Abstract
Proteins incorporating iron–sulfur (Fe-S) co-factors are required for a plethora of metabolic processes. Their maturation depends on three Fe-S cluster assembly machineries in plants, located in the cytosol, mitochondria, and chloroplasts. After de novo formation on scaffold proteins, transfer proteins load Fe-S clusters onto client proteins. Among the plastidial representatives of these transfer proteins, NFU2 and NFU3 are required for the maturation of the [4Fe-4S] clusters present in photosystem I subunits, acting upstream of the high-chlorophyll fluorescence 101 (HCF101) protein. NFU2 is also required for the maturation of the [2Fe-2S]-containing dihydroxyacid dehydratase, important for branched-chain amino acid synthesis. Here, we report that recombinant Arabidopsis thaliana NFU1 assembles one [4Fe-4S] cluster per homodimer. Performing co-immunoprecipitation experiments and assessing physical interactions of NFU1 with many [4Fe-4S]-containing plastidial proteins in binary yeast two-hybrid assays, we also gained insights into the specificity of NFU1 for the maturation of chloroplastic Fe-S proteins. Using bimolecular fluorescence complementation and in vitro Fe-S cluster transfer experiments, we confirmed interactions with two proteins involved in isoprenoid and thiamine biosynthesis, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase and 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate synthase, respectively. An additional interaction detected with the scaffold protein SUFD enabled us to build a model in which NFU1 receives its Fe-S cluster from the SUFBC2D scaffold complex and serves in the maturation of specific [4Fe-4S] client proteins. The identification of the NFU1 partner proteins reported here more clearly defines the role of NFU1 in Fe-S client protein maturation in Arabidopsis chloroplasts among other SUF components.
Keywords: iron–sulfur protein, chloroplast, protein assembly, Arabidopsis thaliana, sulfur, maturation, NFU
Introduction
Iron and sulfur are essential elements for all organisms, in particular as building blocks of iron–sulfur (Fe-S) proteins that are themselves essential for several key molecular processes. In plants, Fe-S proteins participate in photosynthesis and respiration, being present in the electron transfer chains found in chloroplasts and mitochondria, but also for sulfur and nitrogen assimilation or chlorophyll and vitamin metabolisms to cite a few examples (1, 2). The most represented Fe-S cluster forms are the [2Fe-2S] and [4Fe-4S] clusters with some proteins such as the glutamate synthases (GOGAT) incorporating [3Fe-4S] clusters (2, 3). The maturation of Fe-S proteins is not a spontaneous process and relies on dedicated machineries that exist in all kingdoms, although with some variations in the type and number of machineries present and in the molecular actors implicated (4). In plants, three machineries exist: the iron–sulfur cluster (ISC)5 machinery is found in mitochondria; the cytosolic iron–sulfur protein assembly (CIA) machinery provides Fe-S clusters for the maturation of both cytosolic and nuclear Fe-S proteins and depends on the ISC machinery; and the sulfur mobilization (SUF) machinery works independently and is involved in the maturation of plastidial proteins (3, 4). Regardless of the machinery, the Fe-S cluster biogenesis can be divided into several steps. In the early steps, an Fe-S cluster is built de novo on so-called scaffold proteins, necessitating a multiprotein assembly complex for the mobilization, reduction, and assembly of iron and sulfur atoms. The preformed Fe-S cluster will be conveyed to a set of transfer proteins, eventually with the help of chaperones. After possible conversion and exchange among these Fe-S cluster transfer proteins/complexes, the Fe-S cluster is delivered to final targets (4).
In the current model of the mitochondrial ISC machinery, a [2Fe-2S] cluster is assembled on ISU/ISCU scaffold proteins prior to its transfer to a glutaredoxin (GRX), referred to as Grx5 in yeast. Whereas these steps should be sufficient for the maturation of [2Fe-2S] proteins (5), the maturation of [4Fe-4S] proteins also requires a conversion from two [2Fe-2S] clusters to a [4Fe-4S] cluster by reductive coupling, probably occurring in the course of the interaction between GRX5 and ISCA1/2 heterodimer (6). The BOLA1 and IBA57 maturation factors may be involved at this step. Further late-acting Fe-S cluster transfer proteins, NFU1, IND1/INDH (when present), and BOLA3, participate in the maturation of some [4Fe-4S] proteins but not all (7–9).
In the current model of the chloroplastic SUF machinery, the cysteine desulfurase NFS2, assisted by SUFE proteins, provides the required sulfur atoms for the de novo synthesis of an Fe-S cluster onto the SUFBC2D scaffold complex. However, how the system is supplied with iron atoms and electrons is less clear. Accordingly, all these genes are essential as confirmed by the embryonic lethality of the corresponding Arabidopsis thaliana loss–of–function mutants (10–16). Then, only scarce information is available in the following stages, although proteins belonging to the same family as the mitochondrial counterparts are present, i.e. GRXS14/16, BOLA1/4, IBA57.2, SUFA1, NFU1/2/3, and HCF101 (2, 3, 17–19). The first relevant functional information is that, except for IBA57.2, each gene/protein complements the yeast mutants for the corresponding mitochondrial orthologs (20, 21). The physiological analysis of hcf101, nfu2, and nfu3 knockdown or knockout A. thaliana mutants, which exhibit dwarf phenotypes, pointed to their involvement in the maturation of the [4Fe-4S] clusters found in photosystem I (PSI), with NFU2 and NFU3 acting directly upstream of HCF101 (22–25). HCF101 is also required for the maturation of the [4Fe-4S] cluster in the ferredoxin–thioredoxin reductase (26, 27). In accordance with its capacity to bind both [2Fe-2S] and [4Fe-4S] clusters, NFU2 is required for the maturation of the [2Fe-2S] cluster present in the dihydroxyacid dehydratase (DHAD), an enzyme implicated in the synthesis of branched-chain amino acids (23, 28, 29). A valid biochemical characterization of NFU3 was hampered so far by the impossibility to express sufficient amounts of soluble, nonaggregated recombinant protein for a proper spectroscopic characterization, but it was suggested to bind both [3Fe-4S] and [4Fe-4S] clusters (22). However, no firm conclusion has been gained from the study of individual grxS14, grxS16, and sufa1 Arabidopsis mutants except that these proteins are dispensable, at least in the growth conditions tested (17, 23, 30, 31).
Unlike nfu2 and nfu3 mutants, an Arabidopsis nfu1 mutant has no phenotype when grown under standard conditions (23). This result suggests that NFU1 has either an accessory or very specific function or a functional redundancy with other Fe-S transfer protein(s), rendering difficult the determination of its role in planta. Hence, to tackle the role of NFU1 in Arabidopsis, we have characterized the biochemical and spectroscopic properties of the recombinant protein, before identifying its plastidial partner proteins using co-immunoprecipitation (co-IP) experiments and a binary yeast-two hybrid (Y2H) screen with other SUF components and many candidate [4Fe-4S]-containing proteins. The in planta interaction was validated for a few proteins using bimolecular fluorescence complementation (BiFC) assays before assessing the capacity of NFU1 to transfer its Fe-S cluster to two selected client proteins, the 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase (ISPG/GcpE/HDS) and the 4-amino-2-methyl-5-hydroxymethylpyrimidine phosphate synthase (THIC). The identification of these NFU1 partners allowed us to position NFU1 in the nebula of SUF components and Fe-S client proteins.
Results
AtNFU1 binds a [4Fe-4S] cluster into a homodimer
The plastidial NFU1/2/3 isoforms have two NFU domains repeated in tandem, but only the N-terminal domain possesses the conserved CXXC motif participating in Fe-S cluster ligation. They are all able to restore the growth defect of the yeast mutant for the mitochondrial Nfu1 isoform (21). This suggested that they share some common properties such as the capacity to bind a [4Fe-4S] cluster, but this has not yet been explored for any plant NFU1 isoforms. Moreover, it is important to assess whether NFU1 can bind other types of Fe-S clusters as observed for A. thaliana NFU2 (29).
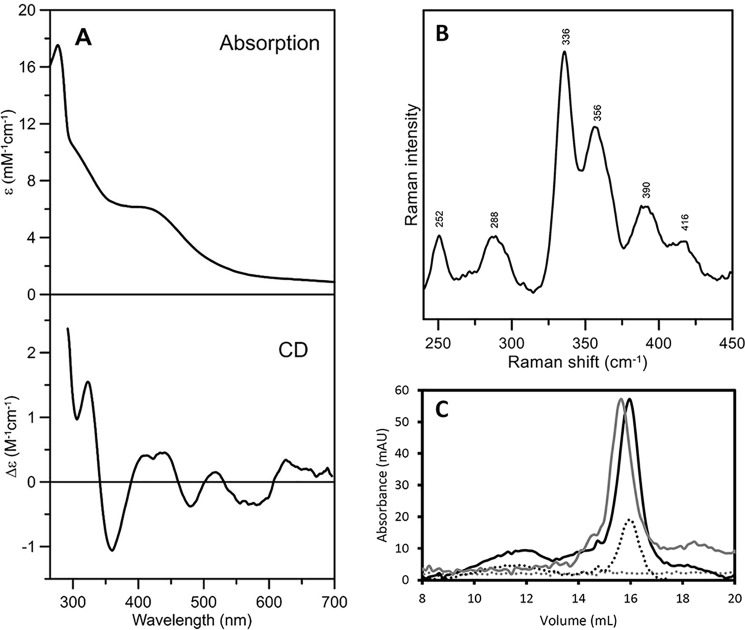
The mature form of Arabidopsis NFU1 was expressed in Escherichia coli as an untagged recombinant protein. Unlike NFU2, which contains [2Fe-2S] clusters as purified (25, 29), no color typical of the presence of Fe-S cluster was visible in the lysed cell extract, and purified NFU1 was devoid of Fe-S clusters, based on a UV-visible absorption spectrum showing only a single absorption peak at 280 nm. To evaluate the ability of NFU1 to bind Fe-S clusters, in vitro enzymatic Fe-S cluster reconstitution experiments were performed under anaerobic conditions in the presence of E. coli cysteine desulfurase (EcIscS). The UV-visible absorption and CD spectra of reconstituted NFU1 are shown in Fig. 1A. The absorption spectrum, comprising broad shoulders centered at ∼400 and ∼320 nm, is characteristic of a [4Fe-4S]2+ cluster and is very similar to that reported for the reconstituted [4Fe-4S] cluster-bound form of NFU2 (Fig. S1) (29). Resonance Raman provides a more definitive assessment of cluster type, based on the observed Fe-S stretching modes (32), and the spectrum obtained with 457.9-nm laser excitation is uniquely indicative of a [4Fe-4S]2+ cluster (Fig. 1B). Interestingly, the UV-visible CD and resonance Raman spectra of the [4Fe-4S] centers in NFU1 and NFU2 are quite distinct (Fig. S1). For example, the dominant symmetric breathing mode of the [4Fe-4S] core shifts from 336 cm−1 in NFU1 to 344 cm−1 in NFU2. The structural origins of such differences may be important for explaining why only NFU2 is capable of binding both [2Fe-2S] and [4Fe-4S] clusters at the subunit interface. X-ray crystal structures will be required for meaningful interpretation.
Figure 1.
