Abstract
Sterile alpha motif and HD domain-containing protein 1 (SAMHD1) is a deoxynucleoside triphosphohydrolase (dNTPase) with a nuclear localization signal (NLS). SAMHD1 suppresses innate immune responses to viral infection and inflammatory stimuli by inhibiting the NF-κB and type I interferon (IFN-I) pathways. However, whether the dNTPase activity and nuclear localization of SAMHD1 are required for its suppression of innate immunity remains unknown. Here, we report that the dNTPase activity, but not nuclear localization of SAMHD1, is important for its suppression of innate immune responses in differentiated monocytic cells. We generated monocytic U937 cell lines stably expressing WT SAMHD1 or mutated variants defective in dNTPase activity (HD/RN) or nuclear localization (mNLS). WT SAMHD1 in differentiated U937 cells significantly inhibited lipopolysaccharide-induced expression of tumor necrosis factor α (TNF-α) and interleukin-6 (IL-6) mRNAs, as well as IFN-α, IFN-β, and TNF-α mRNA levels induced by Sendai virus infection. In contrast, the HD/RN mutant did not exhibit this inhibition in either U937 or THP-1 cells, indicating that the dNTPase activity of SAMHD1 is important for suppressing NF-κB activation. Of note, in lipopolysaccharide-treated or Sendai virus–infected U937 or THP-1 cells, the mNLS variant reduced TNF-α or IFN-β mRNA expression to a similar extent as did WT SAMHD1, suggesting that SAMHD1-mediated inhibition of innate immune responses is independent of SAMHD1's nuclear localization. Moreover, WT and mutant SAMHD1 similarly interacted with key proteins in NF-κB and IFN-I pathways in cells. This study further defines the role and mechanisms of SAMHD1 in suppressing innate immunity.
Keywords: SAM domain and HD domain-containing protein 1 (SAMHD1), innate immunity, NF-kB transcription factor, interferon, cell differentiation, monocyte, inflammation, NF-kappa B (NF-KB), dNTPase activity, innate immune responses, nuclear localization, SAMHD1, NF-κB activation, type I interferon (IFN-I), monocytic cells, viral restriction factor
Introduction
SAMHD1 is a dNTPase3 that reduces intracellular deoxynucleoside triphosphate (dNTP) levels (1, 2). The enzymatic sites responsible for dNTP hydrolysis are in the HD domain, and the SAMHD1 active site mutant (H206R and D207N) known as HD/RN was demonstrated to lack dNTPase activity (3). The majority of SAMHD1 is localized in the nucleus (4), with only a small portion detectable in the cytoplasm (5, 6). The nuclear localization signal (NLS) of SAMHD1 at the N terminus has been identified as 11KRPR14 (5, 7, 8). The SAMHD1 mutation of three amino acids of this motif except Pro-13 to alanine results in SAMHD1 localization only in the cytoplasm although retaining catalytic activity, and hence can also deplete the intracellular dNTP pool (7).
SAMHD1 is a broad viral restriction factor in nondividing immune cells. HIV-1 infection is restricted by SAMHD1 in nondividing myeloid cells (9, 10) and resting CD4+ T cells (6, 11). SAMHD1 can also block infections of other retroviruses, such as feline immunodeficiency virus, murine leukemia virus, equine infectious anemia virus, and human T cell leukemia virus type 1 (12, 13). Furthermore, SAMHD1 can block the infections of several DNA viruses, including hepatitis B virus, vaccinia virus, and herpes simplex virus type I in noncycling myeloid cells (14–17). Interestingly, viral proteins are able to counteract SAMHD1-mediated restriction of viral infections. The Vpx protein from HIV-2 and Vpx/Vpr of certain lineages of simian immunodeficiency viruses target SAMHD1 for degradation through the ubiquitin proteasome pathway (9, 10, 18). SAMHD1-mediated restriction of Epstein-Barr virus infection is also antagonized by a conserved viral protein kinase (BGLF4) encoded by all herpesviruses (19). These results indicate that SAMHD1 plays a significant role in host-virus interactions during viral infections.
Upon viral infection, the innate immune response is the first line of defense. The NF-κB family of transcription factors and interferon regulatory factors (IRFs) are major regulators of innate immune responses (20, 21). NF-κB inhibitor α (IkBα) is phosphorylated and then degraded to release the p105/p65 complex, which is subsequently processed into p50/p65 and translocated to the nucleus to activate the transcription of NF-κB-responsive genes (22). IRF3/7 can be phosphorylated by inhibitor of NF-κB kinase subunit epsilon (IKKϵ), and then forms homodimers to translocate into the nucleus to activate the interferon-sensitive responsive element (ISRE) (23). The activation of NF-κB and IFN-I pathway leads to the expression of proinflammatory cytokines such as TNF-α and IL-6, and IFN-I, which may inhibit viral infections.
SAMHD1 acts as a negative regulator of innate immunity (24). Homozygous mutations of SAMHD1 can cause autoimmune diseases such as Aicardi-Goutières syndrome and systemic lupus erythematosus (4, 25, 26). These patients can have increased levels of IFN-I or IFN-inducible gene expression (4, 27), suggesting that SAMHD1 is a negative regulator of innate immunity. Moreover, SAMHD1 acts at stalled replication forks to restart DNA replication resulting in prevention of IFN-I induction (28). We have reported that SAMHD1 suppresses innate immune responses to viral infection and inflammatory stimuli by inhibiting the NF-κB and IFN-I pathways (29). Knockdown of SAMHD1 by siRNA in primary human macrophages increased TNF-α, IL-6, IFN-α, and IFN-β mRNA levels after SeV infection, indicating SAMHD1 inhibits NF-κB activation and IFN-I induction (29).
To analyze the correlation between SAMHD1-mediated suppression of innate immune responses and its dNTPase activity, we overexpressed WT SAMHD1 and the HD/RN mutant in dividing HEK293T cells and found that both can inhibit IL-1β-induced NF-κB activation and IRF7-mediated ISRE activity (29). These results suggest that SAMHD1 inhibition of NF-κB activation and IFN-I induction is independent of its dNTPase activity in dividing cells. However, it remains unknown whether the dNTPase activity and nuclear localization of SAMHD1 are important for its suppression of innate immune responses in nondividing cells.
In this study, we found that WT SAMHD1, but not mutant HD/RN, suppressed NF-κB activation and IFN-I induction in differentiated monocytic U937 and THP-1 cells in response to lipopolysaccharide (LPS) treatment or SeV infection. These new data suggest that dNTPase activity, but not nuclear localization of SAMHD1, is important for its suppression of innate immunity in response to inflammatory stimulation and viral infection in nondividing monocytic cells. Our findings provide new insights into better understanding the immunoregulation function of SAMHD1.
Results
Inhibition of LPS-induced NF-κB activation by SAMHD1 requires its dNTPase activity
We reported that WT SAMHD1-mediated NF-κB activation is independent of its dNTPase activity in dividing HEK293T cells (29). However, it remains unclear whether SAMHD1-mediated suppression of NF-κB activation is specific for nondividing cell types. SAMHD1 is responsible for down-regulation of dNTP levels by its dNTPase activity in nondividing cells (1, 2). Monocytic U937 cells lacking endogenous SAMHD1 protein have been extensively used in functional and mechanistic studies of WT SAMHD1 or other SAMHD1 variants upon exogenous expression (1, 10, 29–34).
