olson W. K. (2008) 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson K. A. (2008) Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J. Biol. Chem. 283, 26297–26301 10.1074/jbc.R800034200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koag M. C., and Lee S. (2018) Insights into the effect of minor groove interactions and metal cofactors on mutagenic replication by human DNA polymerase β. Biochem. J. 475, 571–585 10.1042/BCJ20170787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., and Wilson S. H. (2005) Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate. Structure 13, 1225–1233 10.1016/j.str.2005.05.010 [DOI] [PubMed] [Google Scholar]
- 41. Smith M. R., Shock D. D., Beard W. A., Greenberg M. M., Freudenthal B. D., and Wilson S. H. (2019) A guardian residue hinders insertion of a Fapy·dGTP analog by modulating the open-closed DNA polymerase transition. Nucleic Acids Res. 47, 3197–3207 10.1093/nar/gkz002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Freudenthal B. D., Beard W. A., and Wilson S. H. (2012) Structures of dNTP intermediate states during DNA polymerase active site assembly. Structure 20, 1829–1837 10.1016/j.str.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eckenroth B. E., Towle-Weicksel J. B., Sweasy J. B., and Doublié S. (2013) The E295K cancer variant of human polymerase β favors the mismatch conformational pathway during nucleotide selection. J. Biol. Chem. 288, 34850–34860 10.1074/jbc.M113.510891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Su Y., Egli M., and Guengerich F. P. (2016) Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem. 291, 3747–3756 10.1074/jbc.M115.706226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirsch P. D., and Ekerdt J. G. (2000) KaleidaGraph: Graphing and Data Analysis. Version 3.5 for Windows Synergy Software, 2457 Perkiomen Ave., Reading, PA 19606–2049. www.Synergy.com. $155.00. J. Am. Chem. Soc. 122, 11755–11755 10.1021/ja004775j [DOI] [Google Scholar]
- 46. Göksenin A. Y., Zahurancik W., LeCompte K. G., Taggart D. J., Suo Z., and Pursell Z. F. (2012) Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 287, 42675–42684 10.1074/jbc.M112.422733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bolin C., and Cardozo-Pelaez F. (2009) Characterization of oxidized guanosine 5′-triphosphate as a viable inhibitor of soluble guanylyl cyclase. Free Radic. Biol. Med. 46, 828–835 10.1016/j.freeradbiomed.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishibashi T., Hayakawa H., Ito R., Miyazawa M., Yamagata Y., and Sekiguchi M. (2005) Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res. 33, 3779–3784 10.1093/nar/gki682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sassa A., Yasui M., and Honma M. (2019) Current perspectives on mechanisms of ribonucleotide incorporation and processing in mammalian DNA. Genes Environ. 41, 3 10.1186/s41021-019-0118-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hayakawa H., Hofer A., Thelander L., Kitajima S., Cai Y., Oshiro S., Yakushiji H., Nakabeppu Y., Kuwano M., and Sekiguchi M. (1999) Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry 38, 3610–3614 10.1021/bi982361l [DOI] [PubMed] [Google Scholar]
- 51. Malfatti M. C., Balachander S., Antoniali G., Koh K. D., Saint-Pierre C., Gasparutto D., Chon H., Crouch R. J., Storici F., and Tell G. (2017) Abasic and oxidized ribonucleotides embedded in DNA are processed by human APE1 and not by RNase H2. Nucleic Acids Res. 45, 11193–11212 10.1093/nar/gkx723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carreras-Puigvert J., Zitnik M., Jemth A.-S., Carter M., Unterlass J. E., Hallström B., Loseva O., Karem Z., Calderón-Montaño J. M., Lindskog C., Edqvist P.-H., Matuszewski D. J., Ait Blal H., Berntsson R. P. A., Häggblad M., et al. (2017) A comprehensive structural, biochemical and biological profiling of the human NUDIX hydrolase family. Nat. Commun. 8, 1541 10.1038/s41467-017-01642-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beard W. A., and Wilson S. H. (1995) Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 262, 98–107 10.1016/0076-6879(95)62013-3 [DOI] [PubMed] [Google Scholar]
- 54. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 55. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 57. Liebschner D., Afonine P. V., Moriarty N. W., Poon B. K., Sobolev O. V., Terwilliger T. C., and Adams P. D. (2017) Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 10.1107/S2059798316018210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.