Spectroscopic and oligomeric state characterization of reconstituted Arabidopsis NFU1. UV-visible absorption and CD spectra (A), resonance Raman spectrum (B), and analytical gel filtration studies (C) of reconstituted Arabidopsis NFU1. The ϵ and Δϵ values for the UV-visible absorption and CD spectra (A) are based on NFU1 monomer concentration. The resonance Raman spectrum (B) was recorded using a droplet of NFU1 (∼2 mm in clusters) frozen at 17 K, using 457.9 nm laser excitation. The spectrum is the sum of 100 scans with each scan involving counting protons for 1 s every 0.5 cm−1, with 7 cm−1 spectral resolution. Bands due to the frozen buffer solution have been subtracted. For analytical gel filtration (C), apo-NFU1 (gray line) and reconstituted holo-NFU1 (black line) proteins were loaded onto a Sephadex S200 10/300 column. Absorbance of the eluted fractions was recorded at 280 nm (solid line) and 420 nm (dashed line), a wavelength characteristic of Fe-S cluster absorption. The apparent molecular masses of apo- and holo-NFU1 were determined from the elution volumes relatively to those of standard proteins, as described under “Experimental procedures.”
Quantification of both iron and acid-labile sulfur atoms bound to the protein showed that the sample contained 1.54 ± 0.29 Fe and 1.68 ± 0.11 acid-labile S per NFU1 monomer. Hence, analytical data indicate that the reconstituted samples contain ∼80% of [4Fe-4S] cluster-loaded NFU1. This is in accord with the UV-visible absorption values of ϵ400 = 6.2 mm−1 cm−1 based on the NFU1 monomer. This translates to ϵ400 = 12.4 mm−1 cm−1 based on the NFU1 dimer and ∼80% of NFU1 with a subunit bridging the [4Fe-4S] cluster, based on typical values of ϵ400 = 15 ± 2 mm−1 cm−1 for one [4Fe-4S]2+ cluster. Moreover, analytical gel-filtration experiments have been performed using both apo- and holo-NFU1 (Fig. 1C). Both forms eluted as a single peak corresponding to an estimated molecular mass of 30 kDa for apo-NFU1 and 25 kDa for holo-NFU1, respectively (Fig. 1B). From the theoretical molecular mass of a mature NFU1 (17 kDa), we concluded that both protein forms exist as homodimers. Taken together, these results indicated that the Fe-S cluster is not required for dimerization and that a reconstituted NFU1 homodimer contained ∼80% [4Fe-4S] cluster.
AtNFU1 physically interacts with SUFD, SUFA1, and various [4Fe-4S]-containing proteins
So far, there are no known NFU1 partners in plants, and only negative results have been obtained using binary Y2H, either with HCF101 or with two putative targets, 5′-adenylyl-phosphosulfate reductase 1 (APR1) and the PSI subunit PsaC (23, 29). As a first approach to determine what are the SUF partners and client Fe-S proteins of NFU1, co-IP experiments using anti-GFP antibodies have been performed on 2-week-old transgenic lines expressing either a NFU1–GFP fusion (ProNFU1::gNFU1–GFP construct) or a GFP alone fused downstream of the chloroplastic targeting peptide (CTP) of the NFU3 protein (Pro35S::CTPNFU3-GFP construct). The latter construct allows a specific targeting of GFP into the stroma of chloroplasts. After verifying that the constructs allow the specific expression of both fusion proteins in chloroplasts (Fig. S2), four replicates have been performed using distinct transgenic lines. Table 1 lists the 49 proteins present in at least three replicates originating from the co-IP performed with the NFU1–GFP-expressing lines and absent in the four replicates performed with the control lines. It also comprises 5 additional proteins (labeled with an asterisk) representing Fe-S proteins or subunits associated with the Fe-S proteins found using a slightly less-stringent cutoff, i.e. also present in at least three replicates but found in one replicate of control experiments. Using the latter filter, a total of 89 additional proteins were retrieved (Table S1). Among these 138 proteins, we identified 6 known Fe-S proteins. HCF101 and GRXS16 are SUF components. Recovering HCF101 was surprising, because no interaction was previously detected by Y2H and BiFC (23). It may be that it has been pulled down as part of a complex formed with other Fe-S proteins. Concerning GRXS16, the question of a direct physical interaction remains to be asked. Although no interaction was observed between NFU1 and GRXS16 by binary Y2H (see below), the NFU2 paralog has proven to be able to efficiently transfer a [2Fe-2S] cluster to GRXS16 (29). A role of GRXS16 in Fe-S cluster biogenesis has yet to be validated in planta, but in vitro the recombinant protein incorporates either a [2Fe-2S] or a [4Fe-4S] cluster (30). Four putative Fe-S client proteins have been isolated. The 5′-adenylyl-phosphosulfate reductase 2 (APR2), one of the three APR isoforms in Arabidopsis, catalyzes the second step of sulfate assimilation and is thus implicated in cysteine and methionine synthesis. The quinolinate synthase SUFE3 is involved in NAD synthesis (12). The glutamate synthase 2 (GLU2) participates to nitrogen assimilation by catalyzing the conversion of glutamine into glutamate at the expense of ferredoxin. The last Fe-S protein firmly identified is THIC, an enzyme involved in the synthesis of vitamin B1 (thiamine) (33). APR2, SUFE3, and THIC are known to bind [4Fe-4S] clusters whereas GLU2 binds a [3Fe-4S] cluster (2). Another likely candidate partner is the isopropyl malate isomerase (IIL1/IPMI), an Fe-S enzyme catalyzing the isomerization between 2-isopropylmalate and 3-isopropylmalate and thus implicated in the synthesis of leucine and glucosinolates (34). It forms a protein complex involving a large subunit and a small subunit, and the three isoforms corresponding to the small subunits have been retrieved from these experiments. Despite being found in the four co-IP replicates, the large subunit, bearing the Fe-S cluster, was not listed here because it was also found in two of the control experiments. Then, what is noticeable among the set of the 45 other identified proteins is the presence of a set of proteins performing redox reactions. It includes four sulfurtransferases (STR9, -10, -12, and -14), enzymes containing a rhodanese domain with a conserved cysteine involved in trans-persulfidation reactions (35). It also comprises several thioredoxin (TRX)-like proteins, TRX z, TRX-lilium2, also referred to as the atypical Cys–His-rich thioredoxin 2, and a novel putative TRX superfamily member (At5g65840). Having reactive cysteines, it may be that these proteins formed covalent bonds with the reactive cysteines of an apo-NFU1. In line with such a possible interaction, we have recently observed that mitochondrial TRXs o have the ability to reduce an intramolecular disulfide formed in the NFU domain of mitochondrial NFU4 and NFU5 (36). Whether this is also true for chloroplastic NFU1/TRX couples remains to be investigated and the physiological relevance assessed. TRX z is known to be part of the plastid-encoded RNA polymerase complex, likely explaining why fructokinase-like 2 and plastid transcriptionally-active 5 and 17 proteins have also been retrieved (37). To conclude on that approach, there are quite a few other proteins that have never been biochemically characterized and for which there is no attributed function. Consequently, it is difficult to extrapolate whether they could or could not be novel Fe-S–containing proteins or involved in the maturation process. For instance, several chloroplastic chaperones are present in the expanded list, and it may be that they are required for Fe-S cluster exchange as documented for the assembly step of the mitochondrial ISC machinery. Although usually powerful, it is worth pointing that the co-IP approach may not be the best for supposedly weak or transient interactions or interactions that would rely on the presence of an oxygen-labile Fe-S cluster.
Table 1.
Potential NFU1 partners obtained by co-immunoprecipitation experiments and their assigned or presumed function
Proteins were considered as interactors if they were identified in co-IP experiments in at least three replicates using ProNFU1::gNFU1-GFP lines and not identified in the four replicates using Pro35S::CTPNFU3-GFP lines. The five hits labeled with an asterisk are Fe-S proteins or Fe-S–associated proteins found in only one replicate using Pro35S::CTPNFU3-GFP lines. Additional non-Fe-S proteins selected using this criterion are listed in Table S1.
| Gene IDs | moy_intensity Pro35S::CTPNFU3-GFP | moy_intensity ProNFU1::gNFU1-GFP | No. of peptides | Symbol | Protein name | Function | Localization | Fe-S cluster type | Sequence coverage |
|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||
| AT5G50210 | 0.00 | 1.87E + 07 | 8 | SUFE3 | Quinolinate synthase SUFE3 | NAD synthesis | Plastid | 4Fe-4S | 17.3 |
| AT1G62180 | 0.00 | 1.09E + 07 | 7 | APR2 | 5′-Adenylyl-phosphosulfate reductase 2 | Sulfate assimilation | Plastid | 4Fe-4S | 30 |
| AT2G29630* | 7.80E + 04 | 1.05E + 07 | 5 | THIC | Thiamine C biosynthesis | Thiamine biosynthesis | Plastid | 4Fe-4S | 11.3 |
| AT2G41220* | 1.17E + 05 | 1.10E + 06 | 8 | GLU2 | Glutamate synthase 2 | Nitrogen assimilation | Plastid | 3Fe-4S | 6.4 |
| AT2G38270* | 8.86E + 04 | 2.64E + 06 | 2 | GRXS16 | Glutaredoxin S16 | Fe-S cluster assembly machinery | Plastid | 2Fe-2S, 4Fe-4S | 16.7 |