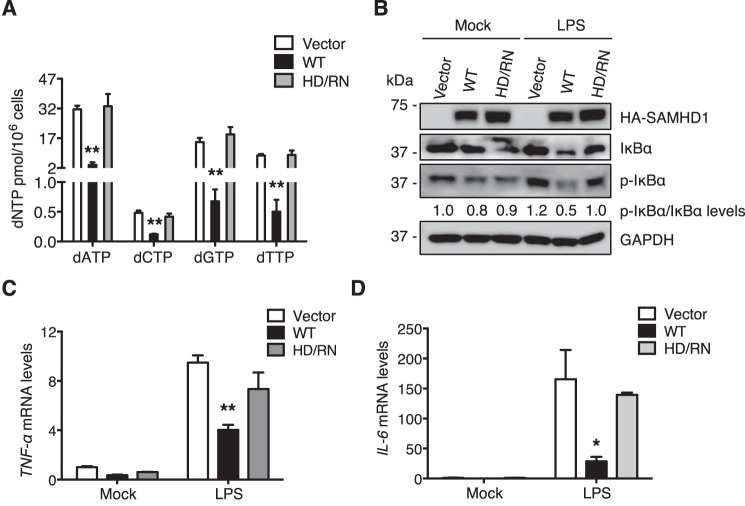
To investigate the correlation between SAMHD1-mediated inhibition of NF-κB activation and its dNTPase activity in nondividing cells, we first generated monocytic U937 cells stably expressing WT SAMHD1 or the HD/RN mutant by lentiviral transduction. SAMHD1 mutant HD/RN is defective for dNTPase activity (3). To differentiate U937 cells and induce expression of WT SAMHD1 or the HD/RN mutant, we treated cells with phorbol 12-myristate 13-acetate (PMA). To validate the dNTPase activity of WT SAMHD1 and HD/RN in differentiated U937 cells, intracellular dNTP levels were measured. We found that WT SAMHD1, but not HD/RN, significantly reduced intracellular dNTP levels compared with vector control cells (Fig. 1A), confirming that HD/RN has no dNTPase activity.
Figure 1.
Inhibition of LPS-induced NF-κB activation by SAMHD1 requires its dNTPase activity in differentiated U937 cells. U937 cells stably expressing WT SAMHD1 or HD/RN mutant were differentiated with PMA (100 ng/ml) for 24 h and then recovered in fresh medium for another 24 h. A, after differentiation, intracellular dNTP levels were measured. WT SAMHD1 but not HD/RN reduces cellular dNTP levels in differentiated U937 cells. Results are shown as mean ± S.D. Statistical significance was determined by unpaired Student's t test; **, p < 0.01 compared with the vector control. B–D, differentiated U937 cells were treated with LPS (100 ng/ml) or mock treated for 6 h. Cells were collected, and cell lysates were analyzed by immunoblotting (IB) to test the expression levels of HA-tagged SAMHD1, total and phosphorylated IkBα (p-IkBα). GAPDH was a loading control when quantifying relative p-IkBα/IkBα levels (B). Relative mRNA levels of TNF-α (C) and IL-6 (D) in cell pellets were measured by RT-PCR. Results are shown as mean ± S.D. Statistical significance was determined using unpaired Student's t test; *, p < 0.05; **, p < 0.01 compared with vector controls. The results are representative of three independent experiments.
To examine whether the dNTPase activity of SAMHD1 is important for its inhibition of NF-κB pathway in nondividing cells, we treated PMA-differentiated U937 cells with LPS for 6 h. Immunoblotting confirmed comparable WT SAMHD1 and HD/RN expression levels independently of LPS treatment (Fig. 1B). We analyzed expression level of phosphorylated IkBα (p-IkBα) as a marker of NF-κB activation. The expression levels of p-IkBα were normalized to total IkBα, and increased slightly after LPS treatment in vector control U937 cells (Fig. 1B), indicating activation of the NF-κB pathway. Of note, p-IkBα expression level was reduced by WT SAMHD1 in LPS-treated cells relative to vector control cells (Fig. 1B), which is consistent with our previous study (29). In contrast, HD/RN mutant did not significantly reduce p-IkBα levels after LPS treatment relative to vector control cells (Fig. 1B).
Next, we measured mRNA levels of the NF-κB-dependent genes TNF-α and IL-6 in cells treated with LPS or mock treated. We found that after LPS treatment WT SAMHD1 reduced TNF-α and IL-6 mRNA levels by ∼2.4-fold and ∼6-fold, respectively, compared with vector control cells (Fig. 1, C and D). These data suggest that WT SAMHD1 suppresses LPS-induced NF-κB activation, in line with our previous study (29). However, the HD/RN mutant was not able to inhibit TNF-α and IL-6 mRNA levels after LPS treatment compared with vector control cells (Fig. 1, C and D, respectively), indicating that dNTPase activity of SAMHD1 is important for its suppression of NF-κB activation induced by LPS in nondividing monocytic cells.
SAMHD1-mediated suppression of SeV infection–induced NF-κB and IFN-I activation requires its dNTPase activity
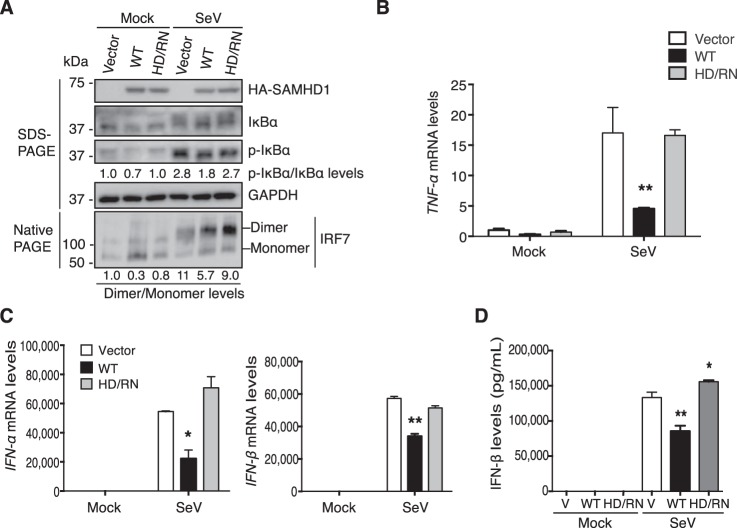
To examine the importance of dNTPase activity of SAMHD1 in its suppression of NF-κB and IFN-I activation induced by viral infection in nondividing cells, we infected PMA-differentiated U937 cells with SeV, an RNA virus that activates both NF-κB and IFN-I pathways (29). We observed p-IkBα expression levels increased in vector control cells at 6 h post SeV infection (Fig. 2A), indicating the activation of the NF-κB pathway. We also tested the dimerization of IRF7 in cells as a marker of activation of the IFN-I pathway. We found significant increases in the dimerization of IRF7 after SeV infection in vector control cells (Fig. 2A), indicating the activation of the IFN-I pathway. Interestingly, WT SAMHD1 significantly reduced p-IkBα expression levels and IRF7 dimerization after SeV infection compared with vector control cells (Fig. 2A), indicating that WT SAMHD1 inhibits NF-κB and IFN-I activation after viral infection in differentiated monocytic cells. Of note, expression levels of p-IkBα and dimerization of IRF7 were not inhibited significantly by HD/RN mutant relative to vector control after SeV infection (Fig. 2A).
Figure 2.
SAMHD1-mediated suppression of SeV infection–induced NF-κB activation and IFN-I induction requires its dNTPase activity in differentiated U937 cells. Differentiated U937 cells stably expressing WT SAMHD1 or HD/RN were infected with SeV at the m.o.i. of 10 for 6 h or mock infected. A, Cell lysates were used to test the expression levels of HA-tagged SAMHD1, total and phosphorylated IkBα, and GAPDH by SDS-PAGE. Dimerization of IRF7 was also tested in native PAGE separately. B and C, dNTPase activity of SAMHD1 is needed for suppressing NF-κB activation and IFN-I induction after SeV infection. Relative mRNA levels of TNF-α (B), IFN-α (C), and IFN-β (C) in cell pellets were measured by RT-PCR. D, IFN-β protein levels in the supernatants were tested after 24 h SeV infection. Results are shown as mean ± S.D. Statistical significance was determined by unpaired Student's t test; *, p < 0.05; **, p < 0.01 compared with vector controls. The results are representative of three independent experiments.