| AT3G24430* | 1.61E + 05 | 1.83E + 07 | 9 | HCF101 | High-chlorophyll fluorescence 101 | Fe-S cluster assembly machinery | Plastid | 4Fe-4S | 27.3 |
| AT2G43100 | 0.00 | 6.92E + 06 | 6 | IPMI2 | Isopropylmalate isomerase 2 | Glucosinolate and leucine biosynthesis | Plastid | None | 29.3 |
| AT3G58990 | 0.00 | 5.76E + 06 | 4 | IPMI1 | Isopropylmalate isomerase 1 | Glucosinolate and leucine biosynthesis | Plastid | None | 23.3 |
| AT2G43090* | 1.54E + 05 | 1.18E + 07 | 8 | IPMI3 | Isopropylmalate isomerase 3 | Glucosinolate and leucine biosynthesis | Plastid | None | 42.6 |
| AT5G10920 | 0.00 | 1.64E + 06 | 2 | none | Argininosuccinate lyase | Amino acid biosynthesis (arginine) | Plastid | None | 6.6 |
| AT1G58080 | 0.00 | 8.08E + 06 | 3 | ATP-PRT1 | ATP phosphoribosyltransferase 1 | Amino acid biosynthesis (histidine) | Plastid | None | 12.9 |
| AT5G05590 | 0.00 | 1.40E + 06 | 3 | PAI2 | Phosphoribosylanthranilate isomerase 2 | Amino acid biosynthesis (tryptophan) | Plastid | None | 18.2 |
| AT4G14210 | 0.00 | 7.17E + 06 | 3 | PDS3 | Phytoene desaturase 3 | Carotenoid biosynthesis | Plastid | None | 6.4 |
| AT5G59370 | 0.00 | 2.63E + 07 | 6 | ACT4 | Actin4 | Cellular architecture | Cytosol | None | 17 |
| AT4G16390 | 0.00 | 1.14E + 07 | 9 | SVR7 | Suppressor of variegation 7 | Chloroplast biogenesis | Plastid | None | 15.7 |
| AT1G69200 | 0.00 | 1.54E + 06 | 2 | FLN2 | Fructokinase-like 2 | Chloroplast organization | Plastid | None | 4.9 |
| AT1G79050 | 0.00 | 5.65E + 06 | 6 | RECA1 | Homolog of bacterial RecA | DNA metabolism | Plastid | None | 19.6 |
| AT2G43710 | 0.00 | 1.19E + 06 | 5 | SSI2 | Suppressor of SA insensitive 2 | Fatty acid desaturation | Plastid | None | 18.7 |
| AT4G15560 | 0.00 | 1.44E + 06 | 2 | DXS | 1-Deoxy-d-xylulose 5-phosphate synthase | Isoprenoid biosynthesis | Plastid | None | 5.9 |
| AT4G21210 | 0.00 | 8.84E + 06 | 5 | RP1 | PPDK regulatory protein 1 | Kinase activity | Plastid | None | 16.1 |
| AT1G70070 | 0.00 | 1.65E + 06 | 3 | ISE2 | Increased size-exclusion limit 2 | Plasmodesmata formation | Plastid, cytosol, nucleus | None | 4 |
| AT2G21280 | 0.00 | 4.09E + 06 | 2 | GC1 | Giant chloroplast 1 | Plastid division | Plastid | None | 7.8 |
| AT5G24020 | 0.00 | 1.54E + 06 | 2 | ARC11 | Accumulation and replication of chloroplasts 11 | Plastid division | Plastid | None | 7.1 |
| AT1G80480 | 0.00 | 1.49E + 07 | 7 | PTAC17 | Plastid transcriptionally active 17 | Plastid gene expression | Plastid | None | 26.1 |
| AT4G13670 | 0.00 | 3.57E + 07 | 10 | PTAC5 | Plastid transcriptionally active 5 | Plastid gene expression | Plastid | None | 39.5 |
| AT5G13630 | 0.00 | 7.04E + 06 | 5 | GUN5 | Genomes uncoupled 5 | Plastid-to-nucleus signal transduction | Plastid | None | 6.1 |
| AT1G02560 | 0.00 | 1.96E + 07 | 4 | CLPP5 | Nuclear-encoded CLP protease 5 | Protease | Plastid | None | 15.4 |
| AT4G20960 | 0.00 | 3.19E + 06 | 4 | PYRD | Pyrimidine deaminase | Riboflavin biosynthesis | Plastid | None | 16 |
| ATCG00810 | 0.00 | 3.44E + 06 | 2 | RPL22 | Ribosomal protein L22 | Ribosomal protein | Plastid | None | 17.5 |
| AT1G78630 | 0.00 | 9,60E + 06 | 2 | emb1473 | Embryo-defective 1473 | Ribosomal protein | Plastid, cytosol, mitochondrion | None | 13.3 |
| AT3G12930 | 0.00 | 4.72E + 06 | 2 | DG238 | Delayed greening 238 | Ribosome availability | Plastid | None | 10.9 |
| AT3G62910 | 0.00 | 2.19E + 06 | 3 | CPRF1 | Chloroplast ribosome release factor 1 | Ribosome availability | Plastid | None | 10.4 |
| ATCG00180 | 0.00 | 1.48E + 06 | 2 | RPOC1 | RNA polymerase β′ subunit-1 | RNA polymerase | Plastid | None | 3.8 |
| ATCG00190 | 0.00 | 2.78E + 06 | 3 | RPOB | RNA polymerase subunit β | RNA polymerase | Plastid | None | 3.8 |
| ATCG00740 | 0.00 | 2.87E + 06 | 3 | RPOA | RNA polymerase subunit α | RNA polymerase | Plastid | None | 10.9 |
| AT5G13030 | 0.00 | 5.78E + 06 | 7 | SELO | Selenoprotein O | ROS regulation | Plastid | None | 12 |
| AT1G32900 | 0.00 | 5.81E + 06 | 8 | GBSS1 | Granule-bound starch synthase 1 | Starch biosynthesis | Plastid | None | 19.7 |
| AT5G24300 | 0.00 | 7.06E + 06 | 5 | SS1 | Starch synthase 1 | Starch biosynthesis | Plastid | None | 10.1 |
| AT5G43780 | 0.00 | 2.67E + 06 | 7 | ATPS4 | ATP sulfurylase 4 | Sulfate assimilation | Plastid | None | 19 |
| AT3G21200 | 0.00 | 2.47E + 06 | 2 | PGR7 | Proton gradient regulation 7 | Tetrapyrrole biosynthesis | Plastid | None | 11.4 |
| AT1G22940 | 0.00 | 5.07E + 05 | 2 | TH1 | Thiamine-requiring 1 | Thiamine biosynthesis | Plastid | None | 5 |
| AT3G06730 | 0.00 | 2.07E + 06 | 2 | TRXZ | Thioredoxin Z | Thioredoxin | Plastid | None | 15.8 |
| AT4G29670 | 0.00 | 4.71E + 06 | 2 | ACHT2 | Atypical Cys–His-rich thioredoxin 2/TRX-lilium2 | Thioredoxin | Plastid | None | 16.6 |
| AT5G65840 | 0.00 | 6.78E + 06 | 4 | none | None | Thioredoxin | Plastid | None | 14.9 |
| AT2G42220 | 0.00 | 8.84E + 06 | 5 | STR9 | Sulfurtransferase 9 | Thiosulfate metabolism | Plastid | None | 26.1 |
| AT3G08920 | 0.00 | 3.14E + 06 | 3 | STR10 | Sulfurtransferase 10 | Thiosulfate metabolism | Mitochondrion | None | 17.8 |
| AT5G19370 | 0.00 | 1.46E + 07 | 9 | STR12 | Sulfurtransferase 12 | Thiosulfate metabolism | Plastid | None | 37.8 |
| AT4G27700 | 0.00 | 8.26E + 06 | 4 | STR14 | Sulfurtransferase 14 | Thiosulfate metabolism | Plastid | None | 24.1 |
| AT1G03160 | 0.00 | 1.58E + 06 | 3 | FZL | FZO-like | Thylakoid organization | Plastid | None | 4.8 |
| AT4G33760 | 0.00 | 3.88E + 06 | 4 | OKI | Okina kuki | tRNA synthesis | Plastid, cytosol, mitochondrion | None | 8.9 |
| AT1G23180 | 0.00 | 1.97E + 06 | 3 | None | ARM repeat superfamily protein | Unknown | Plastid, nucleus | None | 6.6 |
| AT3G62530 | 0.00 | 7.67E + 06 | 4 | None | ARM repeat superfamily protein | Unknown | Plastid, mitochondrion | None | 23.5 |
| AT2G13440 | 0.00 | 1.59E + 06 | 2 | None | Glucose-inhibited division family A protein | Unknown | Plastid, mitochondrion | None | 4.8 |
| AT5G07190 | 0.00 | 5.14E + 06 | 3 | ATS3 | Seed gene 3 | Unknown | extracellular | None | 27.6 |
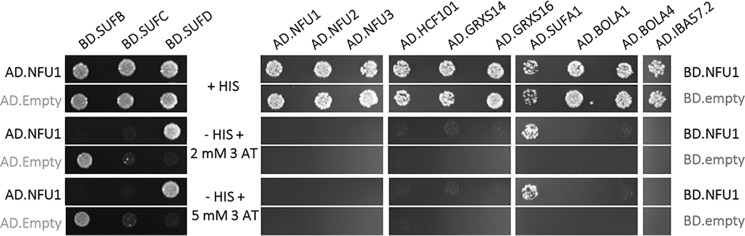
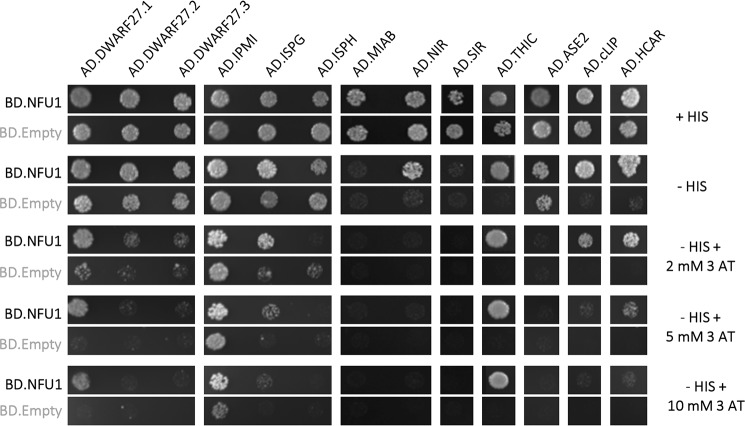
Hence, as a second more systematic approach, we sought to analyze the capacity of NFU1 to interact with the presumed scaffold and transfer proteins of the SUF machinery, i.e. SUFB, SUFC, SUFD, SUFA1, BOLA1, BOLA4, IBA57.2, GRXS14, GRXS16, NUF2, NFU3, and HCF101 using binary Y2H assays. An interaction was only observed with SUFD and SUFA1 (Fig. 2). Then, a large set of [4Fe-4S]-containing proteins involved in various metabolic pathways was tested. This included nitrite reductase (NIR) and sulfite reductase (SIR), two siroheme-containing proteins involved in nitrogen and sulfur assimilation. This also included several radical S-adenosylmethionine enzymes, i.e. THIC, the chloroplastic lipoate synthase (cLIP), which participates in the formation of lipoic acid and should incorporate two [4Fe-4S] clusters by analogy to the bacterial and mitochondrial orthologs and the tRNA-modifying enzyme, MIAB. Additional targets are the glutamine phosphoribosyl pyrophosphate amidotransferase 2 (ASE2) implicated in the de novo synthesis of purine, the ISPG/GcpE/HDS and the 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, ISPH, participating in the synthesis of isoprenoids, the β-carotene isomerase DWARF27.1 notably required for strigolactone synthesis and its two homologs (DWARF27.2/3), the IIL1/IPMI large subunit described above, and the 7-hydroxymethyl chlorophyll a reductase (HCAR), a protein involved in the chlorophyll metabolism and binding two [4Fe-4S] clusters. Among these 13 candidate partners, interactions have been detected with NIR, cLIP, THIC, HCAR, DWARF27.1, and ISPG when NFU1 was fused to the GAL4-binding domain (Fig. 3). In these experiments, we could not firmly confirm the existence of an interaction with the large subunit of IPMI because the GAL4 auto-activation caused by this protein is strong, and conclusions on the growth differences in the presence of NFU1 remain questionable. Looking at the strength of the interactions, the strongest ones were observed with THIC and DWARF27.1, then with ISPG, HCAR, and cLIP, and finally with NIR, with the latter being visible only in the absence of 3-aminotriazole (3-AT).
Figure 2.
Binary Y2H assays between Arabidopsis NFU1 and SUF components. The co-transformed yeast cells were plated at an OD600 of 0.05 on a control plate containing histidine (+His) and on test plates without histidine (−His) and containing 2 or 5 mm 3-AT. Yeast cells were grown for 5 days at 30 °C. Empty pGAD/pGBK-NFU1 and pGAD-NFU1/empty pGBK co-transformed yeast cells do not grow without histidine (data not shown). The interaction between NFU1 and SUFD is not visible when the other combination of chimeric constructs is used.
Figure 3.
Binary Y2H assays between Arabidopsis NFU1 and putative client proteins known to incorporate [4Fe-4S] clusters. The co-transformed yeast cells were plated at an OD600 of 0.05 on a control plate containing histidine (+His) and on test plates without histidine (−His), eventually containing 2, 5, or 10 mm 3-AT. Yeast cells were grown for 5 days at 30 °C. Empty pGAD/pGBK-NFU1 co-transformed yeast cells do not grow without histidine (data not shown).