We then measured TNF-α mRNA levels in cells infected with SeV or mock infected. We found that WT SAMHD1 reduced TNF-α mRNA levels by ∼4-fold after SeV infection relative to vector control cells, whereas the HD/RN mutant did not (Fig. 2B). These results suggest that dNTPase activity of SAMHD1 is necessary for its suppression of NF-κB activation induced by viral infection in differentiated monocytes.
We further measured mRNA levels of IFN-α and IFN-β after SeV infection as indicators of the activation of IFN-I pathway. Compared with vector control cells, we observed after SeV infection WT SAMHD1, but not HD/RN, reduced IFN-α and IFN-β mRNA levels by ∼2.5-fold and ∼2-fold, respectively (Fig. 2C). Moreover, WT SAMHD1, but not HD/RN, significantly decreased IFN-β protein levels in the supernatants from SeV-infected differentiated U937 cells relative to vector control cells (Fig. 2D). These data suggest that the dNTPase activity of SAMHD1 is important for its suppression of IFN-I induction after viral infection in differentiated monocytic cells.
Nuclear localization of SAMHD1 is not required for its suppression of NF-κB activation and IFN-I induction in differentiated U937 cells
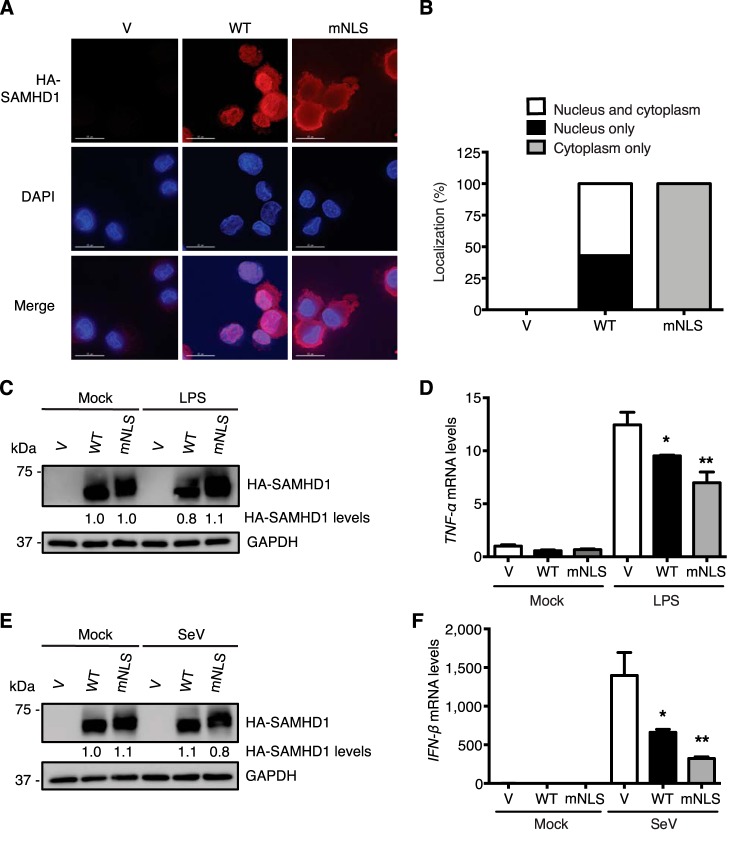
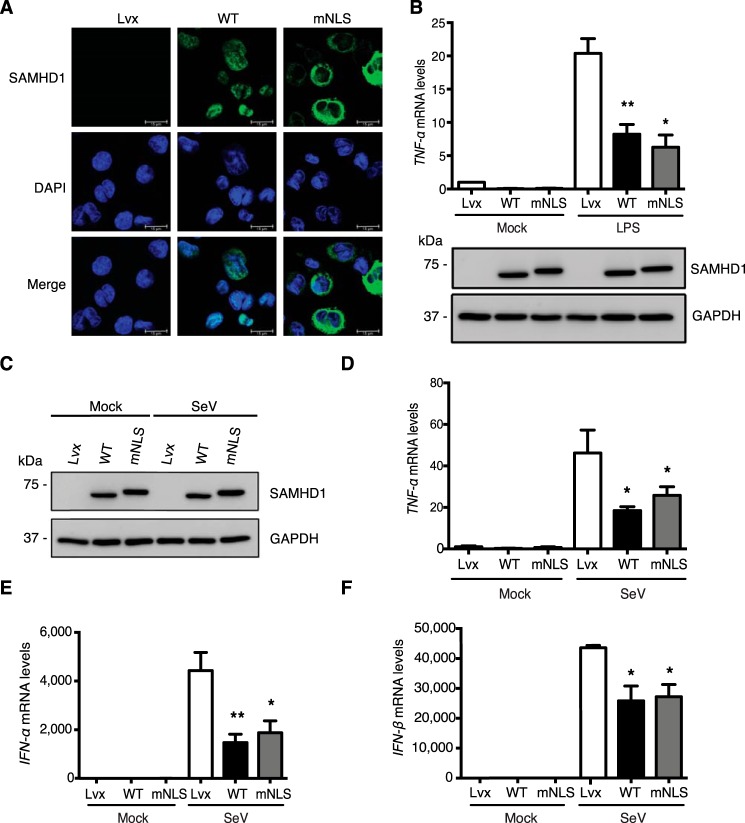
Our previous studies suggest that the mechanism of the suppression of innate immune responses by SAMHD1 is through the interactions with the key proteins in the NF-κB pathway (such as IkBα) or in the IFN-I pathway (such as IRF7), resulting in the inhibition of the phosphorylation of IkBα and IRF7 (29). The majority of endogenous SAMHD1 is located in the nucleus and cytoplasmic SAMHD1 is also reported (5, 6). Our previous studies have not defined whether nuclear or cytoplasmic SAMHD1 is responsible for its suppression of innate immune responses. We hypothesize that SAMHD1-mediated suppression of innate immune responses occurs in the cytoplasm, and nuclear localization of SAMHD1 is not required for the suppressive effect. To test our hypothesis and to better understand the underlying mechanism driving the suppression, we mutated the NLS from 11KRPR14 in WT SAMHD1 to 11AAPA14 (mNLS) which results in cytoplasmic localization (5). We then generated U937 cells stably expressing mNLS, and examined its localization in PMA-differentiated cells by immunofluorescence microscopy. We observed that the mNLS was only localized in the cytoplasm without overlapping with DAPI (Fig. 3A). However, some WT SAMHD1 signal overlapped with DAPI, indicating nuclear localization only, and others throughout the whole cells (Fig. 3A). We then counted 100 cells and analyzed SAMHD1 localization. The mNLS was exclusively cytoplasmic, whereas about half (43%) WT SAMHD1 was only nuclear, and the remaining (57%) was in both nucleus and cytoplasm (Fig. 3B).
Figure 3.
Nuclear localization of SAMHD1 is not required for its suppression of NF-κB activation and IFN-I induction in differentiated U937 cells. U937 cells stably expressing WT SAMHD1 or mNLS variant were differentiated with PMA (100 ng/ml) for 24 h and then recovered in fresh medium for another 24 h. A, mNLS is only localized in the cytoplasm in differentiated U937 cells. Differentiated U937 cells were fixed and stained with HA antibody and DAPI staining. The images shown are from one representative experiment. Red, HA-tagged SAMHD1; blue, DAPI. Scale bars, 15 μm. B, one hundred WT SAMHD1- or mNLS-expressing cells were examined, counted, and scored according to the visual localization. C and D, nuclear localization of SAMHD1 is not required for its suppression of LPS-induced NF-κB activation. Differentiated U937 cells were treated with 100 ng/ml LPS or mock treated for 6 h. C, cell lysates were used to test expression levels of HA-tagged SAMHD1, total and phosphorylated IkBα by IB. GAPDH was a loading control. D, relative mRNA levels of TNF-α were measured by RT-PCR. Results are shown as mean ± S.D. E and F, nuclear localization of SAMHD1 is not required for its suppression of SeV infection–induced IFN-I induction. Differentiated U937 cells were infected with SeV for 6 h (m.o.i. of 10). After infection, cells were collected. E, cell lysates were used to test expression levels of HA-tagged SAMHD1, total and phosphorylated IkBα by IB. GAPDH was a loading control. F, relative IFN-β mRNA levels were quantified by RT-PCR. Results are shown as mean ± S.D. Statistical significance in D and F was determined by unpaired Student's t test; *, p < 0.05; **, p < 0.01 compared with vector controls with LPS treatment or SeV infection. The results are representative of three independent experiments.
To examine whether nuclear localization of SAMHD1 is required for its suppression of LPS-induced NF-κB activation in nondividing cells, we treated PMA-differentiated U937 cells with LPS to activate NF-κB signaling. Comparable expression levels of WT SAMHD1 and mNLS were obtained with or without LPS treatment (Fig. 3C). TNF-α mRNA levels were reduced by both WT SAMHD1 and mNLS after LPS treatment relative to vector control cells (Fig. 3D), suggesting that nuclear localization of SAMHD1 is not required for its suppression of LPS-induced NF-κB activation in differentiated U937 cells.
To demonstrate whether NLS of SAMHD1 is required for its suppression of SeV infection–induced IFN-I induction in nondividing cells, we infected PMA-differentiated U937 cells with SeV. WT SAMHD1 and mNLS had comparable expression levels with or without SeV infection (Fig. 3E). Both WT SAMHD1 and mNLS reduced IFN-β mRNA levels after SeV infection relative to vector control cells (Fig. 3F), suggesting that nuclear localization of SAMHD1 is not required for its suppression of IFN-I induction induced by viral infection in differentiated U937 cells.
Reconstitution of WT SAMHD1, but not HD/RN, in THP-1/KO cells suppresses NF-κB activation
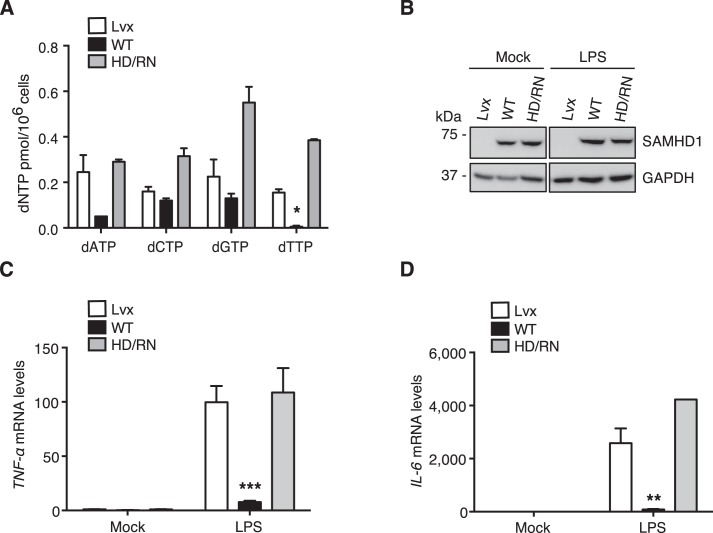
We previously generated SAMHD1-knockout monocytic THP-1 cell lines (THP-1/KO) and characterized their phonotypes (34). Reconstitution of WT SAMHD1 or SAMHD1 mutants in THP-1/KO cells is an important approach to further validate our above results from differentiated U937 cells. Therefore, we reconstituted WT SAMHD1 or HD/RN in THP-1/KO cells by retroviral transduction. To verify the reconstituted cells, we first tested the intracellular dNTP levels and observed that reconstitution of WT SAMHD1 but not HD/RN reduced intracellular dNTP levels in differentiated THP-1/KO cells compared with vector control cells (Fig. 4A), indicating that HD/RN has no dNTPase activity. We then examined the expression levels of WT SAMHD1 and HD/RN after reconstitution in the differentiated THP-1/KO cells. We obtained comparable expression levels of WT SAMHD1 and HD/RN with or without LPS treatment (Fig. 4B).
Figure 4.
Reconstitution of WT SAMHD1 in differentiated THP-1/KO cells suppresses NF-κB activation. THP-1/KO cells were transduced with a Lvx vector (Lvx), or SAMHD1 WT or HD/RN expressing lentiviruses. The reconstituted THP-1/KO cells were differentiated with PMA (30 ng/ml) for 24 h to induce WT SAMHD1 or HD/RN expression. A, after differentiation, intracellular dNTP levels were measured. Reconstitution of WT SAMHD1, but not HD/RN, in THP-1/KO cells reduces intracellular dNTP levels. Results are shown as mean ± S.D. Statistical significance was determined by unpaired Student's t test; *, p < 0.05 compared with the vector control. B–D, the differentiated cells were treated with LPS (100 ng/ml) or mock treated. After 6-h treatment, cells were collected and cell lysates were analyzed by IB to measure the expression levels of WT SAMHD1 or HD/RN. GAPDH was a loading control (B). Relative mRNA levels of TNF-α (C) and IL-6 (D) in the cell pellets were also measured by RT-PCR. Results are shown as mean ± S.D. Statistical significance was determined using unpaired Student's t test; **, p < 0.01; ***, p < 0.001 compared with vector controls. The results are representative of three independent experiments.
To examine whether dNTPase activity of SAMHD1 also correlates with its suppression of NF-κB activation in nondividing monocytic cells, we treated PMA-differentiated THP-1 cells with LPS to activate NF-κB signaling. Compared with vector control cells, a 13- and 32-fold reduction of TNF-α and IL-6 mRNA levels was observed by reconstituting WT SAMHD1 after LPS treatment, respectively (Fig. 4, C and D). However, reconstitution of HD/RN had no effect on TNF-α and IL-6 mRNA levels compared with vector control cells (Fig. 4, C and D, respectively). These results indicate that dNTPase activity of SAMHD1 is important for its suppression of LPS-induced NF-κB activation in differentiated monocytic cells.
Reconstitution of WT SAMHD1 or mNLS suppresses innate immune responses in differentiated THP-1/KO cells
To validate our results observed in differentiated U937 cells using a different monocytic cell line, we reconstituted SAMHD1 WT or mNLS in THP-1/KO cells by retroviral transduction. Immunofluorescence microscopy was conducted to verify SAMHD1 localization. The results showed that WT SAMHD1 was in the nucleus, whereas mNLS was in the cytoplasm of PMA-differentiated THP-1/KO cells after SAMHD1 reconstitution (Fig. 5A).
Figure 5.
Reconstitution of WT SAMHD1 or mNLS variant suppresses innate immune responses in differentiated THP-1/KO cells. THP-1/KO cells were transduced with Lvx (empty vector), WT SAMHD1, or mNLS expressing lentiviruses. The reconstituted THP-1/KO cells were differentiated with PMA (30 ng/ml) for 24 h. A, SAMHD1-reconstituted THP-1/KO cells were fixed and immunofluorescence assay was conducted with SAMHD1 and DAPI staining. Green, SAMHD1; blue, DAPI. Scale bars, 15 μm. B, the differentiated cells were treated with LPS (100 ng/ml) or mock treated. After 6-h treatment, cells were collected, and cell lysates were used to analyze expression levels of WT SAMHD1 or mNLS with a SAMHD1-specific antibody (bottom blots). GAPDH was the loading control. TNF-α mRNA levels in the cells were measured by RT-PCR. C–F, the differentiated cells were infected with SeV for 6 h (m.o.i. of 10). After infection, cells were collected, and cell lysates were analyzed for expression levels of SAMHD1. GAPDH was used as loading control (C). TNF-α (D), IFN-α (E), and IFN-β (F) mRNA levels in the cells were measured by RT-PCR. Results are shown as mean ± S.D. Statistical significance was determined using unpaired Student's t test; *, p < 0.05; **, p < 0.01 compared with vector controls. The results are representative of three independent experiments.