To challenge the relevance of these interactions in plant cells, we additionally performed BiFC assays in Arabidopsis protoplasts. We have first validated NFU1 homodimerization as revealed by the BiFC signal in Arabidopsis chloroplasts in cells co-expressing NFU1–YFP fusions (i.e. NFU1 fused at the N terminus of the N-terminal (AA1–155) or C-terminal (AA156–239) regions of YFP (Fig. 4A and Figs. S3 and S4). We next tested combinations of NFU1 with 5 selected partners isolated by one or the other above-described approach, i.e. SUFA1, ISPG, THIC, SUFE3, and cLIP (Fig. 4B and Figs. S3 and S4). Transfections with combinations of NFU1 fused to the C-terminal region of the YFP protein (NFU1-C) and of the partners fused to the N-terminal region of the YFP (protein-N) revealed positive BiFC in all cases, exclusively in the chloroplasts. These experiments revealed that NFU1 cooperates in a close environment with all the proteins tested in this plant reporter system.
Figure 4.
BiFC assays between Arabidopsis NFU1 and its putative interactors in Arabidopsis leaf protoplasts. Arabidopsis protoplasts were transfected with combinations of two vectors expressing NFU1 with itself (A) or with its potential interactors upstream of the N- or C-terminal halves of YFP (B). Results shown are representative of at least two independent transfection assays for at least 20 cells per transfection. Scale bar, 10 μm. A, replacement of one of these constructs by an empty vector as negative controls gives no signal. B, NFU1-C was assayed individually with THIC-N, ISPG-N, SUFA1-N, SUFE3-N, and cLIP-N. Negative controls using an empty-C vector are shown separately in Fig. S4. The mid-values of argon laser intensities used are indicated. Protoplast transfections with vectors expressing the same combinations of proteins fused to the other YFP halves (NFU1-N and selected protein-C fusions) provided the same results with similar laser intensities (data not shown).
NFU1 transfers its [4Fe-4S] cluster to apo-ISPG and -THIC in vitro
Based on the identified interactions, we tested whether we could obtain evidence for an Fe-S cluster transfer in vitro using two client proteins, ISPG and THIC, as their Fe-S cluster content was characterized previously (38, 39). Moreover, both proteins have a single [4Fe-4S] cluster per monomer, unlike HCAR and cLIP, and they are formed by a single domain and do not rely on additional subunits, unlike SUFE3 and IPMI.
Hence, we produced the respective His-tagged recombinant proteins. After an aerobic purification, ISPG and THIC were both a mixture of apo- and holo-forms. By treating the proteins with an excess of EDTA and TCEP, we could obtain stable apo-proteins. Thus, we first assessed the ability of both apo-forms to incorporate [4Fe-4S] clusters by performing in vitro IscS-mediated Fe-S cluster reconstitution experiments (Fig. S5). Both reconstituted proteins had broad shoulders at ∼400 and ∼320 and nm characteristic of a [4Fe-4S]2+ center. This was quantitatively confirmed by analytical measurements of both iron and acid-labile sulfur atoms bound to the proteins as ISPG incorporated 3.99 ± 0.14 iron and 3.91 ± 0.11 labile sulfur per monomer, whereas THIC contained 3.81 ± 0.43 iron and 3.63 ± 0.63 labile sulfur per monomer after reconstitution.
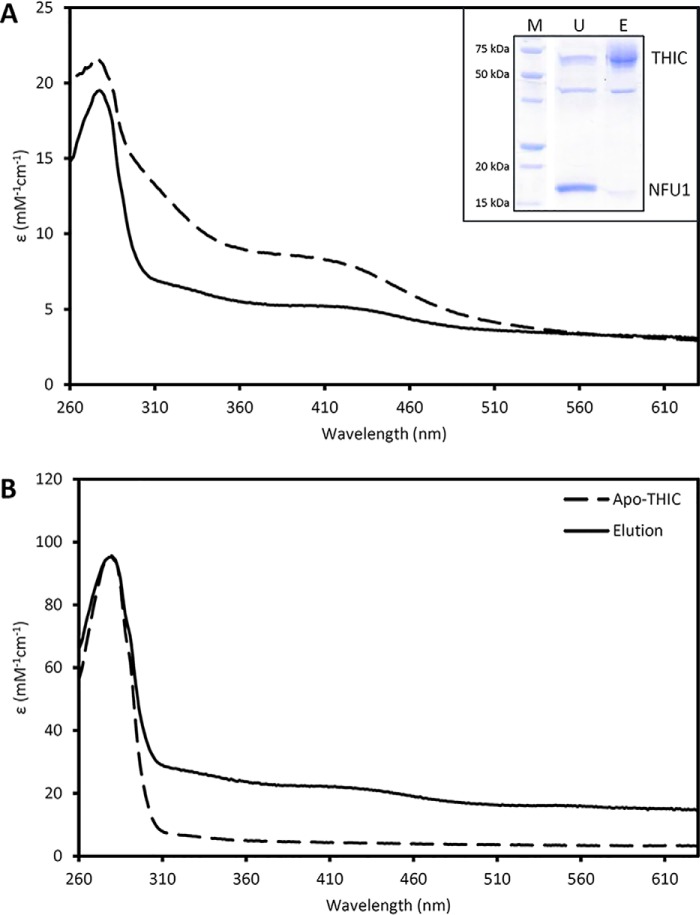
In a second step, in vitro Fe-S cluster transfer experiments were performed using a 2-fold molar excess of untagged holo-NFU1 (reconstituted as before) as compared with apo-acceptors. After a 1-h incubation, donor and acceptor proteins were separated on a nickel affinity chromatography column. The unbound fractions contained NFU1 and eventually tiny amounts of the acceptor proteins (Figs. 5 and 6). The UV-visible absorption spectra indicated that the absorption bands typical of the Fe-S cluster in NFU1 were considerably diminished, but not totally, due to the 2-fold excess of NFU1 [4Fe-4S] clusters. In contrast, the imidazole-eluted fractions, which contained ISPG or THIC, exhibited a brown-red color. The UV-visible absorption spectra showed broad bands centered around 400 and 320 nm (Figs. 5 and 6), similar to the ones observed in the spectra of reconstituted proteins (Fig. S5). To assess transfer efficiency, we titrated the Fe and acid-labile S contents into acceptor proteins (Table S4) and compared these values to the theoretical amounts expected based on the presence of a [4Fe-4S] cluster per monomer of ISPG and THIC. After the 1-h incubation time, we recovered ∼50 and ∼75% of holo-ISPG and holo-THIC, respectively. Altogether, these results pointed to an intact [4Fe-4S] cluster transfer from NFU1 to ISPG or THIC in vitro.
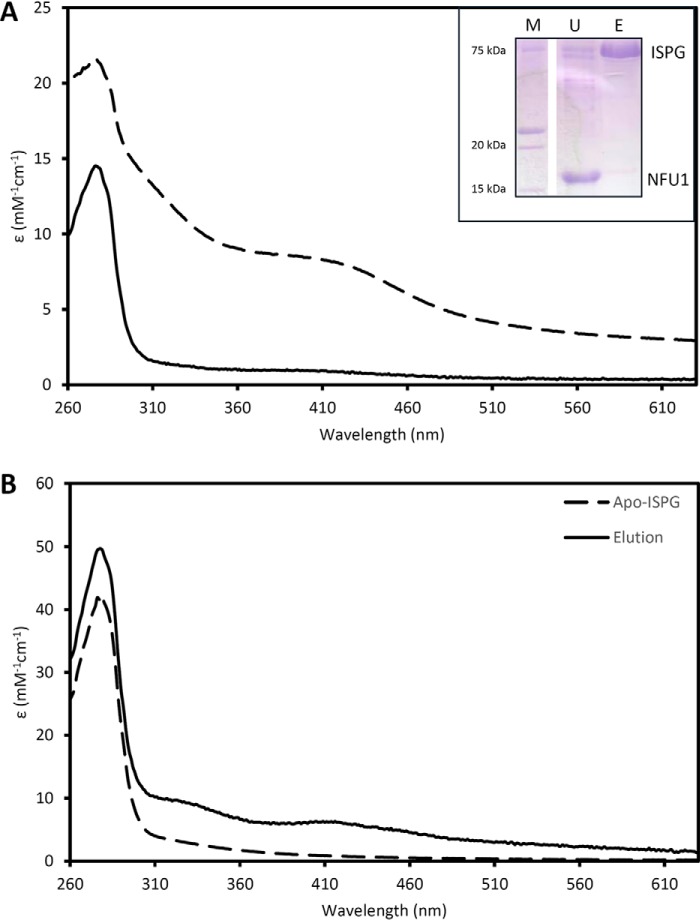
Figure 5.
In vitro Fe-S cluster transfer from holo-NFU1 to apo-ISPG monitored by UV-visible absorption spectroscopy. The transfer reaction was initiated by mixing 2 molar eq of holo-NFU1 (50 or 100 μm) relative to a reduced apo-ISPG (25 or 50 μm). After a 1-h incubation, the untagged NFU1 was separated from the His-tagged ISPG on IMAC. Inset at top shows an SDS-PAGE of the unbound (U) and eluted (E) fractions. M, molecular weight marker. A, UV-visible absorption spectra of IscS-reconstituted holo-NFU1 before (dashed line) and after incubation with apo-ISPG and IMAC separation (solid line). B, UV-visible absorption spectra of apo-ISPG prior (dashed line) and after incubation with holo-NFU1 and IMAC separation (solid line). The ϵ values are based on NFU1 and ISPG monomer concentrations.
Figure 6.
In vitro Fe-S cluster transfer from holo-NFU1 to apo-THIC monitored by UV-visible absorption spectroscopy. The transfer reaction was initiated by mixing 2 molar eq of holo-NFU1 (50 or 100 μm) relative to a reduced apo-THIC (25 or 50 μm). After a 1-h incubation, the untagged NFU1 was separated from the His-tagged THIC on IMAC. Inset at top shows an SDS-PAGE of the unbound (U) and eluted (E) fractions. M, molecular weight marker. A, UV-visible absorption spectra of IscS-reconstituted holo-NFU1 before (dashed line) and after incubation with apo-THIC and IMAC separation (solid line). B, UV-visible absorption spectra of apo-THIC prior (dashed line) and after incubation with holo-NFU1 and IMAC separation (solid line). The ϵ values are based on NFU1 and THIC monomer concentrations.
Discussion
The maturation of all chloroplastic Fe-S proteins depends on the SUF machinery. The late-acting Fe-S cluster transfer proteins, including NFU1/2/3 and HCF101, are, in principle, in direct contact with client proteins. Unlike NFU1, some of the NFU2, NFU3, and HCF101 targets have been unveiled (11, 22–24, 40). The fact that an A. thaliana nfu1 mutant has no macroscopic phenotype when grown under standard conditions prevented to delineate NFU1 function(s) and associated partners using a physiological approach (23).