To validate that SAMHD1-mediated inhibition of NF-κB activation is independent of its nuclear localization in nondividing cells, we treated PMA-differentiated THP-1/KO cells with LPS for 6 h, and observed comparable expression levels of reconstituted WT SAMHD1 and mNLS (Fig. 5B, bottom blots). We found that reconstitution of WT SAMHD1 or mNLS in THP-1/KO cells decreased TNF-α mRNA levels relative to vector control cells (Fig. 5B, top bar chart), indicating that nuclear localization of SAMHD1 is not required for its suppression of NF-κB activation in differentiated THP-1 cells.
Next, we infected the PMA-differentiated THP-1/KO cells expressing WT SAMHD1 or mNLS with SeV and examined the effect on NF-κB activation and IFN-I induction. After SeV infection, we observed comparable expression levels of WT SAMHD1 and mNLS (Fig. 5C). TNF-α and IFN-I mRNA levels were examined as indicators of NF-κB activation and IFN-I induction by SeV infection, respectively. The results showed that reconstituted WT SAMHD1 or mNLS significantly inhibited expression of TNF-α, IFN-α, and IFN-β mRNA in differentiated THP-1/KO cells compared with vector control cells (Fig. 5, D–F, respectively). These results further demonstrated that nuclear localization of SAMHD1 is not required for its suppression of innate immune responses to viral infection in differentiated monocytic cells.
SAMHD1 suppresses NF-κB activation and IFN-I induction independently of its nuclear localization in dividing HEK293T cells
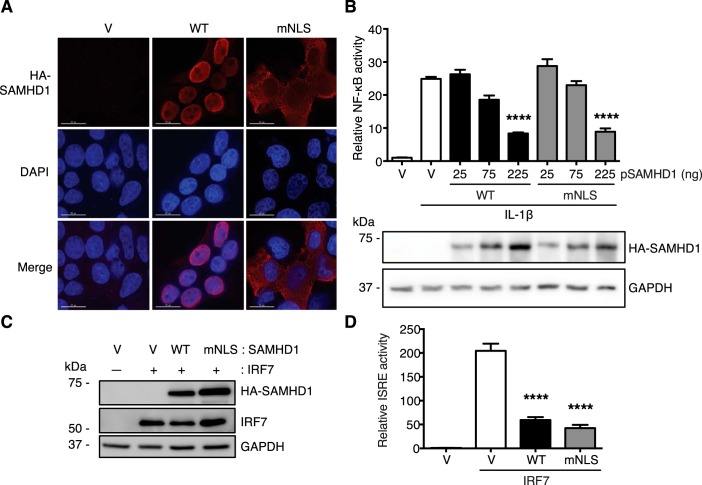
In our previously published studies, we found that SAMHD1 inhibits NF-κB activation and IFN-I induction independently of its dNTPase activity in dividing HEK293T cells (29). However, in this study we showed that dNTPase activity of SAMHD1 correlates with its suppression of NF-κB activation and IFN-I induction in differentiated monocytic cells. This suggests that the correlation between dNTPase activity of SAMHD1 and its suppression of innate immunity differs in dividing cells relative to nondividing cells. We also showed that nuclear localization of SAMHD1 is not required for its suppression of NF-κB activation and IFN-I induction in differentiated monocytic cells. However, the requirement of nuclear localization of SAMHD1 in dividing HEK293T cells remains unknown. Because HEK293T cells express a very low level of endogenous SAMHD1 (35), WT SAMHD1 or mNLS was overexpressed in HEK293T cells to examine whether nuclear localization of SAMHD1 is required for suppressing the innate immune responses. Immunofluorescence results showed that WT SAMHD1 was exclusively localized in the nucleus, whereas the mNLS was only in the cytoplasm of HEK293T cells (Fig. 6A).
Figure 6.
SAMHD1 suppresses NF-κB activation and IFN-I induction independently of its nuclear localization in dividing HEK293T cells. A, mNLS is only localized in the cytoplasm of HEK293T cells. HEK293T cells were transfected with HA-tagged WT SAMHD1 or mNLS for 24 h. Cells were fixed, and immunofluorescence was performed with HA antibody and DAPI staining. Red, HA-tagged SAMHD1; blue, DAPI. Scale bars, 15 μm. B, both WT SAMHD1 and mNLS inhibit IL-1β–induced NF-κB activation. HEK293T cells were transfected with HA-tagged SAMHD1 WT or mNLS, together with NF-κB–luciferase reporter. After 24-h transfection, cells were treated with 10 ng/ml IL-1β for 6 h. Cells were collected and lysed. Cell lysates were used to quantify the relative NF-κB activity, and to detect WT SAMHD1 and mNLS expression levels by IB. Results are shown as mean ± S.D. Statistical significance was determined using one-way ANOVA; ****, p < 0.0001 compared with the vector control with IL-1β treatment. The result is representative of three independent experiments. C and D, both WT SAMHD1 and mNLS inhibit IRF7-mediated ISRE activation in HEK293T cells. HEK293T cells were transfected with HA-tagged SAMHD1 WT or mNLS, together with IRF7 and ISRE-luc reporter. After 24-h transfection, cells were collected and lysed. C, cell lysates were used to detect expression levels of HA-tagged SAMHD1 and IRF7 by IB. D, cell lysates were also used to quantify the relative ISRE activity. Results are shown as mean ± S.D. Statistical significance was determined by unpaired Student's t test; ****, p < 0.0001 compared with the vector control with IRF7 co-expression. The result is representative of three independent experiments.
To test whether nuclear localization of SAMHD1 is required for its suppression of NF-κB activation in HEK293T cells, we co-expressed HA-tagged WT SAMHD1 or mNLS, together with an NF-κB-luciferase reporter in HEK293T cells. The cells were then treated with IL-1β to activate NF-κB signaling. We observed comparable expression levels of WT SAMHD1 and mNLS after IL-1β treatment (Fig. 6B). We also found that high levels of WT SAMHD1 and mNLS reduced NF-κB activity induced by IL-1β treatment relative to vector control cells (Fig. 6B), suggesting that nuclear localization of SAMHD1 is not required for its inhibition of NF-κB activation in dividing HEK293T cells.
Furthermore, to test the requirement of nuclear localization of SAMHD1 in its inhibition of IFN-I induction in dividing cells, we co-expressed WT SAMHD1 or mNLS, together with IRF7 and an ISRE reporter in HEK293T cells. Comparable expression levels of WT SAMHD1 and mNLS were observed as well as IRF7 expression levels (Fig. 6C). We also examined ISRE activity and found that mNLS variant and WT SAMHD1 similarly reduced IRF7-mediated ISRE activity relative to vector control cells (Fig. 6D), suggesting that nuclear localization of SAMHD1 is not required for its suppression of IFN-I induction in dividing HEK293T cells.
SAMHD1 mNLS and HD/RN variants interact with key proteins in both NF-κB and IFN-I pathways in HEK293T cells
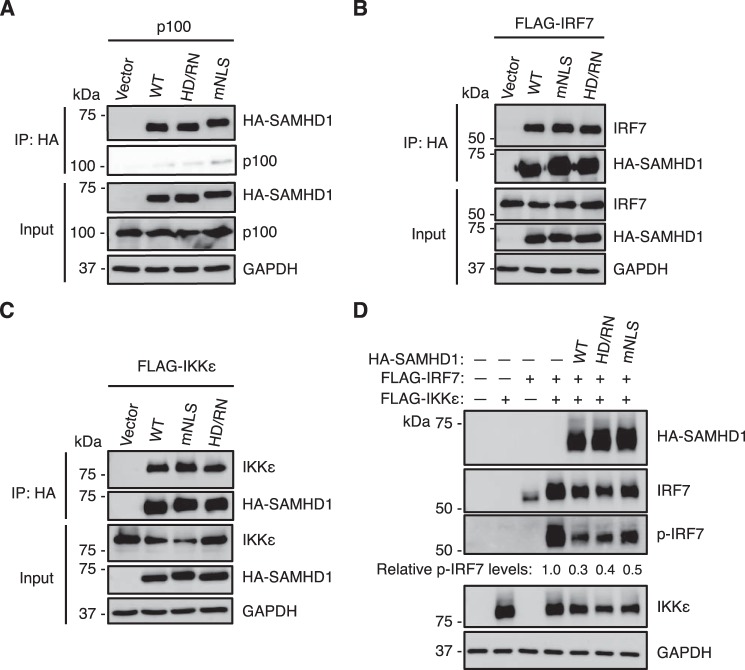
We reported interactions between WT SAMHD1 and key proteins in both the NF-κB and IFN-I pathways, such as p100, IRF7, and IKKϵ (29). Our previous and current results showed that SAMHD1 suppresses NF-κB activation and IFN-I induction independently of its dNTPase activity and nuclear localization in dividing HEK293T cells (29). We hypothesize the independence of dNTPase activity and nuclear localization of SAMHD1 in dividing HEK293T cells is because of the interactions between SAMHD1 variant HD/RN or mNLS and the key proteins, such as p100, IRF7, and IKKϵ, in both NF-κB and IFN-I pathways. To test whether overexpressed HD/RN and mNLS are able to interact with p100, IRF7 and IKKϵ, we co-expressed HA-tagged SAMHD1 WT, variant HD/RN or mNLS, and p100 or IRF7 or IKKϵ in HEK293T cells, and performed co-immunoprecipitation assay. We observed that both SAMHD1 variants HD/RN and mNLS were able to interact with overexpressed p100, IRF7, and IKKϵ similarly to WT SAMHD1 in HEK293T cells (Fig. 7, A–C, respectively). Furthermore, because WT SAMHD1 inhibits IKKϵ-mediated IRF7 phosphorylation in HEK293T cells (29), we hypothesized that SAMHD1 variants HD/RN and mNLS can also inhibit IRF7 phosphorylation. To test this hypothesis, we co-expressed HA-tagged WT SAMHD1, variant HD/RN or mNLS, together with IRF7 and IKKϵ in HEK293T cells. Both the HD/RN and mNLS variants inhibited IKKϵ-mediated phosphorylation of IRF7 (reduced by ∼50–60%) similarly to WT SAMHD1 (reduced by ∼70%) in HEK293T cells (Fig. 7D). These data further help explain why the dNTPase activity and nuclear localization of SAMHD1 are not required to inhibit IFN-I induction in dividing HEK293T cells.
Figure 7.
SAMHD1 mNLS and HD/RN variants interact with key proteins in both NF-κB and IFN-I pathways in HEK293T cells. A–C, overexpressed HA-tagged SAMHD1 WT, mNLS or HD/RN interacts with overexpressed p100 (A), FLAG-tagged IRF7 (B), and FLAG-tagged IKKϵ (C) in HEK293T cells. HEK293T cells were co-transfected with each plasmid. At 30 h post transfection, cells were lysed and analyzed using HA-agarose beads by IP assay. The indicated protein levels were detected by IB. D, SAMHD1 mNLS and HD/RN variants inhibit IKKϵ-mediated phosphorylation of IRF7. HEK293T cells were co-transfected with each plasmid for 24 h. Cells were collected and lysed to analyze expression levels of the indicated proteins by IB. The results are representative of three independent experiments.
Discussion
Upon viral infection, we reported that SAMHD1 suppresses innate immune responses by inhibiting the NF-κB and IFN-I pathways, in which SAMHD1 interacts with the key proteins in both NF-κB and IFN-I signaling (24, 29). In this study, we demonstrated that in nondividing monocytic cells, the dNTPase activity, but not nuclear localization, of SAMHD1 is important for its inhibition of NF-κB activation and IFN-I induction in response to inflammatory stimulation and viral infection. This study facilitates better understanding the function of SAMHD1 in suppressing the innate immune responses to inflammatory stimuli and viral infections. A recent study suggests that SAMHD1 restricts the infection of human cytomegalovirus by inhibiting NF-κB activation (36). They showed that the SAMHD1 dNTPase-defective mutant (HD/AA) loses the suppression of NF-κB activation compared with WT SAMHD1 after human cytomegalovirus infection in U937 cells (36), which is similar to what we observed in U937 cells during SeV infection. The correlation between dNTPase activity of SAMHD1 and its suppression of NF-κB activation is conserved in DNA virus and RNA virus infections in monocytic cells.
Our new data suggest that SAMHD1 suppresses NF-κB activation and IFN-I induction dependent on its dNTPase activity in nondividing monocytic cells, although in dividing HEK293T cells, dNTPase activity of SAMHD1 is not required for its suppression (29). There may be various factors causing the cell-type dependence. We previously reported that SAMHD1 has little effect on intracellular dNTP levels in dividing HEK293T cells (35), although it reduces intracellular dNTP levels in differentiated U937 cells. The difference in intracellular dNTP levels may be important in SAMHD1-mediated suppression of innate immune responses. As to nondividing monocytic cells stimulated by LPS, the intracellular dNTP levels were lower, and the suppression of NF-κB activation by WT SAMHD1 was stronger in THP-1 cells relative to U937 cells (Figs. 1 and 4). The precise mechanism of the dependence of dNTPase activity of SAMHD1 in suppressing innate immune responses in nondividing monocytic cells remains to be examined.
The importance of dNTPase activity of SAMHD1 implies its balanced regulation of innate immune responses in monocytic cells during viral infections. SAMHD1 is a well-studied restriction factor to viral infections that reduces intracellular dNTP levels through its dNTPase activity (13, 15–17, 30). The lack of viral replication blocks leads to the accumulation of viral nucleic acids in cells (35), which may result in the activation of innate immune responses. To avoid too strong innate immune responses, SAMHD1 inhibits the secretion of inflammatory cytokines and IFN-I by interacting with key proteins in NF-κB and IFN-I pathways (29). Thus, the balanced regulation of the antiviral function and negative effect on innate immunity by SAMHD1 may be critical for host cells.
In dividing HEK293T cells, SAMHD1 suppression of NF-κB activation and IFN-I induction is independent of its dNTPase activity (29) and nuclear localization in this study. SAMHD1 dNTPase–defective mutant HD/RN and nuclear localization–defective mutant mNLS can interact with the key proteins in both NF-κB and IFN-I pathways similarly to WT SAMHD1. These suggest that the interactions between SAMHD1 and key proteins in both NF-κB and interferon pathways correlate with its suppression of innate immune responses.
SAMHD1-mediated inhibition of NF-κB activation might occur at the transcriptional level in nondividing monocytic cells. WT SAMHD1 expression in PMA-differentiated U937 and THP-1 cells efficiently inhibited LPS-induced IL-6 mRNA expression (Figs. 1D and 4D, respectively). However, using ELISA, we did not observe SAMHD1-mediated inhibition of release of IL-6 protein from PMA-differentiated U937 cells at 12, 24, and 36 h post LPS treatment (data not shown). Previous studies indicated that LPS-induced production of cytokines, including IL-6, in monocytes and macrophages is highly regulated by intracellular signal transduction pathways (37). It is therefore conceivable that nuclear SAMHD1 may not be able to directly inhibit LPS-induced IL-6 protein production or secretion from differentiated monocytes.
Studying the role of SAMHD1 in suppressing NF-κB and IFN-I pathways to inflammatory stimuli and viral infection is important not only for better understanding autoimmune diseases caused by homozygous mutations of SAMHD1, but also for exploring the underlying mechanisms of innate immune responses to viral infections. Overall, our current study fills the knowledge gap and lays the foundation for future research.