In this work, we purified the Arabidopsis NFU1 protein to investigate which type of Fe-S cluster(s) is bound to the protein in vitro. Previous studies showed that the large diversity in the domain organization of NFU proteins is associated with differences in their biochemical properties. The Helicobacter pylori Nfu, formed by a single NFU domain, and the Arabidopsis NFU2, formed by two NFU domains (only the N-terminal one containing the ligating cysteine residues), were shown to bind either one [2Fe-2S] cluster as purified from E. coli cells or one [4Fe-4S] cluster upon in vitro reconstitution (29, 41). The E. coli and Azotobacter vinelandii NfuA that contain two domains, a C-terminal NFU domain fused to an N-terminal ISCA-type domain lacking the three conserved cysteine residues, were reported to bind only [4Fe-4S] clusters into homodimers (42, 43). In mitochondrial NFUs, the NFU domain is coupled to a domain of unknown function at its N terminus. The human NFU1 was shown to incorporate a [4Fe-4S] cluster upon reconstitution in a homodimer but a small-angle x-ray scattering–derived structural model highlighted the existence of a trimer of dimers (44, 45). These in vitro observations are in agreement with the in vivo investigations, which showed that only the maturation of [4Fe-4S] proteins, including respiratory complexes, aconitase and lipoate synthase, is affected in a yeast nfu1 mutant or in human patients (8, 45–47). Hence, the best and not to say the sole-documented example where both in vitro and in vivo results support the requirement of an NFU protein in the maturation of [2Fe-2S] proteins is for the plant NFU2–DHAD couple (23, 28). Concerning Arabidopsis NFU1, we have observed that it was purified as an apo-form after heterologous expression in E. coli and that a [4Fe-4S] cluster holo-dimeric-loaded form of NFU1 was uniquely formed after an in vitro anaerobic IscS-mediated reconstitution. NFU1 homodimerization was also visible in plant cells using BiFC but not in the yeast nuclear context during Y2H assays. Overall, this suggests that NFU1 should not participate in the maturation of [2Fe-2S] clusters, but only to the maturation of [4Fe-4S] and possibly of [3Fe-4S] clusters.
The question of the NFU1 partners among SUF components was unsolved so far. We have obtained evidence for possible interactions with SUFD, SUFA1, GRXS16, and HCF101. As already discussed, the detection in the co-IP experiments of HCF101 and GRXS16 as putative NFU1 partners, although they were not found to physically interact by Y2H and/or BiFC, raises some doubt about the existence of a direct contact. For this reason, these interactions are not represented in the updated model proposed in Fig. 7. However, the interaction with SUFD and SUFA1 seen in Y2H and in Y2H and BiFC experiments, respectively, suggests that NFU1 directly receives its cluster from the SUFBCD scaffold complex, before exchanging its cluster with SUFA1, the sole A-type transfer protein in chloroplasts. Accordingly, E. coli NfuA is able to receive its Fe-S cluster from both the ISCU and SUFBC2D scaffold and to interact with all A-type transfer proteins, having the ability to transfer its cluster in vitro to SufA, IscA, and ErpA, and forming a complex with ErpA stabilizing its Fe-S cluster (43, 48, 49). However, the model is that ErpA, and NfuA to a lesser extent, are the final Fe-S cluster donors to client apo-proteins (49). A [4Fe-4S] cluster transfer from the NFU domain of A. vinelandii NifU to NIFIscA is also documented (50). Noteworthy, E. coli SufA also receives an Fe-S cluster from SUFBC2D (51) and is competent, for instance, for the maturation of both [2Fe-2S]- and [4Fe-4S]-containing proteins as shown using ferredoxin and biotin synthase (52, 53). This raises the possibility that, in chloroplasts, SUFA1 may obtain its cluster independently of NFU1 and, on the contrary, deliver it to NFU1. This sequence order would fit with the mitochondrial model in which NFUs likely act downstream of the A-type transfer proteins, ISCA1/ISA1 and ISCA2/ISA2. In vitro experiments provided evidence that these A-type transfer proteins exist as homodimers, as ISCA1/2 heterodimers, but also as ISCA-IBA57 heterodimers (6, 54, 55). In fact, it seems that an even higher level of flexibility and adaptability exists for A-type transfer proteins as most of the characterized homo- or heterodimers are able to incorporate either [2Fe-2S] or [4Fe-4S] clusters. Hence, whereas the interaction between NFUs and A-type transfer proteins is a robust observation made in both prokaryote and eukaryote organisms (8, 43, 49, 56), which of the NFU1 or SUFA1 acts upstream remains uncertain in the chloroplastic SUF system. So far, SUFA1 was characterized as a [2Fe-2S] cluster–containing protein able to receive in vitro an Fe-S cluster from GRXS14 and to transfer it to a ferredoxin (17, 31, 57). Hence, an Fe-S cluster exchange from NFU1 to SUFA1 would necessitate an oxidative conversion from the [4Fe-4S]-loaded NFU1 into [2Fe-2S]-loaded SUFA1 forms, unless SUFA1 also binds [4Fe-4S] clusters. Alternatively, a reductive Fe-S cluster conversion from a [2Fe-2S]-loaded SUFA1 to a [4Fe-4S]-loaded NFU1 might be possible. Further experiments are needed to address the direction in which these exchanges occur.
Figure 7.
Positioning of NFU1 and its partner proteins in the current model of the SUF machinery. The previous model (23) for the plastidial SUF machinery was implemented with recent results, including those reported in this paper for NFU1. The approaches used for defining these interactions are depicted with a color code, as visible on the scheme. A question mark remains for the interaction between the NFU1 and IPMI large subunit, because we could not firmly establish it. For the sake of clarity, other candidate SUF components i.e. BOLA1/4, GRXS14/16, and IBA57.2, have not been represented here because their position is unclear.
The question of whether NFU1 has direct client Fe-S proteins was also the purpose of this study. By combining several complementary approaches, we have identified 9 NFU1 partners, if we consider the interaction with IPMI as too uncertain. They incorporate either a [3Fe-4S] cluster in the case of GLU2 or one or several [4Fe-4S] clusters in the case of SUFE3, APR2, cLIP, DWARF27.1, HCAR, ISPG, NIR, and THIC. Although we could not test or obtain evidence for all of these interactions by all methods, most of the interactions have been observed using at least two different approaches. Whereas the chloroplastic lipoate synthase cLIP has not been investigated much so far, the requirement of NFU1 as a maturation factor is consistent with NFU-type proteins being required for the maturation or repair of Fe-S clusters in lipoate synthase present in bacteria, or in yeast and human mitochondria (8, 58). The interactions of NFU1 with THIC and ISPG were evidenced both in vivo by Y2H and BiFC and in vitro from the observation of an intact [4Fe-4S] cluster transfer from a holodimeric NFU1. The fact that THIC was also retrieved from the co-IP experiments shows little doubt about the validity of this interaction. Concerning ISPG, the result is consistent with the observation that E. coli NfuA is also a required maturation factor for bacterial IspG/H enzymes as recently demonstrated by a genetic approach (49). Taken together, these results indicate that the plastidial NFU1 of Arabidopsis is involved in maturation pathways that are conserved throughout evolution. For other proteins specific to plants, such as DWARF27.1 and HCAR, obtaining data about their maturation will be crucial as no extrapolation is possible from other model organisms. Only HCAR was analyzed before by Western blottings in sufb, sufc, sufd, nfu2, and hcf101 Arabidopsis mutants (11). The protein level was diminished in sufb, sufc, and sufd RNAi lines, although it was not totally absent, but it did not vary much in nfu2 and hcf101. This leads us to discuss the physiological consequences of the described interactions and possible redundancies with other SUF maturation factors. The sufa1 and nfu1 Arabidopsis mutants have no or only weak growth phenotypes under standard conditions, which is also the case for apr2 or hcar mutants (59, 60). Nevertheless, mutants for other identified proteins have either strong(er) growth phenotypes, for instance the albino phenotype of an ispg mutant (61), or even embryo- or seedling-lethal phenotypes as observed for sufe3 and thic mutants, respectively (12, 33). These phenotypic differences indicate indeed the existence of back-up system(s) for NFU1, meaning a certain level of redundancy with other maturation factors. This would be totally in line with the E. coli model, in which several factors serve for the maturation of a single target, and their nature may depend on the growth conditions (62). For instance, in E. coli, depending on stress conditions, either NfuA or ErpA is involved in the maturation of ISPG (49). Hence, determining whether the functions of NFU1 are restricted to stress conditions or are more prominent in this context is likely now required. In plant chloroplasts, the most likely candidates for ensuring functions similar to NFU1 are obviously NFU2, NFU3, but also HCF101, because they assemble the same type of cluster and they are critical for plant growth (22, 29, 40). From a phylogenetic point of view, NFU1 and NFU2/3 form two separate phylogenetic clades with the gene duplication generating NFU2 and NFU3 having occurred in an ancestor of angiosperms. For instance, a single NFU2/3 representative is present in chlorophyceae, bryophytes, and lycophytes whereas there are two in monocots and dicots (3, 23). Assuming that NFU1/2/3 originate from a single ancestral gene, these proteins may have diverged to some extent but also conserved common biochemical and structural properties. In support of this conclusion, all three plastidial NFUs were able to restore the growth defect of a yeast mutant for the mitochondrial Nfu1, despite the difference in protein organization between NFUs present in both organelles (21). However, the functions of NFU1 and NFU2/3 diverged and are clearly not fully overlapping. Unlike NFU1, NFU2 and NFU3 are involved in PSI maturation together with HCF101 but are also biochemically competent for the maturation of the [2Fe-2S] cluster in DHAD (23). Additional functions of NFU2/3 may actually have been masked by the strong effect on PSI and by their redundancy (a double nfu2 nfu3 mutant is lethal (23). Hence, to address the question of redundancy among NFUs, it would be mandatory to perform similar experiments with NFU2 and NFU3 in order to identify their set of client proteins.
Experimental procedures
Heterologous expression in E. coli and purification of recombinant proteins
The sequences coding for the presumed mature forms (i.e. devoid of N-terminal targeting sequences) of Arabidopsis NFU1, THIC, and ISPG were cloned, respectively, into the NdeI and BamHI restriction sites of pET12a, the NcoI–XhoI and NdeI–XhoI restriction sites of pET28a (Table S2), in order to produce an untagged NFU1, an N-terminal His-tagged THIC, and a C-terminal His-tagged ISPG.
NFU1 was expressed in the E. coli BL21 (DE3) strain containing the pSBET plasmid (63). Protein expression in 3.2 liters was induced by adding of 100 μm isopropyl β-d-thiogalactopyranoside (IPTG) during exponential growth phase. After 4 h at 37 °C, cells were collected by centrifugation for 20 min at 6,318 × g, and the cell pellets were resuspended in about 25 ml of TN buffer (30 mm Tris-HCl, pH 8.0, 200 mm NaCl). Bacterial cells were lysed by sonication (three times for 1 min), and the cell debris were removed at 4 °C by centrifugation for 30 min at 27,216 × g. The soluble fraction was sequentially precipitated by ammonium sulfate to 40% and then to 80% of the saturation. NFU1 was recovered mainly in the 0–40% ammonium sulfate fraction. This fraction was subjected to gel-filtration chromatography (ACA44) equilibrated with TN buffer. NFU1-containing fractions were pooled, concentrated, and dialyzed against 30 mm Tris-HCl, pH 8.0, buffer by ultrafiltration (YM10 membrane) under nitrogen pressure using an Amicon cell. The sample was loaded on an ion-exchange chromatography (DEAE-Sepharose column) equilibrated in 30 mm Tris-HCl, pH 8.0, buffer before applying a linear 0–0.4 m NaCl gradient. The purest fractions containing NFU1 as judged by SDS-PAGE analysis were pooled and dialyzed against 30 mm Tris-HCl, pH 8.0, buffer by ultrafiltration. Finally, the fractions were concentrated and stored at −20 °C until further use.