Experimental procedures
Plasmids
The plasmids encoding HA-tagged empty pLenti-puro vector and WT SAMHD1 (30) were gifts from Dr. Nathaniel Landau (New York University). The plasmid encoding SAMHD1 mutant HD/RN has been reported (29). The plasmid encoding SAMHD1 mNLS was generated using a QuikChange mutagenesis kit (Agilent Technologies) according to the manufacturer's protocol. To generate mNLS, the following primer was used: 5′-GAG CAG CCC TCC GCG GCT CCC GCT TGC GAT GAC AGC-3′. The plasmids encoding FLAG-IRF7 and ISRE-Luciferase (29, 38) were gifts from Dr. Deyin Guo (Wuhan University, China). The plasmids encoding FLAG-IKKϵ and NF-κB–Luciferase have been described (29, 39). The plasmid encoding p100 was purchased from Addgene (catalogue no. 23287).
Cell culture
Monocytic U937 parental cells and HEK293T cells were from the American Type Culture Collection (ATCC) (31, 40). THP-1/KO cells were described previously (34). U937 cells stably expressing empty vector, WT SAMHD1, SAMHD1 mutant HD/RN, or mNLS were generated by spinoculation of monocytic U937 parental cells with concentrated lentiviral vectors followed by 1 μg/ml puromycin containing media selection as described (7, 31, 41). THP-1/KO cells expressing Lvx vector, WT SAMHD1, HD/RN, or mNLS were generated as described (29). All transduced U937 and THP-1/KO cell lines were cultured in RPMI 1640 (ATCC), supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1 μg/ml puromycin. HEK293T cells were cultured as reported (34). All cell lines were maintained at 37 °C, 5% CO2 and confirmed free from mycoplasma contamination using the universal mycoplasma detection kit (ATCC 30-101-2K).
LPS, IL-1β treatment, and SeV infection of cells
Monocytic U937 or THP-1 cells were treated with 100 ng/ml LPS for 6 h. HEK293T cells were treated with 10 ng/ml IL-1β for 6 h. SeV strain was generated as described previously (29). Viral titer was determined (42) and SeV infection was conducted at a multiplicity of infection (m.o.i.) of 10 for 6 h.
Intracellular dNTP quantification
For intracellular dNTP quantification, cells were harvested and prepared for the single nucleotide incorporation assay as reported (34, 43).
Antibodies and immunoblotting
Cells were lysed with cell lysis buffer (Cell Signaling Technology) containing protease inhibitor mixture (Sigma-Aldrich) and phosphatase inhibitor mixture (Sigma-Aldrich) after harvest. Cell lysates were used to conduct BCA assay for quantifying protein concentrations, and then same amounts of protein were loaded for SDS-PAGE. Immunoblotting was performed as described (29, 31). Blots were detected with antibodies specific against HA (BioLegend, no. 901501), IkBα (Abnova, no. MAB0057), p-IkBα (Cell Signaling Technology, no. 9246S), GAPDH (Bio-Rad, no. AHP1628), IRF7 (Cell Signaling Technology, no. 4920), phosphorylated IRF7 (Cell Signaling Technology, no. 5184), SAMHD1 (Abcam, no. 67820), p100 (Millipore, no. 05-361) and IKKϵ (Cell Signaling Technology, no. 2690). Detection of GAPDH expression acted as the loading control. The detection of IRF7 dimerization using native gel electrophoresis was performed as described (29).
RNA extraction and real-time quantitative RT-PCR assay
Cells were collected and total RNA was extracted using RNeasy Mini Kit from Qiagen. RNA concentrations were measured and equal amounts of RNA were reverse transcribed into cDNA with First-Strand Synthesis kit from Invitrogen. Synthesized cDNAs were used for SYBR Green–based qPCR analyses as described (29). Cellular 18S rRNA was used as a control and relative mRNA levels were calculated using the 2-ΔΔCT method. Primer sequences for target genes were reported (29).
Immunofluorescence microscopy assay
Immunofluorescence microscopy was conducted in PMA-differentiated U937 cells, THP-1/KO cells or dividing HEK293T cells using method described previously (32).
Transfection and luciferase assay
Transfection of HEK293T cells and luciferase assay were conducted as reported (29).
Co-immunoprecipitation assay
Co-immunoprecipitation assay was conducted as described in HEK293T cells (29). Monoclonal anti-HA-agarose beads (Sigma-Aldrich) were used to precipitate samples. IP samples were analyzed by immunoblotting with specific antibodies indicated in the figure and legends.
IFN-β ELISA in the supernatant of SeV-infected differentiated U937 cells
PMA-differentiated U937 cells were infected with SeV as described. The supernatants were collected after 24-h infection. VeriKine Human IFN Beta ELISA Kit (PBL Assay Science, no. 41410) were used to test IFN-β protein levels according to the instructions.
Statistical analyses
As to statistical analyses, p value less than 0.05 is considered significant. GraphPad Prism 6 (GraphPad Software) was used to analyze all data. Statistical tests and biological replicates are in the figure legends.
Author contributions
Z. Q., S.B., S.-H. K., J. M. A., and B. M. data curation; Z. Q., S. B., C. S. G., and L. W. formal analysis; Z. Q., S. B., C. S. G., and T.-W. L. validation; Z. Q., S. B., C. S. G., S.-H. K., J. M. A., and L. W. investigation; Z. Q. visualization; Z. Q., S. B., T.-W. L., J. M. A., B. M., J. S. Y., Y. X., and B. K. methodology; Z. Q. and L. W. writing-original draft; Z. Q., S. B., C. S. G., J. S. Y., and L. W. writing-review and editing; T.-W. L., J. S. Y., and Y. X. resources; B. K. and L. W. supervision; L. W. conceptualization; L. W. funding acquisition; L. W. project administration.
Acknowledgments
We thank the Wu lab members for helpful discussions and suggestions. We thank Drs. Nathaniel Landau and Deyin Guo for sharing constructs.
This work was supported in part by National Institutes of Health grants R01AI141495 and R01AI150343 (to L. W.), R01AI130110 (to J. S. Y.), R21AI136737 (to Y. X.), and R01AI136581 and R01AI150451 (to B. K.). This work was also partially supported by C. Glenn Barber Fund from the Ohio State University and a China Scholarship Council fellowship (to Z. Q.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- dNTPase
- deoxynucleoside triphosphohydrolase
- dNTP
- deoxy-NTP
- NLS
- nuclear localization signal
- IRF
- interferon regulatory factor
- IKKϵ
- inhibitor of NF-κB subunit epsilon
- ISRE
- interferon-sensitive responsive element
- SeV
- Sendai virus
- LPS
- lipopolysaccharide
- PMA
- phorbol 12-myristate 13-acetate
- IkBα
- NF-κB inhibitor alpha
- DAPI
- 4′,6-diamidino-2-phenylindole
- m.o.i.
- multiplicity of infection
- IB
- immunoblotting.