The His-tagged ISPG and THIC were expressed in the E. coli Rosetta2 (DE3) strain. Protein expression was achieved in a 3.2-liter culture. After an initial growth phase at 37 °C up to the exponential phase (OD600 = 0.6–0.8), flasks were placed for 2 h at 4 °C in the presence of 0.5% ethanol before induction by 100 μm IPTG. The cultures were further grown for about 18 h at 20 °C. Cells were collected by centrifugation for 20 min at 6,318 × g and resuspended in about 25 ml of TN buffer plus 20 mm imidazole (TNI20). Bacterial cells were lysed by sonication (three times for 1 min), and soluble and insoluble cell fractions were separated by centrifugation for 30 min at 27,216 × g at 4 °C. The soluble fraction was then loaded onto a Ni-NTA affinity column (Qiagen) equilibrated in TNI20 buffer. After extensive washing, recombinant proteins were eluted in TN buffer containing 250 mm imidazole. Proteins were then concentrated and dialyzed against TN buffer by ultrafiltration under nitrogen pressure (Amicon, YM10 membrane) and stored at −20 °C. The purity of each recombinant protein was checked on SDS-PAGE. The concentrations of apo-proteins were determined spectrophotometrically using the theoretical molecular extinction coefficient at 280 nm of 4595 m−1 cm−1 for NFU1, 41,425 m−1cm−1 for ISPG, and 92,290 m−1 cm−1 for THIC.
In vitro IscS-mediated reconstitution of Fe-S cluster
All experiments were done at room temperature under an anaerobic atmosphere using a Jacomex glovebox (O2 <2 ppm). Reduced apo-NFU1, ISPG, and THIC were obtained by treating purified proteins with a 20-fold excess of TCEP and a 50-fold excess of EDTA for 1 h or overnight in the case of NFU1, which allowed us to get rid of residual polysulfide visible from the presence of a shoulder at 320 nm (64), before desalting on a G-25 column pre-equilibrated with 30 mm Tris-HCl, pH 8.0, buffer. The Fe-S cluster reconstitution was performed in 500 μl of 37.5 mm Tris-HCl, pH 8.0, 37.5 mm NaCl buffer, using 50 μm protein, a 20-fold excess of l-cysteine and ammonium iron(II) sulfate hexahydrate, and a catalytic amount of E. coli cysteine desulfurase IscS purified as described previously (36). The reaction was initiated by the addition of IscS and followed by monitoring the UV-visible absorption spectrum. After a 1-h reaction, the Fe-S cluster-loaded proteins were desalted on a G-25 column equilibrated with TN buffer.
Determination of the oligomerization state of NFU1
The oligomerization state of apo- and holo-NFU1 was determined by size-exclusion chromatography. Samples containing about 200 μg of protein were loaded onto a Sephadex S200 10/300 column equilibrated in TN buffer and connected to an Akta purifier system (GE Healthcare). Proteins were detected by recording absorption at 280 and 420 nm. The column was calibrated using a molecular weight standard from Sigma. Elution volume, protein name, and molecular mass are as follows: 8.47 ml of thyroglobulin, 669 kDa; 10.51 ml of apo-ferritin, 443 kDa; 11.92 ml of β-amylase, 200 kDa; 13.89 ml of BSA, 66 kDa; 15.74 ml of carbonic anhydrase, 29 kDa; 17.01 ml of cytochrome c, 12.4 kDa; and 18.28 ml of aprotinin, 6.5 kDa.
In vitro Fe-S cluster transfer experiments
Under strictly anaerobic conditions in a Jacomex glovebox (O2 <2 ppm), 2 molar eq of reconstituted holo-NFU1 (50 or 100 μm) respective to reduced apo-ISPG or apo-THIC (25 or 50 μm) were mixed for 1 h in the absence or presence of a 10-fold excess of EDTA to ensure that the Fe-S cluster is transferred intact and not upon degradation and reassembly. Untagged NFU1 and tagged acceptor proteins (ISPG or THIC) were then separated on a Ni-NTA affinity column (IMAC-Qiagen) equilibrated in TNI20 buffer. The column was then washed with 6 column volumes of TNI20 buffer, and elution was performed using TN buffer containing 250 mm imidazole. Fractions corresponding to washing and elution steps were concentrated to a volume of 500 μl using Vivaspin 500 centrifugal filters, before recording UV-visible absorption spectra and quantifying the contents in iron and acid-labile sulfide. Aliquots of each fraction were also analyzed by SDS-PAGE.
Spectroscopic methods
UV-visible absorption spectra were recorded using Shimadzu UV-3101 PC scanning or Agilent Cary 60 spectrophotometers. CD spectra were recorded using a JASCO J-715 spectropolarimeter. Septa-sealed quartz cuvette cells with either a 1-mm or a 1-cm path length were used for both absorption and CD spectroscopies. Resonance Raman spectra were acquired using a Raman or U1000 scanning spectrometer (Instruments SA, Edison, NJ) fitted with a cooled photomultiplier tube and photon-counting electronics (Instruments SA, Edison, NJ), using excitation lines from a Sabre argon laser (Coherent, Santa Clara, CA). A droplet of concentrated sample (∼2 mm in Fe-S clusters) was frozen at 17 K on a gold-plated sample holder mounted to a cold finger of a Displex Model CSA-202E closed cycle helium refrigerator (Air Products, Allentown, PA).
Iron and acid-labile sulfide quantification
Protein concentrations were determined using the colorimetric bicinchoninic acid assay kit as recommended (Interchim). For iron quantification, different volumes of protein (25, 50, and 100 μl) were diluted in 130 μl of water. Proteins were precipitated by adding 90 μl of 70% (v/v) perchloric acid for 10 min at room temperature after strong shaking. After centrifugation (10 min, 11,600 × g), 180 μl of supernatant were mixed with 144 μl of 3.2 mm bathophenanthroline disulfate, 72 μl of 192 mm sodium ascorbate, and 152 μl of 6.2 m ammonium acetate. The mixture was homogenized with 30 s of vortex before 30 min of incubation at room temperature. Iron amounts were determined by subtracting the nonspecific absorbance at 680 nm from the specific absorbance of the ferrous iron–chelator complex measured at 535 nm relative to a standard curve obtained with ammonium iron(II) sulfate (0–20 nmol).
For acid-labile sulfide quantification, 25, 50, and 100 μl of proteins (eventually brought to 100 μl with water) were mixed with 300 μl of 1% (w/v) zinc acetate and 15 μl of 3 m NaOH. The mixture was incubated for 10 min at room temperature before adding 50 μl of 20 mm N,N-dimethyl-p-phenylenediamine (prepared in 7.2 m HCl) and 50 μl of 30 mm FeCl3 (prepared in 1.2 m HCl). After 30 s of shaking and incubation at 4 °C for 3 h, the mixture was centrifuged for 5 min at 11,600 × g, and the presence of methylene blue in the supernatant was recorded at 670 nm. Lithium sulfide (0–20 nmol) was used for the calibration curve.
Binary yeast two-hybrid assays
The experiments have been performed with the Gal4-based yeast two-hybrid reporter strain CY306 (65). The sequences coding for proteins devoid of their chloroplastic targeting sequences were cloned in both pGADT7 and pGBKT7 vectors (Clontech) using NcoI-, NdeI-, BamHI-, or XhoI-containing primers (Table S2). Binary interactions were tested using both the activator domain for Gal4 (AD) and the DNA-binding domain for Gal4 (BD) fusion combinations. Cells co-transformed with pGAD- and pGBK-based constructs were selected on minimal YNB medium (0.7% yeast extract without amino acids, 2% glucose, and 2% agar) containing the required amino acids and bases (histidine, adenine, lysine, uracil, and methionine). Interactions were assessed through cell growth on selective YNB media, in the absence of histidine and in the absence or presence of 2–10 mm 3-AT, to get rid of some trans-activating constructs and estimate the strength of each interaction. Negative controls were performed using co-transformations of AD or BD fusions with empty vectors. Each dot is a 7-μl drop of cultures adjusted at an OD600 = 0.05. Representative images shown here were taken 5 days post-dotting.
Bimolecular fluorescence complementation
All proteins selected for BiFC analyses were cloned as full-length open reading frames upstream of the C- and N-terminal regions of the YFP protein into the pUC-SPYCE and pUC-SPYNE vectors (abbreviated as -C and -N in the figures, respectively), using a restriction site-based strategy and the primers listed in Table S3 (66). Leaf protoplasts were prepared from 21- to 28-day-old Arabidopsis plantlets grown in growth chambers in short-day conditions (8 h light/23 °C/217 μmol·m−2·s−1; 16 h dark/20 °C, 65% humidity) and transfected according to Ref. 67 using 10 μg of each pUC-SPYCE and pUC-SPYNE construct expressing selected proteins. YFP fluorescence in Arabidopsis protoplast cells was recorded 20–24 h post-transfection by using a Leica TCS SP8 confocal laser-scanning microscope. YFP was excited with an argon laser at 514 nm and detected at 520–550 nm whereas chlorophyll autofluorescence was monitored at 680–720 nm after excitation at 561 nm. Images were obtained using LAS X software and treated with Adobe Photoshop CS3 at high resolution. Results are representative of at least two independent transfection experiments, including the analysis of around 20 cells per transformation event.
DNA constructs
To generate ProNFU1::gNFU1-GFP–expressing plants, the NFU1 locus (2000 bp prior to the start codon until the end of the coding sequence without the STOP) was amplified with AttB1ProNFU1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGCAGTACCCTAAACCATTG-3′) and AttB2NFU1 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGCTTGTAAAGGTTAC-3′) primers, cloned in pDONR207 vector, and recombined in pGWB4 (68). To generate plants expressing a stroma-targeted GFP (Pro35S::CTPNFU3-GFP), the NFU3 chloroplastic peptide signal was amplified with AttB1NFU3 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGGTTCTGTTTCGGGTC-3′) and AttB2-PS-NFU3 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGCAGCTCACGTGACCAAATAC-3′) primers, cloned in pDONR207, and recombined in pGWB505 (derived from pGWB series (68)). All the PCR products were obtained using high-fidelity Phusion DNA polymerase, and each construct in pDONR207 was sequenced to ensure its integrity.
Co-immunoprecipitation experiments
ProNFU1::gNFU1–GFP- and Pro35S::CTPNFU3-GFP–expressing seedlings were germinated and grown under long day conditions (16/8 h light/dark) on half-strength Murashige and Skoog medium (MS/2) with 0.05% MES, 1% sucrose, 0.7% agar. One g of aerial tissues from 2-week-old seedlings was cross-linked in 1% formaldehyde in PBS buffer two times (7 min under vacuum). The reaction was blocked by adding 300 mm glycine (30 min under vacuum). Fixed tissues were rinsed three times with water, dried, and frozen in liquid nitrogen prior to grinding. Powder was resuspended in 2 ml of RIPA buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate) and centrifuged two times for 10 min at 14,000 × g to remove cell debris. 50 μl of antibodies raised against GFP coupled to magnetic beads (Miltenyi Biotec®) were added to the supernatant and mixed (wheel rotation) for 30 min at 4 °C. Tubes were then placed on a magnetic rack, and after four washes with RIPA buffer, proteins were eluted with 100 μl of 1× Laemmli solution (65 mm Tris-HCl, pH 7.5, 5% glycerol, 2% SDS, 125 mm DTT). These manipulations were done on four biological replicates per genotype.
Mass spectrometry analysis
To analyze co-IP samples by MS, eluted proteins were loaded on a 10% precast acrylamide gel (Bio-Rad) for a short run (15 min, 100 V). The whole band was manually excised from the gel and cut into small pieces. After sequential washes with 25 mm ammonium bicarbonate, 50% acetonitrile in 25 mm ammonium bicarbonate, and 100% acetonitrile, thiol groups of cysteines were reduced with 10 mm DTT for 45 min and alkylated for 30 min with 55 mm iodoacetamide. The bands were then sequentially washed with 50% acetonitrile in 25 mm ammonium bicarbonate and with 100% acetonitrile. The proteins were then digested with 0.25 μg of trypsin (Sequencing Grade Modified, Promega) in 25 mm ammonium bicarbonate overnight at 37 °C. Peptides were eluted first with 2% formic acid and twice with 80% acetonitrile in 2% formic acid. Supernatants were pooled and evaporated in a vacuum centrifuge. Peptides were resuspended in 8 μl of 2% formic acid, and 6 μl were injected for LC-MS/MS analyses. They were performed using an Ultimate 3000 RSLC nano system (Thermo Fisher Scientific, Waltham, MA) interfaced on line with a nano easy ion source and a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific). The samples were analyzed in data-dependent acquisition (DDA). Protein digests were first loaded onto a pre-column (Thermo Fisher Scientific, PepMap 100 C18, 5-μm particle size, 100-Å pore size, 300-μm inner diameter × 5-mm length) at a flow rate of 10 μl/min for 3 min.
The peptides were separated on a reverse-phase column (Thermo Fisher Scientific, PepMap C18, 2-μm particle size, 100-Å pore size, 75-μm inner diameter × 50-cm length) at a flow rate of 300 nl/min. Loading buffer (solvent A) was 0.1% formic acid in water, and elution buffer (solvent B) was 0.1% formic acid in 80% acetonitrile. The linear gradient employed was 2–25% of solvent B for 103 min, then 25–40% of solvent B from 103 to 123 min, and finally 40–90% of solvent B from 123 to 125 min. The total run time was 150 min, including a high organic wash step and re-equilibration step. The Q Exactive Plus mass analyzer was operated in positive electrospray ionization mode at 1.8 kV. In DDA, the top 10 precursors were acquired between 375 and 1500 m/z with a 2-Thomson selection window, dynamic exclusion of 40 s, normalized collision energy of 27, and resolutions of 70,000 for MS and 17,500 for MS2. Raw mass spectrometric data were analyzed in the MaxQuant environment (69), version 1.5.0.0, and Andromeda was employed for database search (70). The MS/MS data were matched against the TAIR10 + GFP database (35,417 entries). The “Trypsin/P” criterion was chosen as digestion enzyme. Up to two missed cleavages were allowed for protease digestion. For protein identification and quantification, cysteine carbamidomethylation was set up as a fixed modification and oxidation of methionine as a variable modification. Mass tolerance for precursor ions was 20 and 4.5 ppm for the first and the main searches, respectively, and it was 20 ppm for the fragment ions. At least two peptides are necessary for protein identification and quantification. A peptide–spectrum match false discovery rate (FDR) and a protein FDR below 0.01 were required. Using the above criteria, the rates of false peptide sequence assignment and false protein identification were lower than 1%. For the other characteristics, MaxQuant default parameters were used. Intensities without normalization were considered, and a protein was considered as a putative interactant if it was found in at least three replicates in ProNFU1::NFU1g-GFP lines and not in three or four replicates of Pro35S::CTPNFU3-GFP lines. The values of Pearson correlation coefficients calculated between each biological replicate were between 0.75 and 0.85. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (71) partner repository with the dataset identifier PXD015295.
Author contributions
M. R., J. P.-T., F. V., N. B., L. C., T. D., M. K. J., J. C., and N. R. conceptualization; M. R., J. P.-T., F. V., N. B., and J. C. data curation; M. R., J. P.-T., F. V., N. B., T. A., L. C., H.-C. W., and T. D. methodology; M. R., J. C., and N. R. writing-original draft; J. P.-T., F. V., N. B., H.-C. W., and T. D. formal analysis; V. S., M. K. J., C. D., J. C., and N. R. supervision; V. S., M. K. J., C. D., J. C., and N. R. validation; M. K. J. and C. D. writing-review and editing; J. C. and N. R. funding acquisition; N. R. project administration.
Supplementary Material
Acknowledgments
We thank Carine Alcon and the Montpellier Rio-Imaging and PHIV platforms for expertise and assistance in confocal microscopy.
This work was supported by Agence Nationale de la Recherche as part of the “Investissements d'Avenir” Program Grant ANR-11-LABX-0002-01, Laboratory of Excellence ARBRE, and Grant ANR-2013-BSV6-0002-01 and by National Institutes of Health Grant R37GM62524 (to M. K. J.). The authors declare that they have no conflicts of interest with the contents of this article.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier accession no. PXD015295.
This article contains Figs. S1–S5 and Tables S1–S4.
- ISC
- iron–sulfur cluster
- 3-AT
- 3-aminotriazole
- Y2H
- yeast-two hybrid
- BiFC
- bimolecular fluorescence complementation
- YFP
- yellow fluorescent protein
- CTP
- chloroplastic targeting peptide
- ISPG
- 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase
- IMAC
- immobilized metal-affinity chromatography
- THIC
- 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate synthase
- SUF
- sulfur mobilization
- Ni-NTA
- nickel-nitrilotriacetic acid
- cLIP
- chloroplastic lipoate synthase
- GRX
- glutaredoxin
- IPTG
- isopropyl β-d-thiogalactopyranoside
- PSI
- photosystem I
- DHAD
- dihydroxyacid dehydratase
- IPMI
- isopropyl malate isomerase
- TRX
- thioredoxin
- NIR
- nitrite reductase
- HCAR
- hydroxymethyl chlorophyll a reductase
- AA
- amino acid
- AD
- activator domain
- BD
- binding domain
- DDA
- data-dependent acquisition
- FDR
- false discovery rate
- co-IP
- co-immunoprecipitation
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1. Balk J., and Schaedler T. A. (2014) Iron co-factor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153 10.1146/annurev-arplant-050213-035759 [DOI] [PubMed] [Google Scholar]
- 2. Przybyla-Toscano J., Roland M., Gaymard F., Couturier J., and Rouhier N. (2018) Roles and maturation of iron–sulfur proteins in plastids. J. Biol. Inorg Chem. 23, 545–566 10.1007/s00775-018-1570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Couturier J., Touraine B., Briat J.-F., Gaymard F., and Rouhier N. (2013) The iron–sulfur cluster assembly machineries in plants: current knowledge and open questions. Front. Plant Sci. 4, 259 10.3389/fpls.2013.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lill R. (2009) Function and biogenesis of iron–sulphur proteins. Nature 460, 831–838 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- 5. Braymer J. J., and Lill R. (2017) Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 292, 12754–12763 10.1074/jbc.R117.787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brancaccio D., Gallo A., Mikolajczyk M., Zovo K., Palumaa P., Novellino E., Piccioli M., Ciofi-Baffoni S., Banci L. (2014) Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J. Am. Chem. Soc. 136, 16240–16250 10.1021/ja507822j [DOI] [PubMed] [Google Scholar]
- 7. Bych K., Kerscher S., Netz D. J., Pierik A. J., Zwicker K., Huynen M. A., Lill R., Brandt U., and Balk J. (2008) The iron–sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27, 1736–1746 10.1038/emboj.2008.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melber A., Na U., Vashisht A., Weiler B. D., Lill R., Wohlschlegel J. A., and Winge D. R. (2016) Role of Nfu1 and Bol3 in iron–sulfur cluster transfer to mitochondrial clients. Elife 5, e15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheftel A. D., Wilbrecht C., Stehling O., Niggemeyer B., Elsässer H.-P., Mühlenhoff U., and Lill R. (2012) The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell. 23, 1157–1166 10.1091/mbc.e11-09-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hjorth E., Hadfi K., Zauner S., and Maier U.-G. (2005) Unique genetic compartmentalization of the SUF system in cryptophytes and characterization of a SufD mutant in Arabidopsis thaliana. FEBS Lett. 579, 1129–1135 10.1016/j.febslet.2004.12.084 [DOI] [PubMed] [Google Scholar]
- 11. Hu X., Kato Y., Sumida A., Tanaka A., and Tanaka R. (2017) The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe-S proteins. Plant J. 90, 235–248 10.1111/tpj.13483 [DOI] [PubMed] [Google Scholar]
- 12. M N. M., Ollagnier-de-Choudens S., Sanakis Y., Abdel-Ghany S. E., Rousset C., Ye H., Fontecave M., Pilon-Smits E. A., and Pilon M. (2007) Characterization of Arabidopsis thaliana SufE2 and SufE3 functions in chloroplast iron–sulfur cluster assembly and NAD synthesis. J. Biol. Chem. 282, 18254–18264 10.1074/jbc.M701428200 [DOI] [PubMed] [Google Scholar]
- 13. Nagane T., Tanaka A., and Tanaka R. (2010) Involvement of AtNAP1 in the regulation of chlorophyll degradation in Arabidopsis thaliana. Planta 231, 939–949 10.1007/s00425-010-1099-8 [DOI] [PubMed] [Google Scholar]
- 14. Van Hoewyk D., Abdel-Ghany S. E., Cohu C. M., Herbert S. K., Kugrens P., Pilon M., and Pilon-Smits E. A. (2007) Chloroplast iron–sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc. Natl. Acad. Sci. U.S.A. 104, 5686–5691 10.1073/pnas.0700774104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu X. M., and Møller S. G. (2006) AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 25, 900–909 10.1038/sj.emboj.7600968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu X. M., and Møller S. G. (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 9143–9148 10.1073/pnas.0400799101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdel-Ghany S. E., Ye H., Garifullina G. F., Zhang L., Pilon-Smits E. A., and Pilon M. (2005) Iron–sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 138, 161–172 10.1104/pp.104.058602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Léon S., Touraine B., Ribot C., Briat J.-F., and Lobréaux S. (2003) Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem. J. 371, 823–830 10.1042/bj20021946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waller J. C., Ellens K. W., Alvarez S., Loizeau K., Ravanel S., and Hanson A. D. (2012) Mitochondrial and plastidial COG0354 proteins have folate-dependent functions in iron–sulphur cluster metabolism. J. Exp. Bot. 63, 403–411 10.1093/jxb/err286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bandyopadhyay S., Gama F., Molina-Navarro M. M., Gualberto J. M., Claxton R., Naik S. G., Huynh B. H., Herrero E., Jacquot J. P., Johnson M. K., and Rouhier N. (2008) Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe–2S] clusters. EMBO J. 27, 1122–1133 10.1038/emboj.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uzarska M. A., Przybyla-Toscano J., Spantgar F., Zannini F., Lill R., Mühlenhoff U., and Rouhier N. (2018) Conserved functions of Arabidopsis mitochondrial late-acting maturation factors in the trafficking of iron–sulfur clusters. Biochim. Biophys. Acta 1865, 1250–1259 10.1016/j.bbamcr.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 22. Nath K., Wessendorf R. L., and Lu Y. (2016) A nitrogen-fixing subunit essential for accumulating 4Fe-4S-containing photosystem I core proteins. Plant Physiol. 172, 2459–2470 10.1104/pp.16.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Touraine B., Vignols F., Przybyla-Toscano J., Ischebeck T., Dhalleine T., Wu H.-C., Magno C., Berger N., Couturier J., Dubos C., Feussner I., Caffarri S., Havaux M., Rouhier N., and Gaymard F. (2019) Iron–sulfur protein NFU2 is required for branched-chain amino acid synthesis in Arabidopsis roots. J. Exp. Bot. 70, 1875–1889 10.1093/jxb/erz050 [DOI] [PubMed] [Google Scholar]