References
- 1. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 10.1038/nature10623 [DOI] [PubMed] [Google Scholar]
- 2. Powell R. D., Holland P. J., Hollis T., and Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 10.1074/jbc.C111.317628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., and Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 10.1038/nsmb.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., et al. (2009) Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandariz-Nuñez A., Valle-Casuso J. C., White T. E., Laguette N., Benkirane M., Brojatsch J., and Diaz-Griffero F. (2012) Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49 10.1186/1742-4690-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofmann H., Logue E. C., Bloch N., Daddacha W., Polsky S. B., Schultz M. L., Kim B., and Landau N. R. (2012) The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86, 12552–12560 10.1128/JVI.01657-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaller T., Pollpeter D., Apolonia L., Goujon C., and Malim M. H. (2014) Nuclear import of SAMHD1 is mediated by a classical karyopherin alpha/beta1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation. Retrovirology 11, 29 10.1186/1742-4690-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., and Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., and Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W., Hofmann H., Kim B., Lindemann D., and Landau N. R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26 10.1186/1742-4690-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sze A., Belgnaoui S. M., Olagnier D., Lin R., Hiscott J., and van Grevenynghe J. (2013) Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14, 422–434 10.1016/j.chom.2013.09.009 [DOI] [PubMed] [Google Scholar]
- 14. Hollenbaugh J. A., Gee P., Baker J., Daly M. B., Amie S. M., Tate J., Kasai N., Kanemura Y., Kim D. H., Ward B. M., Koyanagi Y., and Kim B. (2013) Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481 10.1371/journal.ppat.1003481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim E. T., White T. E., Brandariz-Núñez A., Diaz-Griffero F., and Weitzman M. D. (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87, 12949–12956 10.1128/JVI.02291-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z., Zhu M., Pan X., Zhu Y., Yan H., Jiang T., Shen Y., Dong X., Zheng N., Lu J., Ying S., and Shen Y. (2014) Inhibition of hepatitis B virus replication by SAMHD1. Biochem. Biophys. Res. Commun. 450, 1462–1468 10.1016/j.bbrc.2014.07.023 [DOI] [PubMed] [Google Scholar]
- 17. Sommer A. F., Rivière L., Qu B., Schott K., Riess M., Ni Y., Shepard C., Schnellbächer E., Finkernagel M., Himmelsbach K., Welzel K., Kettern N., Donnerhak C., Münk C., Flory E., Liese J., Kim B., Urban S., and König R. (2016) Restrictive influence of SAMHD1 on hepatitis B virus life cycle. Sci. Rep. 6, 26616 10.1038/srep26616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lim E. S., Fregoso O. I., McCoy C. O., Matsen F. A., Malik H. S., and Emerman M. (2012) The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 10.1016/j.chom.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang K., Lv D. W., and Li R. (2019) Conserved herpesvirus protein kinases target SAMHD1 to facilitate virus replication. Cell Rep. 28, 449–459.e5 10.1016/j.celrep.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q., Lenardo M. J., and Baltimore D. (2017) 30 years of NF-κB: A blossoming of relevance to human pathobiology. Cell 168, 37–57 10.1016/j.cell.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taniguchi T., Ogasawara K., Takaoka A., and Tanaka N. (2001) IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 22. Rahman M. M., and McFadden G. (2011) Modulation of NF-κB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306 10.1038/nrmicro2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ning S., Pagano J. S., and Barber G. N. (2011) IRF7: Activation, regulation, modification and function. Genes Immun. 12, 399–414 10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S., Bonifati S., Qin Z., St. Gelais C., and Wu L. (2019) SAMHD1 suppression of antiviral immune responses. Trends Microbiol. 27, 254–267 10.1016/j.tim.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crow M. K., Kirou K. A., and Wohlgemuth J. (2003) Microarray analysis of interferon-regulated genes in SLE. Autoimmunity 36, 481–490 10.1080/08916930310001625952 [DOI] [PubMed] [Google Scholar]
- 26. Ramantani G., Häusler M., Niggemann P., Wessling B., Guttmann H., Mull M., Tenbrock K., and Lee-Kirsch M. A. (2011) Aicardi-Goutières syndrome and systemic lupus erythematosus (SLE) in a 12-year-old boy with SAMHD1 mutations. J. Child Neurol. 26, 1425–1428 10.1177/0883073811408310 [DOI] [PubMed] [Google Scholar]
- 27. Baechler E. C., Batliwalla F. M., Karypis G., Gaffney P. M., Ortmann W. A., Espe K. J., Shark K. B., Grande W. J., Hughes K. M., Kapur V., Gregersen P. K., and Behrens T. W. (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. U.S.A. 100, 2610–2615 10.1073/pnas.0337679100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coquel F., Silva M. J., Técher H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A. L., Promonet A., Cribier A., Sarrazin A., Niedzwiedz W., Lopez B., et al. (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- 29. Chen S., Bonifati S., Qin Z., St. Gelais C., Kodigepalli K. M., Barrett B. S., Kim S. H., Antonucci J. M., Ladner K. J., Buzovetsky O., Knecht K. M., Xiong Y., Yount J. S., Guttridge D. C., Santiago M. L., and Wu L. (2018) SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways. Proc. Natl. Acad. Sci. U.S.A. 115, E3798–E3807 10.1073/pnas.1801213115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., and Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. St. Gelais C., de Silva S., Hach J. C., White T. E., Diaz-Griffero F., Yount J. S., and Wu L. (2014) Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 88, 5834–5844 10.1128/JVI.00155-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. St. Gelais C., Kim S. H., Ding L., Yount J. S., Ivanov D., Spearman P., and Wu L. (2016) A putative cyclin-binding motif in human SAMHD1 contributes to protein phosphorylation, localization, and stability. J. Biol. Chem. 291, 26332–26342 10.1074/jbc.M116.753947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antonucci J. M., Kim S. H., St. Gelais C., Bonifati S., Li T. W., Buzovetsky O., Knecht K. M., Duchon A. A., Xiong Y., Musier-Forsyth K., and Wu L. (2018) SAMHD1 impairs HIV-1 gene expression and negatively modulates reactivation of viral latency in CD4+ T cells. J. Virol. 92, e00292–18 10.1128/JVI.00292-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonifati S., Daly M. B., St. Gelais C., Kim S. H., Hollenbaugh J. A., Shepard C., Kennedy E. M., Kim D. H., Schinazi R. F., Kim B., and Wu L. (2016) SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495, 92–100 10.1016/j.virol.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. St. Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., and Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim E. T., Roche K. L., Kulej K., Spruce L. A., Seeholzer S. H., Coen D. M., Diaz-Griffero F., Murphy E. A., and Weitzman M. D. (2019) SAMHD1 modulates early steps during human cytomegalovirus infection by limiting NF-κB activation. Cell Rep. 28, 434–448.e436 10.1016/j.celrep.2019.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossol M., Heine H., Meusch U., Quandt D., Klein C., Sweet M. J., and Hauschildt S. (2011) LPS-induced cytokine production in human monocytes and macrophages. Crit. Rev. Immunol. 31, 379–446 10.1615/CritRevImmunol.v31.i5.20 [DOI] [PubMed] [Google Scholar]
- 38. Li S., Zhu M., Pan R., Fang T., Cao Y. Y., Chen S., Zhao X., Lei C. Q., Guo L., Chen Y., Li C. M., Jokitalo E., Yin Y., Shu H. B., and Guo D. (2016) The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 17, 241–249 10.1038/ni.3311 [DOI] [PubMed] [Google Scholar]
- 39. Prinarakis E., Chantzoura E., Thanos D., and Spyrou G. (2008) S-glutathionylation of IRF3 regulates IRF3-CBP interaction and activation of the IFNβ pathway. EMBO J. 27, 865–875 10.1038/emboj.2008.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Antonucci J. M., St. Gelais C., de Silva S., Yount J. S., Tang C., Ji X., Shepard C., Xiong Y., Kim B., and Wu L. (2016) SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat. Med. 22, 1072–1074 10.1038/nm.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang F., St. Gelais C., de Silva S., Zhang H., Geng Y., Shepard C., Kim B., Yount J. S., and Wu L. (2016) Phosphorylation of mouse SAMHD1 regulates its restriction of human immunodeficiency virus type 1 infection, but not murine leukemia virus infection. Virology 487, 273–284 10.1016/j.virol.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yount J. S., Kraus T. A., Horvath C. M., Moran T. M., and López C. B. (2006) A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 177, 4503–4513 10.4049/jimmunol.177.7.4503 [DOI] [PubMed] [Google Scholar]
- 43. Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., and Kim B. (2004) Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553 10.1074/jbc.M408573200 [DOI] [PMC free article] [PubMed] [Google Scholar]