- 24. Touraine B., Boutin J.-P., Marion-Poll A., Briat J.-F., Peltier G., and Lobréaux S. (2004) Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 40, 101–111 10.1111/j.1365-313X.2004.02189.x [DOI] [PubMed] [Google Scholar]
- 25. Yabe T., Morimoto K., Kikuchi S., Nishio K., Terashima I., and Nakai M. (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron–sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007 10.1105/tpc.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lezhneva L., Amann K., and Meurer J. (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37, 174–185 10.1046/j.1365-313X.2003.01952.x [DOI] [PubMed] [Google Scholar]
- 27. Stöckel J., and Oelmüller R. (2004) A novel protein for photosystem I biogenesis. J. Biol. Chem. 279, 10243–10251 10.1074/jbc.M309246200 [DOI] [PubMed] [Google Scholar]
- 28. Gao H., Azam T., Randeniya S., Couturier J., Rouhier N., and Johnson M. K. (2018) Function and maturation of the Fe–S center in dihydroxyacid dehydratase from Arabidopsis. J. Biol. Chem. 293, 4422–4433 10.1074/jbc.RA117.001592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao H., Subramanian S., Couturier J., Naik S. G., Kim S.-K., Leustek T., Knaff D. B., Wu H.-C., Vignols F., Huynh B. H., Rouhier N., and Johnson M. K. (2013) Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry 52, 6633–6645 10.1021/bi4007622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rey P., Becuwe N., Tourrette S., and Rouhier N. (2017) Involvement of Arabidopsis glutaredoxin S14 in the maintenance of chlorophyll content. Plant Cell Environ. 40, 2319–2332 10.1111/pce.13036 [DOI] [PubMed] [Google Scholar]
- 31. Yabe T., and Nakai M. (2006) Arabidopsis AtIscA-I is affected by deficiency of Fe–S cluster biosynthetic scaffold AtCnfU-V. Biochem. Biophys. Res. Commun. 340, 1047–1052 10.1016/j.bbrc.2005.12.104 [DOI] [PubMed] [Google Scholar]
- 32. Czernuszewicz R. S., Macor K. A., Johnson M. K., Gewirth A., and Spiro T. G. (1987) Vibrational mode structure and symmetry in proteins and analogs containing Fe4S4 clusters: resonance Raman evidence that HiPIP is tetrahedral while ferredoxin undergoes a D2d distortion. J. Am. Chem. Soc. 109, 7178–7187 10.1021/ja00257a045 [DOI] [Google Scholar]
- 33. Kong D., Zhu Y., Wu H., Cheng X., Liang H., and Ling H.-Q. (2008) AtTHIC, a gene involved in thiamine biosynthesis in Arabidopsis thaliana. Cell Res. 18, 566–576 10.1038/cr.2008.35 [DOI] [PubMed] [Google Scholar]
- 34. Knill T., Reichelt M., Paetz C., Gershenzon J., and Binder S. (2009) Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. Plant Mol. Biol. 71, 227–239 10.1007/s11103-009-9519-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Selles B., Moseler A., Rouhier N., and Couturier J. (2019) Rhodanese domain-containing sulfurtransferases: multifaceted proteins involved in sulfur trafficking in plants. J. Exp. Bot. 70, 4139–4154 10.1093/jxb/erz213 [DOI] [PubMed] [Google Scholar]
- 36. Zannini F., Roret T., Przybyla-Toscano J., Dhalleine T., Rouhier N., and Couturier J. (2018) Mitochondrial Arabidopsis thaliana TRXo isoforms bind an iron–sulfur cluster and reduce NFU proteins in vitro. Antioxidants 7, 142 10.3390/antiox7100142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arsova B., Hoja U., Wimmelbacher M., Greiner E., Ustün S., Melzer M., Petersen K., Lein W., and Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22, 1498–1515 10.1105/tpc.109.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fenwick M. K., Mehta A. P., Zhang Y., Abdelwahed S. H., Begley T. P., and Ealick S. E. (2015) Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase. Nat. Commun. 6, 6480 10.1038/ncomms7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seemann M., Wegner P., Schünemann V., Bui B. T., Wolff M., Marquet A., Trautwein A. X., and Rohmer M. (2005) Isoprenoid biosynthesis in chloroplasts via the methylerythritol phosphate pathway: the (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase (GcpE) from Arabidopsis thaliana is a [4Fe–4S] protein. J. Biol. Inorg Chem. 10, 131–137 10.1007/s00775-004-0619-z [DOI] [PubMed] [Google Scholar]
- 40. Schwenkert S., Netz D. J., Frazzon J., Pierik A. J., Bill E., Gross J., Lill R., and Meurer J. (2009) Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425, 207–214 10.1042/BJ20091290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benoit S. L., Holland A. A., Johnson M. K., and Maier R. J. (2018) Iron–sulfur protein maturation in Helicobacter pylori: identifying a Nfu-type cluster carrier protein and its iron–sulfur protein targets. Mol. Microbiol. 108, 379–396 10.1111/mmi.13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bandyopadhyay S., Naik S. G., O'Carroll I. P., Huynh B.-H., Dean D. R., Johnson M. K., and Dos Santos P. C. (2008) A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron–sulfur cluster carrier. J. Biol. Chem. 283, 14092–14099 10.1074/jbc.M709161200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Py B., Gerez C., Angelini S., Planel R., Vinella D., Loiseau L., Talla E., Brochier-Armanet C., Garcia Serres R., Latour J.-M., Ollagnier-de Choudens S., Fontecave M., and Barras F. (2012) Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol. Microbiol. 86, 155–171 10.1111/j.1365-2958.2012.08181.x [DOI] [PubMed] [Google Scholar]
- 44. Cai K., Liu G., Frederick R. O., Xiao R., Montelione G. T., and Markley J. L. (2016) Structural/functional properties of human NFU1, an intermediate [4Fe-4S] carrier in human mitochondrial iron–sulfur cluster biogenesis. Structure 24, 2080–2091 10.1016/j.str.2016.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tong W.-H., Jameson G. N., Huynh B. H., and Rouault T. A. (2003) Subcellular compartmentalization of human Nfu, an iron–sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767 10.1073/pnas.1732541100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cameron J. M., Janer A., Levandovskiy V., Mackay N., Rouault T. A., Tong W.-H., Ogilvie I., Shoubridge E. A., and Robinson B. H. (2011) Mutations in iron–sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet. 89, 486–495 10.1016/j.ajhg.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Navarro-Sastre A., Tort F., Stehling O., Uzarska M. A., Arranz J. A., Del Toro M., Labayru M. T., Landa J., Font A., Garcia-Villoria J., Merinero B., Ugarte M., Gutierrez-Solana L. G., Campistol J., Garcia-Cazorla A., et al. (2011) A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am. J. Hum. Genet. 89, 656–667 10.1016/j.ajhg.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Angelini S., Gerez C., Ollagnier-de Choudens S., Sanakis Y., Fontecave M., Barras F., and Py B. (2008) NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283, 14084–14091 10.1074/jbc.M709405200 [DOI] [PubMed] [Google Scholar]
- 49. Py B., Gerez C., Huguenot A., Vidaud C., Fontecave M., Ollagnier de Choudens S., and Barras F. (2018) The ErpA/NfuA complex builds an oxidation-resistant Fe-S cluster delivery pathway. J. Biol. Chem. 293, 7689–7702 10.1074/jbc.RA118.002160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., and Johnson M. K. (2012) Spectroscopic and functional characterization of iron–sulfur cluster-bound forms of Azotobacter vinelandii Nif IscA. Biochemistry 51, 8071–8084 10.1021/bi3006658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chahal H. K., Dai Y., Saini A., Ayala-Castro C., and Outten F. W. (2009) The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48, 10644–10653 10.1021/bi901518y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chahal H. K., and Outten F. W. (2012) Separate FeS scaffold and carrier functions for SufB2C2 and SufA during in vitro maturation of [2Fe2S] Fdx. J. Inorg. Biochem. 116, 126–134 10.1016/j.jinorgbio.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ollagnier-de-Choudens S., Sanakis Y., and Fontecave M. (2004) SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg Chem. 9, 828–838 10.1007/s00775-004-0581-9 [DOI] [PubMed] [Google Scholar]
- 54. Banci L., Brancaccio D., Ciofi-Baffoni S., Del Conte R., Gadepalli R., Mikolajczyk M., Neri S., Piccioli M., and Winkelmann J. (2014) [2Fe-2S] cluster transfer in iron–sulfur protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 6203–6208 10.1073/pnas.1400102111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gourdoupis S., Nasta V., Calderone V., Ciofi-Baffoni S., and Banci L. (2018) IBA57 recruits ISCA2 to form a [2Fe-2S] cluster-mediated complex. J. Am. Chem. Soc. 140, 14401–14412 10.1021/jacs.8b09061 [DOI] [PubMed] [Google Scholar]
- 56. Beilschmidt L. K., Ollagnier de Choudens S., Fournier M., Sanakis I., Hograindleur M.-A., Clémancey M., Blondin G., Schmucker S., Eisenmann A., Weiss A., Koebel P., Messaddeq N., Puccio H., and Martelli A. (2017) ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo. Nat. Commun. 8, 15124 10.1038/ncomms15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mapolelo D. T., Zhang B., Randeniya S., Albetel A.-N., Li H., Couturier J., Outten C. E., Rouhier N., and Johnson M. K. (2013) Monothiol glutaredoxins and A-type proteins: partners in Fe–S cluster trafficking. Dalton Trans. 42, 3107–3115 10.1039/c2dt32263c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCarthy E. L., and Booker S. J. (2017) Destruction and reformation of an iron–sulfur cluster during catalysis by lipoyl synthase. Science 358, 373–377 10.1126/science.aan4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grant K., Carey N. M., Mendoza M., Schulze J., Pilon M., Pilon-Smits E. A., and van Hoewyk D. (2011) Adenosine 5′-phosphosulfate reductase (APR2) mutation in Arabidopsis implicates glutathione deficiency in selenate toxicity. Biochem. J. 438, 325–335 10.1042/BJ20110025 [DOI] [PubMed] [Google Scholar]
- 60. Meguro M., Ito H., Takabayashi A., Tanaka R., and Tanaka A. (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23, 3442–3453 10.1105/tpc.111.089714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gutiérrez-Nava Mde L., Gillmor C. S., Jiménez L. F., Guevara-García A., and León P. (2004) Chloroplast biogenesis genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 135, 471–482 10.1104/pp.103.036996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roche B., Aussel L., Ezraty B., Mandin P., Py B., and Barras F. (2013) Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469 10.1016/j.bbabio.2012.12.010 [DOI] [PubMed] [Google Scholar]
- 63. Schenk P. M., Baumann S., Mattes R., and Steinbiss H. H. (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare argtRNAs. BioTechniques 19, 196–198, 200 [PubMed] [Google Scholar]
- 64. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., and Johnson M. K. (2012) Spectroscopic and functional characterization of iron-bound forms of Azotobacter vinelandii Nif IscA. Biochemistry 51, 8056–8070 10.1021/bi300664j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vignols F., Bréhélin C., Surdin-Kerjan Y., Thomas D., and Meyer Y. (2005) A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 16729–16734 10.1073/pnas.0506880102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., and Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 10.1111/j.1365-313X.2004.02219.x [DOI] [PubMed] [Google Scholar]
- 67. Yoo S.-D., Cho Y.-H., and Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- 68. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., and Kimura T. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- 69. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 70. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 71. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yılmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.